Significance
Specific gene regulation in organ development remains poorly understood. Here, we report that skin-specific ectodysplasin A (Eda) signaling triggers the formation of a protein complex that includes a BAF complex, an NF-kB dimer of p50/RelB, and a specific “linker” protein, Tfg. We further find that Eda-activated RelB recruits BAF complex to specific gene loci for local chromatin remodeling of target genes. These findings may exemplify a more general model for specific gene regulation involving unique ligand–receptor complexes leading to selective activation of transcription factors, specific linkers, and tissue-specific chromatin-remodeling complex.
Keywords: ectodysplasin, RelB, SWI/SNF, BAF, chromatin remodeling
Abstract
Ectodysplasin A (Eda) signaling activates NF-κB during skin appendage formation, but how Eda controls specific gene transcription remains unclear. Here, we find that Eda triggers the formation of an NF-κB–associated SWI/SNF (BAF) complex in which p50/RelB recruits a linker protein, Tfg, that interacts with BAF45d in the BAF complex. We further reveal that Tfg is initially induced by Eda-mediated RelB activation and then bridges RelB and BAF for subsequent gene regulation. The BAF component BAF250a is particularly up-regulated in skin appendages, and epidermal knockout of BAF250a impairs skin appendage development, resulting in phenotypes similar to those of Eda-deficient mouse models. Transcription profiling identifies several target genes regulated by Eda, RelB, and BAF. Notably, RelB and the BAF complex are indispensable for transcription of Eda target genes, and both BAF complex and Eda signaling are required to open chromatin of Eda targets. Our studies thus suggest that Eda initiates a signaling cascade and recruits a BAF complex to specific gene loci to facilitate transcription during organogenesis.
Gene regulation in development is orchestrated by signaling pathways, transcription factors (TFs), and epigenetic changes in accessibility of genomic loci (1–3). In mammals, skin appendages include hair, teeth, and several exocrine glands, and their exquisite differentiation requires the ectodysplasin A (Eda) signaling pathway. Mutations in this pathway result in ectodermal dysplasia (EDA), characterized by defective formation of hair follicles (HFs), sweat glands (SWs), teeth and Meibomian glands (MGs) (4). Ectodermal dysplasia is an X-linked hereditary genetic disorder. Notably, the X-linked skin phenotype was first described by Charles Darwin in 1875 (5). More than 100 y later, people have identified three genes from patients with ectodermal dysplasia, Eda, Edar, and Edaradd, which constitute a specific TNF ligand–receptor–adaptor family that is restricted to skin appendages (6). Edar, like other TNF family receptors, activates NF-κB (7). However, Eda activates a gene-expression profile that is distinct from that stimulated by classic TNF signaling in the immune system (8). How Eda activates a selective cohort of NF-κB–mediated genes remains an open question.
One feature of selective activation of gene expression is genome accessibility to TFs at specific sites in chromatin (9). Accessibility of genomic DNA can be epigenetically modulated by several chromatin-remodeling complexes, including the SWI/SNF (BAF) complex (10). In mammals, BAF complexes contain 12–15 subunits (11). The assembly of complexes containing mutually exclusive subunits is highly divergent and may function in a cell-specific manner (12). Two distinct BAF complexes have been identified, BAF (13) and polybromo-associated BAF (PBAF) (14). In addition, some tissue-specific BAFs have been reported, including embryonic stem cell BAF (esBAF) (15, 16), neuron-specific BAF (nBAF) (17), and l leukemic BAF (eukBAF) (18). However, it has not been determined whether tissue- or stage-specific cell signals during organ development induce distinct BAF complexes to modulate gene expression.
Here, we report that skin-specific Eda signaling triggers the formation of a large BAF-containing complex that includes a BAF complex, an NF-κB dimer of p50/RelB, and a specific linker protein, Tfg (TRK-fusion gene). Thus, Eda/NF-κB signaling operates through a BAF complex to regulate specific gene expression in organ development, which may exemplify a more general paradigm for gene-specific regulation in many other systems.
Results
Eda Signaling Triggers Linkage of RelB to a BAF Complex.
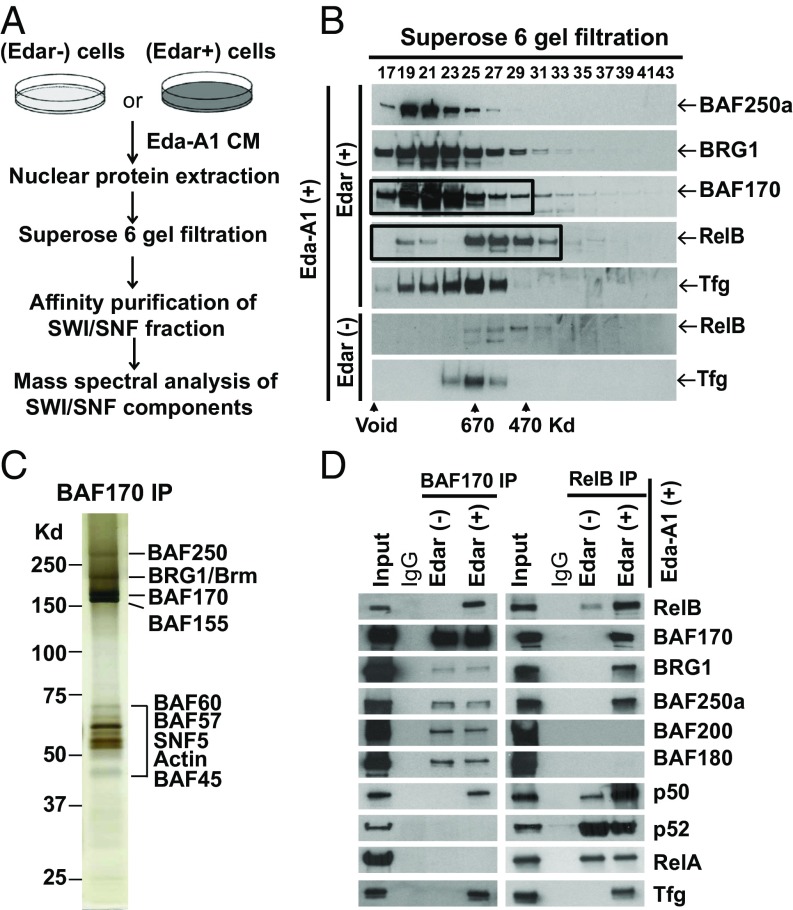
We hypothesized that tissue-specific Eda signaling might require NF-κB activation to specify gene targets and a chromatin-remodeling complex to open chromatin at those sites. To test this hypothesis, we designed a workflow for the purification of BAF complexes from HaCaT keratinocytes before and after Eda stimulation (Fig. 1A), using a successful purification method described previously (13, 19). We first generated a derivative of the HaCaT human keratinocyte stable cell line that expresses the Eda receptor (Edar+). Immunoblotting demonstrated the expected expression of Edar, and when conditioned medium (CM) containing active Eda-A1 protein (20) was added to the cultures, NF-κB luciferase activity increased 15.2-fold in Edar+ cells compared with that in untransfected (Edar−) HaCaT cells (SI Appendix, Fig. S1A).
Fig. 1.
SWI/SNF (BAF) complexes with NF-κB upon Eda signaling in keratinocytes. (A) Schematic overview shows the workflow of BAF complex purification. (B) Immunoblotting shows gel-filtration profiles in the NE from the indicated cells and treatments. Black rectangles indicate fractions collected for IP-MS analysis. (C) A silver-stained gel shows the BAF complexes purified by BAF170 IP from the NE of Edar+ cells. (D) BAF170 (Left) and RelB (Right) IP with total NE was followed by immunoblotting with the indicated antibodies. Five percent total NE from Edar+ cells was used as input. IP with IgG antibody was the negative control.
We next fractionated nuclear extracts (NEs) from Edar− and Edar+ cells treated with Eda-A1 CM. After gel-filtration chromatography, peak fractions containing BAF components—BRG1 and other key proteins including BAF250a and BAF170 (Fig. 1B)—were pooled and immunoprecipitated with a BAF170 antibody. Polyacrylamide gel silver staining (Fig. 1C) and MS analyses (SI Appendix, Table S1) showed the presence of most BAF components including BAF and PBAF in the BAF170 immunoprecipitate. Interestingly, a fraction of BAF complex associates with NF-κB subunits, observed in the BAF170 immunoprecipitate from the NE of Edar+ but not Edar− cells. Of the three NF-κB subunits with DNA-binding capacity, RelA and RelB were both observed, but RelB was ∼10-fold more abundant (SI Appendix, Table S1). To further assess the interaction of RelB and a BAF complex after Eda/Edar stimulation, we examined the size of the RelB-containing complex(es). In contrast to the gel-filtration profile of RelB in Edar− cells, much more RelB was seen in Edar+ cells, and a portion was shifted to a higher molecular weight position (Fig. 1B), consistent with the incorporation of RelB protein into a larger protein complex after Eda/Edar stimulation. Reverse immunoprecipitation (IP)-MS data using a RelB antibody found 10 components of the BAF complex (with neither of the alternative PBAF complex components BAF180 or BAF200) coimmunoprecipitated with RelB from Edar+ but not from Edar− cells (SI Appendix, Table S1). Notably, an additional protein, Tfg, was coprecipitated, that had been suggested as possibly involved in NF-κB action (21) but had not been previously found in any BAF complex. It appeared in both BAF170 and RelB immunoprecipitates from Edar+ cells (SI Appendix, Table S1), and the gel-filtration profile of Tfg was similar to that of RelB (Fig. 1B). Fully ECL-exposed immunoblots excluded the possibility that the absence of Tfg and RelB bands in the higher molecular weight position in the gel filtration was due to their lower protein levels in Edar− cells (SI Appendix, Fig. S1B). BAF170 and RelB IP-Western blotting using total NE further verified that the BAF complex, but not PBAF, had indeed interacted with the NF-κB dimer RelB/p50 and did so only after activation of Eda signaling (Fig. 1D). In further urea denaturation experiments, either p50/RelB or Tfg was still stable in its association with the BAF complex after treatment with more than 2 M urea (SI Appendix, Fig. S1C). To rule out the possibility that NF-κB may indirectly associate with the BAF complex through DNA, IP from NE of Edar+ cells was performed in the presence of ethidium bromide (EtBr), a DNA-interacting drug that dissociates proteins from DNA. The amount of RelB in the BAF170 immunoprecipitate was not affected by the presence of EtBr (SI Appendix, Fig. S1D), indicating that RelB complexes with SWI/SNF through protein interaction.
Direct protein–protein binding experiments in vitro further explored the possibility that Tfg might link the BAF complex to NF-κB. We used a cell-free protein synthesis system to produce individual proteins, including 10 components of the RelB-associated BAF complex, five NF-kBs, and Tfg, detected by immunoblotting (SI Appendix, Fig. S1 E–G). Gel silver staining confirmed the production and purity of key proteins (SI Appendix, Fig. S1H). Protein–protein binding assays showed that Tfg directly bound only RelB among the five NF-kBs and bound BAF45d but no other tested BAF (SI Appendix, Fig. S2 A and B). Reverse IP-Western blotting confirmed the direct binding of Tfg to both RelB and BAF45d (SI Appendix, Fig. S2 C and D).
Thus, biochemical data indicated that skin-specific Eda signaling activated a large protein complex containing BAF complex, p50/RelB, and the linker protein Tfg.
Eda Signaling Is Mediated Mainly by the p50/RelB Subclass of NF-κB.
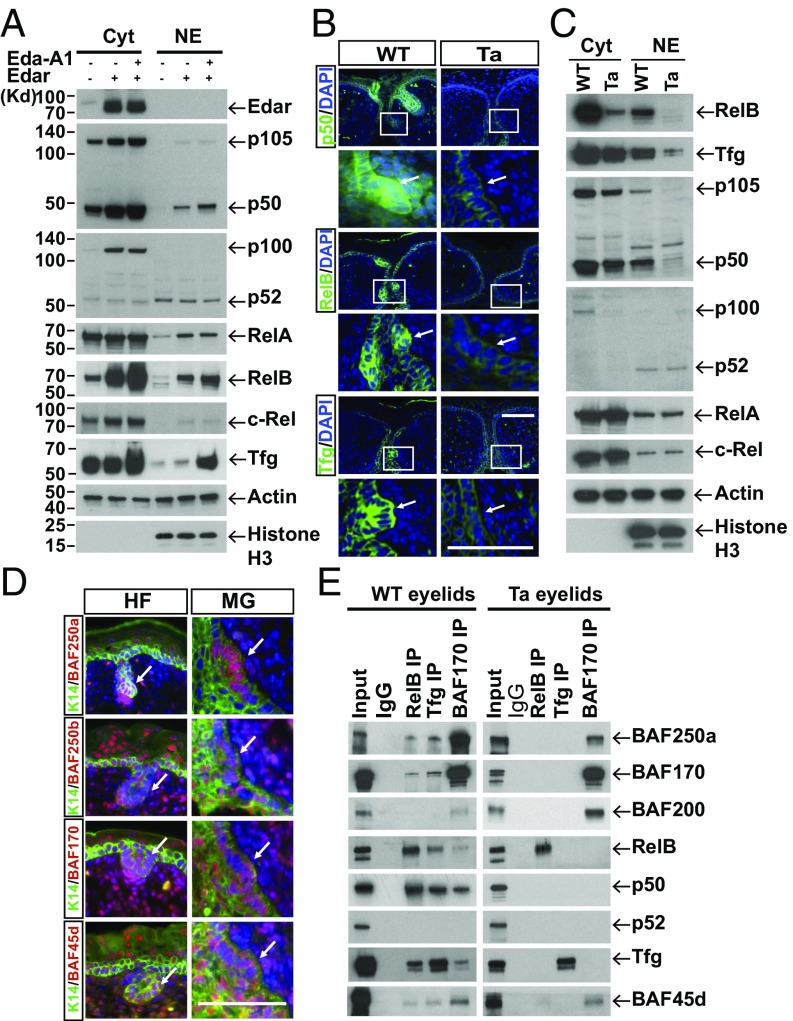
NF-κB mediates Eda/Edar signaling in cell cultures and in animal models (22, 23), but the selective binding of RelB to BAF complex (Fig. 1) suggested that refined specificity of NF-κB action might be gained by the activation of a subclass of components. We therefore checked the protein levels of all five NF-kBs in cell cultures. In the cytoplasm, Eda/Edar action up-regulated the levels of p50, p105 (p50 precursor), p100 (p52 precursor), and RelB, while other NF-kBs, including p52, RelA, and c-Rel, showed no apparent change in levels (Fig. 2A). In the nucleus, upon Edar expression, p52 was unaffected, and p50, RelA, and c-Rel were somewhat up-regulated, consistent with the previous finding that Edar itself can activate NF-κB (24, 25); however, RelB accumulated to a strikingly higher level, and the application of Eda CM further augmented RelB among three Rels (Fig. 2A). We further performed NF-κB oligonucleotide (oligo) IP experiments and found that Eda/Edar strongly activated the binding of p50 and RelB to an oligo with the NF-κB consensus sequence but not to oligos with a mutated binding sequence. Slight oligo binding to RelA was also seen (SI Appendix, Fig. S3 A and B).
Fig. 2.
Eda signaling specifically activates p50/RelB and recruits BAF complex during skin appendage development. (A) Immunoblotting shows levels of the NF-κB subunit as well as Tfg in the cytosol (Cyt) and NE of HaCaT cells with (+) or without (−) the indicated treatment. Actin and histone H3 (a nuclear protein) served as loading controls. (B) IHC staining (green) of p50, RelB, and Tfg in WT or Tabby (Ta) mouse eyelids at E16.5. Enlarged views of the areas within the white rectangles are shown below each image. Arrows indicate MG germs. (C) Immunoblotting shows protein levels of NF-κB subunits and Tfg in WT and Tabby eyelids at E16.5. (D) IHC of K14 (green) and indicated BAF proteins (red) in back skin (Left) and eyelids (Right) from WT mice at E16.5. Arrows indicate HFs (Left) or MGs (Right). (E) Total NE from WT (Left) or Tabby (Right) eyelids was used for RelB, Tfg, and BAF170 IP. Immunoblotting with the indicated antibodies is shown. Five percent NE from WT eyelids was used as input. IgG served as the negative control. (Scale bars, 50 μm.)
We extended studies of NF-κB subunit activation in skin appendage development using mouse Meibomian glands (MGs) as a model (25). At an early stage (E16.5), immunohistochemistry (IHC) images showed that p50, RelB, and RelA were expressed throughout the basal epidermal layer in WT mouse skin, while p52 was undetectable and c-Rel was restricted to the mucus layer. In MG germs, the RelA level was similar to that in epidermal layers, but p50 and RelB were considerably elevated (Fig. 2B, Left and SI Appendix, Fig. S3C). Consistent with these findings, the up-regulation of p50 and RelB in WT MGs was diminished in the MGs of Eda-deficient [Tabby (Ta)] mice (Fig. 2B, Right). Immunoblotting of each NF-κB subunit in developing MGs further confirmed the activation patterns of NF-kBs (Fig. 2C). Notably, Tfg, as a linker protein for RelB–BAF complex formation, was also up-regulated at the protein level and accumulated in nuclei upon Eda/Edar stimulation (Fig. 2 A–C and SI Appendix, Fig. S3D), consistent with the possible importance of this protein in Eda signaling.
Eda-Activated p50/RelB Recruits BAF Complex in Developing Skin Appendages.
To substantiate the function of the Eda-induced BAF complex in vivo, we studied two mouse skin appendage models: HFs and MGs. First, we analyzed the expression pattern of BRG1, a key component of both BAF and PBAF complexes. BRG1 IHC staining showed ubiquitous expression in whole-skin tissues for both HFs and MGs (SI Appendix, Fig. S4A). To further compare the expression of BAF and PBAF complexes, we checked for BAF250a and BAF250b as selective components of BAF (26) and for BAF180 and BAF200 as idiosyncratic PBAF subunits (14). BAF250a was widely expressed in whole skin, including the epidermal layer, labeled by the marker keratin 14 (K14), but BAF250a staining was markedly up-regulated within HFs and MGs (Fig. 2D). Unlike BAF250a, other BAF components (BAF250b, BAF170, and BAF45d) were all expressed uniformly in both dermis and epidermis (Fig. 2D). In contrast to other BAF proteins, IHC staining showed no expression of either BAF180 or BAF200 in HFs or MGs but showed their sporadic expression outside the basal epidermal layers (SI Appendix, Fig. S4B). We infer that BAF, but not PBAF, is expressed in developing skin appendages at E16.5 and that BAF250a is highly up-regulated in this stage.
To test whether Eda activates BAF formation in developing skin appendages, we carried out RelB, Tfg, and BAF170 IP using NEs from WT or Tabby eyelids at E16.5. Indeed, p50/RelB, together with Tfg, formed a complex with BAF but not with PBAF in WT eyelids (Fig. 2E, Left). Similarly, in Eda-deficient (Tabby) eyelids, RelB failed to complex with p50, Tfg, or any BAF protein tested (Fig. 2E, Right). These data thus provide evidence that Eda mainly activates p50/RelB, which then recruits the BAF complex through Tfg in development.
Epidermal Knockout of BAF250a Inhibits Skin Appendage Formation.
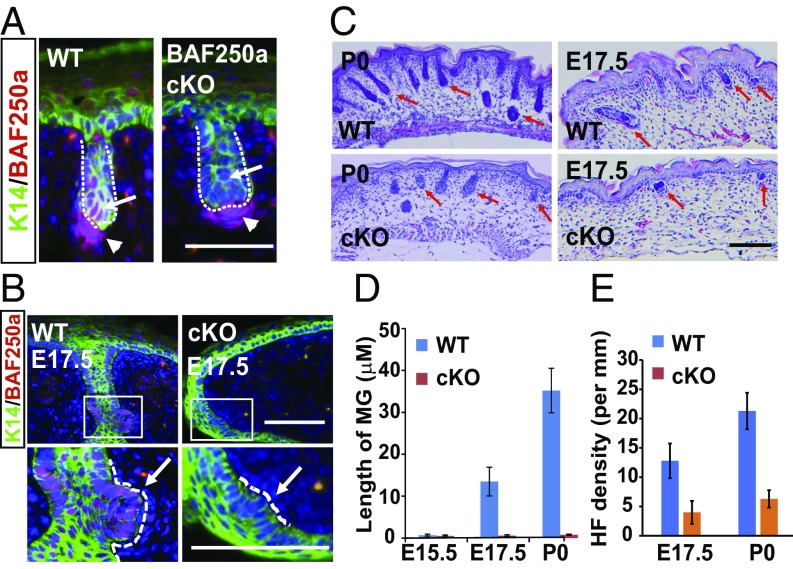
BAF250a showed specific up-regulation in developing appendages (Fig. 2D), which might reflect its functional importance. We thus generated skin-specific BAF250a knockout (cKO) mice by crossing BAF250a loxP/loxP mice with K14-Cre mice. In BAF250a cKO mice at E17.5, BAF250a expression was successfully ablated in K14+ cells (Fig. 3A). We noted that the cKO mice were born with open eyes and died within a few hours (SI Appendix, Fig. S5A).
Fig. 3.
Skin-specific cKO of BAF250a inhibits skin appendage formation. (A) IHC shows that BAF250a expression (red) in HFs from WT (Left) and cKO (Right) mice. The arrowheads indicate dermal condensation. Arrows indicate HF germs. (B) K14 (green) and BAF250a (red) IHC staining of WT and cKO eyelids at E17.5. Dotted lines separate epidermis and dermis in HFs and MGs. Arrows indicate MG germs. (C) H&E staining of WT and cKO back skin at E17.5 and P0. Arrows indicate HFs. (D) Quantitation of MG length at E15.5, E17.5, and P0. (E) HF density in WT and cKO in back skin at E17.5 and P0. Error bars indicate the mean ± SEM from at least 15 random images of three mice. (Scale bar, 50 μm.)
We next examined eyelids from WT and cKO mice collected at birth. Results in H&E-stained tissues were consistent with a previous study (25) with an average length of MGs of 35.2 μm in WT mice. By contrast, in the cKO mice, no nascent MGs progressed to invade the dermis (Fig. 3D). To test whether BAF250a affected the pregerm initiation stage of MGs, we evaluated the expression of a pregerm marker, Lef1, at E15.5, when MG is initiated by the WNT/β-catenin pathway (6). Lef1 was appreciably expressed in both WT and cKO mice, suggesting that BAF250a did not perturb the early initiation of MG germs (SI Appendix, Fig. S5B). MG morphology at E17.5 confirmed that knockout of BAF250a blocked the further growth of MG germs (Fig. 3 B and D). H&E staining during HF development in cKO mice showed a sharp reduction in HF density and length (Fig. 3 C and E), suggesting the loss of guard (tylotrich) HFs. The BAF250a cKO phenotypes thus overlap with findings in Eda-deficient Tabby or Edar-KO mice (27, 28).
Tfg Up-Regulation Is Mediated by Eda-Induced RelB Activation.
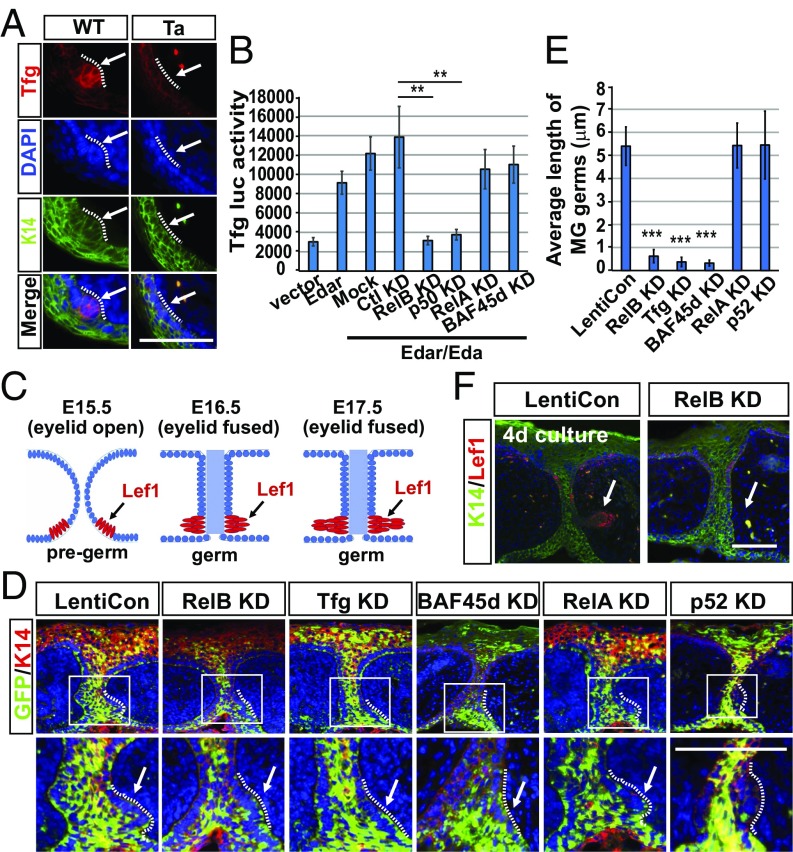
To clarify how Eda signaling induces the formation of the RelB–BAF complex, we first checked the expression of the linker protein Tfg in MGs at the initiation stage. Consistent with data in Fig. 2 A–C, IHC staining showed a specific increase of Tfg in WT MG pregerms, with some accumulation in nucleus, but the increase was blocked in Tabby mice (Fig. 4A). To further characterize the sequential molecular cascade, we used lentivirus encoding shRNA against NF-kBs, Tfg, and BAF45d, respectively. Knockdown (KD) efficacy was validated as more than 80% of each target protein down-regulated (SI Appendix, Fig. S6A). Promoter luciferase activity assays in Kera308 cells showed that Eda indeed activated Tfg transcription through p50 and RelB but independently of RelA and BAF45d (Fig. 4B). Thus, Tfg, induced by Eda-mediated RelB activation, then bridges RelB and BAF45d to form the Eda-induced BAF complex.
Fig. 4.
Eda-induced Tfg recruits the BAF complex and promotes MG growth. (A) IHC shows the staining of Tfg (red) and K14 (green) in WT eyelids at E15.5. (B) Luciferase activity of the Tfg promoter in Kera308 cells with indicated treatment. Data are mean ± SEM for triplicate samples. **P ≤ 0.01, Student’s t test. (C) A schematic showing the development of an MG germ (red, Lef1+) from E15.5 (eyelid open) to E17.5 (eyelid fused). (D) IHC staining of GFP (green) and K14 (red) in cultured WT eyelids transduced by lentivirus coding a GFP marker and scrambled shRNA (LentiCon) or shRNA against each indicated protein. Enlarged views of the areas marked by white rectangles are shown below each image. (E) Average length of MG germs in D. Error bars indicate mean ± SEM from at least 15 MGs of total three cultures. ***P ≤ 0.001, Student’s t test. (F) IHC of K14 (green) and Lef1 (red) in LentiCon- or RelB KD-treated eyelids cultured for 4 d. Arrows, MG germs; dotted lines separate MG germs and dermal cells. (Scale bars, 50 μm.)
RelB, Tfg, and BAF45d All Promote MG Growth During Development.
To test for the indispensability of the three linker proteins in skin appendage development, we used the MG growth model previously described (25). A schematic shows the MG development from E15.5 to E17.5 (Fig. 4C). We then used lentivirus and transfected the shRNAs into cultured eyelids and examined MG morphology. Compared with control shRNA (LentiCon), RelB KD arrested MG growth, leaving pregerm morphology (Fig. 4D), the same phenotype seen in Tabby mice (25). Furthermore, the same repression of MG germs was observed with KD of Tfg or BAF45d (Fig. 4D). The defect was highly significant: MGs transfected with LentiCon showed an average length of 5.6 μm, whereas the length was reduced to 1.5 μm by RelB KD and to 0.6–0.8 μm by Tfg or BAF45d KD (Fig. 4E). As negative controls, neither RelA KD nor p52 KD inhibited MG growth (Fig. 4 D and E). As confirmation, in 4-d cultures, Lef1+ MG germs invaded deeply into dermis, but RelB KD inhibited further MG progression (Fig. 4F). We thus conclude that each of the triad of linked proteins is required for MG germ growth during early development.
Genome-Wide Screen Identifies Gene Targets Regulated by Both Eda Signaling and BAF Complex.
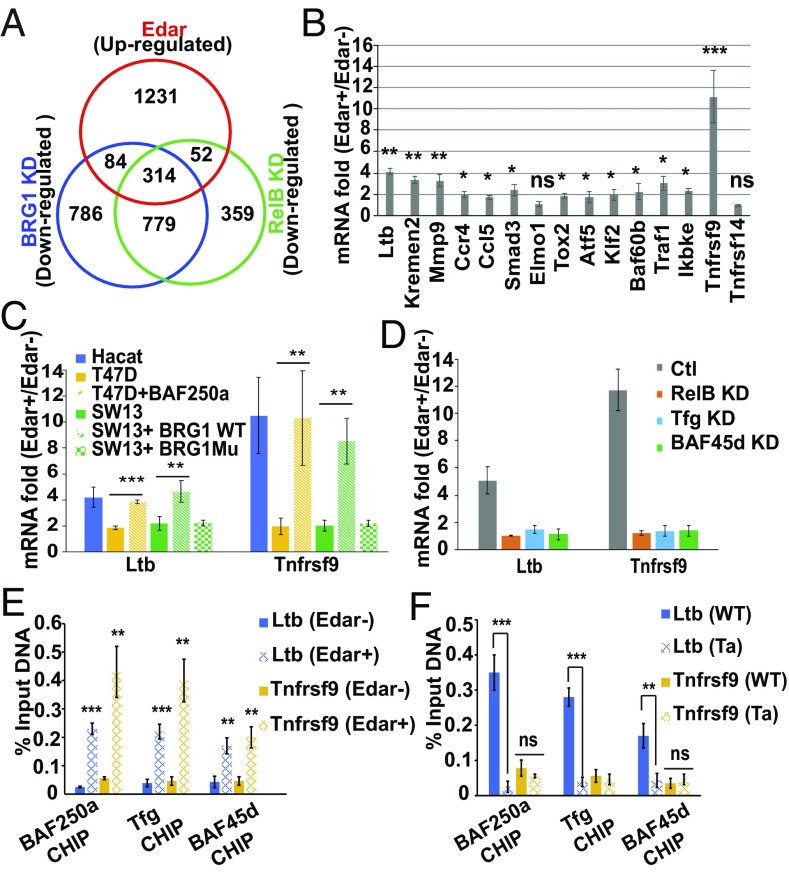
To identify Eda targets regulated by both RelB and BAF, we profiled gene expression in HaCaT cells exposed to Eda-A1 CM. Three comparisons were carried out: (i) Edar− cells vs. Edar+ cells; (ii) Edar+ cells with control siRNA vs. Edar+ cells with RelB siRNA; and (iii) Edar+ cells with control siRNA vs. Edar+ cells with BRG1 siRNA. The efficacy of siRNAs was verified in HaCat cells (SI Appendix, Fig. S6B). A large group of 1,681 genes was significantly up-regulated by Eda/Edar signaling. Among these genes, 314 were also notably down-regulated by both RelB KD and BRG1 KD, making them stronger candidates for up-regulation by Eda-induced BAF (Fig. 5A). The 314 genes included the known Eda targets Ltb, Kremen2, and Mmp9 (29, 30). Consistent with recent findings that Eda may regulate the chemokine pathway and cell migration (31, 32), we also observed some gene targets implicated in chemokine/cell motility pathways (SI Appendix, Table S2). Interestingly, BAF60b, some TFs, and genes in the TNF pathway were also observed (SI Appendix, Table S2), hinting at more possible components regulated by Eda-induced BAF complex. qPCR confirmed the expression levels of most genes selected as markedly up-regulated by Eda/Edar signaling. Among them, Ltb and Tnfrsf9 were the most strikingly augmented, by 4.1-fold and 11.1-fold, respectively (Fig. 5B). Again, these Eda targets identified in cell culture were also verified in eyelids from WT and Tabby mice (SI Appendix, Fig. S7G).
Fig. 5.
Identification of gene targets regulated by both the BAF complex and Eda signaling. (A) Venn diagram showing the number of genes significantly up-regulated by Eda/Edar stimulation (red), down-regulated by BRG1 KD (blue), or down-regulated by RelB KD (green). (B) qPCR assays show the mRNA fold (Edar+/Edar−) of selected genes. (C) qPCR assays show the levels of Ltb and Tnfrsf9 in cell lines with or without indicated vector transfection. BRG1Mu, mutated Brg1. (D) qPCR assays show the levels of Ltb and Tnfrsf9 in HaCaT cells transfected with scrambled siRNA (Ctl) or siRNA (KD) against RelB, Tfg, or BAF45d, respectively. (E) CHIP-qPCR assays show the Ltb or Tnfrsf9 promoter-binding activity of BAF250a, Tfg, and BAF45d. Chromatin from Edar− or Edar+ cells treated with Eda CM was used. (F) As described in E, except that chromatin from eyelids of E16.5 WT or Tabby mice were used. Two percent chromatin DNA was used as input. Error bars indicate mean ± SEM for triplicates. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 by Student’s t test; ns, no significant change.
To further determine the involvement of BAF complex in the regulation of these Eda targets, we used T47D cells (BAF250a-deficient mammary gland cells) and SW13 cells (BRG1-deficeint adrenal gland cells). qPCR showed that the mRNA levels of Ltb and Tnfrsf9 did not change in T47D or SW13 cells with Eda/Edar stimulation but were sharply stimulated in T47D cells transfected with BAF250a vector or in SW13 cells transfected with BRG1 WT vector but not BRG1 Mu (an ATPase-inactive mutant version) (Fig. 5C and SI Appendix, Fig. S6C). Consistent with the action of BAF250a and BRG1, KD of RelB, Tfg, or BAF45d all blocked Eda-induced up-regulation of Ltb and Tnfrsf9 (Fig. 5D).
Eda-Induced BAF Components Bind to the Promoter Regions of Eda Targets.
To test whether RelB, Tfg, and BAF proteins bind to the promoter region of Eda targets, we analyzed the core promoter regions of Ltb and Tnfrsf9 across species using ConTra v2 software (SI Appendix, Fig. S7 A and D). Using CHIP-qPCR, we confirmed that RelB bound to the promoter region of Ltb and Tnfrsf9 in Edar+ cells and that Eda augmented this binding activity. As expected, RelB bound to the Ltb promoter only in WT skin tissue but not to the Tnfrsf9 promoter in mouse skin tissues, due to the lack of the NF-κB–binding site in mouse (SI Appendix, Fig. S7 B, C, E, and F).
Similar to RelB, the other Eda-induced BAF components BAF250a, Tfg, and BAF45d all bound to both Ltb and Tnfrsf9 promoters stimulated by Eda/Edar treatment in human HaCat cells (Fig. 5E). Again, all three bound to the Ltb promoter only in WT but not in Tabby mice, and none bound to the Tnfrsf9 promoter in mouse tissue (Fig. 5F). The data thus suggest that, for gene regulation, Eda-activated RelB can recruit the BAF complex to gene loci containing NF-κB binding sites.
Eda-Induced BAF Complex Increases Chromatin Accessibility of Eda Targets.
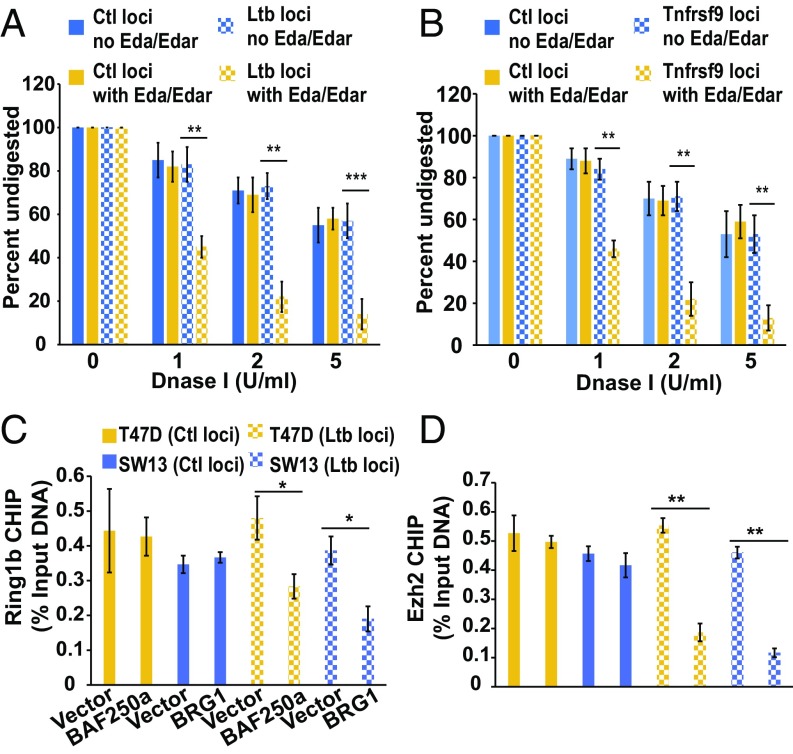
To test whether the Eda-triggered RelB–Tfg–BAF protein complex indeed modulates the chromatin structure of target genes, we evaluated the chromatin status of the distal control (Ctl) loci and proximal promoter loci of Ltb and Tnfrsf9 (SI Appendix, Fig. S7 B and E). In HaCaT cells treated with Eda/Edar, the DNase I sensitivity of both the Ltb and Tnfrsf9 loci, but not of each Ctl locus, was notably increased in Edar+ compared with Edar− cells, suggesting that a more “open” chromatin state is induced by Eda signaling (Fig. 6 A and B). Next, we chose Ltb loci for the following study in several conditions. RelB KD, as expected, blocked the increase of DNase I sensitivity in Edar+ cells (SI Appendix, Fig. S8A). We also checked the Ltb chromatin status in T47D and SW13 cells. Consistent with data in Fig. 5C, the Ltb region of chromatin remained “closed,” as indicated by low DNase I sensitivity, in the both T47D and SW13 cells, irrespective of Eda/Edar stimulus (SI Appendix, Fig. S8 B and D). Reexpression of BAF250a in T47D cells or of WT BRG1 (but not mutated BRG1) in T47D cells restored DNase I sensitivity of the Ltb promoter in response to Eda (SI Appendix, Fig. S8 C, E, and F). Thus, Eda-induced expression of Ltb requires both BAF250a and BRG1 for chromatin remodeling.
Fig. 6.
Both Eda signaling and the BAF complex contribute to chromatin remodeling of Eda target genes. (A) Chromatin was extracted from Edar− or Edar+ cells treated with Eda CM, and then digested with DNase I (from 0 to 5 U/mL). Undigested DNA was amplified by qPCR with primers targeting Ltb loci. A distal Ctl locus was an inner control. Signals for DNase-untreated chromatin are normalized to 100%. (B) As in A, except primers targeting Ctl and Tnfrsf9 loci were used. (C) CHIP-qPCR assays show the binding activity of Ring1b in Ltb and Tnfrsf9 loci in the indicated cells and treatments. (D) As in C, except that Ezh2 CHIP was carried out. Error bars indicate mean ± SEM for triplicates. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 by Student’s t test.
To further confirm the function of the Eda-induced BAF complex in gene regulation of Ltb, we performed CHIP-qPCR assays using antibodies against H3K27me3 (a marker of inactive genes) and H3K27ac (a marker of active genes). Indeed, anti-H3K27me3 bound to the Ltb promoter in HaCaT cells without Eda/Edar stimulation; by contrast, anti-H3K27ac bound to the Ltb promoter after Eda/Edar stimulus (SI Appendix, Fig. S8G). Using anti-H3K27ac CHIP-qPCR assays, we confirmed the requirement for BAF250a and BRG1 as well as for RelB, Tfg, and BAF45d for Eda-mediated Ltb transcription (Fig. 6C and SI Appendix, Fig. S8 H and I). Consistent with recent findings of the BAF-mediated polycomb repressive complex (PRC) eviction mechanism (33, 34), Ring1b and Ezh2 CHIP assays showed that the activation of two Eda targets, Ltb and Tnfrsf9, involved the removal of both the PRC1 and PRC2 complexes (Fig. 6 C and D).
Discussion
How cells and developing tissues select genes for expression in a precise spatiotemporal manner remains largely unknown. In the example of Eda signaling, Eda activates NF-κB for downstream gene expression (7). However, the powerful NF-κB can be activated by many stimuli and may in turn regulate many hundreds of genes (8). One source of specificity in the Eda paradigm was determined early on. The action of Eda in initiating skin appendage growth is restricted to cells that express its receptor Edar and its adaptor protein Edaradd (6). This study focuses on the steps at which specific gene activation is implemented in the regulatory cascade. The results delineate three features: selective activation of NF-κB subunits, transcriptional induction of a linker protein, and recruitment of a chromatin-remodeling complex to NF-κB–binding loci to facilitate gene transcription (SI Appendix, Fig. S9).
The expression pattern of NF-κB subunits in normal epidermis has been reported (35), but which subunits are involved in Eda signaling was unclear. Here, we find that Eda signaling slightly activated RelA and c-Rel, which may have specific transcription targets, but predominant activation of RelB was seen (Fig. 2). Consistently, down-regulation of RelB arrested MG growth (Fig. 4D), producing a phenotype similar to that of the Eda-ablated Tabby mouse.
We stared from unbiased protein purification and found that Eda induced an NF-κB–associated BAF complex, which comprises most, if not all, BAF components and in addition contains the p50/RelB dimer and a linker protein, Tfg (Fig. 1). Thus, tissue-specific Eda signaling may co-opt the chromatin remodeler to regulate organ development. In support of that possibility, the BAF component BAF250a was up-regulated in developing skin appendages, and skin-specific KO of BAF250a led to the malformation of HFs and arrested MG growth (Fig. 3); again, the BAF-mediated process downstream of Eda is consistent with the striking similarity of these phenotypes to those seen in Eda-deficient mice.
Using unbiased expression profiling, we identified several genes activated by Eda, RelB, and the BAF core ATPase BRG1 (Fig. 5 A and B). Some known Eda targets, including Ltb, Kremen2, and Mmp9, as well as previously unidentified candidate targets, were activated. Interestingly, the most discriminated gene identified from our human cell model was Tnfrsf9, which had never appeared as an Eda target in several mouse models. Tnfrsf9 had been previously implicated in NF-κB signaling and function in T cell development (36), and its possible specific function in human skin appendage development remains to be investigated.
In the context of Eda- and BAF complex-coordinated gene regulation (SI Appendix, Fig. S9), there appear to be two successive phases of gene transcription: a first phase in which RelB and Tfg are up-regulated and a second phase in which the RelB recruits BAF complex through Tfg and activates subsequent NF-κB targets. Our study has identified a critical protein, Tfg, which links the two phases, and we have further focused on BAF complex-mediated NF-κB target activation. It remains unknown how other epigenetic mechanisms are dynamically controlled. However, in a larger context, our results are in line with convergent thinking that reciprocal modulation between NF-κB and chromatin organization may regulate gene expression at the right times and places (37). Given the critical roles of both NF-κB and BAF complexes in cancer development (38, 39), our study may also provide a model for the mechanism of gene expression coordinated by NF-κB and BAF complexes in tumorigenesis.
Materials and Methods
All animal study protocols were approved by the National Institute on Aging Animal Care and Use Committee. The materials and methods used in this study, including cell culture, molecular and biochemical assays, histology, mice, and organotypic culture are described in detail in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Marc Michel and Ienglam Lei for technical help on mouse handling and Guobing Chen for critical reading. This work was supported by the Intramural Research Program of the National Institute on Aging, NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE97783).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800930115/-/DCSupplemental.
References
- 1.Emerson BM. Specificity of gene regulation. Cell. 2002;109:267–270. doi: 10.1016/s0092-8674(02)00740-7. [DOI] [PubMed] [Google Scholar]
- 2.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 3.Suganuma T, Workman JL. Chromatin and signaling. Curr Opin Cell Biol. 2013;25:322–326. doi: 10.1016/j.ceb.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Mikkola ML. Genetic basis of skin appendage development. Semin Cell Dev Biol. 2007;18:225–236. doi: 10.1016/j.semcdb.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Darwin C, Gray A. The Variation of Animals and Plants Under Domestication. Orange Judd & Co.; New York: 1868. [Google Scholar]
- 6.Cui CY, Schlessinger D. EDA signaling and skin appendage development. Cell Cycle. 2006;5:2477–2483. doi: 10.4161/cc.5.21.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikkola ML. Molecular aspects of hypohidrotic ectodermal dysplasia. Am J Med Genet A. 2009;149A:2031–2036. doi: 10.1002/ajmg.a.32855. [DOI] [PubMed] [Google Scholar]
- 8.Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 9.Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 10.Kasten MM, Clapier CR, Cairns BR. SnapShot: Chromatin remodeling: SWI/SNF. Cell. 2011;144:310.e1. doi: 10.1016/j.cell.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 14.Xue Y, et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci USA. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan Z, et al. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu JI, et al. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 18.Buscarlet M, et al. Essential role of BRG, the ATPase subunit of BAF chromatin remodeling complexes, in leukemia maintenance. Blood. 2014;123:1720–1728. doi: 10.1182/blood-2013-02-483495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan Z, et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mustonen T, et al. Ectodysplasin A1 promotes placodal cell fate during early morphogenesis of ectodermal appendages. Development. 2004;131:4907–4919. doi: 10.1242/dev.01377. [DOI] [PubMed] [Google Scholar]
- 21.Miranda C, et al. The TFG protein, involved in oncogenic rearrangements, interacts with TANK and NEMO, two proteins involved in the NF-kappaB pathway. J Cell Physiol. 2006;208:154–160. doi: 10.1002/jcp.20644. [DOI] [PubMed] [Google Scholar]
- 22.Yan M, et al. Two-amino acid molecular switch in an epithelial morphogen that regulates binding to two distinct receptors. Science. 2000;290:523–527. doi: 10.1126/science.290.5491.523. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt-Ullrich R, et al. Requirement of NF-kappaB/Rel for the development of hair follicles and other epidermal appendices. Development. 2001;128:3843–3853. doi: 10.1242/dev.128.19.3843. [DOI] [PubMed] [Google Scholar]
- 24.Kumar A, Eby MT, Sinha S, Jasmin A, Chaudhary PM. The ectodermal dysplasia receptor activates the nuclear factor-kappaB, JNK, and cell death pathways and binds to ectodysplasin A. J Biol Chem. 2001;276:2668–2677. doi: 10.1074/jbc.M008356200. [DOI] [PubMed] [Google Scholar]
- 25.Sima J, Piao Y, Chen Y, Schlessinger D. Molecular dynamics of Dkk4 modulates Wnt action and regulates meibomian gland development. Development. 2016;143:4723–4735. doi: 10.1242/dev.143909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei I, et al. BAF250a protein regulates nucleosome occupancy and histone modifications in priming embryonic stem cell differentiation. J Biol Chem. 2015;290:19343–19352. doi: 10.1074/jbc.M115.637389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui CY, et al. Inducible mEDA-A1 transgene mediates sebaceous gland hyperplasia and differential formation of two types of mouse hair follicles. Hum Mol Genet. 2003;12:2931–2940. doi: 10.1093/hmg/ddg325. [DOI] [PubMed] [Google Scholar]
- 28.Vielkind U, Hardy MH. Changing patterns of cell adhesion molecules during mouse pelage hair follicle development. 2. Follicle morphogenesis in the hair mutants, Tabby and downy. Acta Anat (Basel) 1996;157:183–194. doi: 10.1159/000147880. [DOI] [PubMed] [Google Scholar]
- 29.Voutilainen M, et al. Ectodysplasin/NF-κB promotes mammary cell fate via Wnt/β-catenin pathway. PLoS Genet. 2015;11:e1005676. doi: 10.1371/journal.pgen.1005676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui C, et al. Ectodysplasin regulates lymphotoxin, Wnt, BMP and SHH pathways for hair follicle formation. J Invest Dermatol. 2006;126:96. [Google Scholar]
- 31.Ahtiainen L, et al. Directional cell migration, but not proliferation, drives hair placode morphogenesis. Dev Cell. 2014;28:588–602. doi: 10.1016/j.devcel.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Lefebvre S, Fliniaux I, Schneider P, Mikkola ML. Identification of ectodysplasin target genes reveals the involvement of chemokines in hair development. J Invest Dermatol. 2012;132:1094–1102. doi: 10.1038/jid.2011.453. [DOI] [PubMed] [Google Scholar]
- 33.Stanton BZ, et al. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet. 2017;49:282–288. doi: 10.1038/ng.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadoch C, et al. Dynamics of BAF-polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet. 2017;49:213–222. doi: 10.1038/ng.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takao J, et al. Expression of NF-kappaB in epidermis and the relationship between NF-kappaB activation and inhibition of keratinocyte growth. Br J Dermatol. 2003;148:680–688. doi: 10.1046/j.1365-2133.2003.05285.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang C, Lin GHY, McPherson AJ, Watts TH. Immune regulation by 4-1BB and 4-1BBL: Complexities and challenges. Immunol Rev. 2009;229:192–215. doi: 10.1111/j.1600-065X.2009.00765.x. [DOI] [PubMed] [Google Scholar]
- 37.Natoli G. Control of NF-kappaB-dependent transcriptional responses by chromatin organization. Cold Spring Harb Perspect Biol. 2009 doi: 10.1101/cshperspect.a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia Y, Shen S, Verma IM. NF-κB, an active player in human cancers. Cancer Immunol Res. 2014;2:823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.