Significance
Deciphering the mechanisms of how cells restrict viral pathogens is imperative for understanding disease and seeding new therapies. Millions of people suffer with liver disease as a result of chronic infection with hepatitis C virus (HCV). MicroRNA-122, a target of phase II clinical trials, is a microRNA that is beneficial to HCV, in part by binding to viral transcripts and protecting them from XRN exonucleases. As XRNs are specific to 5′ monophosphate transcripts, how XRNs restrict the 5′ triphosphate products of the viral polymerase is unknown. Here, we reveal that the 5′ RNA triphosphatase DUSP11 colludes with XRNs to restrict HCV. These findings implicate DUSP11 as a component of HCV restriction relevant for understanding an emerging class of therapeutics.
Keywords: microRNA, miR-122, restriction factor
Abstract
Seventy percent of people infected with hepatitis C virus (HCV) will suffer chronic infection, putting them at risk for liver disease, including hepatocellular carcinoma. The full range of mechanisms that render some people more susceptible to chronic infection and liver disease is still being elucidated. XRN exonucleases can restrict HCV replication and may help to resolve HCV infections. However, it is unknown how 5′ triphosphorylated HCV transcripts, primary products of the viral polymerase, become susceptible to attack by 5′ monophosphate-specific XRNs. Here, we show that the 5′ RNA triphosphatase DUSP11 acts on HCV transcripts, rendering them susceptible to XRN-mediated attack. Cells lacking DUSP11 show substantially enhanced HCV replication, and this effect is diminished when XRN expression is reduced. MicroRNA-122 (miR-122), a target of current phase II anti-HCV drugs, is known to protect HCV transcripts against XRNs. We show that HCV replication is less dependent on miR-122 in cells lacking DUSP11. Combined, these results implicate DUSP11 as an important component of XRN-mediated restriction of HCV.
Of the tens of millions of people currently infected with hepatitis C virus (HCV), ∼70% will go on to chronic infection, a major cause of cirrhosis and hepatocellular carcinoma. There is limited understanding of why some people naturally clear the virus or of the factors that dictate the outcome to disease progression. There are no licensed vaccines, but recently available drugs can lead to clearance of the virus. However, patients with advanced liver disease remain at increased risk for developing subsequent disease even after HCV clearance (1). Variations in host factors have been associated with both HCV clearance and disease progression (2–8). Defining the mechanisms of HCV restriction by host factors is imperative to optimizing therapeutic strategies and understanding variations in disease outcome.
5′ exonucleases, such as the XRN enzymes, are an emerging class of viral restriction factors (9). Both XRN1 and XRN2 have been implicated as potential HCV restriction factors by attacking viral transcripts (10–12). The abundant liver microRNA (miRNA) miR-122 promotes HCV infection, in part by binding directly to the 5′ end of HCV transcripts and preventing XRN-mediated degradation (10, 11, 13–15). As such, molecules that block miR-122 are currently in phase II clinical trials for treatment of chronic HCV hepatitis (16). Despite this, it is unclear how miR-122 protects against XRN and how XRN enzymes restrict HCV, since XRN activity is specific to 5′ monophosphates and HCV transcripts are a product of a viral RNA-dependent RNA polymerase (RdRP) that leaves a 5′ triphosphate (10). It is important to decipher the activities of XRN and miR-122 on HCV, as this may relate to understanding drug effectiveness and viral escape from anti–miR-122 drugs.
Recently, we demonstrated that the RNA triphosphatase DUSP11 is active on diverse viral small RNAs (17). The result of this activity is the loading of noncanonical retroviral and adenoviral miRNAs into the RNA silencing machinery. Here, we asked whether DUSP11 is also active on longer viral RNAs produced by HCV. Our findings reveal DUSP11 acts on HCV transcripts and promotes XRN restriction of HCV infection. The relevance of these findings and how they relate to miR-122 activity and HCV-associated disease are discussed.
Results
Reducing DUSP11 Levels Enhances HCV Replicon Expression.
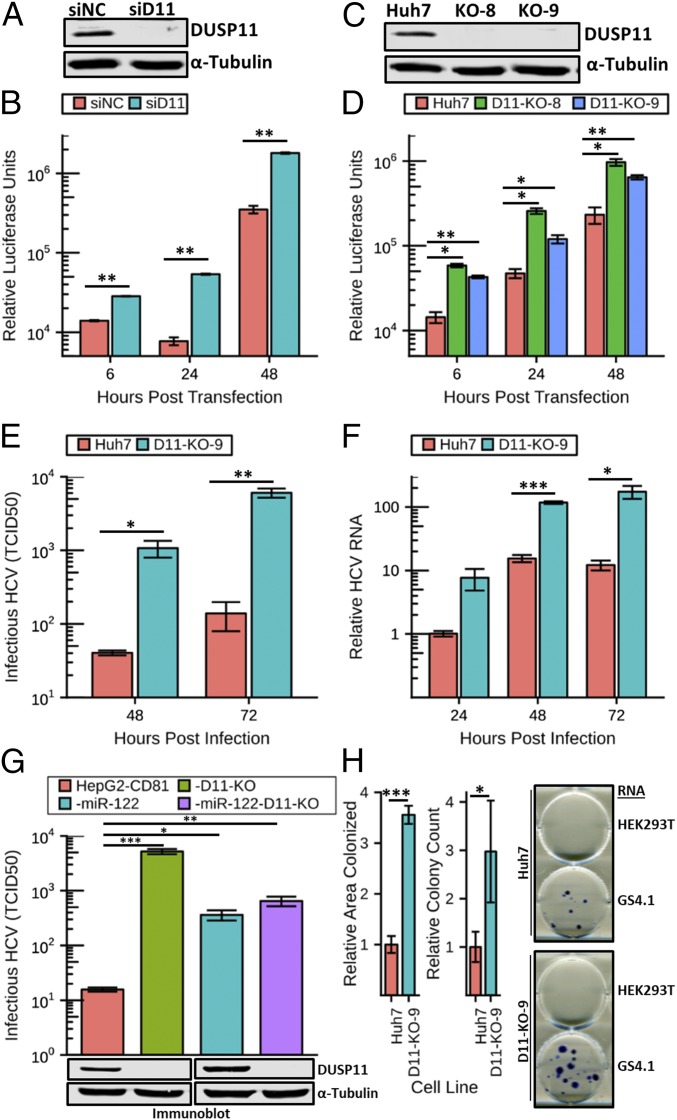
Several studies have established that XRN enzymes restrict HCV replication (10, 11), but how the 5′ triphosphate HCV transcripts are susceptible to the 5′ monophosphate-specific XRNs was unknown. One possibility that has been proposed is that an unknown RNA triphosphatase targets HCV transcripts (18, 19). We have previously reported that DUSP11 acts as a 5′ RNA triphosphatase on viral transcripts produced during bovine leukemia virus and human adenovirus infections, suggesting that DUSP11 could act on other viral transcripts as well (17, 20). We hypothesized that DUSP11 could convert the 5′ triphosphate end structure of HCV RNAs to a monophosphate and promote their turnover. To begin to test this hypothesis, we performed siRNA-mediated knockdown of DUSP11 in Huh7 cells and assayed HCV replication using the genotype 2a sgJFH1-Rluc replicon (SI Appendix, Fig. S1A) (21). We pretransfected Huh7 cells with siRNAs targeting DUSP11 or irrelevant negative-control siRNAs. Forty-eight hours later, we confirmed effective reduction of DUSP11 levels by immunoblot (Fig. 1A) and cotransfected sgJFH1-Rluc replicon RNA along with an additional dose of siRNAs. Luciferase assays performed at 6, 24, and 48 h post transfection (hpt) of replicon RNA demonstrated significantly increased luciferase expression (∼2.5- to 5.4-fold) in the cells treated with the DUSP11 siRNA versus an irrelevant control siRNA sequence (Fig. 1B). These results are consistent with DUSP11 having a restrictive effect on HCV replication.
Fig. 1.
Reducing DUSP11 levels enhances HCV replication and infection. (A) Confirmation of siRNA knockdown of DUSP11 in Huh7 cells by immunoblot. Huh7 were transfected with either an irrelevant control siRNA (siNC) or DUSP11-specific siRNA (siD11). Cell lysates were harvested 48 h post transfection and assayed by immunoblot with the indicated antibodies. (B) sgJFH1-Rluc replicon luciferase assay in Huh7 cells treated with siNC or siD11. Luciferase assays were performed at the indicated times post replicon RNA transfection. The mean ± SEM of three individual experiments is presented. (C) Confirmation of CRISPR/Cas9 disruption of DUSP11 in Huh7 cells by immunoblot. Lysates from parental Huh7 cells and two independent DUSP11 knockout clones (D11-KO-8 and D11-KO-9) were assayed by immunoblot with the indicated antibodies. (D) sgJFH1-Rluc replicon luciferase assay in parental Huh7 cells and two independent DUSP11 knockout clones (D11-KO-8 and D11-KO-9). Luciferase assays were performed at the indicated times post replicon RNA transfection. The mean ± SEM of three individual experiments is presented. (E) HCV cell culture (HCVcc) infectious virus production [multiplicity of infection (MOI) = 1] in Huh7 and D11-KO-9 cells. Data presented are the mean of three replicates ± SEM. (F) HCVcc RNA replication time course (MOI = 1) in Huh7 and D11-KO-9 cells. Data presented are the mean of three replicates ± SEM. (G) HCVcc infectious virus production (MOI = 10) in HepG2-CD81, HepG2-CD81-D11, CD81-miR-122, and HepG2-CD81-miR-122-D11. Data presented are the mean of three replicates ± SEM. (G, Lower) Immunoblot of lysates from the corresponding cell lines with the indicated antibodies. (H) GS4.1 replicon colony formation assays in parental Huh7 cells and DUSP11 knockout cells (D11-KO-9). Quantitation of both relative area colonized and relative colony counts with transfection of GS4.1 RNA is presented (Left). The mean ± SEM of five individual experiments is presented. Example wells with stained colonies from the indicated cell lines transfected with the indicated RNA are displayed (Right). Statistical significance was assessed by Student’s t test and is indicated as follows: *P < 0.05, **P < 0.01, ***P < 0.001.
To further confirm these findings, we assayed sgJFH1-Rluc replicon activity in cells lacking any detectable DUSP11. Our previous work demonstrated the generation of viable DUSP11 knockout (KO) cells using CRISPR/Cas9 technology (17). We applied this same approach to the Huh7 cell line that supports HCV replication. Individual clones lacking detectable DUSP11 were identified by immunoblot (Fig. 1C). Consistent with the above knockdown studies, in the DUSP11 KO clones compared with parental cells, transfection of HCV replicon sgJFH1-Rluc RNA gave rise to statistically significant increased luciferase activity at all time points assayed (6, 24, and 48 hpt) (Fig. 1D). The consistent phenotypes observed between the DUSP11 KO clones and the siRNA knockdown studies identify DUSP11 as an inhibitory factor of HCV replication.
The Absence of DUSP11 Promotes Enhanced HCV Infection.
We next determined if DUSP11 affects HCV infection. HCV infection of Huh7 DUSP11 KO cells gave rise to significantly enhanced secreted virus yield (∼26-fold at 48 h and ∼43-fold at 72 h) compared with parental Huh7 cells (Fig. 1E). Consistent with higher virus replication accounting for the increases in infectious virus yield, DUSP11 KO cells also displayed increased viral RNA (Fig. 1F). Immunoblot analysis revealed no overt differences in DUSP11 levels in HCV-infected Huh7 cells at 24 or 48 hpt (SI Appendix, Fig. S2). To determine if the absence of DUSP11 enhances HCV infection in a different hepatocyte cell line, we also generated DUSP11 knockout cells in the HepG2-CD81 (22) background (HepG2-CD81-DUSP11-KO), which have low miR-122 expression and thus have limited permissivity to HCV replication. The yields of infectious HCV in HepG2-CD81 cells lacking DUSP11 were substantially higher (∼331-fold) compared with the parental cells. Compared with HepG2-CD81 cells undergoing forced expression of miR-122, HepG2-CD81-miR-122–expressing DUSP11 KO cells showed no significant enhancement of HCV infection (Fig. 1G). This suggests that miR-122 and DUSP11 function antagonistically within the same pathway of HCV restriction (the role of DUSP11 in HCV miR-122 dependence is further addressed below). These results demonstrate that in independent cell lineages lacking DUSP11, HCV infection is enhanced.
Stable HCV Replicon Expression Is Enhanced in DUSP11 KO Cells.
HCV replicons that express a selectable antibiotic resistance protein can be used to measure stable replicon expression in cell culture as an independent assay for replicon replication. We hypothesized that since cells with lower DUSP11 levels have enhanced HCV replication, DUSP11 KO cells should also support longer-term replicon expression. To test this, we transfected Huh7 DUSP11 KO cells and parental Huh7 cells with either RNA from GS4.1 cells containing HCV genotype 1b-derived neomycin-resistant replicon RNA (23) or negative-control RNA from parental HEK293T (nonreplicative control RNA source) (SI Appendix, Fig. S1B). As the GS4.1 replicon is genotype 1b, this assay also affords us the ability to measure the activity of DUSP11 on a different genotype of HCV. We applied selection with G418 for 2 wk, fixed and stained the cells, and quantitated both the number of colonies and the relative area of each well that was colonized. Cells lacking DUSP11 displayed significantly higher colonized area (∼3.6-fold) and colony counts (∼3-fold) compared with the parental Huh7 cells (Fig. 1H and SI Appendix, Fig. S3). This result is consistent with DUSP11 having a restrictive effect on stable HCV replication in cell culture. Furthermore, these results demonstrate that the absence of DUSP11 provides a fitness advantage to multiple HCV genotypes.
The HCV Restriction Activity of XRNs Is Reduced in Cells Lacking DUSP11.
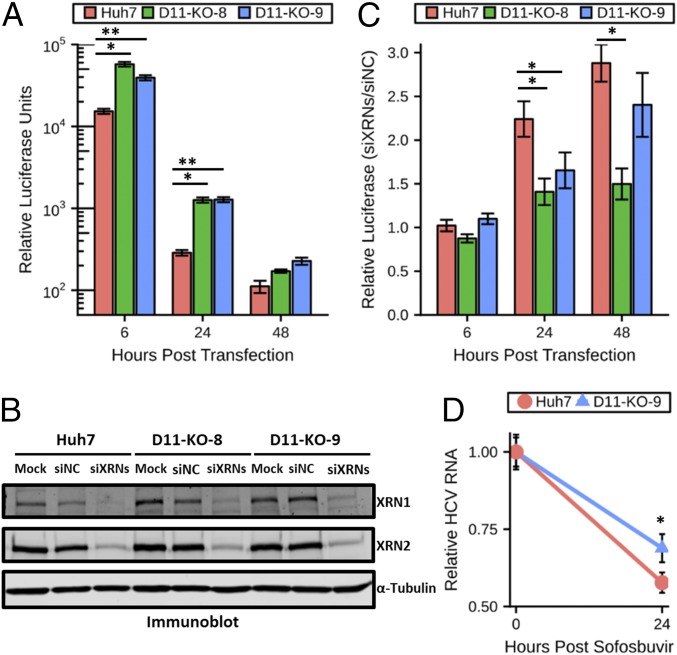
XRNs are 5′-to-3′ exonucleases that are specific to transcripts bearing a 5′ monophosphate (24). If a major reason DUSP11 KO cells are more permissive to HCV replication and infection is due to HCV transcripts being resistant to XRN-mediated exonucleolytic attack, then gene expression from triphosphorylated HCV transcripts should be enhanced irrespective of viral genome replication. Therefore, we tested if the advantage to HCV replicon expression in cells lacking DUSP11 depends on HCV replication. Huh7 DUSP11 KO cells transfected with sgJFH1-Rluc-GND, a mutant replicon incompetent for replication (21), displayed significantly enhanced luciferase activity at both 6 and 24 hpt (levels approached background for all cell lines at the 48-h time point) (Fig. 2A). These results establish that the advantage provided by reduced DUSP11 levels does not depend entirely on replication of the HCV genome.
Fig. 2.
HCV restriction activity of XRNs is reduced in cells lacking DUSP11. (A) sgJFH1-Rluc-GND (nonreplicative polymerase mutant) replicon luciferase assay in parental Huh7 cells and two independent DUSP11 knockout clones (D11-KO-8 and D11-KO-9). Luciferase assays were performed at the indicated times post replicon RNA transfection. The mean ± SEM of three individual experiments is presented. (B) Confirmation of siRNA knockdown of XRN1 and XRN2 in parental Huh7 cells and two independent DUSP11 knockout clones (D11-KO-8 and D11-KO-9) by immunoblot. Huh7 were either mock transfected (Mock) or transfected with either an irrelevant control siRNA or a pool of XRN1- and XRN2-specific siRNAs (siXRNs). Cell lysates were harvested 48 h post transfection and assayed by immunoblot with the indicated antibodies. (C) sgJFH1-Rluc replicon luciferase assay in parental Huh7 cells and two independent DUSP11 knockout clones (D11-KO-8 and D11-KO-9) treated with siNC or a pool of siXRNs. Luciferase assays were performed at the indicated times post replicon RNA transfection. The mean ± SEM of three experiments is presented. (D) Huh7 and D11-KO-9 cells were infected with HCV for 96 h and treated with 5 μM sofosbuvir, and HCV RNA levels were quantified at the indicated times post treatment. The mean ± SEM of eight samples is presented. Statistical significance was assessed by Student’s t test and is indicated as follows: *P < 0.05, **P < 0.01.
We hypothesized that if DUSP11 sensitizes HCV RNAs to XRN-mediated degradation, then knockdown of XRNs should have a diminished effect in DUSP11 KO cells. To test this hypothesis, we pretransfected Huh7 DUSP11 KO cells and parental Huh7 cells with siRNAs targeting both XRNs 1 and 2 or irrelevant negative-control siRNAs. Forty-eight hours later, we confirmed knockdown of XRNs 1/2 by immunoblot (Fig. 2B) and cotransfected sgJFH1-Rluc replicon RNA along with an additional dose of siRNAs. We then performed luciferase assays at 6, 24, and 48 hpt. At both the 24- and 48-h time points, we observed larger relative increases in luciferase expression (2.24-fold vs. 1.41 and 1.65 at 24 hpt; 2.88-fold vs. 1.50 and 2.40 at 48 hpt) from the parental cells treated with the XRN siRNAs versus the DUSP11 KO cells treated with the XRN siRNAs (Fig. 2C and SI Appendix, Fig. S4). We also tested whether HCV RNA decay is reduced in infected DUSP11 KO cells. We treated infected cells with sofosbuvir (NS5B polymerase inhibitor) to attenuate new viral RNA production. Under these conditions, a moderate decrease in HCV RNA decay was observed in the Huh7 DUSP11 KO cells, consistent with a role for DUSP11 in HCV RNA degradation pathways during infection (Fig. 2D). Combined, these results strongly suggest that DUSP11 sensitizes HCV transcripts to XRN-mediated turnover.
HCV Genome Replication Is Less Dependent on miR-122 in Cells Lacking DUSP11.
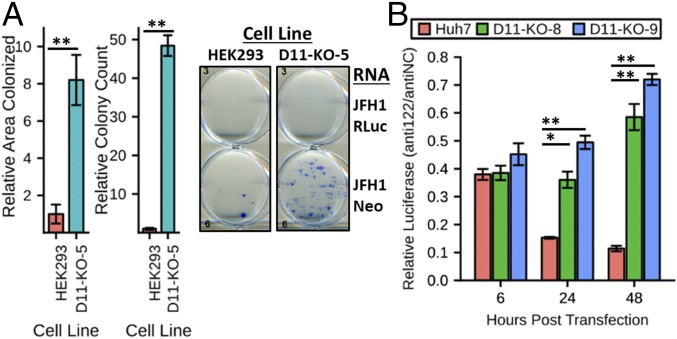
HCV replication is highly dependent on miR-122, and this partly accounts for the liver tropic replication of the virus (25). To test whether knockout of DUSP11 could enhance replicon activity in the absence of miR-122, we performed HCV replicon colony assays in the background of nonliver human embryonic kidney (HEK293) cells [which lack miR-122 expression (26, 27)] with in vitro transcribed sgJFH1-Neo (28) replicon RNA (Fig. 3A and SI Appendix, Figs. S1C and S5). We observed significantly higher colonized area (∼8.2-fold) and colony counts (∼48-fold) in the DUSP11 KO cells. These results are consistent with our infection data of HepG2 cells that differentially express miR-122 (Fig. 1G), and suggest that the absence of DUSP11 relaxes the dependence of HCV replication on miR-122.
Fig. 3.
HCV genome replication is less dependent on miR-122 in cells lacking DUSP11. (A) sgJFH1-Neo replicon colony formation assays in parental HEK293 cells and DUSP11 knockout cells (D11-KO-5). Quantitation of both the relative area colonized and relative colony counts with transfection of sgJFH1-Neo RNA is presented (Left). The mean ± SEM of three individual experiments is presented. Example wells with stained colonies from the indicated cell lines transfected with the indicated RNA are displayed (Right). (B) sgJFH1-Rluc replicon luciferase assay in parental Huh7 cells and two independent DUSP11 knockout clones (D11-KO-8 and D11-KO-9) treated with irrelevant control anti-miR (antiNC) or miR-122–specific anti-miR (anti122). Luciferase assays were performed at the indicated times post replicon RNA transfection. The mean of three individual experiments ± SEM is presented. Statistical significance was assessed by Student’s t test and is indicated as follows: *P < 0.05, **P < 0.01.
To test if the absence of DUSP11 reduces the dependence of HCV genome replication on miR-122, we examined sgJFH1-Rluc replicon activity in cells in which miR-122 was specifically inhibited. Huh7 DUSP11 KO cells or parental Huh7 cells were transfected with either an miR-122 anti-miR or an irrelevant negative-control anti-miR. Forty-eight hours later, we cotransfected sgJFH1-Rluc replicon RNA along with an additional dose of anti-miRs. Luciferase assays performed at multiple times post transfection revealed significantly larger decreases in replicon activity in the miR-122 anti-miR–treated parental cells versus the similarly treated DUSP11 KO cells (0.15 vs. 0.36 and 0.49 at 24 hpt; 0.11 vs. 0.59 and 0.72 at 48 hpt; for parental vs. D11-KO-8 and D11-KO-9) (Fig. 3B and SI Appendix, Fig. S6). These results strongly suggest that miR-122 protects HCV transcripts from XRN attack, which is enabled by the activity of DUSP11.
DUSP11 Acts Directly on the 5′ End of HCV Transcripts and Renders Them Susceptible to XRN-Mediated Cleavage.
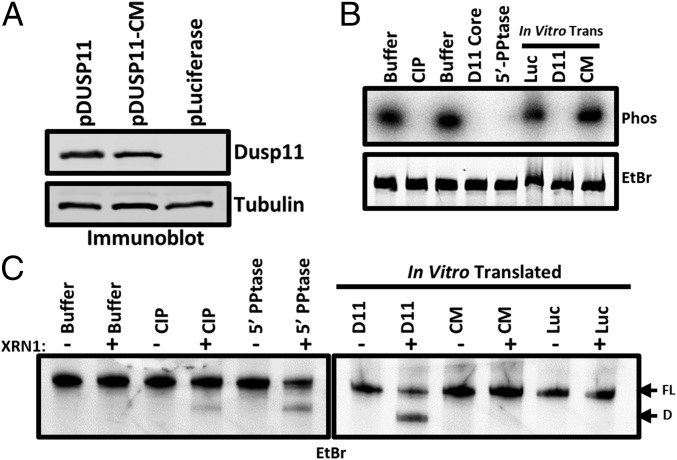
The above genetic and knockdown studies support the model that DUSP11 acts directly on HCV transcripts to render them susceptible to XRN-mediated cleavage. To directly test the activity of DUSP11 on the 5′ end of HCV RNAs, we in vitro transcribed the 5′ UTR of HCV type 1 (NC_004102) in the presence of [γ-32P]GTP to produce 5′ triphosphate end-labeled RNA. Incubation of this RNA with positive-control calf intestinal phosphatase (CIP), positive-control bacterial RNA 5′ polyphosphatase (5′-PPtase), purified DUSP11 core protein (DUSP11 core), or lysate enriched for full-length in vitro translated DUSP11 (D11) (Fig. 4A) all reduced the radioactive signal (Fig. 4B). However, incubation with lysate containing negative-control irrelevant in vitro translated luciferase protein (Luc) or lysate containing in vitro translated catalytically inactive mutant DUSP11 (CM) had little effect (Fig. 4B). These results demonstrate that the enzymatic activity of DUSP11 can act directly on the 5′ end of HCV RNA.
Fig. 4.
DUSP11 directly dephosphorylates HCV 5′ UTR RNA and sensitizes it to XRN-mediated degradation. (A) Confirmation of in vitro translation products by immunoblot. Membrane was incubated with the indicated antibodies. “pLuciferase” indicates reactions programmed to express luciferase as a negative control. (B) In vitro phosphatase assay. 5′ γ-32P–radiolabeled HCV 5′ UTR RNA was incubated with the indicated enzymes (calf intestinal phosphatase, purified DUSP11 core protein, and bacterial 5′ RNA polyphosphatase), or in vitro translated products from A (pDUSP11, pDUSP11-CM, and negative-control pLuciferase). Products were separated by urea/PAGE and stained with ethidium bromide. Products were then transferred to a nitrocellulose membrane and exposed to a phosphor storage screen (Phos). (C) In vitro XRN susceptibility assay. In vitro phosphatase reactions were performed as in B [calf intestinal phosphatase, bacterial 5′ RNA polyphosphatase, or in vitro translated products from A (pDUSP11, pDUSP11-CM, and negative-control pLuciferase)], but products were recovered and incubated ± recombinant XRN1. Products were separated by urea/PAGE and stained with EtBr. “FL” arrow points to the position of the full-length HCV 5′ UTR. “D” arrow points to the position of a faster-migrating degradation product.
To directly test if DUSP11 renders the 5′ end of HCV transcripts susceptible to XRNs, we performed in vitro phosphatase assays, recovered the RNA, and then incubated in reactions with or without recombinant XRN1. We separated the products on a polyacrylamide gel and visualized the RNA by ethidium bromide (EtBr) staining. These results indicated that DUSP11-containing lysate, but not lysate expressing catalytically inactive DUSP11, promoted the formation of XRN-mediated cleavage products (Fig. 4C). Notably, these cleavage products were nearly identical to what had previously been reported for XRN activity on 5′ monophosphate HCV 5′ UTRs (15, 29). Combined, these findings demonstrate that DUSP11 catalytic activity acts directly on the 5′ end of HCV transcripts, rendering them susceptible to XRN-mediated cleavage.
Discussion
The extraordinary role of miR-122 in promoting HCV infection stands as one of the most exciting findings in the field of viral–host miRNA interactions (13). Despite this, the exact protective activities of miR-122 are still being elucidated (25). Independent laboratories have converged on the model that one major proviral activity of miR-122 is binding directly to the 5′ end of the HCV genome and protecting against XRN-mediated degradation (18, 19). Exonucleases act as restriction factors against diverse RNA viruses (9). Current consensus indicates that XRN enzymes are restriction factors against HCV; however, the mechanism for how this occurs is unknown. Because the HCV RdRP generates 5′ triphosphate viral transcripts, there must be either an endonuclease or triphosphatase activity that serves along with the monophosphate-specific XRNs to effect restriction (18, 19). Here, we identify the RNA triphosphatase DUSP11 as being responsible for converting the 5′ end of HCV transcripts to monophosphates and enabling XRN-mediated restriction.
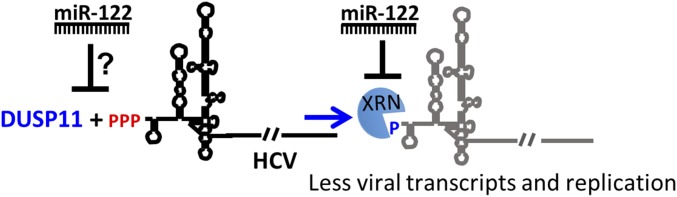
Our work reveals that cells lacking DUSP11 have enhanced HCV replication and virus infection (Fig. 1). The mechanism accounting for this is linked to enhanced abundance of viral transcripts (Fig. 1F). Consistent with this, even replication-defective replicons show increased gene expression in cells lacking DUSP11, suggesting enhanced RNA stability (Fig. 2A). Three independent lines of experimentation link DUSP11 activity to promoting XRN attack of HCV transcripts. First, jointly knocking down XRN1/2 promotes HCV replication in wild-type cells to a greater magnitude than in DUSP11 KO cells (Fig. 2B). Second, miR-122 is known to protect HCV transcripts against XRN, and the magnitude of HCV dependence on miR-122 is diminished in cells lacking DUSP11 (Figs. 1G and 3B). In fact, deleting DUSP11 expression can even promote low levels of HCV replicon replication in nonliver HEK293 cells that do not express any detectable miR-122 (26, 27) (Fig. 3A). Third, DUSP11 directly dephosphorylates the 5′ end of HCV transcripts, and these transcripts are resistant to XRN cleavage in in vitro assays unless treated with catalytically active DUSP11 (Fig. 4C). Combined, the above results support a model whereby DUSP11 forms an integral component of the XRN-mediated anti-HCV restriction pathway (Fig. 5).
Fig. 5.
Model: DUSP11 converts 5′ triphosphorylated HCV RNAs to 5′ monophosphate substrates for XRNs. This results in reduced HCV RNA levels and reduced virus yield.
Our findings raise several interesting questions. Previous in vitro biochemical studies established that miR-122 can directly block XRN activity against HCV transcripts in vitro (15). However, it remains possible in cells that RNA-induced silencing complex-bound miR-122 may also occlude DUSP11. Answering this question will be important for fully understanding the complete mechanisms of action of anti–miR-122 HCV drugs. Subgenomic flaviviral RNAs (sfRNAs) are known to result in “stalling” XRNs and inhibition of their function (29). It is not clear what the 5′ end structures of the RNAs that ultimately produce sfRNAs are, and it will be interesting to determine what role, if any, DUSP11 has in sfRNA production in the related flaviviruses. Finally, in addition to our previous work on small viral RNAs in retroviruses and adenoviruses, this study expands the known viral substrates of DUSP11 to larger viral RNAs. We have previously shown that structured host RNAs, some of which are known to be proinflammatory, are negatively modulated by DUSP11 (17). It therefore remains possible that DUSP11 activity may actually be proviral to those viruses whose transcripts are exceptionally sensitive to the triphosphatase-sensing RIG-I host defense response (30). Although HCV is sensed by RIG-I (31), which is functional in Huh7 and HepG2 cells, the antiviral activity associated with XRN exonucleases is dominant over any proviral RIG-I–associated activity. It will be interesting to further probe the repertoire of viruses and their transcripts that are affected by DUSP11.
In summary, we have identified DUSP11 as a component of XRN-mediated restriction of HCV. DUSP11 activity is linked to the magnitude of miR-122 pro-HCV activities and XRN anti-HCV activities. These findings suggest that DUSP11 is a previously unknown player in the restriction of HCV infection.
Materials and Methods
Cell Lines and Virus.
Huh7 cells were obtained from Charles Rice, Rockefeller University, New York, NY, while HepG2-CD81 and HepG2-CD81-miR-122 cells (22) were obtained from Matthew Evans, Icahn School of Medicine at Mount Sinai. HEK293 cells were obtained from ATCC. HEK293-D11-KO-5 cells have been previously described (17). They were maintained in DMEM (Cellgro) supplemented with FBS (10% vol/vol) (Life Technologies). GS4.1 cells were a gift from Christoph Seeger, Fox Chase Cancer Center, Philadelphia, PA, and maintained in DMEM supplemented with FBS (10% vol/vol) and G418 (500 μg/mL) (Sigma-Aldrich) (23). Infectious HCV was a genotype 2a J6-JFH1 chimera infectious clone pJFHxJ6-CNS2C3, Arash Grakoui, Emory University, Atlanta, GA, that was previously described (32).
Generation of DUSP11 Knockout Cell Lines and Rescue Cell Lines.
DUSP11 knockout cell lines were derived as previously described (17).
Plasmids.
pSG-JFH1-Rluc and pSG-JFH1-Rluc-GND plasmids were previously described (21). Plasmid pSG-JFH1-Rluc has been previously described (28). PISK-T7-HCV-5′UTR was constructed by cloning a synthetic gBlock (see SI Appendix, Table S1 for the sequence; HCV_5UTR_gBlock) (Integrated DNA Technologies) with the sequence of the 5′ UTR of HCV type 1 (NC_004102) into the XhoI and XbaI sites of a pIDT-SMART-kan vector (Integrated DNA Technologies).
Immunoblots.
Cells were lysed directly in Laemmli sample buffer (33) by boiling for 5 min. Equal volumes of lysates were size-fractionated by SDS/PAGE and transferred to nitrocellulose membranes (Bio-Rad). DUSP11 antibody (10204-2-AP) was purchased from Proteintech and used at a 1:2,000 dilution. α-Tubulin antibody (66031-1-Ig) was purchased from Proteintech and used at a dilution of 1:10,000. α-Tubulin antibody (T6199) was purchased from Sigma-Aldrich and used at a dilution of 1:10,000 (Fig. 4A only). XRN1 antibody (A300-443A) was purchased from Bethyl Laboratories and used at a dilution of 1:1,000. XRN2 antibody (H-300) (sc-99237) was obtained from Santa Cruz Biotechnology and used at a dilution of 1:1,000. IRDye 800CW and IRDye 680LT secondary antibodies were purchased from LI-COR and used at a 1:10,000 dilution. Membranes were imaged with an Odyssey CLx imaging system (LI-COR).
In Vitro Transcription of HCV Replicon RNA.
Preparative restriction enzyme digests containing 50 μg of either pSG-JFH1-Rluc, pSG-JFH1-Rluc-GND (21), or pSG-JFH1-Neo (28) plasmid and 10 μL XbaI (New England Biolabs) in 500 μL 1× CutSmart buffer were incubated for 30 min at 37 °C. The reactions were then incubated at 30 °C for 30 min with the addition of 5 μL mung bean nuclease (New England Biolabs). The linearized plasmid DNA was then purified by phenol/chloroform extraction. In vitro transcription was performed with the AmpliScribe T7-Flash Transcription Kit according to the manufacturer’s instructions (Epicentre).
Replicon Luciferase Assays.
Fifty percent confluent Huh7, Huh7-D11-KO-8, and Huh7-D11-KO-9 cells in 12-well plates were transfected in triplicate with 100 ng of the sgJFH1-Rluc or sgJFH1-Rluc-GND in vitro transcribed RNA using Lipofectamine 2000 reagent (Thermo Fisher Scientific). Luciferase assays were then performed at the indicated times post replicon RNA transfection using the Dual-Luciferase Reporter Assay System (Promega) and measured with a GloMax 96 microplate luminometer (Promega). Reported values were relative mean-normalized and derived from three independent experiments with each condition assayed in triplicate.
DUSP11 siRNA Replicon Luciferase Assays.
Silencer Select Negative Control 1 siRNA (4390843) and DUSP11 siRNA (AM16708; ID 105842) were obtained from Ambion. T25 flasks of 50% confluent Huh7 cells were transfected with either 100 pmol of the negative-control or DUSP11 siRNA using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific). Twenty-four hours later, cells were replated and seeded in triplicate at a density of 105 cells per well in 24-well plates. Approximately 24 h later, cells were cotransfected with 100 ng of the in vitro transcribed sgJFH1-Rluc RNA and either 5 pmol of negative-control or DUSP11 siRNA using Lipofectamine 2000 reagent (Thermo Fisher Scientific). Luciferase assays were then performed at the indicated times post replicon RNA transfection using the Dual-Luciferase Reporter Assay System (Promega) and measured with a GloMax 96 microplate luminometer (Promega). Reported values are derived from three independent experiments with each condition assayed in triplicate.
HCV Growth Curves.
RNA was extracted from infected cells using the RNeasy 96 Kit (Qiagen). RNA copy number was determined via quantitative real-time RT-PCR as previously described (34). Relative HCV copy numbers were calculated by the ΔΔCt method and normalized to the 24-h HCV-infected Huh7 value. Infectious HCV tissue culture infectious dose 50% was quantified by titrating infection of Huh7.5 cells and NS5A (9E10; provided by Charles Rice) immunohistochemistry as previously described (35).
HCV RNA Decay.
Huh7 and D11-KO-9 cells were infected with HCV for 96 h and treated with 5 μM sofosbuvir, and HCV RNA levels were quantified as above at the indicated times post treatment. The mean ± SEM of eight samples is presented. Statistical significance was assessed by Student’s t test.
XRN1/2 siRNA Replicon Luciferase Assays.
Silencer Select Negative Control 1 siRNA (4390843), Silencer Select XRN1 siRNA (4392420; ID s29015), and Silencer Select XRN2 siRNA (4392420; ID s22412) were obtained from Ambion. T75 flasks of 50% confluent Huh7, Huh7-D11-KO-8, and Huh7-D11-KO-9 were transfected with either 300 pmol of the negative-control siRNA or a mixture containing 150 pmol of both XRN1 and XRN2 siRNAs using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific). Twenty-four hours later, cells were replated and seeded in triplicate at a density of 105 cells per well in 24-well plates. Approximately 24 h later, cells were cotransfected with 100 ng of the in vitro transcribed sgJFH1-Rluc RNA and either 5 pmol of negative-control siRNA or a mixture containing 2.5 pmol of both XRN1 and XRN2 siRNAs using Lipofectamine 2000 reagent (Thermo Fisher Scientific). Luciferase assays were then performed at the indicated times post replicon RNA transfection using the Dual-Luciferase Reporter Assay System (Promega) and measured with a GloMax 96 microplate luminometer (Promega). Reported values are derived from three independent experiments with each condition assayed in triplicate.
miR-122 Anti-miR Replicon Luciferase Assays.
Anti-miR Negative Control 1 (AM17010) and Anti-miR miRNA Inhibitor hsa-miR-122-5p (AM17000; ID AM11012) were obtained from Ambion. T75 flasks of 50% confluent Huh7, Huh7-D11-KO-8, and Huh7-D11-KO-9 were transfected with either 300 pmol of the negative-control anti-miR or hsa-miR-122-5p anti-miR using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific). Twenty-four hours later, cells were replated and seeded in triplicate at a density of 105 cells per well in 24-well plates. Approximately 24 h later, cells were cotransfected with 100 ng of the in vitro transcribed sgJFH1-Rluc RNA and either 5 pmol of the negative-control anti-miR or hsa-miR-122-5p anti-miR using Lipofectamine 2000 reagent (Thermo Fisher Scientific). Luciferase assays were then performed at the indicated times post replicon RNA transfection using the Dual-Luciferase Reporter Assay System (Promega) and measured with a GloMax 96 microplate luminometer (Promega). Reported values are derived from three independent experiments with each condition assayed in triplicate.
GS4.1 Replicon Colony Formation Assays.
Total RNA for transfection was obtained by PIG-B (36) extraction from T75 plates of subconfluent HEK293T and GS4.1 cells. Six-well plates of either Huh7, Huh7-D11-KO-9, HEK293, or HEK293-D11-KO-5 were transfected with 3 μg of total RNA per well in triplicate from either HEK293T (negative control) or GS4.1 cells using Lipofectamine 2000 reagent (Thermo Fisher Scientific). Media were replaced 24 h post transfection with selective media containing 500 μg/mL G418 (Sigma-Aldrich). Selective media were then replaced every 2 to 3 d for 3 wk. Cells were then stained with a solution of 50% methanol containing methylene blue (0.5% wt/vol). Plates were imaged with an Epson Perfection 4490 scanner, and quantification of the area colonized was performed with ImageJ software (NIH). Reported values are derived from five independent experiments with each condition assayed in triplicate.
sgJFH1-Neo Replicon Colony Formation Assays.
Six-well plates of either HEK293 or HEK293-D11-KO-5 were transfected with 3 μg of in vitro transcribed sgJFH1-Neo RNA or, as a negative control, sgJFH1-Rluc RNA per well in triplicate using Lipofectamine 2000 reagent (Thermo Fisher Scientific). Media were replaced 24 h post transfection with selective media containing 500 μg/mL G418 (Sigma-Aldrich). Selective media were then replaced every 2 to 3 d for 3 wk. Cells were then stained with a solution of 50% methanol containing methylene blue (0.5% wt/vol). Plates were imaged with an Epson Perfection 4490 scanner, and quantification of the area colonized was performed with ImageJ software (NIH). Reported values are derived from five independent experiments with each condition assayed in triplicate.
In Vitro Transcription of HCV 5′ UTR.
Template for in vitro transcription was produced by PCR amplification (see SI Appendix, Table S1 for primers HCVT7F and HCVT7R) of a fragment containing the T7 promoter followed by the 5′ UTR of the HCV type 1 reference sequence from the PISK-T7-HCV-5′UTR plasmid using Phusion polymerase (Thermo Fisher Scientific). For unlabeled RNA, in vitro transcription was performed with the AmpliScribe T7-Flash Transcription Kit according to the manufacturer’s instructions (Epicentre). For γ-32P 5′ end-labeled RNA, in vitro transcription was performed with the AmpliScribe T7-Flash Transcription Kit according to the manufacturer’s instructions with 37.5 mM [γ-32P]GTP (PerkinElmer) added to the reaction.
Purification of DUSP11 Core Protein.
DUSP11 core (residues 29 to 205) was amplified from synthetic gene construct pcDNA3.1-puro-3XFLAG-DUSP11 (17) by PCR and cloned into an expression vector (parental vector pET28a with GST tag). The sequence was confirmed by Sanger DNA sequencing. DUSP11 core construct was expressed in BL21 (DE3) strain Escherichia coli over 12 h of growth at 16 °C after induction with 0.5 mM isopropyl β-d-1-thiogalactopyranoside at an OD600 of 0.8. Cells were lysed by sonication in lysis buffer (50 mM Hepes, pH 7.5, 500 mM NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol) and purified over nickel beads. Protein fractions were collected and dialyzed into ion-exchange buffer (50 mM Tris⋅Cl, pH 7.5, 200 mM NaCl, 5 mM β-mercaptoethanol) overnight, and then purified via ion exchange over a 0.05 to 1 M gradient on a Mono Q 10/100 column (GE Life Sciences). Enzyme activity was verified by p-nitrophenyl phosphate (Sigma-Aldrich) hydrolysis (37).
In Vitro Translation.
In vitro translation of DUSP11, catalytic mutant, and irrelevant luciferase control has been previously described (17).
In Vitro Phosphatase Assay.
In vitro phosphatase assays were performed as previously described (17) with the following modifications. Ten picomoles of labeled HCV 5′ UTR RNA was incubated in a 20-μL reaction [50 mM Tris, 10 mM KCl, 5 mM DTT, 50 mM EDTA, 40 units SUPERaseIn (Thermo Fisher Scientific)] with either 1.3 μL in vitro translated products (DUSP11-3xFlag, DUSP11-CM-3x-Flag, or a negative-control luciferase), 1 μg of calf intestinal phosphatase (New England Biolabs), 1 μL RNA 5′ polyphosphatase (Epicentre), or 1 μg of purified DUSP11 core protein for 10 min at 37 °C. EDTA was omitted from CIP control reactions as it is inhibitory of its enzymatic activity. The reactions were fractionated by 5% urea/PAGE and the RNAs were visualized by EtBr staining. The RNAs were transferred to Amersham Hybond N+ membranes (GE Healthcare). The membranes were exposed to a storage phosphor screen (GE Healthcare) and visualized with a Typhoon biomolecular imager (GE Healthcare).
In Vitro XRN Susceptibility Assay.
In vitro phosphatase reactions were set up as described above but scaled to 100-μL volume and included 5 μg of nonradiolabeled HCV 5′ UTR RNA. Reactions were stopped by extraction of RNA with PIG-B (36). Each sample was then used to set up two 20-μL reactions containing 1 μg of treated RNA in NEBuffer 3 (New England Biolabs) with or without the addition of 1 μL recombinant XRN1 (New England Biolabs) and then incubated for 60 min at 37 °C. Reaction products were separated by 7.5% urea/PAGE and visualized by staining with ethidium bromide.
Note Added in Proof.
While this manuscript was in revision, Amador-Cañizares et al published data demonstrating that knockdown of DUSP11 enhances HCV replication in a manner consistent with our own data (38).
Supplementary Material
Acknowledgments
We gratefully acknowledge Christoph Seeger (Fox Chase Cancer Center) for generously providing the GS4.1 cell line; Arash Grakoui (Emory University) for providing the infectious HCV clone; Charles Rice (Rockefeller University) for providing Huh7 and Huh7.5 cells and the 9E10 antibody; Dr. Jessie Zhang (UT Austin) for providing guidance on the purification of the DUSP11 core; and Dr. Jason Upton for use of laboratory equipment. This work was supported by a Burroughs Wellcome Investigators in Pathogenesis Award and Cancer Prevention and Research Institute of Texas Grant RP140842 (to C.S.S.). G.R. is supported by R01DK102883.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The primary data and code reported in this paper have been deposited in the Zenodo repository (https://doi.org/10.5281/zenodo.1313694).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802326115/-/DCSupplemental.
References
- 1.Kanwal F, et al. Risk of hepatocellular cancer in HCV patients treated with direct-acting antiviral agents. Gastroenterology. 2017;153:996–1005.e1. doi: 10.1053/j.gastro.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Zhou L-Y, Zhang L-L. Host restriction factors for hepatitis C virus. World J Gastroenterol. 2016;22:1477–1486. doi: 10.3748/wjg.v22.i4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You H, et al. Novel host genetic variations associated with spontaneous clearance of a single-source outbreak of HCV1b infections. BMJ Open Gastroenterol. 2015;1:e000010. doi: 10.1136/bmjgast-2014-000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas DL, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Missiha SB, Ostrowski M, Heathcote EJ. Disease progression in chronic hepatitis C: Modifiable and nonmodifiable factors. Gastroenterology. 2008;134:1699–1714. doi: 10.1053/j.gastro.2008.02.069. [DOI] [PubMed] [Google Scholar]
- 6.Ge D, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 7.Suppiah V, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka Y, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 9.Molleston JM, Cherry S. Attacked from all sides: RNA decay in antiviral defense. Viruses. 2017;9:E2. doi: 10.3390/v9010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sedano CD, Sarnow P. Hepatitis C virus subverts liver-specific miR-122 to protect the viral genome from exoribonuclease Xrn2. Cell Host Microbe. 2014;16:257–264. doi: 10.1016/j.chom.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Yamane D, Lemon SM. Dissecting the roles of the 5′ exoribonucleases Xrn1 and Xrn2 in restricting hepatitis C virus replication. J Virol. 2015;89:4857–4865. doi: 10.1128/JVI.03692-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thibault PA, et al. Regulation of hepatitis C virus genome replication by Xrn1 and microRNA-122 binding to individual sites in the 5′ untranslated region. J Virol. 2015;89:6294–6311. doi: 10.1128/JVI.03631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 14.Shimakami T, et al. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci USA. 2012;109:941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mortimer SA, Doudna JA. Unconventional miR-122 binding stabilizes the HCV genome by forming a trimolecular RNA structure. Nucleic Acids Res. 2013;41:4230–4240. doi: 10.1093/nar/gkt075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 17.Burke JM, Kincaid RP, Nottingham RM, Lambowitz AM, Sullivan CS. DUSP11 activity on triphosphorylated transcripts promotes Argonaute association with noncanonical viral microRNAs and regulates steady-state levels of cellular noncoding RNAs. Genes Dev. 2016;30:2076–2092. doi: 10.1101/gad.282616.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Yamane D, Masaki T, Lemon SM. The yin and yang of hepatitis C: Synthesis and decay of hepatitis C virus RNA. Nat Rev Microbiol. 2015;13:544–558. doi: 10.1038/nrmicro3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarnow P, Sagan SM. Unraveling the mysterious interactions between hepatitis C virus RNA and liver-specific microRNA-122. Annu Rev Virol. 2016;3:309–332. doi: 10.1146/annurev-virology-110615-042409. [DOI] [PubMed] [Google Scholar]
- 20.Kincaid RP, Burke JM, Sullivan CS. RNA virus microRNA that mimics a B-cell oncomiR. Proc Natl Acad Sci USA. 2012;109:3077–3082. doi: 10.1073/pnas.1116107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger KL, et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narbus CM, et al. HepG2 cells expressing microRNA miR-122 support the entire hepatitis C virus life cycle. J Virol. 2011;85:12087–12092. doi: 10.1128/JVI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo J-T, Zhu Q, Seeger C. Cytopathic and noncytopathic interferon responses in cells expressing hepatitis C virus subgenomic replicons. J Virol. 2003;77:10769–10779. doi: 10.1128/JVI.77.20.10769-10779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jinek M, Coyle SM, Doudna JA. Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol Cell. 2011;41:600–608. doi: 10.1016/j.molcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Y, et al. miRNA independent hepacivirus variants suggest a strong evolutionary pressure to maintain miR-122 dependence. PLoS Pathog. 2017;13:e1006694. doi: 10.1371/journal.ppat.1006694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panwar B, Omenn GS, Guan Y. miRmine: A database of human miRNA expression profiles. Bioinformatics. 2017;33:1554–1560. doi: 10.1093/bioinformatics/btx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flores O, Kennedy EM, Skalsky RL, Cullen BR. Differential RISC association of endogenous human microRNAs predicts their inhibitory potential. Nucleic Acids Res. 2014;42:4629–4639. doi: 10.1093/nar/gkt1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato T, et al. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 29.Moon SL, et al. XRN1 stalling in the 5′ UTR of hepatitis C virus and bovine viral diarrhea virus is associated with dysregulated host mRNA stability. PLoS Pathog. 2015;11:e1004708. doi: 10.1371/journal.ppat.1004708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burke JM, Sullivan CS. DUSP11—An RNA phosphatase that regulates host and viral non-coding RNAs in mammalian cells. RNA Biol. 2017;14:1457–1465. doi: 10.1080/15476286.2017.1306169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mateu G, Donis RO, Wakita T, Bukh J, Grakoui A. Intragenotypic JFH1 based recombinant hepatitis C virus produces high levels of infectious particles but causes increased cell death. Virology. 2008;376:397–407. doi: 10.1016/j.virol.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Randall G, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Randall G, et al. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology. 2006;131:1584–1591. doi: 10.1053/j.gastro.2006.08.043. [DOI] [PubMed] [Google Scholar]
- 36.Weber K, Bolander ME, Sarkar G. PIG-B: A homemade monophasic cocktail for the extraction of RNA. Mol Biotechnol. 1998;9:73–77. doi: 10.1007/BF02752699. [DOI] [PubMed] [Google Scholar]
- 37.Bergmeyer HU, Gawehn K, Grassl M. Methods of Enzymatic Analysis. 2nd Ed Academic; New York: 1974. [Google Scholar]
- 38.Amador-Cañizares Y, Bernier A, Wilson JA, Sagan SM. miR-122 does not impact recognition of the HCV genome by innate sensors of RNA but rather protects the 5’ end from the cellular pyrophosphatases, DOM3Z and DUSP11. Nucleic Acids Res. 2018;46:5139–5158. doi: 10.1093/nar/gky273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.