Significance
Gram-negative bacteria possess an outer membrane (OM) which reduces antibiotic effectiveness. The OM has a dense outer layer of lipopolysaccharide (LPS), a glycolipid which is directly targeted by the last-resort antibiotic polymyxin B (PmB). However, we lack accurate molecular data on the PmB–OM interaction to inform future drug development. Here we study the PmB–OM interaction in vitro and show that in vivo results are reproduced only when the LPS is in a liquid crystalline phase, which occurs at body temperature. These findings not only explain the temperature dependence of PmB function but also support the notion that bacteria actively control the viscosity of their outer membranes as growth temperatures vary.
Keywords: outer membrane, polymyxin, neutron reflectometry, lipopolysaccharide, phase transition
Abstract
The outer membrane (OM) of Gram-negative bacteria is a robust, impermeable, asymmetric bilayer of outer lipopolysaccharides (LPSs) and inner phospholipids containing selective pore proteins which confer on it the properties of a molecular sieve. This structure severely limits the variety of antibiotic molecules effective against Gram-negative pathogens and, as antibiotic resistance has increased, so has the need to solve the OM permeability problem. Polymyxin B (PmB) represents those rare antibiotics which act directly on the OM and which offer a distinct starting point for new antibiotic development. Here we investigate PmB’s interactions with in vitro OM models and show how the physical state of the lipid matrix of the OM is a critical factor in regulating the interaction with the antimicrobial peptide. Using neutron reflectometry and infrared spectroscopy, we reveal the structural and chemical changes induced by PmB on OM models of increasing complexity. In particular, only a tightly packed model reproduced the temperature-controlled disruption of the asymmetric lipid bilayer by PmB observed in vivo. By measuring the order of outer-leaflet LPS and inner-leaflet phospholipids, we show that PmB insertion is dependent on the phase transition of LPS from the gel to the liquid crystalline state. The demonstration of a lipid phase transition in the physiological temperature range also supports the hypothesis that bacteria grown at different temperatures adapt their LPS structures to maintain a homeoviscous OM.
Gram-negative bacteria lack the thick peptidoglycan cell wall of Gram-positive bacteria; instead, they have an additional semipermeable outer membrane (OM) (1). This unique biological membrane is composed of a phospholipid inner leaflet surrounded by an outer layer of large, peculiar glycolipids termed lipopolysaccharides (LPSs) (Fig. 1A) (2). The selective barrier function of the OM relies heavily on its asymmetric lipid distribution, which is established by the lipopolysaccharide transport (Lpt) protein system (3) and preserved by the specialist maintenance of lipid asymmetry (Mla) family of OM proteins (4), along with the OM enzymes PldA and PagP (2). To kill Gram-negatives, most antimicrobials (e.g., β-lactams) avoid the hydrophobic core of the OM and enter the cell via the water-filled protein pores (Fig. 1A), which normally enable bacterial uptake of water-soluble nutrients (1, 5–7). Consequently, mutations of these pores that prevent antibiotic entry are a significant cause of antibiotic resistance (8). Furthermore, restricted antibiotic influx through the OM indirectly increases the effectiveness of a wide range of other resistance strategies which either degrade, actively pump out, or alter the target of clinically important drugs. Four out of six “ESKAPE” organisms responsible for most hospital-acquired infections are Gram-negative bacteria: Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. A better understanding of the properties of the OM is therefore of primary importance to tackle antibiotic resistance (6).
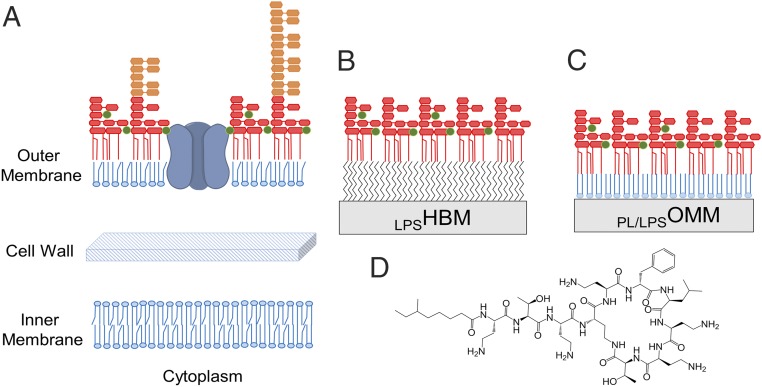
Fig. 1.
Gram-negative outer membrane and models. (A) Schematic representation of the Gram-negative cell envelope. The cytoplasm is enclosed by a phospholipid inner membrane (proteins not shown) surrounded by a thin peptidoglycan cell wall. The outer layer is the asymmetric outer membrane composed of phospholipids in the inner leaflet and lipopolysaccharides in the outer leaflet. It contains water-filled channel proteins (porins) through which most antibiotics enter the cell. The LPS is shown as two forms, a rough LPS (red) containing the lipid A and core oligosaccharide, and variable lengths of O-antigen present in smooth strains of Gram-negative bacteria in orange. Stabilizing divalent cations are shown in green. (B) Simple OM model formed by a hybrid bilayer membrane containing a monolayer of LPS self-assembled on a hydrophobic monolayer. (C) Asymmetric OM model assembled in vitro on a silicon substrate by sequential deposition of synthetic phospholipids and purified rough LPS using a Langmuir trough. (D) Chemical structure of polymyxin B.
Since OM penetration is increasingly recognized as a limiting factor in the development of effective antimicrobials (8), accurate assays to investigate antibiotic–OM interactions are needed. The OM is difficult to precisely address by biophysical methods in vivo, largely because of the close proximity of the inner membrane. This has led to considerable debate concerning its structure and dynamics (9). Although its asymmetric lipid distribution was initially controversial, it is now widely accepted that LPS is the major lipid component of the outer leaflet while phospholipids are confined to the periplasmic side of the membrane (1). However, while plasma membranes are generally accepted to be fluid systems, the physical state of OM lipids remains unclear. While some studies on isolated LPS show a transition to a liquid crystalline phase at physiological temperatures (10), others suggest that the structured OM may remain in a gel-like state on the basis of its observed physicochemical properties (1). Reports of bacterial cells synthesizing lower–melting-point LPS when grown at lower temperatures suggest that homeoviscous adaptation to maintain a liquid crystalline phase is physiologically relevant (11–14).
LPS is a complex glycolipid composed of three sections: (i) the hydrophobic lipid A formed from anionic, phosphorylated diglucosamine that is usually hexaacylated; (ii) the oligosaccharide core region of about 10 sugar units, which also carry anionic groups; and (iii) the distal O-antigen region containing up to 100 oligosaccharide repeats (Fig. 1A) (1). A short LPS, formed only by the lipid A and core oligosaccharide, is typical of most laboratory Escherichia coli termed rough strains (1). LPS with a highly variable number of O-antigens is characteristic of smooth strains but, even here, up to 60% of their LPS contains either no or one O-antigen unit (15).
Polymyxin B (PmB) is a mixture of closely related LPS-binding amphiphiles which combine an acyl tail with a cyclic cationic peptide (Fig. 1D) (16). Its status as last-resort treatment against pan-resistant Gram-negative infections has reinforced interest in the molecular basis of PmB’s unique interaction with the OM. Moreover, its distinctive physicochemical properties place it in a niche isolated from the chemical space occupied by other antibiotics active against Gram-negative bacteria, making it an interesting starting point for the design of new antimicrobials (17, 18).
To better understand LPS–PmB interactions, we have adopted the use of in vitro models which can be studied using a range of biophysical methods, including neutron scattering. Two models are used here. The first, which can be easily created with nonspecialist equipment, simulates only the outer surface of the OM by the simple assembly of an LPS monolayer on a hydrophobic surface (19). The second, developed by us over several years (20–22), permits the formation of both the outer LPS and the inner phospholipid leaflet to form a complete asymmetric OM-like bilayer. In this paper, we report the interaction of PmB with these in vitro OM models (Fig. 1 B and C). We show that the dynamics of the membrane’s hydrophobic core critically determine how PmB binds to the asymmetric OM model, and that in vivo behavior is reproduced by densely packed LPS in a liquid crystalline phase.
Results
PmB Binds to Self-Assembled LPS Monolayers at Room Temperature.
Initially, we created a hybrid bilayer membrane (HBM) consisting of an LPS monolayer (Ra chemotype) deposited via self-assembly onto a hydrophobic surface (LPSHBM) (Fig. 1B) (19). These easily formed HBMs enable the binding of PmB to be probed by a range of biophysical techniques (23, 24). We monitored the formation of the LPSHBM, on a silicon wafer coated with hydrophobic octadecyltrichlorosilane (OTS), by attenuated total internal reflection–Fourier transform infrared spectroscopy (ATR-FTIR) until saturation of the surface (SI Appendix, Fig. S1A). ATR-FTIR yields information on the amount and chemical nature of the species at the interface by measuring the infrared absorbance of the molecules adsorbed on a substrate. Following the addition of PmB to the solution bathing the LPSHBM, a large increase in the amide bond absorbance at 1,650 cm−1 revealed binding of the peptide antibiotic to the lipid-coated surface (Fig. 2A). PmB binding shifted the C–H stretching vibrations toward higher wavenumbers, indicating decreased ordering of the acyl-chain region of the monolayer and suggesting the intercalation of disordered lipopeptide acyl tails into the LPS hydrophobic core. The same interaction studied quantitatively by surface plasmon resonance (SPR) showed a clear relationship between PmB concentration and the amount of bound lipopeptide, which saturated at concentrations above 10 µg/mL (Fig. 2B and SI Appendix, Fig. S1B).
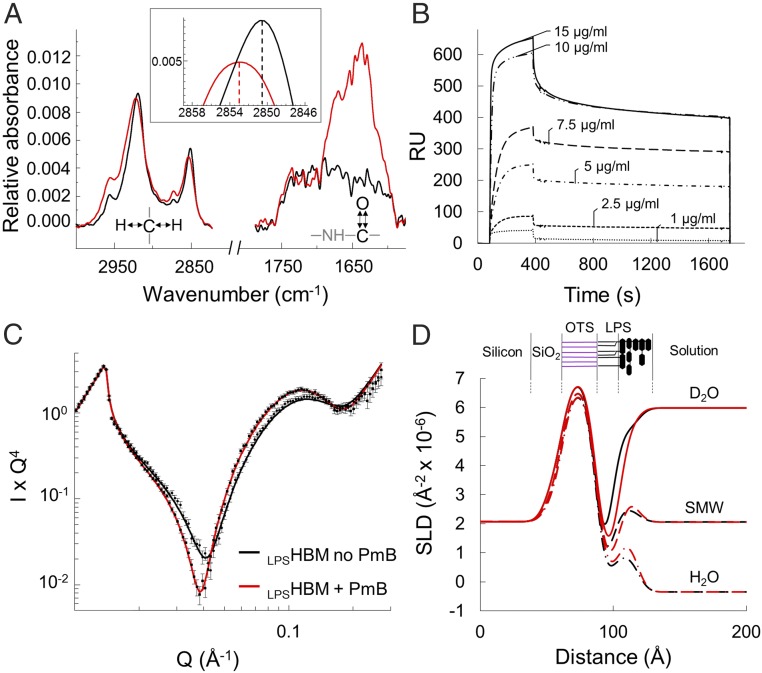
Fig. 2.
PmB binds to LPSHBM at room temperature. (A) Infrared spectra of an LPS monolayer on OTS before (black) and after (red) addition of PmB (100 µg/mL), showing a prominent increase in the amide band at 1,650 cm−1 and the shift of the signal of the C–H stretching vibrations around 2,900 cm−1. Magnified in the Inset, the peak shift of the symmetric CH2 vibration indicating the increased disorder of the hydrocarbon chains. (B) Concentration-dependent binding of PmB to the LPSHBM measured by surface plasmon resonance and reported as resonance units (RU). The antimicrobial concentration during the injection phase (80 to 400 s) is reported in µg/mL next to each binding curve. This is followed by a PmB-free buffer wash to measure the stability of the binding. (C) Neutron reflectivity data points and fit lines obtained before (black) and after (red) the interaction of PmB (100 µg/mL) with the LPSHBM on deuterated OTS in D2O, showing the shift to lower Q of the reflectivity after the binding event. (D) Scattering length density profiles obtained from fitting the reflectivity data in three solution contrasts: D2O (line), silicon-matched water (dashes), and H2O (dash-dot). The three datasets were constrained to fit to a common model of the interface (SI Appendix, Fig. S2 and Table S1; see Materials and Methods for NR data analysis). Results show the change in thickness of the hydrogen-rich LPS tail region before (black) and after (red) interaction with PmB. A representation of the LPSHBM is shown above the profile showing the deuterated OTS in purple and LPS in black.
Next, we went on to examine the structural aspects of this interaction using neutron reflectometry (NR). NR provides structural and chemical information on the distribution of molecules along the axis perpendicular to the surface. NR’s unique strength is in its ability to detect differences in neutron scattering length density (SLD, or “neutron refractive index”), which relate to the chemical composition of the material. In a biological context, the large difference between the SLD of hydrogen and deuterium enables the exploitation of deuterated molecules to generate contrast between otherwise chemically indistinguishable regions. We thus created an LPSHBM on a deuterated OTS layer to clearly resolve the structure of the hydrogen-rich LPS monolayer. Neutron reflectivity profiles were collected at three isotopic solution contrasts by varying the amount of D2O in the aqueous buffer bathing the monolayer (Fig. 2C and SI Appendix, Fig. S2). The constrained fit to the three datasets yielded the structure of the LPSHBM described by its SLD profile perpendicular to the surface (Fig. 2D). The LPS monolayer was clearly resolved from the deuterated OTS and coated the hydrophobic surface covering 92.3 ± 4.5% of the substrate (SI Appendix, Table S1).
Against a D2O solution background, the neutron reflectivity profile for the isotopically asymmetric LPSHBM displays undulations known as “Kiessig fringes,” which are sensitive to subtle structural alterations in the hydrogen-rich acyl tail region of LPS (Fig. 2C) (20). The binding of PmB to the monolayer resulted in a visible shift of the reflectivity profile toward lower values of scattering vector (Q), corresponding to an increased thickness of the LPSHBM. The fit to the data confirmed an increased thickness of the LPS tail region from 14.2 ± 0.7 Å to 18.0 ± 1.3 Å and a displacement of ∼28% of the volume of water associated with the core oligosaccharide (Fig. 2D and SI Appendix, Table S1). The changes in the monolayer structure revealed by NR are consistent with a PmB–LPS model proposed by NMR studies in which the PmB hydrophobic moiety inserts into the hydrophobic acyl-chain region of LPS while the cationic cyclic peptide binds to the inner carbohydrate head group (25). Additionally, the data show a reduction in the thickness of the LPS core oligosaccharide layer of ∼8 Å, which suggests a partial collapse induced by antimicrobial binding. Furthermore, the binding affinity measured by SPR is within the range of the minimal inhibitory concentration (26).
Temperature Regulates PmB Binding to the Asymmetric Bilayer Model OM.
We next wished to examine whether PmB penetrates beyond the LPS layer to interact with the phospholipid inner leaflet of the OM. For this purpose, we used specialized OM models previously developed in our group (20–22). Here an asymmetric phospholipid–LPS outer-membrane model (PL/LPSOMM) is created using Langmuir–Blodgett and Langmuir–Schaefer deposition steps, which sequentially transfer a phospholipid monolayer, followed by an LPS monolayer, from the air–water interface onto a solid substrate (Fig. 1C). The lipid monolayers are compressed to in vivo surface pressures, which maximizes both the lateral packing of the lipids and the stability of the resulting asymmetric model (27–29). To enable NR resolution of the two leaflets, the resulting asymmetric OM models contained deuterated phospholipids in the inner leaflet and natural hydrogenous LPS in the outer leaflet (dPL/LPSOMM) (20). NR measurements were again performed using three isotopic solution contrasts, and the resulting reflectivity datasets were fitted to a single model of the interface as above. The resulting SLD profiles described a highly asymmetric bilayer with less than 1% of the hydrophobic core volume fraction occupied by water, indicating both comprehensive coverage and tight lipid packing (Fig. 3 A and B and SI Appendix, Table S2). The water associated with the hydrophilic phospholipid head groups amounted to ∼50% of the volume occupied by the phosphatidylcholine moiety, a value in line with previously characterized high-coverage asymmetric bilayers displaying inner–head-group hydration values of ∼60% (20). Overall, the membrane structure was found to be in very good agreement with previously published values for solid-supported asymmetric phospholipid–LPS bilayers (20, 28). However, in sharp contrast with what we observed under the same conditions with the LPSHBM, the reflectivity from dPL/LPSOMM was completely unaffected by the addition of 100 µg/mL PmB (Fig. 3C). This lack of interaction led us to hypothesize that the controlled assembly afforded by the Langmuir–Blodgett and –Schaefer depositions yielded a tighter molecular arrangement that prevented the intercalation of PmB within the model membrane. Considering that the action of PmB has been shown to be highly temperature-dependent (30–32), we sought to discover if this effect could be reproduced in our model.
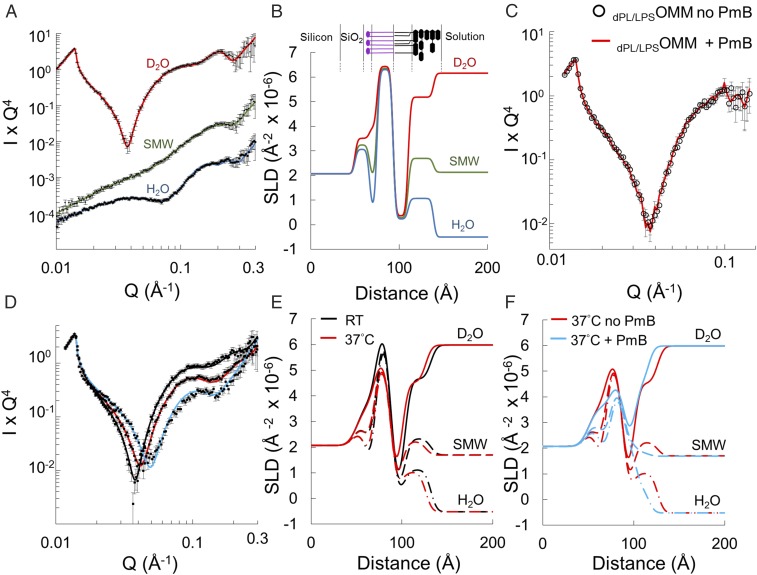
Fig. 3.
Temperature-dependent disruption of dPL/LPSOMM by PmB. (A) Neutron reflectivity data and best fit lines for a dPL/LPSOMM at room temperature (RT) at three solution contrasts: D2O (red), SMW (green), and H2O (blue). The reduced χ2 values of the fits to the three datasets are 1.04, 0.82, and 0.78, respectively. Data are offset vertically for clarity. (B) SLD profile of the dPL/LPSOMM derived from the fit in A. Cartoon of the dPL/LPSOMM is shown above; deuterated PC inner (purple) and LPS outer leaflet (black). (C) NR in D2O before (empty circles) and after (red line) addition of 100 µg/mL PmB to the dPL/LPSOMM at RT. (D) NR data and best fit lines for a dPL/LPSOMM in D2O at RT (black line) and upon heating to 37 °C (red line) and subsequent addition of 100 µg/mL PmB at 37 °C (blue line). PmB at 37 °C significantly alters the reflectivity. Data were collected in H2O and SMW as well as in D2O (shown here) and used in the constrained fit (SI Appendix, Figs. S3A, S4A, and S5A). (E and F) SLD profiles obtained from fits to data collected at three solution contrasts: D2O (solid lines) (shown in D), SMW (dashes), and H2O (dash-dot) (SI Appendix, Figs. S3–S5) at RT (black) and 37 °C before (red) and after (blue) addition of PmB. Red lines are identical in E and F. Lipid asymmetry remains after heating, but 100 µg/mL PmB at 37 °C mixed the leaflets.
Heating the dPL/LPSOMM to 37 °C in the absence of PmB caused the reflectivity profile to shift toward higher values of scattering vector that corresponded to a decreased total thickness of the model membrane, from ∼70 to ∼64 Å (Fig. 3 D and E). The contrast provided by the asymmetric H and D (2H) isotopic distributions in the hydrophobic region of the bilayer allowed us to assign the main thickness change to the LPS outer leaflet, which decreased by ∼6 Å (SI Appendix, Tables S3 and S4). The dPL/LPSOMM maintained a high level of lipid asymmetry upon heating to 37 °C, retaining more than 90% of LPS in its outer leaflet.
Contrary to what we observed at room temperature, the addition of PmB to the dPL/LPSOMM at 37 °C produced a prominent shift of the reflectivity toward higher Q values (Fig. 3D). The comparison between the SLD profiles of the dPL/LPSOMM at 37 °C before and after the addition of PmB indicated a further reduction in total thickness and a significant mixing of the two lipid layers (Fig. 3F). The final isotopic composition of the hydrophobic core of the PmB-treated bilayer, described by its SLD, showed ∼35% of hydrogenous hydrocarbons (LPS and PmB tails) penetrating the inner leaflet while the outer leaflet gained the same amount of deuterated material (phospholipid tails). The inner- and outer-bilayer head groups exhibited a concomitant loss of asymmetry revealed by the convergence in size and SLD of these regions to values intermediate between those of LPS and phospholipids as a result of the leaflet mixing (SI Appendix, Table S5). The temperature effect was confirmed by stepwise heating of the model membrane in the presence of the antibiotic while monitoring the neutron reflectivity from the bilayer in D2O (Fig. 4A and SI Appendix, Fig. S8).
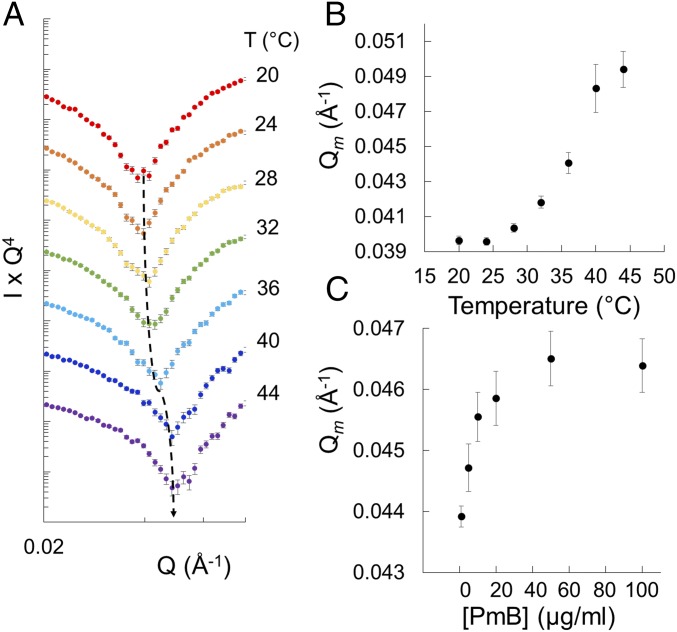
Fig. 4.
Effect of temperature and PmB concentration on dPL/LPSOMM disruption. (A) Temperature-dependent shift of the Kiessig fringe in the neutron reflectivity profile for the dPL/LPSOMM in D2O, heated in the presence of 100 µg/mL PmB. The curves are offset vertically for clarity. The dashed line follows the trough of the Kiessig fringe. Full curves are shown in SI Appendix, Fig. S8. (B) Plot of the shift in Q of the fringe shown in A, quantified as the barycentric mean of the data. (C) Shift in barycentric mean Q of the fringe from a dPL/LPSOMM sample titrated with increasing amounts of PmB at 37 °C (SI Appendix, Fig. S9). The error bars in B and C are associated with the uncertainty of the measured neutron intensity profiles.
Stepwise increases in temperature caused the Kiessig fringe to progressively shift to higher Q values, with a sigmoidal response revealed by the plot of the barycentric mean of the data (Fig. 4B). In the presence of the lipopeptide, minor changes occurred between 20 and 28 °C, while between 32 and 40 °C the reflectivity altered significantly. Heating the dPL/LPSOMM in the presence of PmB led to an overall change in reflectivity identical to that caused by the addition of the antimicrobial to the dPL/LPSOMM at 37 °C. Under the same conditions, the deacylated analog of the lipopeptide, PmB nonapeptide, which lacks bactericidal activity, caused no disruption (SI Appendix, Figs. S6 and S7). The acyl chain of the lipopeptide was thus critical for its interaction with the fluid model membrane. Finally, having so far purposely used a high concentration of PmB, we examined the concentration dependence by adding increasing amounts of PmB to the dPL/LPSOMM at 37 °C (Fig. 4C and SI Appendix, Fig. S9). The effects of the antimicrobial on the model OM were already evident at 2 µg/mL and reached 50% saturation below 10 µg/mL. Increasing the concentration from 50 to 100 µg/mL did not cause any significant further change in reflectivity. Notably, in both the temperature ramp and the titration experiments and at each condition of temperature and PmB concentration, the dPL/LPSOMM reached a stable equilibrium despite a continuous flow of PmB solution in the aqueous phase.
Phase Behavior of OM Models.
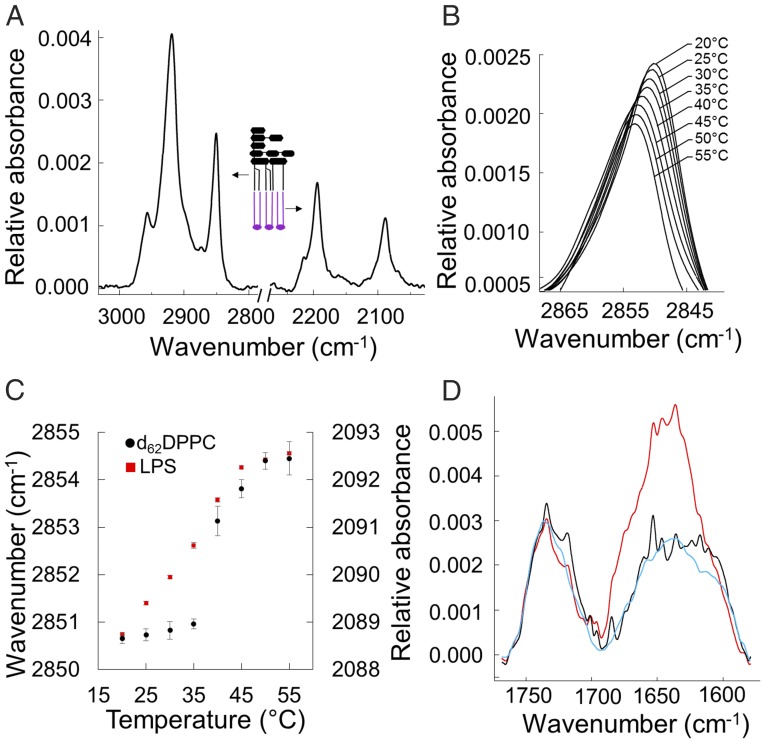
To complement the structural characterization of the model membranes, we investigated the dynamics of the systems using ATR-FTIR. The isotopically asymmetric leaflets [H vs. D (2H)] of the dPL/LPSOMM (Fig. 1C) generate distinct IR signals that enable differentiation between the acyl chains of the phospholipids and those of the LPS. The IR spectrum of the dPL/LPSOMM clearly displays the distinct LPS C–H stretching vibrations between ∼3,000 and ∼2,800 cm−1 and the deuterated phospholipid C–D stretching vibrations between ∼2,250 and ∼2,050 cm−1 (Fig. 5A). The peak absorbance of the H–C–H symmetric bond stretching is strongly affected by the ordering of the aliphatic hydrocarbons and undergoes an abrupt 3- to 4-cm−1 shift as the acyl chains undergo the gel-to-liquid phase transition (10) (Fig. 5B). As we gradually increased the temperature of the dPL/LPSOMM from 20 to 55 °C, both leaflets of the bilayer changed from gel to liquid disordered states (Fig. 5C). As we expected, the synthetic phospholipid displayed a sharp transition while the chemically heterogeneous, natural LPS extract displayed a more gradual peak shift. The midpoints (Tm) calculated from nonlinear regressions of data from two independent samples showed a Tm of 36.2 ± 1.6 °C and 39.3 ± 0.4 °C for LPS and tail-deuterated DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine), respectively, in good agreement with data published for the two separate lipids in suspension (10, 33). Cooling the sample to room temperature caused the IR peaks to return to their original positions, confirming the reversibility of the measured phase transition (SI Appendix, Fig. S10A). The same experiment on the LPSHBM showed a similar gel-to-liquid crystalline phase transition upon heating to physiological temperature (SI Appendix, Fig. S10B). This implies that PmB disruption of the LPSHBM model membrane was independent of the lipid phase, rather than being regulated by the physical state of the membrane, as observed with the dPL/LPSOMM. Last, to further test our hypothesis on the critical role of controlled molecular assembly in regulating PmB penetration of OM models, we examined the interaction of the antibiotic with the dPL/LPSOMM using ATR-FTIR. Unlike with the LPSHBM (Fig. 2A), no PmB binding was detected until the dPL/LPSOMM (Fig. 5D) was taken through its thermal phase transition, in line with the NR data (Fig. 3).
Fig. 5.
Phase transitions of a dPL/LPSOMM and PmB binding measured by ATR-FTIR. (A) ATR-FTIR absorption spectra of a dPL/LPSOMM. The two main peaks on the left are the C–H bond vibrations of the asymmetric (∼2,920 cm−1) and symmetric (∼2,850 cm−1) H–C–H stretching modes, arising from the acyl chains of LPS. On the right, the asymmetric (∼2,193 cm−1) and symmetric (∼2,088 cm−1) D–C–D stretching modes of the tail-deuterated phospholipids. (B) Peak absorption of the LPS H–C–H symmetric stretching vibration measured between 20 and 55 °C. (C) Phase transition of LPS (red squares; left y axis) and d62DPPC (black circles; right y axis) measured by the peak shift of the symmetric H–C–H and D–C–D stretching modes as a function of temperature (mean and SE of five measurements). (D) Infrared spectra of a dPL/LPSOMM before (black) and after (blue) the addition of PmB at RT and after cycling the bilayer once through its phase transition (red). The increase in the amide band signal at 1,650 cm−1, characteristic of the PmB binding, becomes apparent only after the phase transition has occurred.
Discussion
Generally, a drug’s cell-membrane permeability is simply related to its hydrophobicity or LogP (octanol/water partition coefficient), but antibiotics are more polar than other drug categories (6). For Gram-negative bacteria this is because the majority of antibiotics enter via water-filled pores within OM proteins that only transport small (<600-Da), water-soluble molecules (1). Although antibiotics following this route are numerous and chemically varied, their common pathway gives them a shared vulnerability to simple mutations in the pore proteins (8). Gram-positive bacteria, lacking this molecular sieve, are sensitive to molecules with a much wider range of chemistries, sizes, and polarities (6). The only large, polar antibiotic used against Gram-negative infections is polymyxin B (1.2 kDa), which directly targets the OM and currently provides the clearest model for the development of new molecules that can avoid the limitations of the molecular sieve (17). To open up the nonprotein pore route into Gram-negative bacteria requires new molecule families (34) and a better understanding of the physical chemistry of the OM.
We have shown here that, once PmB binding to the dPL/LPSOMM was enabled by heating the membrane through its phase transition, the antibiotic caused a large-scale disruption of the bilayer structure and the loss of membrane asymmetry (Figs. 3 D–F and 4 A and B). The in vivo temperature-dependent activity of PmB against E. coli shows a cooperative rather than linear response to temperature (30–32, 35), characteristic of lipid phase transitions arising from the melting of the acyl chains within the hydrophobic core of the membrane. As exemplified in Fig. 4A, the interaction of PmB with the dPL/LPSOMM above and below its phase transition (Figs. 3–5) reproduced the behavior observed in vivo. Furthermore, the saturation concentration of PmB on the dPL/LPSOMM (Fig. 4C) occurs in the same concentration range that inhibits cell growth in vivo (30–32, 35). Finally, FTIR showed distinct transition temperatures for the two lipid leaflets (Fig. 5C).
Although both the dPL/LPSOMM and LPSHBM achieved high lipid coverages of 92 and 99%, respectively (SI Appendix, Tables S1 and S3), subtle differences in the arrangement of the glycolipids may explain the completely different response to PmB at room temperature. Closer inspection of the NR data revealed that the LPS head-group layer of the LPSHBM is ∼30% thinner (20.9 ± 1.8 Å) than that of the dPL/LPSOMM (29 ± 0.6 Å) (SI Appendix, Tables S1 and S2) (20) and has a substantially higher water content (∼68% compared with ∼42%). This suggests a less efficient assembly of the molecules in the self-assembled monolayer, which would explain the accessibility of proposed PmB binding sites (25). Phospholipid HBM models are widely used in studies of membrane systems (23, 24, 36) and can play a role in shortening the drug development process (37, 38). However, possibly due to poor packing of the complex head groups, LPSHBMs do not fully imitate the behavior of the OM. The lipid phase-dependent binding of PmB displayed by the more accurate dPL/LPSOMM was not reproduced in the LPSHBM, which instead showed significant antibiotic binding in the gel phase at room temperature. Interestingly, in a molecular dynamics simulation of PmB interactions with the OM, penetration by the PmB acyl chain was not observed for a rough LPS outer layer at 40 °C but was achieved for a lipid A bilayer at 50 °C (39).
It is well-established that phospholipid membranes in many organisms are maintained in a liquid crystalline state across varying temperatures by adjusting the lipid’s phase-transition temperature. However, clear observation of a lipid phase transition in the LPS leaflet of the OM has been difficult (1, 10, 40–43). Recently, several lines of evidence have emerged to reconcile a liquid crystalline LPS acyl-chain region with the known impermeability of the OM at mammalian body temperatures (44). The OM is made rigid by divalent cation interactions (45), which elevate the temperature transition (Tm) (44), while the complex oligosaccharide head-group interactions also determine the packing and stability of LPS monolayers (46). The biological relevance of an LPS acyl-chain phase transition is also supported by biological phenomena such as the homeoviscous adaptation of LPS in E. coli grown at 12 or 37 °C (13) or Antarctic bacterial species grown at 4 or 22 °C (14). Meanwhile, X-ray diffraction studies of the liquid phase of LPS indicate that the single–hydrocarbon-chain area (26 Å2) is small, due to both strong acyl-chain van der Waals attractions and head-group attractions (46). In the asymmetric dPL/LPSOMM, the surface pressures and lipid packing found in natural membranes can be reproduced in vitro. X-ray studies of LPS at the air–water interface show that increasing the surface pressure of the monolayer toward the values used here for the fabrication of the dPL/LPSOMM induces a crystalline rearrangement of the glycolipid into a 2D hexagonal lattice (27). Thus, the LPS layer at 37 °C can be considered to be an unusual liquid crystalline lipid phase with gel-like characteristics driven by complex head-group interactions. Interestingly, cold can also increase the action of antibiotics against Gram-negative bacteria, including the large and polar molecule vancomycin, especially if homeoviscous adaptation is prevented by mutation (12). Resistance is regained by mutants, which may improve LPS packing below the phase transition (47).
In vivo, PmB causes a large-scale disruption of the OM (1, 17). This includes the formation of bleb-like protrusions (48), destabilization of the outer leaflet with loss of LPS (49), and various levels of permeabilization of the outer membrane depending upon the dosage and ambient salt concentration (1, 35). PmB has been proposed to undergo self-promoted uptake across the OM, complexing with LPS and forming OM pores which also enable the passage of molecules as large as lysozyme (1, 17, 35). Resistance to PmB correlates with both changes in LPS structure (see below) and reduced physical damage to the OM (35, 50). Since all PmB resistance mutations identified so far involve LPS, it is clear that this interaction is central and essential for the antibiotic action. The basic mechanism is attributed to the displacement by PmB of the divalent cations necessary to bridge the negative charges of neighboring LPS molecules (35, 39). We have previously shown how removal of calcium by EDTA from the asymmetric model OM causes a loss of lipid asymmetry, similar to that reported here (Fig. 3F) (21, 28), corroborating the view that PmB displaces divalent cations from the anionic LPS. The effects shown here by PmB include mixing of leaflets, with PmB intercalating into the bilayer, and increased water penetration, which correlate well with the self-promoted uptake, OM destabilization, and permeability changes observed in vivo (35). The thinning of the model membrane is associated with loss of LPS from the surface, but we do not observe the wholesale removal of LPS observed in some in vivo studies (49).
The primary resistance strategy of Gram-negative bacteria to evade cationic antimicrobial peptide toxicity consists of altering LPS charge by decorating the lipid A phosphate groups with moieties such as positively charged l-4-aminoarabinose and zwitterionic phosphoethanolamine (51). These modifications reduce both the electrostatic binding of cationic antimicrobials and the requirement for divalent cations to provide OM stability. Along with these resistance mechanisms, a second subset of LPS modifications employed by resistant strains alters the hydrophobic interior of the OM by changing lipid A acylation. The alteration of both LPS net charge and acylation pattern is controlled by two interconnected regulatory systems, PmrAB and PhoPQ (51). LPS modification with l-4-aminoarabinose, which protects against PmB, has been shown to require prior myristoylation of lipid A in E. coli and Salmonella typhimurium (52) and directly inhibit lipid A deacylation in Yersinia enterocolitica and S. typhimurium (53, 54). Addition of acyl chains to the lipid A of A. baumannii conferred resistance to PmB by increasing the stability of the OM hydrophobic core (26). Interestingly, while underacylated LPS of K. pneumoniae correlated both with an increased fluidity of the OM and a higher sensitivity to PmB (55), a similar modification in P. aeruginosa lipid A further increased resistance in PmB-resistant strains and altered binding in models similar to those used here (29). Moreover, increased hydrophobicity of PmB synthetic analogs increases their potency and overcomes resistance to the natural lipopeptide (17). These observations and results shown here demonstrate that the LPS acyl matrix plays an important role in regulating the penetration of antibiotics excluded by the OM molecular sieve. In the future, the interface between LPS and outer-membrane proteins will also need to be taken into account, particularly in view of our current understanding of the dynamic nature of protein biogenesis in the OM (7, 56).
Taken together, our results presented here provide clear support for the suggestion that Gram-negative OMs adopt a closely packed liquid crystalline phase at their adapted growth temperature. The comparisons of different in vitro models show that this unusual combination of fluidity and dense packing must be recreated in any study of antibiotic interactions with the outer membrane.
Materials and Methods
Materials.
Ra (rough) chemotype LPS from EH100 E. coli, polymyxin B sulfate, and polymyxin B nonapeptide chloride were from Sigma-Aldrich. Tail-deuterated d-DPPC [1,2-dipalmitoyl(d62)-sn-glycero-3-phosphocholine] was from Avanti Polar Lipids. All phospholipid and LPS samples were used without further purification. Buffer salts, solvents, and deuterium oxide (D2O) were sourced from Sigma-Aldrich or Fisher Scientific. Deuterated octadecyltrichlorosilane (d-OTS) was a kind gift from Peixun Li (STFC, Harwell, United Kingdom) and R. K. Thomas (University of Oxford, Oxford, United Kingdom). A buffer of 5 mM CaCl2 and 20 mM Hepes in D2O or H2O (pD/pH 7.4) was used throughout to provide the various solution contrasts as defined in each section.
Model Membrane Fabrication for Neutron Reflectometry and Infrared Measurements.
The deposition of asymmetric d62DPPC/LPS outer-membrane model bilayers on silicon substrates for NR experiments has been described by us in previous articles (20–22). An identical procedure was used to prepare a dPL/LPSOMM on substrates for IR analysis. For the NR and IR measurements of RaLPS monolayers deposited by vesicle rupture, silicon crystals were coated with a self-assembled monolayer (SAM) of octadecyl-trichlorosilane (perdeuterated for the NR experiments) as described previously (57). Vesicles were prepared by tip sonication of a resuspended dry film of RaLPS in a D2O buffer containing 20 mM Hepes (pD 7.4) and 5 mM CaCl2.
Neutron Reflectometry Measurements.
NR measurements were carried out on the INTER and CRISP reflectometers at the ISIS Pulsed Neutron and Muon Source, Rutherford Appleton Laboratory, as previously described (20) with the following modifications. A range of neutron wavelengths of 1.5 to 17 Å and 0.5 to 6.5 Å was used for INTER and CRISP, respectively. The reflected intensity was measured at two glancing angles on INTER (0.7° and 2.3°) and three angles on CRISP (0.35°, 0.65°, and 1.5°) as a function of the momentum transfer Q. For each isotopic contrast change a total of 18 mL of 20 mM (pH/D 7.4) Hepes, 5 mM CaCl2 buffer solution was pumped through the cell (six cell volumes) at a speed of 1 mL/min, followed by continuous flow of the same solution at 0.1 mL/min during the measurements. The three solution isotopic contrasts used here are D2O, H2O, and silicon-matched water (SMW), composed of 62% H2O and 38% D2O. D2O and H2O, respectively, provide the highest and lowest aqueous-phase SLD achievable and maximize contrast with the different SLD membrane components. With SMW, the contribution from the silicon substrate is null, and reflectivity data result solely from the interfacial bilayer. Temperature control of the solid–liquid flow cell was achieved using a Julabo FP50 circulating water bath and monitoring the temperature with a temperature sensor on the silicon substrate. Chosen PmB concentrations were prepared by pumping an appropriate mixture of two Hepes-buffered solutions through the sample flow cell, one containing no PmB and another with 100 µg/mL. For each concentration, 15 mL of PmB solution was flushed through the solid–liquid flow cells containing the heated bilayer samples at a rate of 1 mL/min. At each condition of temperature and PmB concentration, data were collected until no further changes were observed to ensure that the result represented the equilibrium condition. Shifts in the Kiessig fringes due to temperature or PmB effects were plotted as barycentric means of the reflectivity profile (58).
Neutron Reflectivity Data Analysis.
Reflectivity data were analyzed as previously described (28). Datasets collected on the same silicon substrate under different conditions, as for the LPSHBM before and after PmB addition, were constrained to fit a unique structure of the substrate which remains unaffected by the antimicrobial interaction. The barycentric mean of the Kiessig fringes was calculated in the 0.012 to 0.12 Q range, which extends from the critical edge at low Q to the maximum intensity of the fringe at high Q.
Attenuated Total Reflectance Infrared Spectroscopy.
Measurements were taken on a Nicolet iS50 spectrometer (Thermo Fisher Scientific) equipped with a mercury-cadmium-telluride detector. The instrument and sample chamber were purged of water vapor and CO2 using an air purge system.
The spectrometer was fitted with a Specac liquid attenuated total reflectance accessory. The IR beam was reflected through a trapezoidal silicon crystal (Crystran) with four of its six faces (the beam either traverses across or reflects from) polished to ∼3 Å rms roughness. General information about the use of this setup in solid–liquid interfacial studies can be found in Arteta et al. (59).
Asymmetric d62DPPC (inner leaflet) RaLPS (outer leaflet) was assembled on ATR silicon crystals as described for NR. The solid–liquid cell containing the silicon crystal, with the bilayer deposited on its surface, was then placed into the sample chamber of the spectrometer, and D2O buffer solution was pumped into the cell. Solution flow was controlled by a peristaltic pump. The sample chamber was allowed to purge of H2O vapor and CO2 for at least 1 h before spectra were acquired. For data collection, 128 individual spectra were summed. Once the IR measurements were taken, the cell was cleaned with ethanol and 1% Hellmanex to remove the bilayer from the silicon surface, rinsed with water, and filled with the D2O buffer, and a background measurement was taken which was then subtracted from the measurements.
For the IR measurements of LPS monolayers deposited by vesicle rupture, silicon ATR crystals were coated with a monolayer octadecyltrichlorosilane SAM (57) and placed in the solid–liquid cell. A background measurement was taken once the solid–liquid cell was mounted on the spectrometer and filled with D2O buffer. Vesicles were prepared as described above. The monolayer formation from the vesicle suspension was monitored over time until the IR signal reached saturation, after about 2 h.
Surface Plasmon Resonance.
SPR measurements were performed on a Biacore X100 (GE Healthcare) using gold chips (SIA Kit Au; GE Healthcare) coated with undecane thiol (Sigma). The chip was mounted on the Biacore and a suspension of LPS vesicles (0.25 mg/mL in 5 mM CaCl2, 20 mM Hepes, pH 7.4) was injected for 1,000 s, followed by two brief 30-s washes with 0.1 M NaOH to remove nonruptured vesicles. The deposition procedure was repeated until, upon injection of vesicles, no further increase in the signal was detected, indicating saturation of the hydrophobic surface by a monolayer of LPS. PmB was injected on the monolayer for 400 s followed by 1,200 s of buffer. After each PmB interaction, the monolayer was regenerated with a 0.1 M NaOH solution to remove the bound polymyxin from the surface.
Supplementary Material
Acknowledgments
The technical assistance of Dr. Helen Waller is gratefully acknowledged. Deuterated OTS was a kind gift from Dr. Peixun Li and Prof. R. K. Thomas. We thank Dr. Arwel Hughes for help with the Bayesian error analysis. The critical evaluation of the manuscript by Profs. Rick Lewis, Bert van den Berg, and Waldemar Vollmer is much appreciated. This work was supported by ISIS beam time Awards 1600025, 1510379, and 1400031. N.P. holds a Facility Development Studentship funded by STFC, Newcastle University, and OJ-Bio Ltd.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803975115/-/DCSupplemental.
References
- 1.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henderson JC, et al. The power of asymmetry: Architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu Rev Microbiol. 2016;70:255–278. doi: 10.1146/annurev-micro-102215-095308. [DOI] [PubMed] [Google Scholar]
- 3.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. Lipopolysaccharide transport and assembly at the outer membrane: The PEZ model. Nat Rev Microbiol. 2016;14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abellón-Ruiz J, et al. Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat Microbiol. 2017;2:1616–1623. doi: 10.1038/s41564-017-0046-x. [DOI] [PubMed] [Google Scholar]
- 5.Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta. 2009;1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov. 2015;14:529–542. doi: 10.1038/nrd4572. [DOI] [PubMed] [Google Scholar]
- 7.Arunmanee W, et al. Gram-negative trimeric porins have specific LPS binding sites that are essential for porin biogenesis. Proc Natl Acad Sci USA. 2016;113:E5034–E5043. doi: 10.1073/pnas.1602382113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pagès J-M, James CE, Winterhalter M. The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 9.Brandenburg K, Seydel U. A comment on the preparation of liposomes from and on the beta in equilibrium alpha acyl chain melting behaviour of rough mutant lipopolysaccharide. Biochim Biophys Acta. 1991;1069:1–4. doi: 10.1016/0005-2736(91)90096-q. [DOI] [PubMed] [Google Scholar]
- 10.Brandenburg K, Seydel U. Investigation into the fluidity of lipopolysaccharide and free lipid A membrane systems by Fourier-transform infrared spectroscopy and differential scanning calorimetry. Eur J Biochem. 1990;191:229–236. doi: 10.1111/j.1432-1033.1990.tb19114.x. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, et al. LPS remodeling is an evolved survival strategy for bacteria. Proc Natl Acad Sci USA. 2012;109:8716–8721. doi: 10.1073/pnas.1202908109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vorachek-Warren MK, Carty SM, Lin S, Cotter RJ, Raetz CRH. An Escherichia coli mutant lacking the cold shock-induced palmitoleoyltransferase of lipid A biosynthesis: Absence of unsaturated acyl chains and antibiotic hypersensitivity at 12 °C. J Biol Chem. 2002;277:14186–14193. doi: 10.1074/jbc.M200408200. [DOI] [PubMed] [Google Scholar]
- 13.Carty SM, Sreekumar KR, Raetz CRH. Effect of cold shock on lipid A biosynthesis in Escherichia coli. Induction at 12 °C of an acyltransferase specific for palmitoleoyl-acyl carrier protein. J Biol Chem. 1999;274:9677–9685. doi: 10.1074/jbc.274.14.9677. [DOI] [PubMed] [Google Scholar]
- 14.Kumar GS, Jagannadham MV, Ray MK. Low-temperature-induced changes in composition and fluidity of lipopolysaccharides in the Antarctic psychrotrophic bacterium Pseudomonas syringae. J Bacteriol. 2002;184:6746–6749. doi: 10.1128/JB.184.23.6746-6749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson AA, McGroarty EJ. High-molecular-weight components in lipopolysaccharides of Salmonella typhimurium, Salmonella minnesota, and Escherichia coli. J Bacteriol. 1985;162:738–745. doi: 10.1128/jb.162.2.738-745.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orwa JA, et al. Isolation and structural characterization of polymyxin B components. J Chromatogr A. 2001;912:369–373. doi: 10.1016/s0021-9673(01)00585-4. [DOI] [PubMed] [Google Scholar]
- 17.Velkov T, et al. Teaching ‘old’ polymyxins new tricks: New-generation lipopeptides targeting Gram-negative ‘superbugs.’. ACS Chem Biol. 2014;9:1172–1177. doi: 10.1021/cb500080r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Shea R, Moser HE. Physicochemical properties of antibacterial compounds: Implications for drug discovery. J Med Chem. 2008;51:2871–2878. doi: 10.1021/jm700967e. [DOI] [PubMed] [Google Scholar]
- 19.Schneck E, et al. Calcium ions induce collapse of charged O-side chains of lipopolysaccharides from Pseudomonas aeruginosa. J R Soc Interface. 2009;6(Suppl 5):S671–S678. doi: 10.1098/rsif.2009.0190.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clifton LA, et al. The effect of lipopolysaccharide core oligosaccharide size on the electrostatic binding of antimicrobial proteins to models of the Gram negative bacterial outer membrane. Langmuir. 2016;32:3485–3494. doi: 10.1021/acs.langmuir.6b00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clifton LA, et al. Effect of divalent cation removal on the structure of Gram-negative bacterial outer membrane models. Langmuir. 2015;31:404–412. doi: 10.1021/la504407v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clifton LA, et al. Asymmetric phospholipid: Lipopolysaccharide bilayers; a Gram-negative bacterial outer membrane mimic. J R Soc Interface. 2013;10:20130810. doi: 10.1098/rsif.2013.0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper MA, Try AC, Carroll J, Ellar DJ, Williams DH. Surface plasmon resonance analysis at a supported lipid monolayer. Biochim Biophys Acta. 1998;1373:101–111. doi: 10.1016/s0005-2736(98)00091-1. [DOI] [PubMed] [Google Scholar]
- 24.Besenicar M, Macek P, Lakey JH, Anderluh G. Surface plasmon resonance in protein-membrane interactions. Chem Phys Lipids. 2006;141:169–178. doi: 10.1016/j.chemphyslip.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Mares J, Kumaran S, Gobbo M, Zerbe O. Interactions of lipopolysaccharide and polymyxin studied by NMR spectroscopy. J Biol Chem. 2009;284:11498–11506. doi: 10.1074/jbc.M806587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boll JM, et al. Reinforcing lipid A acylation on the cell surface of Acinetobacter baumannii promotes cationic antimicrobial peptide resistance and desiccation survival. MBio. 2015;6:e00478-15. doi: 10.1128/mBio.00478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Brun AP, et al. Structural characterization of a model Gram-negative bacterial surface using lipopolysaccharides from rough strains of Escherichia coli. Biomacromolecules. 2013;14:2014–2022. doi: 10.1021/bm400356m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clifton LA, et al. An accurate in vitro model of the E. coli envelope. Angew Chem Int Ed Engl. 2015;54:11952–11955. doi: 10.1002/anie.201504287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han ML, et al. Polymyxin-induced lipid A deacylation in Pseudomonas aeruginosa perturbs polymyxin penetration and confers high-level resistance. ACS Chem Biol. 2018;13:121–130. doi: 10.1021/acschembio.7b00836. [DOI] [PubMed] [Google Scholar]
- 30.Teuber M, Bader J. Resistance to polymyxin-B at low temperature: A function of the outer membrane in Gram-negative bacteria. FEMS Microbiol Lett. 1977;1:75–77. [Google Scholar]
- 31.Hodate K, Bito Y. Temperature dependence of bactericidal action of polymyxin B. Microbiol Immunol. 1982;26:737–740. doi: 10.1111/j.1348-0421.1982.tb00217.x. [DOI] [PubMed] [Google Scholar]
- 32.Katsu T, Yoshimura S, Tsuchiya T, Fujita Y. Temperature dependence of action of polymyxin B on Escherichia coli. J Biochem. 1984;95:1645–1653. doi: 10.1093/oxfordjournals.jbchem.a134777. [DOI] [PubMed] [Google Scholar]
- 33.Guard-Friar D, Chen CH, Engle AS. Deuterium isotope effect on the stability of molecules: Phospholipids. J Phys Chem. 1985;89:1810–1813. [Google Scholar]
- 34.Stokes JM, et al. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat Microbiol. 2017;2:17028. doi: 10.1038/nmicrobiol.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daugelavicius R, Bakiene E, Bamford DH. Stages of polymyxin B interaction with the Escherichia coli cell envelope. Antimicrob Agents Chemother. 2000;44:2969–2978. doi: 10.1128/aac.44.11.2969-2978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chia CSB, Gong Y, Bowie JH, Zuegg J, Cooper MA. Membrane binding and perturbation studies of the antimicrobial peptides caerin, citropin, and maculatin. Biopolymers. 2011;96:147–157. doi: 10.1002/bip.21438. [DOI] [PubMed] [Google Scholar]
- 37.Frostell-Karlsson A, et al. Biosensor analysis of the interaction between drug compounds and liposomes of different properties; a two-dimensional characterization tool for estimation of membrane absorption. J Pharm Sci. 2005;94:25–37. doi: 10.1002/jps.20215. [DOI] [PubMed] [Google Scholar]
- 38.Danelian E, et al. SPR biosensor studies of the direct interaction between 27 drugs and a liposome surface: Correlation with fraction absorbed in humans. J Med Chem. 2000;43:2083–2086. doi: 10.1021/jm991156g. [DOI] [PubMed] [Google Scholar]
- 39.Berglund NA, et al. Interaction of the antimicrobial peptide polymyxin B1 with both membranes of E. coli: A molecular dynamics study. PLoS Comput Biol. 2015;11:e1004180. doi: 10.1371/journal.pcbi.1004180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melchior DL, Steim JM. Thermotropic transitions in biomembranes. Annu Rev Biophys Bioeng. 1976;5:205–238. doi: 10.1146/annurev.bb.05.060176.001225. [DOI] [PubMed] [Google Scholar]
- 41.Labischinski H, et al. High state of order of isolated bacterial lipopolysaccharide and its possible contribution to the permeation barrier property of the outer membrane. J Bacteriol. 1985;162:9–20. doi: 10.1128/jb.162.1.9-20.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garidel P, Howe J, Brandenburg K. Thermodynamic analysis of the interaction of lipopolysaccharides with cationic compounds. Eng Life Sci. 2008;8:523–529. [Google Scholar]
- 43.Schneck E, et al. Mechanical properties of interacting lipopolysaccharide membranes from bacteria mutants studied by specular and off-specular neutron scattering. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;80:041929. doi: 10.1103/PhysRevE.80.041929. [DOI] [PubMed] [Google Scholar]
- 44.Garidel P, et al. Divalent cations affect chain mobility and aggregate structure of lipopolysaccharide from Salmonella minnesota reflected in a decrease of its biological activity. Biochim Biophys Acta. 2005;1715:122–131. doi: 10.1016/j.bbamem.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 45.Herrmann M, Schneck E, Gutsmann T, Brandenburg K, Tanaka M. Bacterial lipopolysaccharides form physically cross-linked, two-dimensional gels in the presence of divalent cations. Soft Matter. 2015;11:6037–6044. doi: 10.1039/c5sm01002k. [DOI] [PubMed] [Google Scholar]
- 46.Snyder S, Kim D, McIntosh TJ. Lipopolysaccharide bilayer structure: Effect of chemotype, core mutations, divalent cations, and temperature. Biochemistry. 1999;38:10758–10767. doi: 10.1021/bi990867d. [DOI] [PubMed] [Google Scholar]
- 47.Stokes JM, et al. Cold stress makes Escherichia coli susceptible to glycopeptide antibiotics by altering outer membrane integrity. Cell Chem Biol. 2016;23:267–277. doi: 10.1016/j.chembiol.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 48.Koike M, Iida K, Matsuo T. Electron microscopic studies on mode of action of polymyxin. J Bacteriol. 1969;97:448–452. doi: 10.1128/jb.97.1.448-452.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lounatmaa K, Mäkelä PH, Sarvas M. Effect of polymyxin on the ultrastructure of the outer membrane of wild-type and polymyxin-resistant strains of Salmonella. J Bacteriol. 1976;127:1400–1407. doi: 10.1128/jb.127.3.1400-1407.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaara M, Vaara T. Sensitization of Gram-negative bacteria to antibiotics and complement by a nontoxic oligopeptide. Nature. 1983;303:526–528. doi: 10.1038/303526a0. [DOI] [PubMed] [Google Scholar]
- 51.Needham BD, Trent MS. Fortifying the barrier: The impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol. 2013;11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tran AX, et al. Resistance to the antimicrobial peptide polymyxin requires myristoylation of Escherichia coli and Salmonella typhimurium lipid A. J Biol Chem. 2005;280:28186–28194. doi: 10.1074/jbc.M505020200. [DOI] [PubMed] [Google Scholar]
- 53.Reinés M, et al. Deciphering the acylation pattern of Yersinia enterocolitica lipid A. PLoS Pathog. 2012;8:e1002978. doi: 10.1371/journal.ppat.1002978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawasaki K, Ernst RK, Miller SI. Inhibition of Salmonella enterica serovar Typhimurium lipopolysaccharide deacylation by aminoarabinose membrane modification. J Bacteriol. 2005;187:2448–2457. doi: 10.1128/JB.187.7.2448-2457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Velkov T, et al. Molecular basis for the increased polymyxin susceptibility of Klebsiella pneumoniae strains with under-acylated lipid A. Innate Immun. 2013;19:265–277. doi: 10.1177/1753425912459092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rassam P, et al. Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria. Nature. 2015;523:333–336. doi: 10.1038/nature14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang M, Liechti KM, Wang Q, White JM. Self-assembled silane monolayers: Fabrication with nanoscale uniformity. Langmuir. 2005;21:1848–1857. doi: 10.1021/la048483y. [DOI] [PubMed] [Google Scholar]
- 58.Chalton DA, Lakey JH. Simple detection of protein soft structure changes. Anal Chem. 2010;82:3073–3076. doi: 10.1021/ac902932c. [DOI] [PubMed] [Google Scholar]
- 59.Arteta MY, et al. On the formation of dendrimer/nucleolipids surface films for directed self-assembly. Soft Matter. 2015;11:1973–1990. doi: 10.1039/c4sm02712d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.