Significance
Curcumin is an ancient drug derived from turmeric and has been found to exhibit potent anticancer properties albeit through controversial mechanisms of action. Using a biochemical model, mouse cancer model, and cellular models, we show that curcumin is a highly potent and selective inhibitor of dual-specificity tyrosine-regulated kinase 2 (DYRK2), a positive regulator of the 26S proteasome. Curcumin perturbs 26S proteasome activity via DYRK2 inhibition in various cancer cells and in the mouse cancer model leading to impairment of cell proliferation and reduction of cancer burden in mice. This novel mechanism of action of curcumin opens up new avenues for potential preventative or therapeutic strategies in proteasome-addicted cancers like triple-negative breast cancer and multiple myeloma.
Keywords: triple-negative breast cancer, multiple myeloma, proteasome inhibitor, kinase specificity profiling, DYRK
Abstract
Curcumin, the active ingredient in Curcuma longa, has been in medicinal use since ancient times. However, the therapeutic targets and signaling cascades modulated by curcumin have been enigmatic despite extensive research. Here we identify dual-specificity tyrosine-regulated kinase 2 (DYRK2), a positive regulator of the 26S proteasome, as a direct target of curcumin. Curcumin occupies the ATP-binding pocket of DYRK2 in the cocrystal structure, and it potently and specifically inhibits DYRK2 over 139 other kinases tested in vitro. As a result, curcumin diminishes DYRK2-mediated 26S proteasome phosphorylation in cells, leading to reduced proteasome activity and impaired cell proliferation. Interestingly, curcumin synergizes with the therapeutic proteasome inhibitor carfilzomib to induce apoptosis in a variety of proteasome-addicted cancer cells, while this drug combination exhibits modest to no cytotoxicity to noncancerous cells. In a breast cancer xenograft model, curcumin treatment significantly reduces tumor burden in immunocompromised mice, showing a similar antitumor effect as CRISPR/Cas9-mediated DYRK2 depletion. These results reveal an unexpected role of curcumin in DYRK2-proteasome inhibition and provide a proof-of-concept that pharmacological manipulation of proteasome regulators may offer new opportunities for anticancer treatment.
The medicinal effects of Curcuma longa or turmeric have been known since ancient times with earliest reference found in circa 600 BCE (1, 2). The primary active component of turmeric is curcumin or diferuloylmethane, a hydrophobic polyphenol extracted from the rhizomes (1). In more modern times, the therapeutic potential of curcumin was first reported in 1748 (2, 3) followed by a review article on the medicinal properties of Curcuma in 1815 (2, 4). In 1937, Albert Oppenheimer carried out a clinical study and reported a successful therapeutic application of curcumin on patients with biliary disease (5). Since then, there has been a growing body of literature (>11,500 publications) claiming that curcumin has a myriad of therapeutic efficacies in various diseases including cancer, neurological disorders, topical infections, etc. (6). Curcumin has been reported to induce anticancer and antiproliferative activity via multiple pathways including, but not limited to, induction of apoptosis by caspase activation, down-regulation of essential transcription factors like NFκB, inhibition of c-Jun N-terminal kinase (JNK) and protein tyrosine kinases, and down-regulation of growth factor receptors like Her2 and EGFR (7). Traditionally curcumin has been implicated as a serine-threonine kinase inhibitor by directly inhibiting IKK in the NFκB pathway (8, 9) and also as a potent inhibitor of GSK3β with an IC50 of 66 nM (10). Despite the widespread interest in the therapeutic potential of curcumin, this body of often-controversial literature has led researchers to term curcumin an improbable metabolic panacea the exact biological function of which is very difficult to dissect (6, 11). Although there are various concerns and contradictions regarding curcumin’s mechanism(s) of action, there is very strong evidence in general regarding the anticancer properties of curcumin (12–15). One mechanism of curcumin action that has been reported is the inhibition of the proteasome (16–21). Various groups have reported that curcumin acts as a proteasome inhibitor, resulting in increased p53 levels and induction of apoptosis via mitochondrial caspase activation (16, 17). Despite all these known modes of action, the exact mechanism of curcumin-mediated proteasome inhibition has not been convincingly demonstrated.
The mature 26S proteasome is a complex of 33 distinct subunits that catalyzes 80% of eukaryotic protein degradation (22, 23). Recent studies have shown that both proteasome activity and cellular abundance are dynamically regulated during physiological and pathological processes such as cell differentiation, aging, neurodegenerative diseases, and cancer (24–29). In fact, addiction to the proteasome has been identified to be the “Achilles’ heel” of the aggressive basal-like triple-negative breast cancer (TNBC) (30) and the devastating plasma cell malignancy, multiple myeloma (31). Therapeutic proteasome inhibitors bortezomib (Velcade) (31, 32), carfilzomib (Kyprolis) (33), and ixazomib (Ninlaro) (34) are Food and Drug Administration-approved with proven clinical benefit in treating early stage and refractory multiple myeloma.
Given the proteasome’s biological complexity coupled with the normal cell toxicity of proteasome inhibitor drugs, recent work has focused on inhibiting the proteasome indirectly by identifying and targeting proteasome regulators (35, 36). Recently, our laboratory reported a proteasome regulator, dual-specificity tyrosine-regulated kinase 2 (DYRK2) that directly phosphorylates the conserved Thr25 of the ATPase RPT3 subunit of the proteasome (37). In that study we demonstrated that DYRK2 depletion impairs proteasome activity and results in accumulation of numerous proteins involved in diverse cellular processes (37). These DYRK2-depleted cells exhibited a slower proliferation rate and significantly reduced tumor burden in a mouse xenograft model (37). Taken together, we established that DYRK2 is a molecular target with promising anticancer potential not only for chemosensitive but also for proteasome inhibitor-resistant/adapted cancers.
In the current study, we provide evidence that curcumin is a specific and potent inhibitor of DYRK2 and regulates the proteasome activity via DYRK2 inhibition. Cocrystal structure of curcumin with DYRK2 reveals that curcumin binds potently to the active site of DYRK2 via hydrophobic and hydrogen bonds. Furthermore, curcumin was found to not effect the proteasome activity of cells with DYRK2 deletion. Notably, curcumin treatment significantly reduced tumor volume in a TNBC mouse xenograft model, and the tumor volume was comparable to DYRK2-depleted tumors. The results establish that the inhibition of the DYRK2–proteasome axis is the primary mode of action of curcumin with expanded therapeutic utility in proteasome inhibitor-resistant cancer burdens.
Results
Curcumin Is a Potent and Selective Inhibitor of DYRK2.
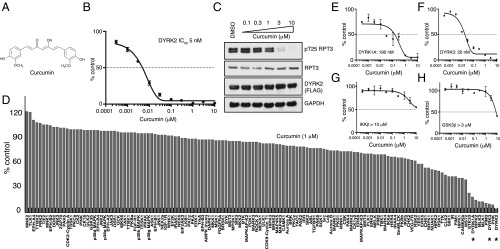
The structure of curcumin is shown in Fig. 1A. Curcumin inhibits DYRK2 with an IC50 of 5 nM (Fig. 1B) in vitro. To evaluate whether curcumin could suppress cellular DYRK2 activity, we treated HEK293T cells with stable DYRK2-FLAG overexpressing with increasing concentrations of curcumin and assessed curcumin-mediated changes of RPT3 phosphorylation at Thr25, the major site of DYRK2 phosphorylation on the proteasome. We observed that curcumin treatment suppressed pT25 RPT3 phosphorylation in a dose-dependent manner, with maximal effects observed at inhibitor concentrations of 3–10 μM (Fig. 1D). To evaluate the specificity of curcumin, we also examined the activity of 140 protein kinases, including CMGC kinase family members that are closely related to DYRKs (Fig. 1C and SI Appendix, Table S1). Curcumin targeted DYRK2 specifically; curcumin treatment was found to also inhibit DYRK1A, DYRK3, PIM, MLK, and PHK kinases albeit to a lesser extent compared with DYRK2. Curcumin had off-target effects on DYRK1A with an IC50 of 190 nM (Fig. 1E) and on DYRK3 with an IC50 of 20 nM (Fig. 1F). Contrary to previous literature, curcumin did not exhibit potent inhibition of purified IKKβ (IC50 > 10 μM) (Fig. 1G) or purified GSK3β (IC50 > 3 μM) (Fig. 1H) in vitro. The results reveal that curcumin is a highly potent inhibitor of DYRK2 with off-target effects on related DYRK isoforms.
Fig. 1.
Curcumin is a potent and selective inhibitor of DYRK2. (A) Chemical structure of curcumin. (B) DYRK2 was assayed using 300 μM of Woodtide in the presence of 50 μM [γ-33P]ATP with the indicated concentrations of curcumin. The IC50 graph was plotted using GraphPad Prism software. The results are presented as the percentage of kinase activity relative to the DMSO-treated control. Results are means ± SD for triplicate reactions with similar results obtained in at least one other experiment. (C) HEK293T cells stably expressing FLAG-DYRK2 were treated with the indicated concentrations of curcumin over 0.5 h. Cells were lysed and immunoblotting was carried out with the indicated antibodies. (D) Kinase profiling of curcumin at 1 μM was carried out against the panel of 140 kinases at the The International Centre for Protein Kinase Profiling (www.kinase-screen.mrc.ac.uk/). DYRK family kinases are indicated with an asterisk. (E) DYRK1A was assayed using the indicated concentrations of curcumin. The IC50 graph was plotted using GraphPad Prism software. (F) DYRK3 was assayed as in E. (G) IKKβ was assayed as in E. (H) GSK3β was assayed as in E.
Structure of DYRK2 in Complex with Curcumin.
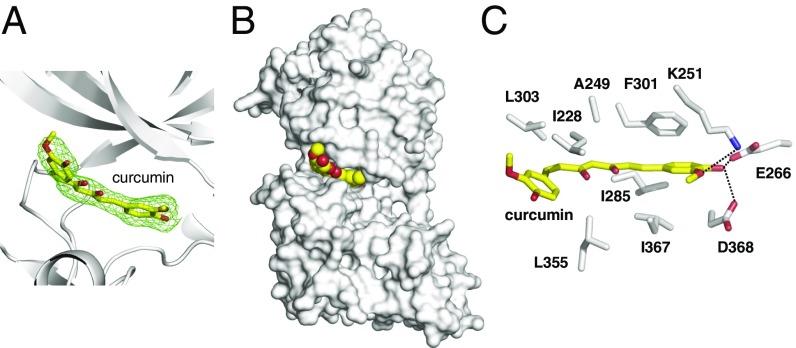
To elucidate how curcumin specifically inhibits DYRK2, we crystallized DYRK2 in the presence of curcumin and determined the structure at 2.5 Å (SI Appendix, Table S2) [Protein Data Bank (PDB) ID code: 5ZTN]. Strong electron densities are present for curcumin, allowing confident interpretation of its position (Fig. 2A). Curcumin occupies the ATP-binding pocket of DYRK2 (Fig. 2B). One of the 4-hydroxy-3-methoxyphenyl groups of curcumin forms hydrogen bonds with the Lys251 (the ion pair Lys), Glu266 (the ion pair Glu), and Asp368 (the DFG Asp) of DYRK2 that anchors curcumin deep within the ATP-binding pocket of DYRK2 (Fig. 2C). Ile228, Ala249, Ile285, Phe301, Leu303, Leu355, and Ile367 are involved in making hydrophobic interactions with curcumin (Fig. 2C). The cocrystal structure of DYRK2-curcumin clearly reveals that curcumin imparts its inhibitory effect on DYRK2 by directly binding to the DYRK2 ATP-binding pocket.
Fig. 2.
Structure of DYRK2 in complex with curcumin. (A) The Fo–Fc difference electron density map (2.5 σ) calculated before curcumin was modeled is shown as a green mesh, revealing the presence of curcumin. (B) Curcumin occupies the ATP-binding pocket of DYRK2. Curcumin atoms are shown as yellow and red spheres. DYRK2 is shown in a surface representation. (C) Detailed interactions between DYRK2 and curcumin. Hydrogen bonds are shown as dashed lines.
Curcumin Inhibits 26S Proteasome Activity.
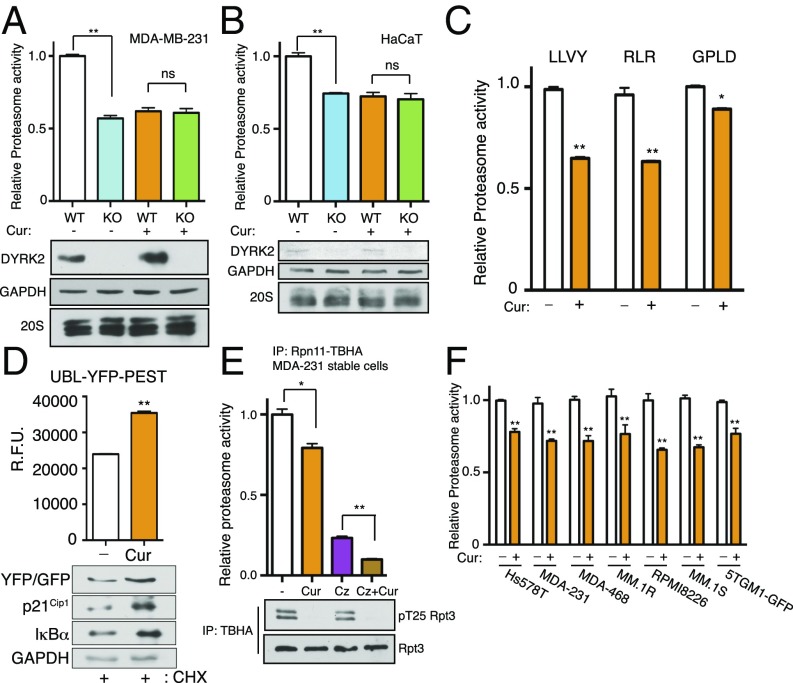
We reasoned that curcumin, by inhibiting DYRK2, may reduce proteasome activity in cells. Indeed, curcumin treatment of MDA-MB-231 and HaCaT cells caused a 25–40% decrease of proteasome activity toward the fluorogenic peptide substrate Suc-LLVY-AMC without altering proteasome abundance as indicated by Western blotting (Fig. 3 A and B). Notably, the same degree of proteasome inhibition was observed with CRISPR/Cas9-mediated DYRK2 knockout (37), and yet the DYRK2-null MDA-MB-231 (Fig. 3A) or HaCaT (Fig. 3B) cells showed no further decrease of proteasome activity with curcumin treatment. These results strongly support that curcumin down-regulates proteasome activity via DYRK2 inhibition. Moreover, in 293T cells, curcumin treatment reduced all three types of proteasome peptidase activities (Fig. 3C) and also stabilized UBL-YFP-PEST (37), a short-lived reporter protein that undergoes rapid ubiquitination and proteasome degradation (Fig. 3D). Remarkable accumulation and stabilization of two other well-established proteasome substrates, p21Cip1 and IκBα, was also evident in curcumin-treated cells compared with control (Fig. 3D). Furthermore, the inhibitory effect of curcumin on proteasome activity was observed across a panel of TNBC and multiple myeloma cell lines (Fig. 3F). Importantly, cotreatment of the TNBC MDA-MB-231 cell line with curcumin and carfilzomib resulted in a much stronger proteasome inhibition than either compound alone (Fig. 3E). Together, these results demonstrate that the antiproteasome activity of curcumin, due to DYRK2 inhibition, is similar in different cancer cell types and exhibits synergistic inhibition with carfilzomib.
Fig. 3.
Curcumin perturbs proteasome activity. (A) Proteasome activity in total cell lysates from MDA-MB-231 cells or (B) HaCaT cells (parental of DYRK2 KO) with or without a 10-μM curcumin treatment for 1 h was measured with Suc-LLVY-AMC. Immunoblotting of the cell lysates was carried out with the indicated antibodies. **P < 0.01, ns: not significant (compared with control treated, ordinary one-way ANOVA, mean ± SD from n = 3 independent experiments). (C) Proteasome activity in total cell lysates from HEK293T cells with or without a 10-μM curcumin treatment for 1 h was measured with Suc-LLVY-AMC (chymotrypsin-like activity) or Ac-RLR-AMC (trypsin-like activity) or Ac-GPLD-AMC (caspase-like activity). *P < 0.05, **P < 0.01 (compared with control treated, two-tailed paired Student’s t test, mean ± SD from n = 3 independent experiments). (D) Curcumin impedes UBL-YFP-PEST degradation. HEK293T cells were transfected with UBL-YFP-PEST. After a 1-h pretreatment of DMSO or 10 μM of curcumin, cycloheximide (CHX, 50 μg/mL) was added for 2 h along with the DMSO/curcumin. YFP fluorescence in live cells was determined, background-subtracted, and normalized to the starting level at time 0. **P < 0.01 (DMSO-treated vs. curcumin-treated, unpaired Student’s t test, mean ± SD from n = 3 independent experiments). Immunoblotting of the cell lysates was carried out with the indicated antibodies. (E) MDA-MB-231 Rpn11-TBHA cells were pretreated with the indicated drugs (Cz: carfilzomib; Cur: curcumin) for 1 h, 26S proteasome was affinity-purified, and proteasome activity was measured using Suc-LLVY-AMC. *P < 0.05; **P < 0.01, ns: not significant (compared with control treated, ordinary one-way ANOVA, mean ± SD from n = 3 independent experiments). Immunoblotting of the cell lysates was carried out with the indicated antibodies. (F) Proteasome activity in total cell lysates from the indicated cells was measured with Suc-LLVY-AMC. **P < 0.01 (compared with control treated for each cell line, two-way ANOVA, mean ± SD from n = 3 independent experiments).
Curcumin Impedes Cell Proliferation and Invasion and Induces Apoptosis.
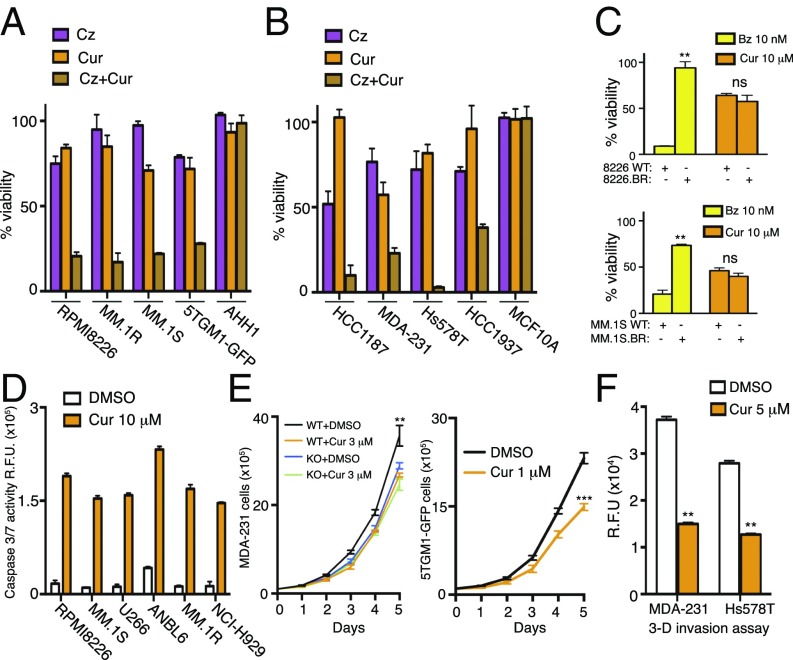
Next, we wanted to determine if curcumin–carfilzomib-mediated synergistic impairment of proteasome activity could have an effect on cancer cell viability. Interestingly, a marked synergistic cytotoxicity was observed across four different multiple myeloma cell lines with a combination index (CI) of <1 (Fig. 4A and SI Appendix, Table S3). However, noncancerous myeloid cell AHH1 exhibited modest cytotoxicity toward the combination (Fig. 4A). Similar data were observed in four different triple-negative breast cancer cell lines with a curcumin-carfilzomib CI <1 for cytotoxicity in all cancer cells with modest to no effect in noncancerous mammary cell line MCF10A (Fig. 4B and SI Appendix, Table S3). We next used bortezomib-resistant cell lines that were generated by adaptation to continuous proteasome inhibition (38). RPMI8226.BR and MM.1S.BR cells were treated with 10 μM curcumin in parallel with their bortezomib-sensitive WT counterparts. RPMI8226.BR and MM.1S.BR cells exhibited comparable cytotoxicity to 10 μM of curcumin although these cells were >20- to 100-fold more resistant to bortezomib than their respective bortezomib-sensitive cells (Fig. 4C). Treatment with curcumin induced apoptotic cell death with a significant increase in caspase3/7 activity in multiple myeloma cell lysates (Fig. 4D). DYRK2-mediated pT25-RPT3 phosphorylation is essential for proteasomal degradation of cell-cycle regulators and timely progression of S-phase (37). Curcumin treatment clearly perturbs cell proliferation that is consistent with DYRK2 inhibition. MDA-MB-231 and 5TGM1-GFP cells treated with curcumin exhibited a markedly slower rate of cell proliferation compared with DMSO-treated controls (Fig. 4E). However, curcumin treatment of MDA-MB-231 DYRK2 KO cells did not exhibit a further decrease in cell proliferation (Fig. 4E), suggesting that the curcumin-mediated impediment of cell proliferation is directly linked to DYRK2 inhibition. Furthermore, curcumin treatment largely blocked the ability of TNBC cells to migrate in transwell chemotaxis and 3D matrigel invasion assays (Fig. 4F). These results support that curcumin-mediated DYRK2-proteasome activity impairment has a direct effect on cytotoxicity, proliferation, and invasion of proteasome-dependent TNBC and multiple myeloma cells. It has been previously shown that curcumin shows synergism with proteasome inhibitors resulting in cytotoxicity in cancer cells (38); however, our data clearly indicate that curcumin-mediated cytotoxicity is predominantly rendered by curcumin-mediated inhibition of DYRK2.
Fig. 4.
Curcumin induces apoptosis and impedes proliferation and invasion of cancer cells. (A) Multiple myeloma cell lines RPMI8226, MM.1R, MM.1S, 5TGM1-GFP, and noncancerous AHH1 cells were treated with carfilzomib alone (RPMI8226 = 3 nM; MM.1R and MM.1S = 1 nM; 5TGM1-GFP and AHH1 = 5 nM) or with curcumin alone (5 μM) or with the combination of carfilzomib and curcumin for 24 h, and cell viability was analyzed by a CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay kit. Viability of DMSO-treated cells was utilized as the control. Data are represented as the percentage of viability of DMSO-treated control cells. (B) Triple-negative breast cancer cell lines HCC1187, MDA-MB-231, Hs578T, and HCC1937 and noncancerous triple-negative MCF10A cells were treated with carfilzomib alone (HCC1937 = 5 nM; HCC1187, MDA-MB-231, Hs578T and MCF10A = 10 nM) or with curcumin alone (10 μM) or with the combination of carfilzomib and curcumin for 24 h, cell viability was analyzed, and data are represented as in A. (C) WT (RPMI8226 and MM.1S) and bortezomib-resistant strains of multiple myeloma cell lines (RPMI8226.BR and MM.1S.BR) were treated with 10 nM of bortezomib (Bz) or 10 μM of curcumin for 24 h, cell viability was analyzed, and data are represented as in A. **P < 0.01; ns: nonsignificant (compared with control treated for each cell line, two-way ANOVA, mean ± SD from n = 3 independent experiments). (D) Caspase 3/7 activity after 16 h of DMSO or curcumin treatment was quantified in the indicated multiple myeloma cell lines using CellEvent Caspase-3/7 Green Detection Reagent. (E) Growth curves of MDA-MB-231 (WT and DYRK2 KO) and 5TGM1-GFP control and curcumin-treated cells. **P < 0.01, ***P < 0.001 (two-way ANOVA, mean ± SD from n = 3 independent experiments). (F) Bar graph depicting cell invasion in a Matrigel transwell migration assay using DMSO-treated or 5 μM of curcumin-treated MDA-MB-231 or Hs578T cells. Data were acquired 24 h after seeding in the upper chamber of a 8-μm pore-size transwells. Cells that invaded the Matrigel were quantified based on DNA content using CyQuant dye; data are represented as R.F.U. (relative fluorescence units). **P < 0.01 (compared with DMSO-treated for each cell line, two-way ANOVA, mean ± SD from n = 2 independent experiments with triplicates in each experiment).
Curcumin Reduces Tumor Burden at a Rate Consistent with DYRK2 Inhibition.
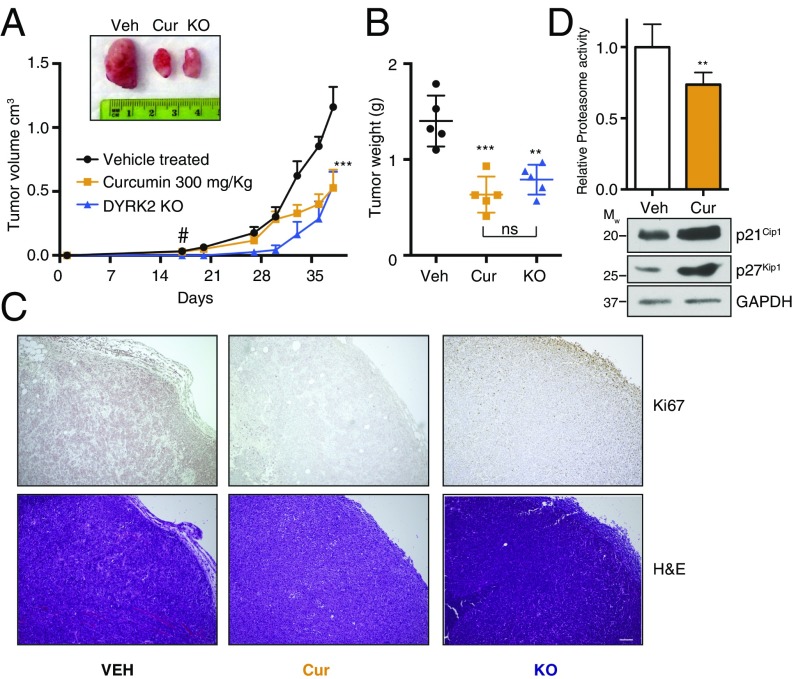
MDA-MB-231 is a basal-like TNBC cell line shown to be resistant to proteasome inhibition-mediated cytotoxicity (30). Our results on cell viability and biochemistry made us wonder whether curcumin could inhibit the tumorigenic growth of these cells in vivo. It was documented previously that curcumin reduces tumor burden in various cancer models at 200 mg/kg–1 gm/kg body weight (12–15). We have previously reported that tumors derived from DYRK2 KO cells grow at a significantly lower rate than those formed by the parental cells (37). To compare the effect of curcumin on tumor growth to DYRK2-depleted tumor growth, parental and DYRK2 KO MDA-MB-231 cells were injected s.c. into NOD scid gamma (NSG) immunocompromised mice to induce tumor formation. Parental MDA-MB-231 bearing mice with palpable tumors were randomized into two groups of n = 5 each, and vehicle control or 300 mg/kg curcumin was injected intraperitoneally every alternate day. Indeed, curcumin treatment significantly reduced tumor burden after 2 wk of treatment (Fig. 5A), and curcumin-treated tumor weights (Fig. 5B) were comparable with tumors derived from MDA-MB-231 DYRK2 KO cells. Histological examination of the xenograft tumors also showed greatly attenuated Ki-67 staining (a cellular marker for proliferation) in both curcumin-treated and DYRK2 KO tumors (Fig. 5C). Furthermore, the total proteasome activity in curcumin-treated tumor lysates were significantly reduced compared with vehicle-treated tumor lysates (Fig. 5D), thus suggesting the direct contribution of DYRK2 inhibition in tumor regression. These data strongly support the role of curcumin-mediated inhibition of the DYRK2–proteasome axis in regulating cell proliferation in vivo and suggest that targeting proteasome regulators (such as DYRK2) in combination with proteasome inhibitors may be a promising approach of anticancer therapy.
Fig. 5.
Curcumin reduces tumor burden to a similar extent as DYRK2 deletion. (A) Tumor xenograft studies were carried out with or without 300 mg/kg of curcumin treatment. MDA-MB-231 parental or genome-edited (DYRK2 KO) cells were injected s.c. into NSG mice. MDA-MB-231 parental cells bearing mice with palpable tumors (16 d postinjection marked by a “#”) were treated with vehicle control of curcumin two to three times a week by i.p. injection, and tumor volume was measure twice a week (n = 5 per condition). (B) Forty-two days postinjection, tumors were resected and tumor weight was measured. **P < 0.01, ***P < 0.001 (compared with vehicle-treated, ordinary one-way ANOVA, mean ± SD from n = 5 mice each). (C) Histological examination of consecutive sections of the tumors with Ki-67 and hematoxylin/eosin staining. (Scale bar, 100 μm.) (D) Proteasome activity in whole-tumor lysates from vehicle or curcumin-treated tumor-bearing mice was measured with Suc-LLVY-AMC. Immunoblotting of the whole-tumor lysates was carried out with indicated antibodies. **P < 0.01 (compared with control treated, two-tailed paired Student’s t test, mean ± SD from n = 3 different tumors for each treatment).
Discussion
This current study is a comprehensive report of a major mechanism of action of curcumin in alleviating proteasome-addicted tumor burden. Results show that curcumin potently binds and inhibits DYRK2 (Figs. 1 and 2), resulting in reduced 26S proteasome activity, (Fig. 3) which leads to impaired cell proliferation with induction of apoptotic cell death, and (Fig. 4) culminating in reduced tumor growth (Fig. 5). We also report a comprehensive kinase specificity profile of curcumin (Fig. 1D and SI Appendix, Table S1), which clearly shows that curcumin is a highly specific and potent inhibitor of DYRK2 with a >10-fold higher potency for DYRK2 compared with other DYRK isoforms (Fig. 1 E and F). Higher concentrations of ATP had a modest effect on the IC50 of curcumin for DYRK2 (SI Appendix, Fig. S1). This suggests that binding of curcumin to DYRK2 is potentially irreversible. Over the past three decades many groups have reported that curcumin exhibits therapeutic efficacy in mouse models for various cancers (12–15) and have shown curcumin to exhibit anti-inflammatory activity via either IKK inhibition and down-regulation of NFκB (8, 9) or inhibition of GSK3β (10). However, in our research, curcumin exhibited an in vitro IC50 of >10 μM for IKKβ (Fig. 1G) and >3 μM for GSK3β, which consequently requires >30-μM concentrations of curcumin in cells to see activity. One possibility is that the down-regulation of NFκB observed by the various groups is brought about by the accumulation of IκBα in cells due to impairment of the proteasome activity by curcumin (Fig. 3C). Moreover, DYRK isoforms have been known to play a general role in priming phosphorylation of GSK3 substrates (39), which is possibly why other researchers have observed changes in GSK3 substrate phosphorylation upon curcumin treatment (10). Various other kinases have been reported as potential targets of curcumin; however, in the absence of proper biochemical kinetics, kinase specificity, or target engagement analyses, the credibility of the reports is dramatically reduced (6). Fig. 1D shows that 1 μM of curcumin has modest to no effect on JNK isoforms, HER isoforms, and VEGFR kinases, which were previously reported to be potential targets (7). In fact, curcumin does not inhibit 139 other kinases tested in vitro to the same extent as DYRK2 (Fig. 1D). Furthermore, curcumin inhibits DYRK2 in cells at 1–10 μM (Fig. 1C), which is much lower than 30–100 μM as reported previously for other targets. It has also been convincingly shown that using curcumin at >20 μM leads to aggregation and rapid degradation of curcumin in standard cell culture and biochemical assay conditions (6).
To clear all doubts regarding the mechanism of target engagement, we solved the cocrystal structure of DYRK2 with curcumin bound at 2.5 Å (Fig. 2 and SI Appendix, Table S2) (PDB ID code: 5ZTN). This cocrystal structure of curcumin with a kinase target reveals that curcumin binds the ATP-binding pocket of DYRK2 via hydrogen bonds and hydrophobic interactions (Fig. 2). To explain the selectivity of curcumin for DYRK2, we identified that the essential curcumin-interacting residues in DYRK2, Ile285 and Ile367, are replaced by valines in DYRK1A and DYRK1B. We hypothesize that this Ile-Val replacement would result in a slightly larger pocket in the DYRK1s and reduce the shape complementarity to curcumin, explaining the selectivity of curcumin for DYRK2 (SI Appendix, Fig. S2). Alignment with representative members of distant and closely related CMGC protein kinases (SI Appendix, Fig. S3) echoes this conclusion. Substitutions of polar residues, which alter the hydrophobicity of the pocket (e.g., see SI Appendix, Fig. S3B, F301:Q), or less bulky hydrophobic residues, which result in a more open pocket (e.g., see SI Appendix, Fig. S3A, F301:L), are indicative of the resistance of a given kinase to curcumin. Substitutions of the residues interacting via hydrogen bonding (Lys251, Glu266, and D368) was not observed in these alignments, but their conservation in even highly resistant kinases (e.g., GSK3β) suggests that they are not major contributors to curcumin specificity. Efforts to generate a curcumin-resistant mutant of DYRK2 have failed, as the binding of curcumin to DYRK2 was quite strong and most single mutations either did not confer drug resistance or dramatically inhibited the kinase activity of DYRK2. Collectively, these analyses demonstrate that the majority of curcumin’s specificity toward DYRK2 is garnered by tight hydrophobic interactions made uniquely possible at the DYRK2 ATP-binding pocket.
We recently reported that DYRK2 directly phosphorylates the 26S proteasome on the Thr25 of the RPT3 subunit that impedes the 26S proteasome activity (37). The Thr25 site on RPT3 is thought to enhance substrate translocation into the 20S proteasome core, leading to protein degradation (37). Various groups have previously reported curcumin-mediated impairment of proteasome in cells (16–21); however, the exact mechanism of curcumin activity has not been not reported. Curcumin treatment inhibits all three peptidase activities of the proteasome by 10–30% (Fig. 3A). This effect, however, is not observed in DYRK2-depleted cells (Fig. 3B), clearly suggesting that curcumin targets the proteasome via inhibition of DYRK2. Interestingly, curcumin at 10 μM had no significant effect in vitro on the activity of affinity-purified proteasome to degrade BODIPY-casein substrate (SI Appendix, Fig. S4). Loss of DYRK2 delays the cell cycle by possible accumulation of proteasome substrates in the cell cycle (37). Consistent with DYRK2 depletion phenotypes, curcumin treatment impedes cell proliferation (Figs. 4E and 5C) and invasion (Fig. 4F) at concentrations <10 μM and reduces tumor burden on a TNBC mouse xenograft model at 300 mg/kg body weight dose (Fig. 5 A and B).
Accumulation of proteins with short half-lives (UBL-YFP-PEST, IκBα, p27Kip1, p21Cip1) (Figs. 3C and 5D) indicates that curcumin treatment leads to a global reduction of proteasome activity and that this activity is synergistic with the proteasome inhibitor carfilzomib (Fig. 3D). The synergism between curcumin and carfilzomib was observed not only at a biochemical level, but also in apoptotic cell death in TNBC and multiple myeloma cells lines (Fig. 4 A, B, and D). Recently, Allegra et al. (38) reported a synergism between carfilzomib and curcumin toward induction of cytotoxicity on U266 myeloma cells. Consistent with their work, we report that eight different “proteasome-adapted” cell lines exhibit strong synergistic cytotoxicity upon treatment with carfilzomib and curcumin combination (Fig. 4 A and B and SI Appendix, Table S3). However, noncancerous cells did not exhibit the similar levels of cytotoxicity (Fig. 4 A and B), thus conveying the fact that cancer cells with acquired proteasome resistance could be selectively targeted by the curcumin–carfilzomib combination. An important point to note is that higher concentrations of curcumin are equally fatal for all cells cancerous or noncancerous (HaCaT and MCF10A) (SI Appendix, Fig. S5). Bortezomib-resistant myeloma cells did not exhibit any significant resistance to curcumin (Fig. 4C). This was expected since curcumin inhibits the upstream proteasome regulator DYRK2 and does not dock into the 20S proteasome peptidase active site that is often mutated in these cell lines (40). Consistent with our cell-based data, we saw a major reduction in the cell proliferation marker Ki67 in the sections of the curcumin-treated tumors (Fig. 5C). Tumor reduction upon curcumin treatment was observed comparable to the volume and weight of DYRK2-depleted tumors (Fig. 5 A and B). Furthermore, proteasome activity in the curcumin-treated mouse tumors was reduced by 30% along with accumulation of p21Cip1 and p27Kip1, suggesting that curcumin-mediated DYRK2 inhibition successfully perturbed proteasome activity in the mouse xenograft model (Fig. 5D). The phenotypic similarity between curcumin treatment and DYRK2 depletion reinforces our hypothesis that curcumin-mediated effects on cell viability, proliferation, invasion, and tumor development are all a direct consequence of DYRK2 inhibition and impairment of the proteasome activity.
Curcumin has been one of the most controversial yet extensively studied natural products over the past three decades, having been implicated in a wide range of diseases, often with contradictory results reported by various groups worldwide (6). Considering the poor pharmacokinetics/pharmacodynamics (PK/PD) of curcumin, there is an unresolved debate on curcumin treatment as a viable therapeutic option (6). Our current work establishes curcumin not only as an excellent research tool for DYRK2 inhibition, but also as a potential therapeutic possibility. Derivatives of curcumin with improved PK/PD could be viable treatment options especially for proteasome-dependent highly refractory TNBC or multiple myeloma in the future. With respect to the vast body of literature on potential targets of curcumin, we do not claim inhibition of the DYRK2–proteasome axis as the only possible mechanism of action of curcumin; nevertheless, we have conclusively established that impairment of proteasome activity by DYRK2 inhibition is a major mechanism of action for curcumin in the context of the alleviation of proteasome-dependent neoplastic malignancies.
Materials and Methods
Details on the general methods, antibodies, reagents, curcumin preparation and treatment, IC50 determination, protein kinase inhibitor specificity screen, cell lines, transfection, lysis, genome editing, protein purification, crystallography, alignment studies, genome editing, proteasome assays, cell proliferation, viability, invasion, and tumor xenograft studies are presented in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Babatunde Oyajobi (University of Texas Health San Antonio) and Dr. Robert Orlowski (University of Texas MD Anderson Cancer Center) for providing cell lines; Dr. Carolyn Worby, Dr. Sandra Wiley, Dr. Adam Pollak, Mandira Mookerjee, and Vibhav Nadkarni for valuable input; and Dr. Hilary McLauchlan and the kinase-screen team (International Centre for Protein Kinase Profiling). This work was supported by National Key Research and Development Program of China Grants 2017YFA0505200 (to J.X.) and 2016YFC0906000 (to J.X.); NIH Grants DK018849-41 (to J.E.D.) and DK018024-43 (to J.E.D.); Mary Kay Ash Breast Cancer Grant 047.16 (to J.E.D.); Natural Science Foundation of China Grant 31671391 (to X.G.); Zhejiang Natural Science Foundation Grant LR18C050001 (to X.G.); and a University of California at San Diego NIH/National Cancer Institute Cancer Training Grant (Ruth L. Kirchstein National Research Service Award T32 CA009523) (to J.E.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5ZTN).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806797115/-/DCSupplemental.
References
- 1.Aggarwal BB, Sundaram C, Malani N, Ichikawa H. Curcumin: The Indian solid gold. Adv Exp Med Biol. 2007;595:1–75. doi: 10.1007/978-0-387-46401-5_1. [DOI] [PubMed] [Google Scholar]
- 2.Bandyopadhyay D. Farmer to pharmacist: Curcumin as an anti-invasive and antimetastatic agent for the treatment of cancer. Front Chem. 2014;2:113. doi: 10.3389/fchem.2014.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loeber CC, Buechner AE. Dissertatio inauguralis medica de curcuma officinarum ejusque genuinis virtutibus [Scientific dissertation on the real medicinal virtues of the rhizomes of curcuma] In: Præs, Büchnero AE, editors. Diss Inaug Halae. HalaeMagdeburgicae; Halle, Germany: 1748. pp. 1–28. [Google Scholar]
- 4.Vogel HA, Pelletier J. Chemische Untersuchung der Gilbwurzel (Kurkume) J Pharm (Cairo) 1815;7:20. [Google Scholar]
- 5.Oppenheimer A. Turmeric (curcumin) in biliary diseases. Lancet. 1937;229:619–621. [Google Scholar]
- 6.Nelson KM, et al. The essential medicinal chemistry of curcumin. J Med Chem. 2017;60:1620–1637. doi: 10.1021/acs.jmedchem.6b00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aggarwal BB, Kumar A, Bharti AC. Anticancer potential of curcumin: Preclinical and clinical studies. Anticancer Res. 2003;23:363–398. [PubMed] [Google Scholar]
- 8.Singh S, Aggarwal BB. Activation of transcription factor NF-kappa B is suppressed by curcumin (diferuloylmethane) J Biol Chem. 1995;270:24995–25000, and erratum (1995) 270:30235. doi: 10.1074/jbc.270.42.24995. [DOI] [PubMed] [Google Scholar]
- 9.Bharti AC, Donato N, Singh S, Aggarwal BB. Curcumin (diferuloylmethane) down-regulates the constitutive activation of nuclear factor-kappa B and IkappaBalpha kinase in human multiple myeloma cells, leading to suppression of proliferation and induction of apoptosis. Blood. 2003;101:1053–1062. doi: 10.1182/blood-2002-05-1320. [DOI] [PubMed] [Google Scholar]
- 10.Bustanji Y, et al. Inhibition of glycogen synthase kinase by curcumin: Investigation by simulated molecular docking and subsequent in vitro/in vivo evaluation. J Enzyme Inhib Med Chem. 2009;24:771–778. doi: 10.1080/14756360802364377. [DOI] [PubMed] [Google Scholar]
- 11.Bisson J, et al. Can invalid bioactives undermine natural product-based drug discovery? J Med Chem. 2016;59:1671–1690. doi: 10.1021/acs.jmedchem.5b01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aggarwal BB, et al. Curcumin suppresses the paclitaxel-induced nuclear factor-kappaB pathway in breast cancer cells and inhibits lung metastasis of human breast cancer in nude mice. Clin Cancer Res. 2005;11:7490–7498. doi: 10.1158/1078-0432.CCR-05-1192. [DOI] [PubMed] [Google Scholar]
- 13.Kunnumakkara AB, et al. Curcumin potentiates antitumor activity of gemcitabine in an orthotopic model of pancreatic cancer through suppression of proliferation, angiogenesis, and inhibition of nuclear factor-kappaB-regulated gene products. Cancer Res. 2007;67:3853–3861. doi: 10.1158/0008-5472.CAN-06-4257. [DOI] [PubMed] [Google Scholar]
- 14.Kawamori T, et al. Chemopreventive effect of curcumin, a naturally occurring anti-inflammatory agent, during the promotion/progression stages of colon cancer. Cancer Res. 1999;59:597–601. [PubMed] [Google Scholar]
- 15.Huang MT, et al. Inhibitory effects of dietary curcumin on forestomach, duodenal, and colon carcinogenesis in mice. Cancer Res. 1994;54:5841–5847. [PubMed] [Google Scholar]
- 16.Bech-Otschir D, et al. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 2001;20:1630–1639. doi: 10.1093/emboj/20.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jana NR, Dikshit P, Goswami A, Nukina N. Inhibition of proteasomal function by curcumin induces apoptosis through mitochondrial pathway. J Biol Chem. 2004;279:11680–11685. doi: 10.1074/jbc.M310369200. [DOI] [PubMed] [Google Scholar]
- 18.Dikshit P, Goswami A, Mishra A, Chatterjee M, Jana NR. Curcumin induces stress response, neurite outgrowth and prevent NF-kappaB activation by inhibiting the proteasome function. Neurotox Res. 2006;9:29–37. doi: 10.1007/BF03033305. [DOI] [PubMed] [Google Scholar]
- 19.Si X, et al. Dysregulation of the ubiquitin-proteasome system by curcumin suppresses coxsackievirus B3 replication. J Virol. 2007;81:3142–3150. doi: 10.1128/JVI.02028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neuss H, et al. The ubiquitin- and proteasome-dependent degradation of COX-2 is regulated by the COP9 signalosome and differentially influenced by coxibs. J Mol Med (Berl) 2007;85:961–970. doi: 10.1007/s00109-007-0197-y. [DOI] [PubMed] [Google Scholar]
- 21.Milacic V, et al. Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and in vivo. Cancer Res. 2008;68:7283–7292. doi: 10.1158/0008-5472.CAN-07-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 23.Collins GA, Goldberg AL. The logic of the 26S proteasome. Cell. 2017;169:792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vilchez D, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang JT, et al. Asymmetric proteasome segregation as a mechanism for unequal partitioning of the transcription factor T-bet during T lymphocyte division. Immunity. 2011;34:492–504. doi: 10.1016/j.immuni.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 28.Vilchez D, et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 29.Tai HC, Schuman EM. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat Rev Neurosci. 2008;9:826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 30.Petrocca F, et al. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell. 2013;24:182–196. doi: 10.1016/j.ccr.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hideshima T, et al. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001;61:3071–3076. [PubMed] [Google Scholar]
- 32.Richardson PG, et al. A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med. 2003;348:2609–2617. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 33.Kuhn DJ, et al. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood. 2007;110:3281–3290. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson PG, et al. Phase 1 study of twice-weekly ixazomib, an oral proteasome inhibitor, in relapsed/refractory multiple myeloma patients. Blood. 2014;124:1038–1046. doi: 10.1182/blood-2014-01-548826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song Y, et al. Blockade of deubiquitylating enzyme Rpn11 triggers apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Oncogene. 2017;36:5631–5638. doi: 10.1038/onc.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, et al. Capzimin is a potent and specific inhibitor of proteasome isopeptidase Rpn11. Nat Chem Biol. 2017;13:486–493. doi: 10.1038/nchembio.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo X, et al. Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat Cell Biol. 2016;18:202–212. doi: 10.1038/ncb3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allegra A, et al. Curcumin ameliorates the in vitro efficacy of carfilzomib in human multiple myeloma U266 cells targeting p53 and NF-kappaB pathways. Toxicol In Vitro. 2017;47:186–194. doi: 10.1016/j.tiv.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Woods YL, et al. The kinase DYRK phosphorylates protein-synthesis initiation factor eIF2Bepsilon at Ser539 and the microtubule-associated protein tau at Thr212: Potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem J. 2001;355:609–615. doi: 10.1042/bj3550609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salem K, McCormick ML, Wendlandt E, Zhan F, Goel A. Copper-zinc superoxide dismutase-mediated redox regulation of bortezomib resistance in multiple myeloma. Redox Biol. 2015;4:23–33. doi: 10.1016/j.redox.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.