Significance
EmrE is a small membrane transporter found in Escherichia coli that exports drug-like molecules from the cell, contributing to antibiotic resistance. In EmrE, as well as in the wider small-multidrug resistance transporter family, a specific anionic amino acid (E14) has been implicated in governing the conformational changes that export drugs. However, due to sparse structural information, the exact interactions remain unidentified. Through interactive molecular dynamics to incorporate existing cryo-electron microscopy data, we create a fully refined atomic model of EmrE. We then embed this model in a lipid bilayer and evaluate the interactions within EmrE under different loading states. We find that E14 makes specific hydrogen bonds to neighboring residues, coupling observed experimental phenomena to interactions at the atomic scale.
Keywords: molecular dynamics, membrane protein, structure refinement, proton-coupled transport
Abstract
EmrE is a small, homodimeric membrane transporter that exploits the established electrochemical proton gradient across the Escherichia coli inner membrane to export toxic polyaromatic cations, prototypical of the wider small-multidrug resistance transporter family. While prior studies have established many fundamental aspects of the specificity and rate of substrate transport in EmrE, low resolution of available structures has hampered identification of the transport coupling mechanism. Here we present a complete, refined atomic structure of EmrE optimized against available cryo-electron microscopy (cryo-EM) data to delineate the critical interactions by which EmrE regulates its conformation during the transport process. With the model, we conduct molecular dynamics simulations of the transporter in explicit membranes to probe EmrE dynamics under different substrate loading and conformational states, representing different intermediates in the transport cycle. The refined model is stable under extended simulation. The water dynamics in simulation indicate that the hydrogen-bonding networks around a pair of solvent-exposed glutamate residues (E14) depend on the loading state of EmrE. One specific hydrogen bond from a tyrosine (Y60) on one monomer to a glutamate (E14) on the opposite monomer is especially critical, as it locks the protein conformation when the glutamate is deprotonated. The hydrogen bond provided by Y60 lowers the of one glutamate relative to the other, suggesting both glutamates should be protonated for the hydrogen bond to break and a substrate-free transition to take place. These findings establish the molecular mechanism for the coupling between proton transfer reactions and protein conformation in this proton-coupled secondary transporter.
EmrE is a membrane transporter found in Escherichia coli that uses the electrochemical proton gradient across the bacterial inner membrane to export a variety of toxic polyaromatic cations (1–4). EmrE acts as a homodimer of 110-residue monomers, each structured into four transmembrane helices (5, 6), a common fold within the small-multidrug resistance (SMR) transporter family (7). Due to this common fold and well-studied biochemistry, EmrE is used as a model to study the transport of drug-like molecules across the SMR family (7).
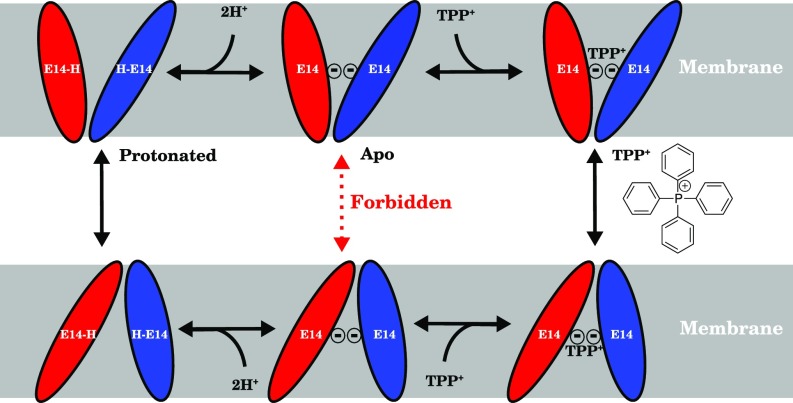
Structural studies of EmrE using NMR spectroscopy (8, 9), cryo-electron microscopy (cryo-EM) (5), and X-ray crystallography (10) have established two conformationally distinguishable monomeric states that are simultaneously populated within each dimer (11). Förster resonance energy transfer (FRET) experiments have conclusively demonstrated the antiparallel orientation of the monomers with respect to one another (8), which may be due to the more favorable helix–helix interactions in antiparallel orientations relative to parallel ones (12). Based on this evidence, proposed transport cycles for EmrE proceed by an alternating access mechanism (13, 14), whereby the monomers swap conformations between these two states, as in other transporters (Fig. 1) (8, 15).
Fig. 1.
Schematic of the transport cycle of EmrE following the alternating access mechanism with a strict 2:1 : stoichiometry. EmrE monomers (red and blue ellipsoids) swap conformations (schematically indicated by the size and angle of the ellipse relative to the membrane) and thus alternate the substrate/proton accessibility to the inside or outside of the cell when loaded with protons (Left) or substrate (Right). When the EmrE dimer is in its apo (Center) form, the transition should be forbidden (red dashed arrow) to preserve the coupling of proton and substrate transport and prevent proton leakage across the membrane. This apo state is in contrast to both the fully protonated and substrate-bound states (Left and Right columns, respectively), whose transition should be allowed (black solid arrows) for EmrE to fulfill its physiological function as a proton-driven polyaromatic cation transporter.
Alternating access transport presents an interesting structural dilemma shared by all secondary antiporters, which use the gradient of one species to drive another against its gradient. In the simplest form of this model, the conformational transition of the empty transporter (apo) ought to be forbidden (Fig. 1); otherwise transporter action would run down the established proton gradient rather than fulfill its physiological function in exporting polyaromatic cations. For antiporters of known structure, such as the glycerol 3-phosphate transporter GlpT, this chemomechanical requirement is reflected in a higher energetic barrier for the apo transition relative to a substrate-bound transition (16). However, due to the rapid transition between conformational states in EmrE (3, 17, 18) and a dearth of complete EmrE structural models, no systematic study exists of how this conformational transition is controlled in EmrE at the atomic level. The glutamate (E14) residues in the transporter lumen that are thought to act as protonation sites as well as drug interaction partners during the transition (19–21) couple proton binding to conformational change. However, the specific interaction partners that may control the transition have not been elucidated due to the prohibitively low resolution and incompleteness of prior structural models.
To answer these questions explicitly, we first construct a complete and refined atomic model of EmrE using molecular dynamics (MD) flexible fitting (MDFF) (22) and interactive biased MD techniques (23) to incorporate existing experimental data into the model. Through this procedure, the model expands upon the positions provided in published crystallographic structures (10) to encompass the full sequence of EmrE, including side chain positions essential to a full structural model. These positions are consistent with experimental electron densities and structural proclivities of individual side chains. Using this rigorously constructed fully atomic refined model (faRM), we carried out a series of MD simulations where the loading state of EmrE was varied to mimic the different intermediates of the transport cycle. The studied transport intermediates include a fully deprotonated state (apo), two singly protonated states where only () or () is protonated, and the doubly protonated state. To represent the drug-bound state, a tetraphenylphosphonium cation () was chosen from among the many EmrE substrates (3) due to crystallographic identification of its binding site (10) and proven binding within the EmrE lumen (21). During simulation design, the best estimate for the : stoichiometry was 2:1 (24), and thus our state is fully deprotonated, whereas newer models permit a 1:1 stoichiometry as well (25).
In addition to conventional equilibrium MD simulations, replica exchange thermodynamic integration (RETI) (26) calculations were performed to determine the differences between the E14 residues in each monomer and to assign specific causal factors to the observed experimental shift of 1.5–2.0 units (27) between E14 residues in each monomer. Nonequilibrium work calculations were also performed for a semiquantitative comparison of the conformational transition rates of different substrate loading states, using the inherent symmetry of EmrE to generate structural models for both inward- and outward-facing states.
Over the course of an aggregate simulation time of 12.5 s, the secondary structure of the faRM remained intact, with a more expansive helical architecture compared with prior models for EmrE (10, 28, 29), similar to other membrane proteins. EmrE is highly dynamic within the simulations, and aromatic residues around the binding site play a pivotal role in controlling the observed conformational heterogeneity. Tyrosine Y60 from monomer B (), in particular, interacted frequently with E14 of the opposing monomer (). When combined with previous mutagenesis data (30) demonstrating a deleterious effect of the Y60F mutation on cell fitness, this finding suggests that the hydrogen bond formed between Y60 and deprotonated E14 locks the conformational transition. In addition, the hydrogen bond lowers the of , thus forcing a second proton-binding event before the interaction is broken and the conformational transition can take place. The identification of this hydrogen bond is a substantial insight into the mechanism of proton coupling to conformational change in EmrE.
Results and Discussion
The faRM generated by the refinement approach is a marked improvement over the initial crystal structure, which features only atoms (10). In a simple static test of the faRM, metrics to assess the overall quality of the model were determined as a first step. Crystallographers frequently use the molprobity score, which considers features such as rotameric states of side chains and interatomic clashes, as a metric to assess their structures (31). The score is scaled such that the score is commensurate with the approximate resolution of the crystal structure (31). The molprobity score for the faRM was 0.89, a result that would be expected from a subångstrom resolution crystal structure (31), compared with the actual crystal structure resolution of 3.8 Å (10). More than 95% of side chains, which were all modeled due to their absence in the previously available structures, are in their favored rotameric and Ramachandran regions of conformational space, with only two outliers each in the 220 total amino acids of the structure. Separate analysis with PROCHECK (32) shows 93% of residues occupy their most favored region, compared with 73% in a recent electron paramagnetic resonance (EPR)-based model (33). Through dynamic simulation, additional features of the modeled structure become apparent, as described below.
Stability of the faRM EmrE Structure.
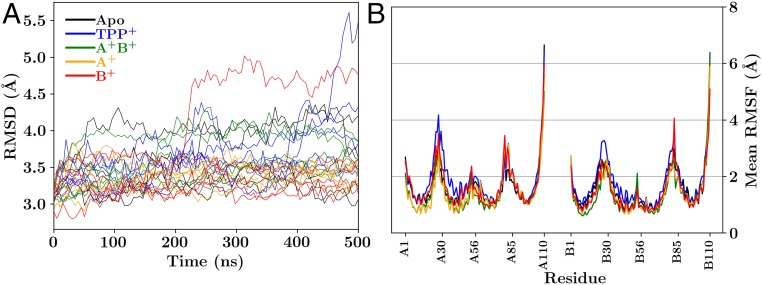
With the extensive structural remodeling that took place before simulation and the poor resolution of the starting structure, the EmrE faRM might be expected to suffer from instability and fall apart over an extended simulation. While the root-mean-square deviation (rmsd) with respect to the crystal structure can be high relative to other simulated membrane proteins (Fig. 2A), the majority of the simulations show rmsds that are comparable to, or below, the crystal structure resolution (3.8 Å), as is generally the case for membrane proteins (34), independent of the reference state (SI Appendix, Fig. S5) or the monomers considered (SI Appendix, Fig. S6). The two notable exceptions are one simulation and one -bound simulation. In the -bound case, lipids intercalate into the dimer interface formed between helix 2 of each monomer and directly interact with . This splits apart the dimer, leading to a large rmsd for this simulation (SI Appendix, Movie S3), but not for the individual monomers themselves (SI Appendix, Fig. S6). For the case, a helix rotation causes the formation of a continuous water channel, which increases the rmsd relative to the crystal (SI Appendix, Movie S4). These conditions were not observed in other simulations.
Fig. 2.
Structural deviation and fluctuation measured over the course of the trajectories. (A) rmsd of each trajectory compared against the 3B5D crystal structure. Due to limitations of the original crystal structure, the comparison strictly involves positions reported in the crystal structure and does not include side chain atoms. The color of the lines indicates the loading state of EmrE: black for the apo state, blue for the -bound state, green for the doubly protonated state (), orange for the state where the proton is bound to monomer A (), and red when the proton is bound to monomer B (). SI Appendix, Fig. S5 is comparable, where the reference state is the first frame of the trajectory, rather than the crystal structure. (B) Mean rmsf per residue, combining the results over all five independent simulations after trajectory alignment for each loading state of EmrE, using the same color scheme as in A. For a detailed trajectory-by-trajectory analysis, see SI Appendix, Fig. S7.
Based on the per-residue root-mean-square fluctuation (rmsf) measurements conducted over these same trajectories (Fig. 2B and SI Appendix, Fig. S7), no large-scale conformational changes indicative of a concerted transport cycle are observed during these simulations. The rmsfs are comparable to what would be expected for a dynamic and diminutive -helical membrane protein, with small fluctuations in the helical regions and larger fluctuations in the connecting loops and termini (Fig. 2B). The spontaneous fluctuations lead the protein away from an inverted faRM (ifaRM) state that would reflect an inward- to outward-facing state transition (SI Appendix, Fig. S8). This finding, coupled with the short simulation timescales (500 ns) relative to measured millisecond timescale turnover rates (17, 18), means that biased simulations are required to drive conformational transitions.
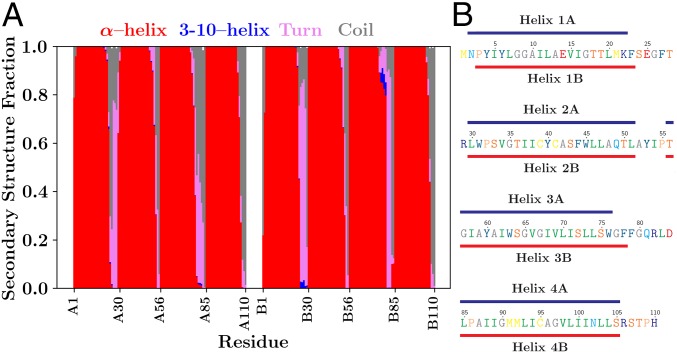
An essential feature of membrane proteins is the secondary structure formed by the polypeptide spanning the low-dielectric environment of the membrane (38, 39). High rmsd can hide the dissolution of protein secondary structure, including the eight transmembrane -helices of EmrE that collectively span the hydrophobic core of the 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) bilayer into which they were embedded (SI Appendix, Fig. S1). By explicitly determining the secondary structure throughout the simulations using Stride (35), we show that the -helical secondary structure is maintained in all four transmembrane helical segments in both monomers (Fig. 3A and SI Appendix, Fig. S9). The identity of the residues belonging to each helix differs slightly, depending on the monomer (Fig. 3B), likely due to the different structure of the connecting loops between the individual helices pulling on the ends of helical segments. Since it is plausible that the conformation at the ends of helices might change during the conformational transitions, further experimental restraints, e.g., those derived from future NMR experiments, would be instrumental in further refining the secondary structure assignment of each residue and improve the veracity of the simulated structure.
Fig. 3.
Representations of the predominant secondary structure over all trajectories. (A) A residue-by-residue breakdown of secondary structures present and their relative predominance throughout the 25 individual simulations as determined by Stride (35). The evolution of secondary structure over time is reported in SI Appendix, Fig. S9. Each secondary structure observed is shown using a different color in the cumulative bar plot above: red for a conventional -helix, blue for a 310-helix, pink for turns, and gray for random coils. (B) Residues that are -helical 80% of the time were identified and grouped into individual -helices. The helices formed in the faRM differ slightly between monomers, as indicated by the lines above (monomer A, blue) and below (monomer B, red) the EmrE sequence, which has been colored according to hydropathy (36) and typeset using TeXshade (37). This mapping includes residues 2–22, 30–51, 56–76, and 85–105 of monomer A and residues 3–23, 30–51, 56–78, and 85–105 of monomer B. As a comparison, the cryo-EM–based model from Fleishman et al. (28) identifies residues 4–21, 34–52, 58–80, and 87–104 as helical. The conservative Fleishman helix definitions are used consistently in other analyses performed on the simulations.
One example of an experimental observable that could be applied comes from recently published measurements of interresidue distances measured by EPR spectroscopy (33). These EPR distance restraints were not considered during model construction in this work; however, the distances measured through EPR suggest substantial conformational change between different loading states for EmrE (33). Over our own simulations, large conformational differences were not immediately apparent between different loading states, as the distances between individual side chains correlate strongly across loading states (SI Appendix, Fig. S11). However, since the correlations are not identically unity, and some spread can be observed, these results cannot rule out that subtle loading-dependent conformational changes have occurred, which we investigate more closely through the nonequilibrium transitions. The EPR experiments focused on two loading states for EmrE: a -bound form for EmrE and EmrE at pH 5, corresponding to our doubly protonated state. The distance between equivalent residues in the two monomers was measured using site-directed spin labeling (33), which we compared against the measured distances from our own simulations (SI Appendix, Fig. S10). The correlation between the EPR spin-label distances and the distance between carbons () is low (SI Appendix, Fig. S10A), and the trendline has a slope below unity. The difference in slope can be attributed to spin labels being larger than individual residues, which results in EPR-measured distances that are typically larger than the actual . The low correlation coefficient comes primarily from residues within loops of the protein with measured distances that are substantially different in each method (SI Appendix, Fig. S10B). Since the cryo-EM densities were weakest between helices, it is not surprising that the loop structures in our model have the weakest agreement with the EPR results. Incorporating spin-label distributions into future EmrE refinements would improve loop structure.
State-Dependent Hydration of the Lumen.
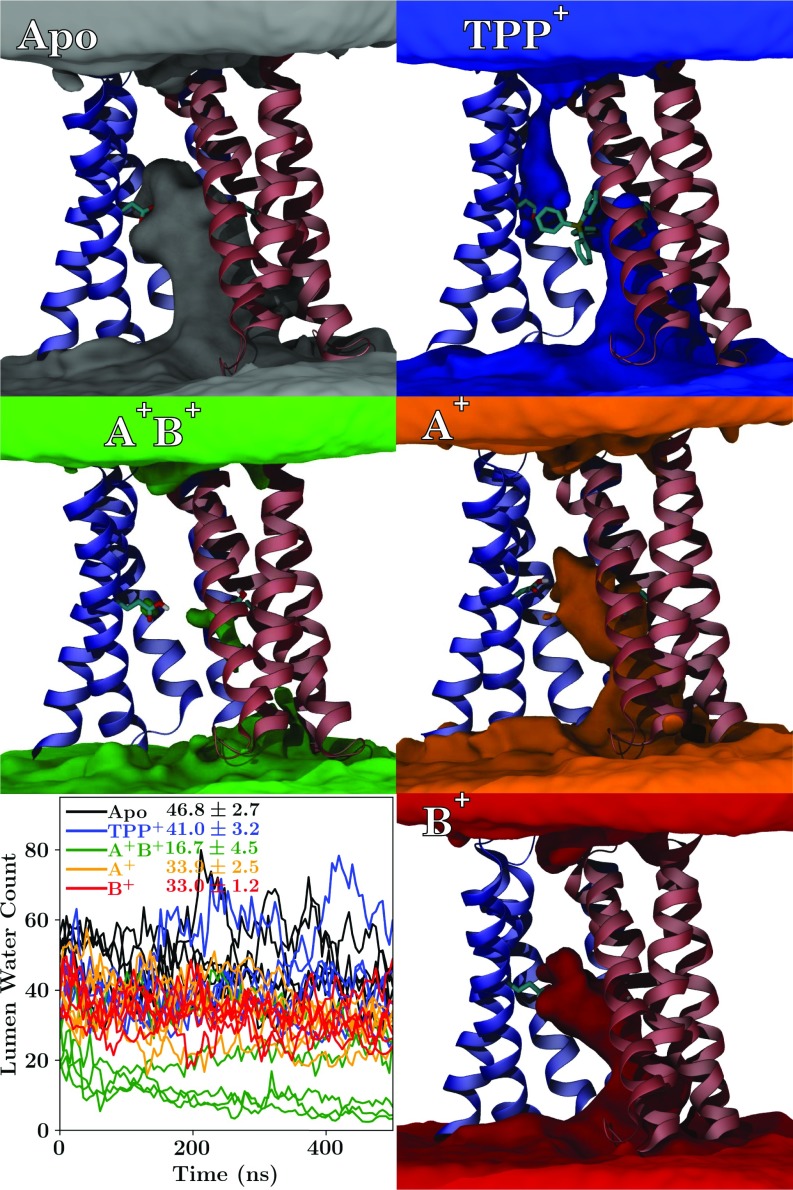
In investigating the different loading states of EmrE, an unexpected event occurred several times within our simulation set—the lumen of the doubly protonated state spontaneously dehydrated (Fig. 4). Other protonation states show typical behavior for a membrane transporter, with lumen water that can exchange directly with bulk water on one side of the membrane. In contrast, the connection from the lumen to bulk water was severed in the state, and the transporter spontaneously transitioned to a new state. Although this state looks occluded because the pocket of water in the lumen is disconnected from bulk solution, no large-scale conformational change has taken place (SI Appendix, Fig. S8). Instead, water interactions with the protein appear too weak to maintain a water channel into the lumen once the glutamates are protonated as in the state, spontaneously breaking the water pathway. The rapid formation of an occluded intermediate is consistent with pH-dependent NMR studies, where low pH facilitates rapid conformational transition (18).
Fig. 4.
Visualization and quantification of the water occupancy across all five replicates within the lumen of EmrE under different loading conditions. Regions where water is frequently present are represented by isosurfaces of different colors (gray for apo, blue for bound, orange for , red for , and green for ), along with a cartoon representation of EmrE for context, including an explicit representation of E14 and where present. Animations of these views during rotation are presented in SI Appendix, Movie S5 A–E. Lower Left shows the number of water molecules within the lumen of EmrE as a function of time for the different loading conditions. All 25 independent trajectories are shown simultaneously and are colored according to the EmrE loading state, with the mean number of luminal waters ( Å) reported beside the key.
The -bound state (Fig. 4) appears weakly leaky, with a visible water pathway between both top and bottom bulk water regions to the -binding site. Water leaks have been reported previously for other membrane transporters (40), and thus it is not surprising for a highly dynamic transporter such as EmrE to exhibit transient leaks. However, water leaks in EmrE are particularly interesting since EmrE transport is driven by a proton gradient. Thus, any significant water leak could potentially serve as a conduit for protons across the membrane, short circuiting the transport cycle (Fig. 1). The leaky states, where a water path exists between the two sides of the membrane, occur only transiently (Table 1), accounting for less than 10% of the total simulation time. These leaky states are inhomogeneously distributed across the simulations conducted (SI Appendix, Figs. S12 and S13). Critically, the leaky states are often not conducive to rapid proton translocation via a water wire, as one or more water molecules within the path orient suboptimally for proton conductance due to interactions with the surrounding residues. The water orientation disrupts the chain of hydrogen bonds needed to form a conductive water wire, resulting in a minimal proton translocation probability (Table 1) that is consistent with the maintenance of a proton gradient in the absence of substrate (25). Thus, the transient water wires observed in our simulations likely would not serve as efficient proton pathways (41), thereby maintaining the proton gradient that drives substrate export.
Table 1.
Formation propensity and lifetime of water pores in different loading states of EmrE
| Water pore existence | conductance | Mean lumen water | |||
| Loading state | Probability, % | Mean duration, ps | Probability, % | Mean duration, ps | Exchange time, ns |
| Apo | 11.6 | 0.3 | |||
| 11.8 | 0.6 | ||||
| 3.9 | 0.7 | ||||
| 0.7 | 0.1 | ||||
| 4.1 | 0.1 | ||||
A water pore exists at a specific time point if water molecules connect on either side of the membrane outside of the transporter lumen (|z|>11:8 Å ). Similarly, we say that EmrE is in a H+ leaky state if a water wire exists across the lumen that might transfer a proton by a concerted Grotthuss-like mechanism. For each of these states, we quantify the percentage of the simulation time where the proton wire exists, as well as the mean duration of the state existing in successive simulation snapshots. The final column measures the mean time taken for luminal water to exchange, as measured by determining the number of frames required for all luminal water molecules to be replaced by bulk water. Reported ranges indicate the SE, not the SD.
Water dynamics are particularly sensitive to the loading state of EmrE. The -bound or states exchange luminal water with bulk solution more slowly than do the other loading states (Table 1). For the case, this delay stems from the irregularity with which the water molecules isolated within the lumen can exchange with bulk water during the sporadic formation of water pathways at the dimer interface, as evidenced by the low number of water molecules in the lumen at any given time (Fig. 4). blocks the exchange of water through a different mechanism—trapping it within the lumen by limiting the accessibility of water molecules nearest to the substrate and thereby retarding their exchange. This observation is consistent with the model in Fig. 1 since slower water disconnected from the bulk would be the first step in forming an occluded state. Thus, viewed from the perspective of water dynamics, the and -bound states are closer to transitioning than the other loading states tested.
Residue-Specific Interactions in EmrE Structure.
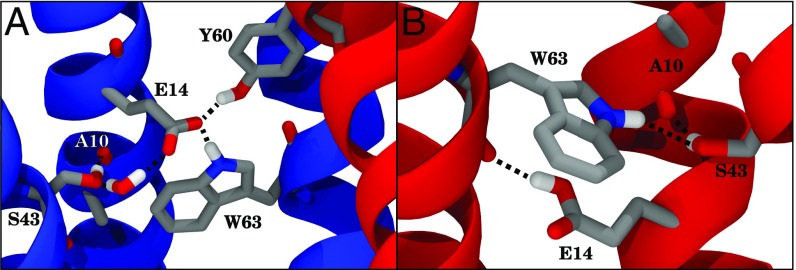
The current model provides information about atomic interactions within EmrE, such as how hydrogen-bonding patterns change during the transport cycle. SI Appendix, Table S1 details the interactions that exist throughout the antisymmetric dimeric faRM structure modeled here. Many interactions, particularly those involving peripheral residues, remain unchanged under the different loading states for EmrE. Other robust interactions include those internal to the structure, such as polar side chains interacting with nearby backbone carbonyl groups like the serine to alanine (S43–A10) interaction exemplified in Fig. 5.
Fig. 5.
Examples of hydrogen bonding (black dashed lines) during simulation and how they change with the protonation of E14. Monomers A and B are drawn in blue and red, respectively. (A) Example interaction network surrounding deprotonated E14, highlighting multiple influential interactions to . These include , , and an example of water-mediated interaction (). Helices 1 and 2 of monomer B have been omitted for clarity, as have hydrogen atoms not directly involved in hydrogen bonds. (B) Example interaction network when is protonated, detailing how E14 can act as a donor to the backbone of rather than as an acceptor as it was in A, which in turn changes into a potential donor for .
The most significant changes in hydrogen bonding occur in the vicinity of E14 as it responds to different loading states (Table 2). Typically, the E14 side chain interacts directly with residues Y40, S43, and W63 of the same monomer and Y60 of the opposite monomer. T18, rather than interacting with the side chain of E14, instead interacts with the backbone, as in Fig. 6B. These interactions are modulated by changing the loading state of EmrE. For example, the proton from W63 interacts with the side chain of E14 from the same monomer if E14 is deprotonated, but stretches across to the backbone or other nearby residues instead if E14 is protonated (Fig. 5). Previous mutagenesis experiments have identified these E14-interaction partners as important to the function of EmrE (30, 42–45). Tryptophan W63, for instance, is highly conserved in the SMR transporter family (SI Appendix, Fig. S15), and its mutation changes EmrE into a uniporter of cationic substrates (42). Mutations of tyrosine Y40 or serine S43 reduce the effectiveness of EmrE in exporting toxic substrates (30, 43), and S43 has been implicated in the specificity of EmrE to its substrates (44). Tyrosine Y60 is one of the few completely conserved amino acids in the SMR family (SI Appendix, Fig. S15), suggesting a role for the hydroxyl of Y60 in regulating the transport cycle. The conservative mutation Y60F renders EmrE nonfunctional (30, 45), while the Y60T mutant remains functional but at a reduced capacity (30). Thus, it is clear that studying these specific interactions is highly relevant to the overall function of EmrE.
Table 2.
Hydrogen bonds between protein side chains and E14, reported as a percentage of the total simulation time the hydrogen bonds existed
| Hydrogen bond propensity | ||||||
| E14 chain | E14 partner | Apo | TPP bound | |||
| A | 63.2 | 65.8 | 75.2 | 60.4 | 63.5 | |
| 14.5 | 10.9 | 0.7 | 1.5 | 7.5 | ||
| 5.4 | 10.7 | — | 0.4 | 1.5 | ||
| 32.8 | 13.8 | 38.6 | 41.0 | 57.1 | ||
| 8.6 | 0.1 | 10.3 | 3.5 | 32.2 | ||
| B | — | 11.7 | 15.9 | 0.1 | 0.1 | |
| 6.1 | 27.2 | — | 0.2 | 0.3 | ||
| 57.8 | 58.7 | 71.4 | 60.1 | 54.2 | ||
| 6.8 | 9.2 | 0.2 | 9.4 | 1.5 | ||
| 22.1 | 16.2 | — | 9.0 | 1.4 | ||
| 15.9 | 8.7 | 71.7 | 44.2 | 51.0 | ||
The results are grouped according to which pair of monomers is involved in the interaction. Note that unlike SI Appendix, Table S1, donor and acceptor interactions to E14 are aggregated here into a single element.
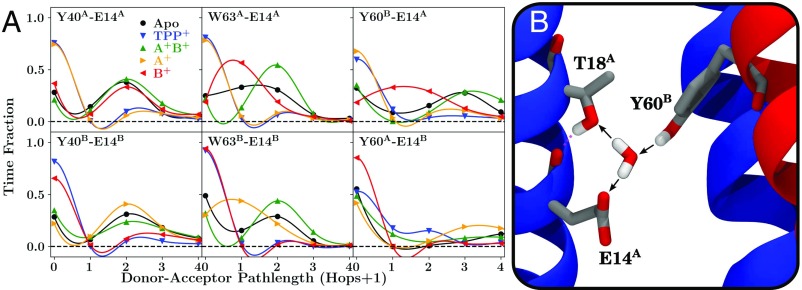
Fig. 6.
(A) Water-mediated hydrogen bonding within EmrE for selected residue pairs. (B) A snapshot exemplifying how these measurements are made. The path length between two residues is determined from a directional hydrogen-bonding network (black arrows in B), and the distribution of this path length over the aggregate trajectories for E14-Y40, E14-W63, and E14-Y60 pairs in each EmrE loading state is reported as labeled at the top of each sub panel in A. Note that since the hydrogen-bonding pathway is directional, we count the interaction only if a donor–acceptor relationship might exist between the residues. Thus, in the example pictured in B, Y60 has a path length of 2 to both T18 and E14, as they are connected via the shown water, while T18 has a path length of 0 to E14, signifying that the two side chains are disconnected since the bridging water acts as the donor to both residues, even though the side chain of T18 does interact with the backbone of E14 (pink dashed interaction). To assist in determining the location of the datapoints for each loading state and to visualize the overall pattern, the datapoints have been connected with an interpolating polynomial in the plots in A.
However, measuring direct hydrogen bonds tells only part of the story. Water-mediated hydrogen bonding, in which water molecules bridge the gap between donor and acceptor, also occurs between E14 and the key residues highlighted previously: Y40, W63, and Y60 (Fig. 6A). The water-mediated hydrogen-bonding pattern observed within a single monomer (Y40 and W63) remained unchanged for equivalent loading states, with similar patterns in both the A and B monomers, indicating that the interactions are retained throughout the transport cycle (Fig. 6A). Critically, the interaction between and (Figs. 5A and 6B) is clearly inequivalent to the interaction between the opposite set of monomers. Additionally, this interaction is strongest when is deprotonated (that is, the , , and apo states), suggesting that the protonation of monomer A might unlock the “electrostatic lock” that couples the monomers together and prevents conformational change when is deprotonated. This mechanistic hypothesis was tested by driven conformational changes, as discussed below.
Dynamics and Membrane Interaction Within EmrE.
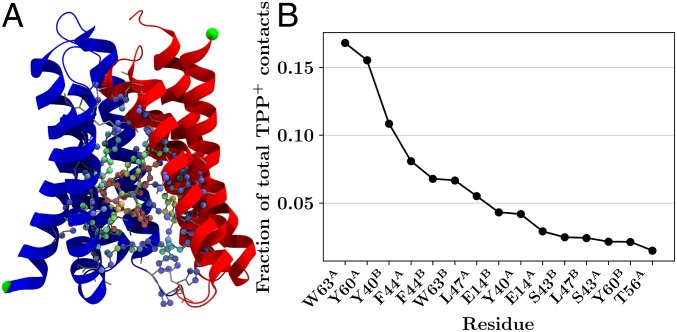
By including an explicit molecule borrowed from the low-resolution crystal structure into the faRM, we are uniquely positioned to investigate the nature of the interactions between and its binding site within EmrE. The observed binding site for was consistent across the five independent trajectories, focusing primarily on the adjacent aromatic residues (Fig. 7). came into contact with a number of other residues as well, including some on the C-terminal helix of monomer A (Fig. 7). The nonspecific nature of the observed interaction aligns well with the polyspecific nature of EmrE transport (3). EmrE requires only two features of its substrates: (i) The substrate must be a cation so it is attracted to the electrostatic potential created by the two E14 residues and (ii) the substrate should have aromatic groups to satisfy the -stacking requirement of neighboring aromatic residues.
Fig. 7.
contact analysis of elements within EmrE. (A) Pictorial representation of the contact data, where each heavy atom that contacted (and itself) is colored according to the number of contacts. Blue atoms had fewer contacts, and red atoms had more contacts. A cartoon representation of the protein and gray licorice representations of every side chain are also included for context. (B) Ordered listing of the top 15 aa with the most contacts with , expressed as a fraction of the total number of contacts, as computed by SI Appendix, Eq. S1 (46, 47), over all trajectories. Together, the 15 residues listed here account for over 92% of the total contacts with .
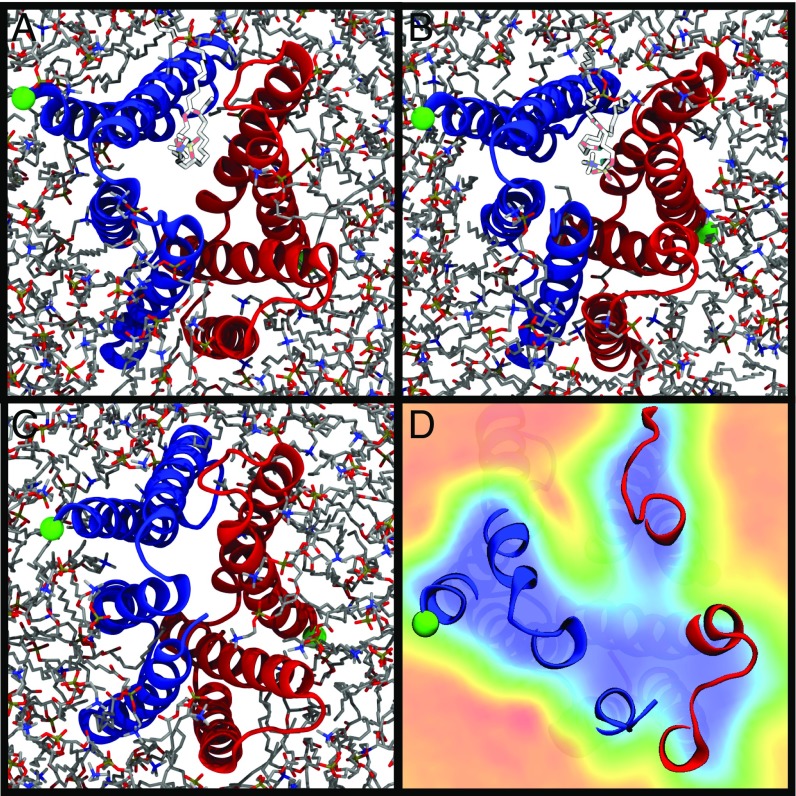
As mentioned previously, lipids play an active role in the structural dynamics of EmrE and consequently may influence the -binding pathway. Lipids wriggle their tails between helical interfaces and can influence protein structure, as observed in some high-rmsd cases (SI Appendix, Movie S3). Indeed, the portal formed by cracking apart the interface between helices 2 from both monomers opens in a subset of simulations (Fig. 8). We propose two explanations for this phenomenon: (i) The residue packing along that interface is nonoptimal in the faRM, resulting in lipids wedging the monomers apart, or (ii) these gaps may form in vivo as well and might serve as a conduit for binding and unbinding as it arrives via a membrane-derived route (48).
Fig. 8.
Lipid intercalation into the interface between helix 2 monomers. (A and B) Snapshots from the trajectories where an intercalated lipid (highlighted lighter-colored lipid) splits the monomers apart (blue for monomer A, red for monomer B, with the N terminus tagged in green). (C) This is a stochastic process, and the intercalation is not always present. (D) The lipid occupancy across all simulation conditions for a specific slice along the membrane normal, with higher lipid occupancies colored in red and lower occupancies in blue, with the protein provided for context. Sequential slices along the membrane normal are displayed in SI Appendix, Movie S7, highlighting the specificity of lipid intercalation to the “open” side of the transporter. The snapshots shown all have the open side of the transporter (the side accessible to solution) oriented toward the viewer.
The lipid-binding regions at the interface between the two helices 2 are large enough to accommodate entry in some cases (Fig. 8 and SI Appendix, Movie S7). As a hydrophobic cation, is enriched in the membrane by a factor of 100 relative to solution (49). EmrE may thus increase its substrate accessibility by allowing direct access of the substrate through the gap between the transmembrane helices, similar to other membrane proteins where substrates or ligands arrive via the membrane (48, 50, 51). However, since the pathway for binding is currently unresolved, further study will be required to distinguish between these hypotheses.
Driving the Transition of EmrE.
The lock formed by the interaction between and would be expected to slow the conformational transition for the apo state relative to the protonated states, similar to how protonation controls conformational change in acrAB-TolC (52), a complementary drug resistance transporter also found in E. coli (1, 53). This hypothesis can be tested through driven simulations, where the average nonequilibrium work needed to induce the transition between an initial “inward-facing” and target “outward-facing” state can be compared between different loading states to deduce which one has the lowest barrier to conformational transition. Our initial states are seeded from the final conformation of the equilibrium trajectories. Exploiting the unique symmetries presented by an antisymmetric homodimer, swapping the conformations of each monomer and exploiting symmetry generate the appropriate target state after conformational transition. A biased simulation then drives the initial structure to the target structure by minimizing the rmsd to the target, with an example transition shown in SI Appendix, Movie S8. Using rmsd as a collective variable is known to generate suboptimal transition pathways (54). For the particular case of EmrE in the absence of a gradient driving conformational change and in a symmetric membrane, we know that the true free energy change must be zero, as the initial and target structures are related by symmetry to one another. Thus, we use the nonequilibrium work, acknowledging that it contains a large dissipative component, as an approximate metric to judge the transition barriers, with less work implying lower transition barriers between inward-facing and outward-facing states. The order of the barrier height computed here can be qualitatively compared with measured transition rates from NMR (8, 18) and the prevailing transport cycle (Fig. 1).
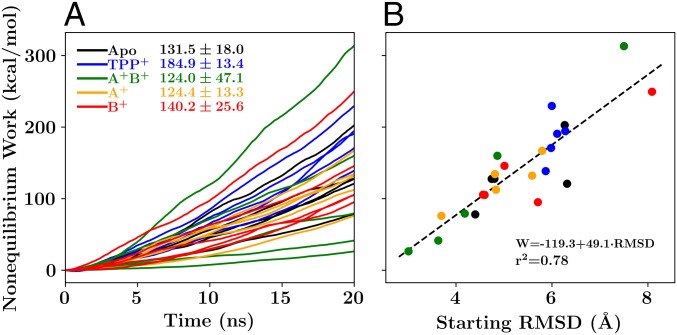
The nonequilibrium work results (Fig. 9) contrast with the transport cycle of EmrE (Fig. 1). A forbidden apo transition should have the highest nonequilibrium work of the states tested, signifying a high barrier to the transition. Instead, the -bound transition requires the most work (Fig. 9A). The higher level of work for the -bound case suggests that the -bound transition should be “forbidden,” in contrast with its physiological role. Although the high barrier for -bound transition is inconsistent with the simple transport model in Fig. 1, it is consistent with measured transition rates from previous NMR studies (8, 18). Protonation of EmrE by lowering the pH experimentally increases the conformational transition rate in the drug-free state from 40 to 220 (18), both of which are much faster than the conformational exchange rate of 5 when is bound to EmrE (8). This trend qualitatively suggests that the transition barrier under the experimental conditions should be apo . Likewise in our simulations, the barrier for the state is lowest, followed by the singly protonated and the deprotonated apo or singly protonated states, which are distantly followed by the -bound state. Critically, these measurements are made in the absence of any electrochemical gradient or applied electric potential, which prevents a direct comparison of our nonequilibrium simulations to the functional state in vivo, where there is a proton gradient as well as an electric potential difference across the bilayer.
Fig. 9.
Nonequilibrium work profiles. (A) Work profiles associated with the conformational transition of EmrE for each loading state. (B) The relationship between the total nonequilibrium work and the starting rmsd difference between the initial states (taken from the equilibrium trajectories) and the final states (which are symmetry related to the initial states). The mean and SD of the nonequilibrium work required for the transition in each of the five independent loading states are reported adjacent to the key in A, using our standard color scheme (black for apo, blue for , green for , orange for , and red for ). Fit parameters describing the linear relationship between the nonequilibrium work and the starting rmsd are presented in B.
Thus, while we cannot say definitively how the results fit into the transport cycle in Fig. 1, the protonation results are in line with the concept of the electrostatic lock. If is protonated (the and states), the transition is easier since the interaction between and is disrupted (Fig. 6). If, however, remains deprotonated, the transition is made more difficult by retention of the interaction with , albeit with large uncertainties as a result of the disparate work values. Thus, the faRM accurately recapitulates available experimental transition rates and allows us to connect these observed rates to specific interactions induced by the protonation state of E14, as well as dehydration of the lumen upon E14 protonation.
The work values can also guide us toward additional insight about the symmetry of the states observed in equilibrium. For example, the state yields both the highest and the lowest nonequilibrium work value (Fig. 9B). In the low-work case, the initial state was close to being symmetric, with only a small rmsd difference between the initial state and the inverted state. In the case where the initial state is highly asymmetric, as measured by a much larger rmsd difference between the initial and inverted states, a much higher work value results. This is again consistent with our examination of the interactions surrounding E14. Most interactions to E14 are intramonomer interactions that are unable to change the relative configurations of the two monomers. The locking interaction between and , however, is different, in that it crosses between monomers and breaks the symmetry between the monomeric interactions. Thus, states where this locking interaction is intact will be more asymmetric, increasing the cost of transition. By contrast, when the locking interaction is broken, as it is in the four lowest-cost transitions, the dimers are more symmetric, lowering the barrier to transition. This intuition is consistent with the recent EPR study discussed previously, which suggested that the doubly protonated state undergoes conformational change to become more symmetric than the apo state (33). Together these results indicate that – interaction changes in response to protonation control conformational transition in EmrE.
Computational Determination of .
As discussed above, the E14 residues form direct and water-mediated interactions with surrounding residues. These interactions change with protonation state and, critically, differ between individual monomers. The asymmetry in interactions should cause a noticeable shift between the two E14 residues. NMR studies have previously reported the shift as units in the temperature range we are simulating in the absence of , with one at 6.8 and the other at 8.5 at C (27). Mechanistically, these values suggest that the E14 residue with 8.5 is almost always protonated at physiological pH whereas the lower E14 would be selectively protonated, leaving open the question of which monomer has which . This question is difficult to answer experimentally, as the symmetry of EmrE makes assignment of to a specific E14 challenging, even as drug-bound NMR experiments have placed the proton on (25). Instead, we leverage our faRM to determine the order of protonation (and thereby assign ) in two ways: (i) analysis of structural elements and the interactions observed and (ii) a RETI calculation of the proton transfer reaction interconverting the and states.
The structural argument for shifted E14 values is that the hydrogen-bonding patterns of E14 to other residues are largely equivalent between monomers A and B, except with respect to the interaction with Y60 from the opposite monomer (Fig. 6). Since could in principle donate a proton to along the formed hydrogen bond, but does not donate a proton to since there is no hydrogen bond between them, should be more willing to lose its own proton (lower ) as it already partially shares the proton from . This general trend of increasing hydrogen bonds leading to lower has been noted previously within many biological contexts (55, 56) Therefore, the free energy change of the proton transfer process from to should be positive, and any singly protonated state should place the proton on the B monomer, creating the state.
We measure this effect explicitly through the alchemical transition of a proton on transferring to . RETI calculations suggest that the movement of the proton from the to is uphill by 1.53 0.12 kcal/mol, equivalent to a shift of 1.08 0.08 units. The positive G is expected based on the asymmetrical interaction between Y60 and E14, confirming that monomer B will be protonated to a greater degree than monomer A. In fact, since the measured 1.7- unit shift at C (27) is equivalent to a 2.4 kcal at room temperature, the overall accuracy of the RETI calculations performed is to within 1 kcal, approximately the limit of our current classical force fields.
Connecting this evidence to the nonequilibrium work measurements, favoring the state over the state would raise the barrier to transition and would delay conformational transition until the second proton binds or the proton hops with small probability to . In the context of a free-exchange model featuring exchanges of singly protonated EmrE (25), our expectation is that only would be protonated, leading to a transport rate closer to that of the Apo state (40 ) where the locking interaction is in place rather than the state where the locking interaction is broken (220 ) (18). Our simulations do not test explicitly whether the – locking interaction would be retained if both and a single proton are bound to EmrE, as is required for a free-exchange model (25). However, since the locking interaction is also in place even when is bound (Fig. 6A), and is the preferred protonation site based on NMR data (25), we hypothesize that the locking – interaction remains when is bound.
Conclusions
After substantial refinement based on the electron density maps provided by previous cryo-EM experiments (5), we have generated a stable dimeric structure of EmrE that is sequence complete and includes side chains of all residues. Based on traditional crystallographic metrics, the quality of the resulting model improves substantially on the initial low-resolution X-ray structure. This high structural quality is in large part due to the interactive refinement used to extend the transmembrane helices to span the full membrane bilayer and position side chains such as proline to be consistent with protein population averages.
With this model in hand, we provide mechanistic insight into the regulation of the EmrE transport cycle that had been unattainable previously, identifying interactions responsible for changes in observed conformational transition rates (8, 18) and their implications for new transport cycle models (25). Our simulations indicate that transferring the proton from the state to create requires approximately enough energy (1.5 kcal) to break a single hydrogen bond (57), definitively indicating that protonates first, and that accepts the second proton. This binding sequence is due to the asymmetry of cross-monomer Y60–E14 interactions, with in a position to hydrogen bond with , but not vice versa. Nonequilibrium work measurements indicate a lower barrier to transition whenever is protonated and the – interaction is broken. This evidence strongly suggests that Y60 is the “electrostatic lock” that couples protonation to conformational change. The mechanistic importance of Y60 may explain its high conservation across the larger SMR family and suggests that novel drugs that mimic the locking action may improve the efficacy of existing antibiotics. In addition to the electrostatic lock, the hydration of the EmrE lumen also responds strongly to changes in the protonation state and transitions spontaneously into an occluded state when both and are protonated. Together with recent studies highlighting similar interactions regulating conformational changes in other drug transporters (52), the evidence presented here suggests that these pH-dependent locking interactions may be a general mechanism by which proton binding can be connected to conformational change in transporters.
Future experimental datasets, particularly from NMR spectra and recent EPR data (33), could be used in conjunction with the current model for further refinement. Future successor models that incorporate these additional datasets may shed light on questions that have not been addressed. These include the relevance of the occasionally observed lipid intercalation into the dimer interface, the ingress and egress pathways of the substrate, and under what protonation conditions substrate binding can take place, such as adding to singly protonated states (25). Another open question is how the behavior of substrate and protonation state changes under in vivo conditions, which include a substantial electrical potential (58) opposing the egress of charged substrate. Answering these questions will be essential to further exploring how drug export is coupled to protonation in EmrE as well as in the broader SMR family.
Materials and Methods
There are two primary accomplishments, the creation of the faRM itself and the simulations conducted once the faRM was in hand. Both are detailed extensively in SI Appendix and briefly described here.
faRM Creation.
The original basis for the faRM is the X-ray structure of a -bound EmrE dimer (10), which was completed using a combination of psfgen (59) and Modeller (60, 61) software. This model was further refined based on fitting the intermediate structure into an electron density derived from cryo-EM experiments (5), using the MDFF (22) to steer atoms to areas of high electron density. The fitting procedure was carried out interactively (23) with the dimer embedded in an implicit solvent representation (62, 63), leveraging GPU acceleration of the implicit solvent model (64) in NAMD (59) to permit interactivity while the fitting was being carried out. Applying interactive forces accelerated structural changes by allowing high-conformational transition barriers to be overcome over a short timescale. Restraints maintained chirality during interactive fitting and adjustment of the structure (65).
After refinement, the model was embedded into a bilayer of DMPC lipids, which was created using CHARMM-GUI (66, 67). The membrane-embedded structure was simulated again with the MDFF restraints retained to equilibrate the lipids around the inserted EmrE dimer. The final product of this process is the faRM, which was simulated using five different loading states: the apo state, a -bound state, two singly protonated states (either or protonated), and a doubly protonated state.
faRM Simulations.
The simulations carried out all use the CHARMM36 force field for proteins (68), lipids (69), and TIP3 water (70). Parameters for the substrate were determined using a combination of CGenFF (71) and reoptimization of the charges near the phosphorus center, using the force field toolkit (72). The parameters are available in SI Appendix.
Each of the five loading states was equilibrated using NAMD (59) for 25 ns under a NPT ensemble to equilibrate the size of the periodic box dimensions. This structure was then converted into a GROMACS-compatible format using TopoGromacs (73). Unbiased equilibrium trajectories of the five different loading states were then repeated five times for 500 ns each using GROMACS 5.0.4 (74–76) under a NVT ensemble, for an aggregate simulation time of 12.5 s. These simulations were used for much of the analysis, which was carried out with the NumPy (77), SciPy, NetworkX (78), and Matplotlib (79) python packages.
The final snapshots from these equilibrium simulations were also used as the starting point for biased simulations to drive the structural transition associated with substrate transport. Since EmrE is an antisymmetric homodimer (8), the target structure for these steered MD calculations was determined by symmetry from the end state of the equilibrium simulations, effectively driving the individual monomers to swap conformations. The COLVARS module (80) of NAMD (59) was used to drive the transition by with the rmsd to the target state as the reaction coordinate. Additional RETI calculations (26) were carried out to determine the relative of and .
Supplementary Material
Acknowledgments
We acknowledge Katherine Henzler-Wildman, Nate Traaseth, Chao Wu, and Reza Dastvan for helpful conversations that guided the research direction. Additionally, we thank Reza Dastvan and Hassane Mchaourab for providing the spin-label distances in a text format for comparison with our own results. J.V.V. and S.B.R. gratefully acknowledge support from the Sandia National Laboratories (SNL) Campus Executive Program, which is funded by the Laboratory Directed Research and Development Program. This project made extensive use of the high-performance computing resources provided by SNL. The project also received support from the Defense Threat Reduction Agency–Joint Science and Technology Office for Chemical and Biological Defense (Interagency Agreement DTRA10027IA-3167, to S.B.R.). SNL is a multimission laboratory managed and operated by National Technology and Engineering Solutions of Sandia LLC, a wholly owned subsidiary of Honeywell International Inc. for the US Department of Energy’s (DOE) National Nuclear Security Administration under Contract DE-NA0003525. This work was performed, in part, at the Center for Integrated Nanotechnologies, an Office of Science User Facility operated for the US DOE’s Office of Science by Los Alamos National Laboratory (Contract DE-AC52-06NA25296) and SNL. This research is also supported by the National Institutes of Health, through Grants P41-GM104601 and U54-GM087519 (to E.T.). The views expressed in this article do not necessarily represent the views of the US DOE or the US government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 8060.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722399115/-/DCSupplemental.
References
- 1. Schuldiner S, The Escherichia coli effluxome. Res Microbiol, in press. [DOI] [PubMed]
- 2.Yerushalmi H, Lebendiker M, Schuldiner S. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem. 1995;270:6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]
- 3.Morrison EA, Henzler-Wildman KA. Transported substrate determines exchange rate in the multidrug resistance transporter EmrE. J Biol Chem. 2014;289:6825–6836. doi: 10.1074/jbc.M113.535328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korkhov VM, Tate CG. Electron crystallography reveals plasticity within the drug binding site of the small multidrug transporter EmrE. J Mol Biol. 2008;377:1094–1103. doi: 10.1016/j.jmb.2008.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ubarretxena-Belandia I, Baldwin JM, Schuldiner S, Tate CG. Three-dimensional structure of the bacterial multidrug transporter EmrE shows it is an asymmetric homodimer. EMBO J. 2003;22:6175–6181. doi: 10.1093/emboj/cdg611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler P, Ubarretxena-Belandia I, Warne T, Tate C. The Escherichia coli multidrug transporter EmrE is a dimer in the detergent-solubilised state. J Mol Biol. 2004;340:797–808. doi: 10.1016/j.jmb.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Bay DC, Rommens KL, Turner RJ. Small multidrug resistance proteins: A multidrug transporter family that continues to grow. Biochim Biophys Acta Biomembr. 2008;1778:1814–1838. doi: 10.1016/j.bbamem.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 8.Morrison EA, et al. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature. 2012;481:45–50. doi: 10.1038/nature10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gayen A, Banigan JR, Traaseth NJ. Ligand-induced conformational changes of the multidrug resistance transporter EmrE probed by oriented solid-state NMR spectroscopy. Angew Chem Int Ed. 2013;52:10321–10324. doi: 10.1002/anie.201303091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen YJ, et al. X-ray structure of EmrE supports dual topology model. Proc Natl Acad Sci USA. 2007;104:18999–19004. doi: 10.1073/pnas.0709387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korkhov VM, Tate CG. An emerging consensus for the structure of EmrE. Acta Crystallogr D Biol Crystallogr. 2009;65:186–192. doi: 10.1107/S0907444908036640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Julius A, Laur L, Schanzenbach C, Langosch D. BLaTM 2.0, a genetic tool revealing preferred antiparallel interaction of transmembrane helix 4 of the dual-topology protein EmrE. J Mol Biol. 2017;429:1630–1637. doi: 10.1016/j.jmb.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell P. Translocations through natural membranes. In: Nord FF, editor. Advances in Enzymology and Related Areas of Molecular Biology. Wiley; Hoboken, NJ: 1967. pp. 33–87. [DOI] [PubMed] [Google Scholar]
- 15.Vergara-Jaque A, Fenollar-Ferrer C, Kaufmann D, Forrest LR. Repeat-swap homology modeling of secondary active transporters: Updated protocol and prediction of elevator-type mechanisms. Front Pharmacol. 2015;6:183. doi: 10.3389/fphar.2015.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moradi M, Enkavi G, Tajkhorshid E. Atomic-level characterization of transport cycle thermodynamics in the glycerol-3-phosphate:phosphate transporter. Nat Commun. 2015;6:8393. doi: 10.1038/ncomms9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho MK, Gayen A, Banigan JR, Leninger M, Traaseth NJ. Intrinsic conformational plasticity of native EmrE provides a pathway for multidrug resistance. J Am Chem Soc. 2014;136:8072–8080. doi: 10.1021/ja503145x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gayen A, Leninger M, Traaseth NJ. Protonation of a glutamate residue modulates the dynamics of the drug transporter EmrE. Nat Chem Biol. 2016;12:141–145. doi: 10.1038/nchembio.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muth GW, Ortoleva-Donnelly L, Strobel SA. A single adenosine with a neutral pka in the ribosomal peptidyl transferase center. Science. 2000;289:947–950. doi: 10.1126/science.289.5481.947. [DOI] [PubMed] [Google Scholar]
- 20.Yerushalmi H, Schuldiner S. An essential Glutamyl residue in EmrE, a multidrug antiporter from Escherichia coli. J Biol Chem. 2000;275:5264–5269. doi: 10.1074/jbc.275.8.5264. [DOI] [PubMed] [Google Scholar]
- 21.Ong YS, Lakatos A, Becker-Baldus J, Pos KM, Glaubitz C. Detecting substrates bound to the secondary multidrug efflux pump EmrE by DNP-enhanced solid-state NMR. J Am Chem Soc. 2013;135:15754–15762. doi: 10.1021/ja402605s. [DOI] [PubMed] [Google Scholar]
- 22.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stone JE, Gullingsrud J, Grayson P, Schulten K. A system for interactive molecular dynamics simulation. In: Hughes JF, Séquin CH, editors. 2001 ACM Symposium on Interactive 3D Graphics. ACM SIGGRAPH; New York: 2001. pp. 191–194. [Google Scholar]
- 24.Rotem D, Schuldiner S. EmrE, a multidrug transporter from Escherichia coli, transports monovalent and divalent substrates with the same stoichiometry. J Biol Chem. 2004;279:48787–48793. doi: 10.1074/jbc.M408187200. [DOI] [PubMed] [Google Scholar]
- 25.Robinson AE, Thomas NE, Morrison EA, Balthazor BM, Henzler-Wildman KA. New free-exchange model of EmrE transport. Proc Natl Acad Sci USA. 2017;114:E10083–E10091. doi: 10.1073/pnas.1708671114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woods CJ, Essex JW, King MA. The development of replica-exchange-based free-energy methods. J Phys Chem B. 2003;107:13703–13710. [Google Scholar]
- 27.Morrison EA, Robinson AE, Liu Y, Henzler-Wildman KA. Asymmetric protonation of EmrE. J Gen Physiol. 2015;146:445–461. doi: 10.1085/jgp.201511404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fleishman SJ, et al. Quasi-symmetry in the cryo-EM structure of EmrE provides the key to modeling its transmembrane domain. J Mol Biol. 2006;364:54–67. doi: 10.1016/j.jmb.2006.08.072. [DOI] [PubMed] [Google Scholar]
- 29.Padariya M, Kalathiya U, Baginski M. Structural and dynamic changes adopted by EmrE, multidrug transporter protein–Studies by molecular dynamics simulation. Biochim Biophys Acta Biomembr. 2015;1848:2065–2074. doi: 10.1016/j.bbamem.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 30.Rotem D, Steiner-Mordoch S, Schuldiner S. Identification of tyrosine residues critical for the function of an ion-coupled multidrug transporter. J Biol Chem. 2006;281:18715–18722. doi: 10.1074/jbc.M602088200. [DOI] [PubMed] [Google Scholar]
- 31.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Cryst D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. Procheck: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 33.Dastvan R, Fischer AW, Mishra S, Meiler J, Mchaourab HS. Protonation-dependent conformational dynamics of the multidrug transporter EmrE. Proc Natl Acad Sci USA. 2016;113:1220–1225. doi: 10.1073/pnas.1520431113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Law RJ, et al. Membrane protein structure quality in molecular dynamics simulation. J Mol Graphics Model. 2005;24:157–165. doi: 10.1016/j.jmgm.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Frishman D, Argos P. Knowledge-based secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 36.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 37.Beitz E. TeXshade: Shading and labeling of multiple sequence alignments using LaTeX2e. Bioinformatics. 2000;16:135–139. doi: 10.1093/bioinformatics/16.2.135. [DOI] [PubMed] [Google Scholar]
- 38.Barrera FN, Fendos J, Engelman DM. Membrane physical properties influence transmembrane helix formation. Proc Natl Acad Sci USA. 2015;109:14422–14427. doi: 10.1073/pnas.1212665109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popot JL, Engelman DM. Helical membrane protein folding, stability, and evolution. Annu Rev Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 40.Li J, et al. Transient formation of water-conducting states in membrane transporters. Proc Natl Acad Sci USA. 2013;110:7696–7701. doi: 10.1073/pnas.1218986110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hassanali A, Giberti F, Cuny J, Kühne TD, Parrinello M. Proton transfer through the water gossamer. Proc Natl Acad Sci USA. 2013;110:13723–13728. doi: 10.1073/pnas.1306642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brill S, Falk OS, Schuldiner S. Transforming a drug/H+ antiporter into a polyamine importer by a single mutation. Proc Natl Acad Sci USA. 2012;109:16894–16899. doi: 10.1073/pnas.1211831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Rath A, Deber CM. Functional response of the small multidrug resistance protein EmrE to mutations in transmembrane helix 2. FEBS Lett. 2014;588:3720–3725. doi: 10.1016/j.febslet.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 44.Brill S, Sade-Falk O, Elbaz-Alon Y, Schuldiner S. Specificity determinants in small multidrug transporters. J Mol Biol. 2015;427:468–477. doi: 10.1016/j.jmb.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 45.Yerushalmi H, Lebendiker M, Schuldiner S. Negative dominance studies demonstrate the oligomeric structure of EmrE, a multidrug antiporter from Escherichia coli. J Biol Chem. 1996;271:31044–31048. doi: 10.1074/jbc.271.49.31044. [DOI] [PubMed] [Google Scholar]
- 46.Vermaas JV, Tajkhorshid E. Conformational heterogeneity of -synuclein in membrane. Biochim Biophys Acta Biomembr. 2014;1838:3107–3117. doi: 10.1016/j.bbamem.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vermaas JV, et al. Mechanism of lignin inhibition of enzymatic biomass deconstruction. Biotechnol Biofuels. 2015;8:217. doi: 10.1186/s13068-015-0379-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayne CG, et al. The cellular membrane as a mediator for small molecule interaction with membrane proteins. Biochim Biophys Acta Biomembr. 2016;1858:2290–2304. doi: 10.1016/j.bbamem.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flewelling R, Hubbell W. Hydrophobic ion interactions with membranes. Thermodynamic analysis of tetraphenylphosphonium binding to vesicles. Biophys J. 1986;49:531–540. doi: 10.1016/S0006-3495(86)83663-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boiteux C, et al. Local anesthetic and antiepileptic drug access and binding to a bacterial voltage-gated sodium channel. Proc Natl Acad Sci USA. 2014;111:13057–13062. doi: 10.1073/pnas.1408710111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hurst DP, et al. A lipid pathway for ligand binding is necessary for a cannabinoid G protein-coupled receptor. J Biol Chem. 2010;285:17954–17964. doi: 10.1074/jbc.M109.041590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yue Z, Chen W, Zgurskaya HI, Shen J. Constant pH molecular dynamics reveals how proton release drives the conformational transition of a transmembrane efflux pump. J Chem Theor Comput. 2017;13:6405–6414. doi: 10.1021/acs.jctc.7b00874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tal N, Schuldiner S. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc Natl Acad Sci USA. 2009;106:9051–9056. doi: 10.1073/pnas.0902400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moradi M, Tajkhorshid E. Computational recipe for efficient description of large-scale conformational changes in biomolecular systems. J Chem Theor Comput. 2014;10:2866–2880. doi: 10.1021/ct5002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shokri A, Abedin A, Fattahi A, Kass SR. Effect of hydrogen bonds on pKa values: Importance of networking. J Am Chem Soc. 2012;134:10646–10650. doi: 10.1021/ja3037349. [DOI] [PubMed] [Google Scholar]
- 56.Huang Y, Yue Z, Tsai CC, Henderson JA, Shen J. Predicting catalytic proton donors and nucleophiles in enzymes: How adding dynamics helps elucidate the structure–function relationships. J Phys Chem Lett. 2018;9:1179–1184. doi: 10.1021/acs.jpclett.8b00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Myers J, Pace C. Hydrogen bonding stabilizes globular proteins. Biophys J. 1996;71:2033–2039. doi: 10.1016/S0006-3495(96)79401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bot CT, Prodan C. Quantifying the membrane potential during E. coli growth stages. Biophys Chem. 2010;146:133–137. doi: 10.1016/j.bpc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 59.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martí-Renom MA, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 61.Eswar N, et al. Comparative protein structure modeling using MODELLER. Current Protocols in Protein Science. 2007;50:2.9.1–2.9.31. doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 62.Still WC, Tempczyk A, Hawley RC, Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J Am Chem Soc. 1990;112:6127–6129. [Google Scholar]
- 63.Onufriev A, Bashford D, Case DA. Exploring protein native states and large-scale conformational changes with a modified generalized Born model. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 64.Tanner DE, Phillips JC, Schulten K. GPU/CPU algorithm for generalized Born/solvent-accessible surface area implicit solvent calculations. J Chem Theor Comput. 2012;8:2521–2530. doi: 10.1021/ct3003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jo S, Kim T, Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comp Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 68.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone , and side-chain and dihedral angles. J Chem Theor Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jorgensen W, Chandrasekhar J, Maudura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 71.Vanommeslaeghe K, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comp Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mayne CG, Saam J, Schulten K, Tajkhorshid E, Gumbart JC. Rapid parameterization of small molecules using the force field toolkit. J Comp Chem. 2013;34:2757–2770. doi: 10.1002/jcc.23422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vermaas JV, Hardy DJ, Stone JE, Tajkhorshid E, Kohlmeyer A. TopoGromacs: Automated topology conversion from CHARMM to GROMACS within VMD. J Chem Inf Model. 2016;56:1112–1116. doi: 10.1021/acs.jcim.6b00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abraham MJ, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- 75.Pronk S, et al. Gromacs 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van der Spoel D, et al. Gromacs: Fast, flexible, and free. J Comp Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 77.Van Der Walt S, Colbert SC, Varoquaux G. The NumPy array: A structure for efficient numerical computation. Comput Sci Eng. 2011;13:22–30. [Google Scholar]
- 78.Hagberg A, Swart P, Chult D. Exploring network structure, dynamics, and function using NetworkX. In: Varoquaux G, Travis V, Millman J, editors. Proceedings of the 7th Python in Science Conference (SciPy2008) Pasedena, CA: 2008. pp. 11–15. [Google Scholar]
- 79.Hunter JD. Matplotlib: A 2d graphics environment. Comput Sci Eng. 2007;9:90–95. [Google Scholar]
- 80.Fiorin G, Klein ML, Hénin J. Using collective variables to drive molecular dynamics simulations. Mol Phys. 2013;111:3345–3362. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.