Summary
Brain microenvironment plays an important role in neurodevelopment and pathology, where the extracellular matrix (ECM) and soluble factors modulate multiple cellular processes. Neural cell culture typically relies on heterologous matrices poorly resembling brain ECM. Here, we employed neurospheroids to address microenvironment remodeling during neural differentiation of human stem cells, without the confounding effects of exogenous matrices. Proteome and transcriptome dynamics revealed significant changes at cell membrane and ECM during 3D differentiation, diverging significantly from the 2D differentiation. Structural proteoglycans typical of brain ECM were enriched during 3D differentiation, in contrast to basement membrane constituents in 2D. Moreover, higher expression of synaptic and ion transport machinery was observed in 3D cultures, suggesting higher neuronal maturation in neurospheroids. This work demonstrates that 3D neural differentiation as neurospheroids promotes the expression of cellular and extracellular features found in neural tissue, highlighting its value to address molecular defects in cell-ECM interactions associated with neurological disorders.
Keywords: 3D culture, neurospheroids, hiPSC, neural progenitors, neural differentiation, neural cell models, microenvironment, ECM, proteomics, transcriptomics
Graphical Abstract
Highlights
-
•
Efficient neural differentiation of hiPSC-derived NPC as neurospheroids
-
•
Neurospheroid transcriptome and proteome landscapes diverge from 2D cultures
-
•
Brain ECM and plasma membrane constituents enriched in neurospheroids
-
•
Prediction of transcriptional regulators driving 3D and 2D microenvironment features
In this article, Brito and colleagues show that differentiation of human neural stem cells as three-dimensional neurospheroids induces significant changes at cell membrane and ECM composition, relative to monolayer cultures, mimicking a neural-like microenvironment. These findings support the potential of neurospheroids to address ECM-affecting neurological disorders.
Introduction
CNS development culminates in the establishment of highly specialized structures, where the architectural and biochemical features of the extracellular space play a pivotal role in maintaining tissue function. Neurons, glia, and other non-neural cells (e.g., endothelial cells) contribute to this microenvironment through the secretion/uptake of extracellular matrix (ECM) components and soluble factors. ECM composition undergoes an intense remodeling process during CNS development, which is essential to support cell migration and promote axonal growth/guidance and synaptogenesis (Bandtlow and Zimmermann, 2000).
Neural ECM differs in composition from other tissues, containing low levels of basement membrane constituents such as collagen, laminin, and fibronectin (Rutka et al., 1988). Instead, the neural ECM is highly enriched in proteoglycans (PGs), including chondroitin sulfate PGs (CSPGs) and heparan sulfate PGs (HSPGs), link proteins (e.g., tenascin-R and tenascin-C) and hyaluronic acid (Soleman et al., 2013). Lecticans are the most important family of CSPGs found in brain tissue, namely the ubiquitously expressed aggrecan and versican and the CNS-specific brevican and neurocan (Howell and Gottschall, 2012). In the final stages of neuronal development these CSPGs accumulate around somatic and dendritic synapses, generating the cartilage-like structures perineuronal nets (PNNs) (Wang and Fawcett, 2012). PNN formation plays a major role in retaining the existing synaptic network and regulating synaptic plasticity. This has been demonstrated by enzymatic digestion of CSPGs, which results in impairment of long-term potentiation/depression and changes the neuronal network activity toward more naive patterns (Bikbaev et al., 2015). Perturbations in neural microenvironment homeostasis have been also associated with pathological events in several neurological disorders, both at a causal or modulatory level. Many studies have suggested a link between ECM alterations and brain injuries (e.g. traumatic brain injury, stroke) (Kim et al., 2016) or Alzheimer's disease (Snow et al., 1994).
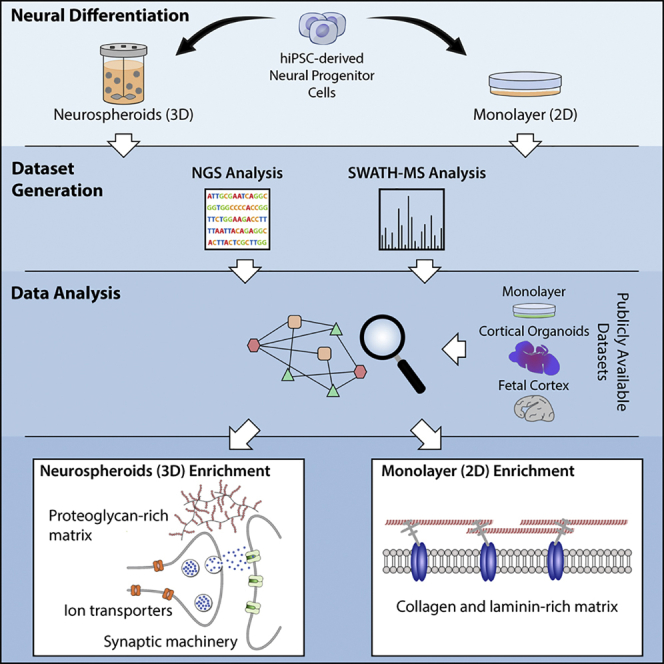
Despite increasing evidence linking alterations of neural microenvironment with neurological pathologies, the availability and validity of in vitro models to address these questions remains uncertain. Organotypical cultures are considered the most accurate model available, preserving ECM composition and tissue architecture. Nevertheless challenges arise from the use of these cultures, namely the scarcity of human tissue, the difficulty in maintaining long-term cultures, and ensuring optimal culture conditions to attain a model that represents a non-inflamed and non-reactive tissue (Humpel, 2015). The use of human induced pluripotent stem cells (hiPSCs) provides a stable and renewable source of human neural cells, also providing access to patient-derived cells with the disease-associated genetic background. Moreover, the combination of hiPSC with three-dimensional (3D) culture systems supports the development of more accurate in vitro cell models, relative to traditional two-dimensional (2D) monolayer systems. Human stem cell-derived 3D in vitro neural models have been shown to recapitulate important features of the target tissue, namely neuron-glia interactions (Simão et al., 2016a) or cellular organization (Lancaster and Knoblich, 2014, Lancaster et al., 2013). However, most of these rely on the use of exogenous matrices, and their ability to mimic neural extracellular space features remains elusive. We hypothesized that 3D differentiation of hiPSC-derived human neural progenitor cells (hNPCs) as neurospheroids, in perfusion stirred-tank bioreactors, could sustain a microenvironment remodeling toward ECM components typical of neural ECM. To address this question, we applied transcriptome sequencing and quantitative proteomics during 3D in vitro differentiation. Here we demonstrate that 3D differentiation as neurospheroids promotes the secretion and accumulation of neural ECM structural components and expression of synaptic machinery, typically found in neural tissue. This study demonstrates that human stem cell-derived 3D neural models are promising tools to address the role of neural microenvironment in healthy and diseased states and are potential tools for the development of novel therapeutic strategies targeting neural ECM.
Results
Transcriptomic and Proteomic Analysis of hiPSC-NPC 3D Differentiation
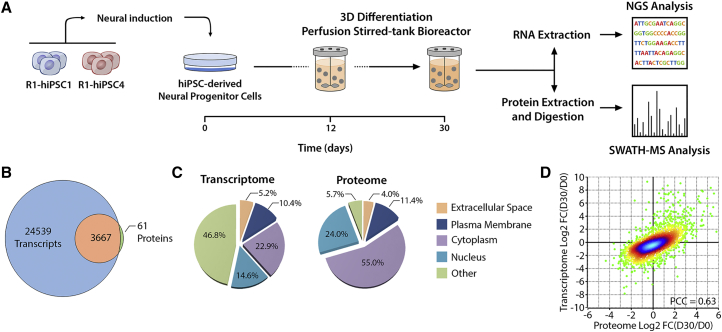
Two NPC lines derived from hiPSCs (Figure S1) were used to address the molecular remodeling process occurring during 3D differentiation, at both transcriptomic and proteomic levels (Figures 1A and S2). We used a 3D culture strategy previously established by our group (Simão et al., 2016b) to generate and differentiate neurospheroids. Three representative time points were selected for analysis, including an initial sampling from the hiPSC-NPC cultures used for inoculation of 3D cultures (day 0), an early differentiation state (day 12), and a late differentiation state (day 30). The initial 7 days of culture correspond to a cell aggregation stage performed in presence of epidermal growth factor and basic fibroblast growth factor. This step promotes self-assembly of cells into compact neurospheres (∼100 μm diameter; Figures S1L and S1M) while maintaining a proliferative phenotype, as demonstrated by the incorporation of 5-ethynyl-2′-deoxyuridine (EdU) in approximately 65% of cells (Figure S1O). Still, the cell density remained almost constant, around 5 × 105 cells/mL (Figure S1N), indicating a considerable decrease of cell growth rate in the 3D cultures. Along differentiation, a drastic decrease in the percentage of actively proliferating cells was observed, with only 2% of cells staining positively for EdU at day 30 (Figure S1O), suggesting differentiation into post-mitotic cell populations. Cell density remained fairly constant despite the reduction in cell proliferation, further supporting the high cell viability observed along the culture time course (Figure S1M).
Figure 1.
Schematic Workflow for hiPSC-Derived NPC 3D Differentiation
(A) NPCs were derived from two hiPSC lines, expanded as monolayer, and differentiated as neurospheroids. Cells harvested at days 0, 12, and 30 were processed for transcriptomic or proteomic analysis.
(B) The coverage and overlap between total identified transcripts and proteins are represented in the Venn diagram.
(C) Cellular localization of the identified transcripts and proteins according to Ingenuity Pathway Analysis (IPA) knowledge base.
(D) Density scatterplot describes the transcriptome-proteome correlation, where each dot indicates the fold change (logarithmized) of an individual transcript/protein (total of 3,667). The Pearson correlation coefficient (PCC) is indicated in the scatterplot. The color code represents the density of dots included in a region of the scatterplot. Data shown represent four pooled independent biological experiments (two independent experiments of two cell lines).
See also Figures S1 and S2.
Transcriptomic (RNA sequencing [RNA-seq]) and proteomic (SWATH-mass spectrometry [MS]) analyses were performed in samples from four independent cultures (two from each cell line), identifying and quantifying 28,206 transcripts and 3,728 proteins. The replicates both at transcriptome and proteome levels revealed good correlation and low inter-cell line variability, as demonstrated by the Pearson correlation coefficient (PCC) higher than 0.9 (Figures S2A and S2B, respectively). For approximately 98% of the identified proteins we could also quantify the transcript abundance (Figure 1B). From these datasets, the most represented cellular compartments were cytoplasm and nucleus, which together consisted of 37.5% and 79% of the identified transcripts and proteins, respectively (Figure 1C). Approximately 15% of the identified transcripts and proteins were classified as associated with the extracellular space and plasma membrane compartments (Figure 1C).
Next, we compared the changes induced by the 3D differentiation in the transcriptome and proteome. Looking to the fold changes between day 0 and day 30, a good correlation between transcript and protein was obtained (PCC of 0.63; Figure 1D). However, when we analyzed the different time points individually, a higher PCC was obtained for day 0 (0.60) in comparison with the samples from 3D differentiation (0.46 and 0.49, respectively, for day 12 and day 30; Figure S2).
Pathway Modulation during 3D Differentiation
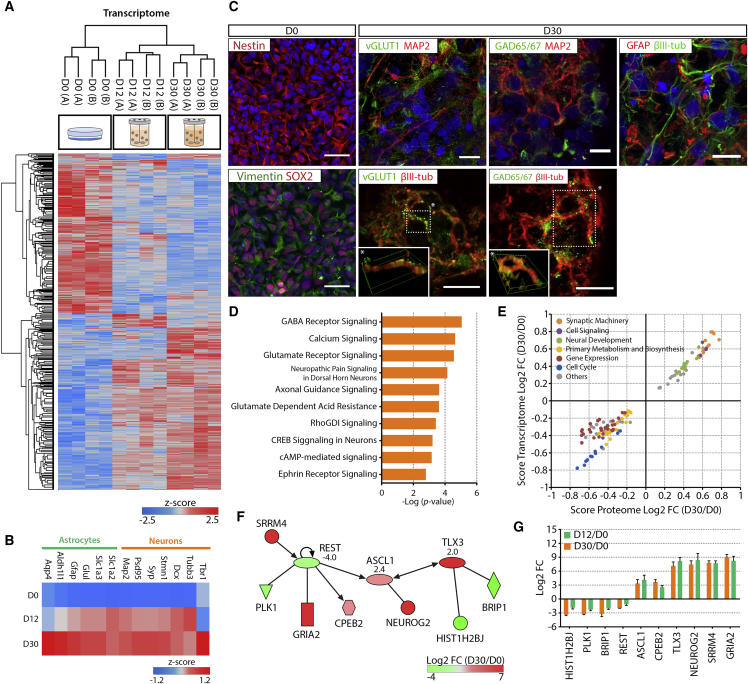
Hierarchical clustering of transcriptomic data revealed that the samples from early (day 12) and late (day 30) differentiation stages clustered together, indicating a higher degree of similarity in gene expression profile relative to non-differentiated cultures (day 0), which were in a more distant cluster (Figure 2A). This suggests that an intense remodeling occurs early in time defining the commitment toward the neuronal and glial lineages, as demonstrated by the upregulation of lineage-specific markers (Figure 2B). The transition from progenitors toward neuronal and astrocytic cells was also evident by immunofluorescence (Figure 2C). Most cells were positive for the NPC markers nestin, vimentin, and SOX2 at day 0, while differentiated neurospheres (day 30) were highly enriched in neuronal (βIII-tubulin-positive) and glial (GFAP-positive) lineage cells (Figure 2C). Concerning neuronal differentiation, vGlut1- and GAD65/67-positive neuronal populations were also detected at day 30, suggesting the presence of glutamatergic and GABAergic neurons, respectively (Figure 2C).
Figure 2.
Transcriptome Dynamics during hiPSC-NPC 3D Differentiation
(A) Heatmap of significantly modulated transcripts between days 0 (D0), 12 (D12), and 30 (D30) (total of 12,116). Hierarchical clustering performed for the biological replicates of each time point and cell line (in columns; A and B refer to R1-hiPSC1-NPC and R1-hiPSC4-NPC, respectively), and for transcripts (in rows). Significantly modulated transcripts identified by multi-sample ANOVA test with a permutation-based false discovery rate (FDR) cutoff of 0.05 applied on the logarithmized intensities. Z-score values were color coded from blue (downregulation) to red (upregulation).
(B) Heatmaps of gene expression profiles of neuronal and astrocytic specific markers at day D0, D12, and D30. Z score values were color coded from blue (downregulation) to red (upregulation).
(C) (Left) Immunocytochemistry of hiPSC-NPC at D0, with the expression of nestin, vimentin, and Sox-2. Scale bars, 50 μm. (Right) Confocal imaging of neurospheroids at D30, with staining of βIII-tubulin or dendritic MAP2 and presynaptic markers of GABAergic (GAD65/67-positive) and glutamatergic (VGlut1-positive) neurons (3D-rendering insets of regions indicated by the asterisks highlight co-localization), βIII-tubulin, and glial fibrillary acidic protein (GFAP). Scale bars, 10 μm.
(D) Top ten canonical pathways identified by IPA to describe the transcriptome modulation between D0 and D30.
(E) Two-dimensional annotation enrichment analysis, correlating the pathways significantly modulated during 3D differentiation at the transcriptome and proteome level (Benjamini-Hochberg FDR < 0.05). Negative score values describe downregulation and positive scores indicate upregulation. Each dot represents a GO-Biological Process (GO-BP) term. Related GO-BP terms are presented with the same color.
(F) Proposed regulatory network responsible for the transcriptome remodeling during differentiation. Different factors were color labeled according to the fold change between D0 and D30, ranging from green (downregulation) to red (upregulation). Predicted activation Z score is indicated by the number bellow each of the three transcription factors (REST, ASCL1, and TLX3), where negative and positive values indicate repression and activation, respectively.
(G) Transcript levels of the different players represented in the regulatory network. Data are presented as mean ± SEM of the log2 of the fold change between D0 and D12 (green bars) or D0 and D30 (orange bars). Data shown in all panels represent four pooled independent biological experiments (two independent experiments of two cell lines).
See also Figures S1 and S3; Table S1.
The main modulated pathways and networks between day 0 and day 30 were identified by analyzing the transcriptomic dataset. The resulting top ten canonical pathways highlight the over-representation of neuronal differentiation pathways, such as cyclic AMP (cAMP) and CREB signaling (Figure 2D). Other pathways related to neuronal morphogenesis were also among the most represented pathways, namely Ephrin and RhoGDI signaling. Genes associated with establishment of excitatory glutamatergic and inhibitory GABAergic synaptic machineries were highly enriched in the obtained datasets, consistent with the immunofluorescence observations (Figure 2C). Concerning regional identity, neurospheroids presented a mixed phenotype with expression of genes associated with development of forebrain, midbrain, and hindbrain (Figures S3A and S3B). Furthermore, correlation analysis with brain tissue data revealed a higher correlation with cortical structures, to the detriment of thalamus and cerebellum (Figure S3C). This was also in line with the upregulation of several telencephalic markers observed during neurospheroid differentiation, in which no clear trend was observed toward dorsal or ventral identity (Figures S3D and S3E).
The correlation between the transcriptome and proteome data in terms of main modulated pathways was evaluated by gene ontology (GO) enrichment analysis. Both datasets were well correlated in terms of significantly modulated biological processes (GO-BP), as shown by the two-dimensional annotation enrichment analysis (Figure 2E). The obtained data demonstrated that synaptic machinery, neural development, and cell signaling related processes were upregulated during 3D differentiation both at transcriptome and proteome level. Most downregulated pathways were associated with cellular proliferation, such as cell cycle, gene expression, and primary metabolism/biosynthesis related processes, suggesting a progression in the developmental pathways toward mature post-mitotic neural cell populations. This is consistent with the EdU proliferation assay data (Figure S1O) and with the modulation of cyclin-dependent kinase (CDK) expression. CDK1, CDK4, and CDK7, which govern cell-cycle progression, were downregulated while CDK5 was upregulated (Figure S1P).
The transcriptomic dataset was used to infer the main transcription factors responsible for the modulation observed during neurospheroid differentiation (Figure 2F). We found the repression of REST and the activation of ASCL1 and TLX3 to play a pivotal role in driving the transcriptional changes observed. The activity modulation of these transcription factors induced the expression of neurogenic genes, such as ASCL1, NEUROG2, and TLX3. Concomitantly, this orchestrated signaling network also included the repression of important cell-cycle modulators, such as PLK1, BRIP1, and HIST1H2BJ. Analyzing the fold changes in the expression levels of this network's players, we observed that these different molecules are regulated early (day 12) in the differentiation process, exerting its downstream effects along the culture time course (Figure 2G).
ECM and Plasma Membrane Remodeling in 3D Cultures
To address our hypothesis that the neurospheroid differentiation would induce a cell microenvironment with higher similarity to what is observed in vivo, we compared the proteome of neurospheroid cultures with 2D cultures after 30 days of differentiation. Principal component analysis (PCA) revealed a clear separation between the neurospheroid and 2D differentiated experimental groups; nonetheless, the 2D differentiated group was closer to neurospheroids at day 12 than at day 30 (Figure S4A). Hierarchical clustering of the significantly modulated proteins between the different experimental groups showed that all of the differentiated groups were clustering together and away from the non-differentiated day 0 (Figure S4B). However, when we filtered the proteome dataset only for proteins annotated as part of the extracellular space and plasma membrane (according to the Ingenuity Pathway Analysis [IPA] knowledge base), we observed a shift in the hierarchical clustering of the different groups: the 2D differentiated group clustered together with day 0, while the 3D differentiated groups were in a separate cluster.
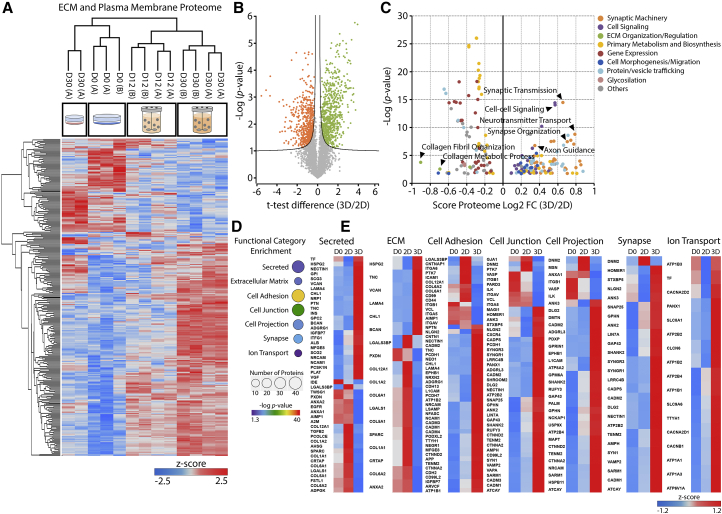
A direct comparison of the total proteome of neurospheroids and 2D cultures at day 30 revealed that there were 663 and 513 proteins specifically enriched in neurospheroids or 2D, respectively (Figure 3B). This differential protein expression corresponded to a significant enrichment in GO-BP terms associated with cell signaling, synaptic machinery, and cell morphogenesis/migration in neurospheroids. On the other hand, 2D differentiation resulted in enrichment of GO-BP terms related to ECM organization/regulation, namely collagen fibril organization and collagen metabolic process (Figure 3C).
Figure 3.
Extracellular Space and Plasma Membrane Proteome Remodeling during hiPSC-NPC 3D Differentiation
(A) Heatmap of proteins significantly modulated at the extracellular or plasma membrane level, during 3D and 2D differentiation (total of 294 proteins). Hierarchical clustering performed for the biological replicates of each time point and cell line (in columns) and for proteins (in rows). Significantly modulated proteins identified by multi-sample ANOVA test with a permutation-based FDR cutoff of 0.05 applied on the logarithmized intensities. Z score values were color coded from blue (downregulation) to red (upregulation).
(B) Volcano plot of proteins identified after 2D or 3D differentiation of hiPSC-NPCs. Significantly enriched proteins after 2D (orange) or 3D (green) differentiation are color labeled. Significantly modulated proteins identified by permutation-based FDR t test applied on the logarithmized intensities, with a threshold value of 0.05 and S0 of 0.1.
(C) GO-BP terms significantly over-represented (Benjamini-Hochberg FDR < 0.02) in 2D (negative score) or 3D (positive score) differentiated cells. The y axis presents the corresponding p values (in negative log scale). Related GO-BP terms are presented with the same color, where some specific terms are highlighted by arrowheads.
(D) Selected functional categories significantly over-represented during 3D differentiation (Benjamini-Hochberg FDR < 0.05) for proteins annotated as extracellular space or plasma membrane components. p values and number of proteins are graphically represented by different colors and sphere sizes, respectively.
(E) Heatmaps of protein abundance profile at day 0 (D0), 2D at day 30, and 3D at day 30 of selected categories. Z score values were color coded from blue (downregulation) to red (upregulation). Data shown in all panels represent four (two independent experiments of two cell lines) or two pooled independent biological experiments, for neurospheroids or 2D cultures, respectively.
Analyzing the extracellular space and plasma membrane proteome, several functional categories were found to be significantly enriched, namely secreted proteins, ECM, cell adhesion, cell junction, cell projection, synapse, and ion transport (Figures 3D and S6A). Closer inspection of these categories showed significant differences in the ECM composition of neurospheroids and 2D differentiated cultures. Neurospheroid cultures presented low levels of collagens and high levels of the proteoglycans BCAN, VCAN, and HSPG2 and the link protein TNC (Figure 3E). LAMA4, the only laminin subunit found to be enriched in neurospheroids, has been predicted to play a relevant role during neural development (Chen et al., 2015). 2D cultures presented a collagen-rich ECM, with the upregulation of different subunits of collagen types I, V, VI, and XII, collagen-interacting proteins (SPARC and CRTAP), and ANXA2, which can mediate collagen secretion (Dassah et al., 2014). Integrin receptors responsible for binding basement membrane ECM components, such as ITGA6, ITGB1, and ITGAV, were also enriched in 2D.
Analyzing the profile of differentially expressed cell adhesion molecules, we observed that most of the proteins enriched in neurospheroids are key players in neural development and morphogenesis. These include several neural cell adhesion molecules (CAMs), known to mediate axon guidance/outgrowth by modulating neuron-ECM interaction and attraction/repulsion properties (PCDH1, NEO1, EPHB1, NRXN2, ADGRG1, CDH13, NRCAM, LSAMP, NCAM1, NFASC, NEGR1, CNTN1, NECTIN1, and CTNNA2) (Figure 3E). Structural synaptic machinery proteins were significantly enriched in neurospheroids, including NECTIN1, NRXN2, NLGN2, SHANK2, HOMER1, GPHN, and CADM1–CADM4. Moreover, proteins related with synaptic vesicle trafficking and neurotransmitter release were also found in higher abundance in neurospheroids, including SYN1, VAMP2, SNAP25, STXBP5, LIN7A, SYNGR1, SYNGR3, CADPS, and AMPH. Similar observations were made for proteins involved in ion transport, as the expression of several Na+/K+-ATPase subunits (ATP1A1, ATP1A3, ATP1B1, ATP1B3, ATP2B2, and ATP2B4) are essential in maintaining the resting potential at the expense of large ATP amounts. Among these different subunits, ATP1B3 was the only one that was present in similar abundance levels between day 0 and day 30 in neurospheroids, which is in line with previous observations in primary murine NPC in vitro neuronal differentiation (Frese et al., 2017).
The majority of the observations on proteomics data were further supported by the transcriptomic analysis, whereby similar trends in fold changes of neurospheroids between day 0 and day 30 were observed at transcript level for most of the differentially expressed proteins (Figure S4D). Moreover, we further confirmed the accumulation of TNC and the CNS-specific proteoglycans, BCAN and NCAN, in neurospheroids by immunostaining and wisteria floribunda agglutinin labeling (Figures S4E and S4F). NCAN, although not quantified in our proteomic dataset, was found to be significantly upregulated at transcript level.
Impact of Culture System on Neuronal Differentiation
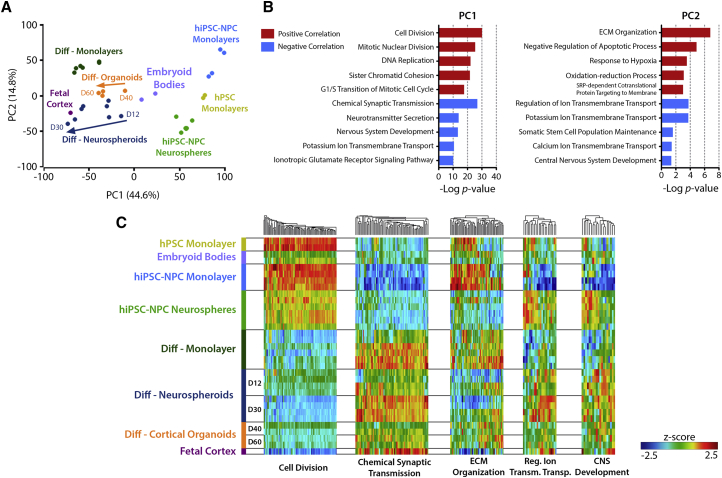
To further validate our findings, we compared our datasets with publicly available data obtained from neural differentiation of human stem cells in different culture systems. The dataset from Srikanth et al. (2015) allowed a comparison with hiPSC-NPC cultured as proliferative neurospheres and their neural differentiation as 2D monolayers. For comparison with human pluripotent stem cell (hPSC) cultures expanded in 2D cultures, embryoid bodies, Matrigel-embedded cortical organoids, and fetal cortex (male, 19 weeks gestation), we used the dataset from Luo et al. (2016). All samples were normalized and compared through PCA, where most of the variance could be explained by PC1 (44.6%) and PC2 (14.8%) (Figure 4A). PC1 revealed a clear separation between the undifferentiated stem cell samples (hPSCs and hiPSC-NPCs) and the differentiated samples. Among the differentiated samples, neurospheroids from day 30 were the samples with a profile closest to that of the fetal cortex.
Figure 4.
Transcriptomic Remodeling during Neural Differentiation in Different 2D and 3D Culture Systems
(A) Principal component analysis of different in vitro culture systems.
(B) Main GO-BP terms significantly over-represented in genes with top 1,000 positive (red) or negative (blue) loadings for principal component 1 (PC1) or PC2. The y axis corresponds to the negative log10 of the p values (FDR corrected).
(C) Heatmap of the genes included in the GO-BP terms of cell division (87 genes), chemical synaptic transmission (68 genes), ECM organization (59 genes), regulation of ion transmembrane transport (22 genes), and CNS development (19 genes). Hierarchical clustering was performed for different genes (columns). Rows represent the different samples. Z score values were color coded from blue (downregulation) to red (upregulation).
See also Table S3.
Next, we analyzed the genes associated with the top PCA loadings contributing to PC1 and PC2 scores (Figure 4B). PC1 positive scores were mostly correlated with the high expression levels of cell-cycle progression genes in proliferating hPSCs and hiPSC-NPCs (Figures 4B and 4C). However, hiPSC-NPCs cultured as proliferative neurospheres present a lower activation of these genes, which is in line with many other cell types that have lower proliferation rates when cultured in 3D systems (Edmondson et al., 2014). All differentiated samples demonstrated a repression of these cell-cycle-associated genes, whereby the fetal cortex presented the lowest expression levels followed by neurospheroids at day 30. PC1 negative scores were highly correlated with nervous system development and synaptic maturation/function. Analyzing the cluster of genes associated with chemical synaptic transmission, we observed that these were highly upregulated in neurospheroids at day 30 and in differentiated monolayers, while displaying lower levels in the cortical organoid samples. Furthermore, these results on chemical synaptic transmission related genes are also supported by the correlation of PC2 negative scores with genes related to regulation of ion transmembrane transport, comprising several potassium and calcium voltage-gated channels critical for synaptic function. These genes were also more enriched in neurospheroids, as compared with monolayers or cortical organoids (Figure 4C). PC2 positive scores were found to be mostly correlated with ECM organization processes, which include several genes encoding for basement membrane components (collagens and laminins) and CAMs (integrins). This gene cluster was more enriched in monolayer cultures of hNPCs and 2D differentiated cells, presenting very low expression levels in fetal cortex. 3D cultures presented an intermediate expression profile for this cluster, still with neurospheroids presenting lower expression of collagen- and laminin-related genes relative to cortical organoids. The CNS development cluster identified in PC2 negative correlation includes some of the main structural neural ECM components, such as the proteoglycans aggrecan, brevican, and neurocan, which were found to have higher expression levels in 3D differentiated cultures (neurospheroids and organoids), relative to differentiation in monolayer (Figure 4C). The CNS-specific hyaluronan binding link proteins HAPLN2 and HAPLN4 were expressed at higher levels on neurospheroids relative to cortical organoids.
To compare and highlight the differences between the expression profiles of genes encoding for extracellular space and plasma membrane proteins in neurospheroids and 2D differentiated samples, we used IPA to predict the most relevant upstream regulators in each dataset. Of the top five transcription factors identified for the neurospheroids dataset, three presented similar activation scores to the 2D dataset (TLX3, REST, and ASCL1), while two were only predicted to be activated on neurospheroids (PTF1A and MITF) (Figure S5).
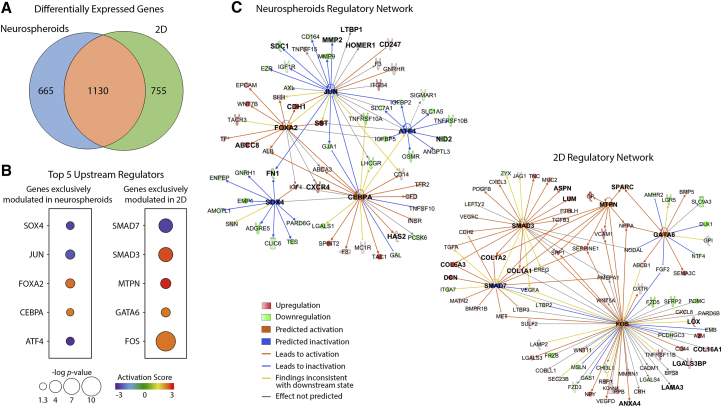
Among the differentially expressed genes associated with extracellular space and plasma membrane in 3D and 2D, 1,130 genes were modulated in both datasets while 665 and 755 genes were exclusively modulated on neurospheroids and 2D differentiation, respectively (Figure 5A). We focused on these two lists of exclusively modulated genes to address in more detail the differences between 3D and 2D differentiation, by identifying the most significant upstream regulators (Figures 5B and 5C). The top five identified regulators in neurospheroids play a relevant role in modulating the expression of ECM genes. CEBPA, which was predicted to be activated, is known to drive the expression of HAS2 in neuronal cells, in agreement with the upregulation observed for this gene in neurospheroids. Conversely, downregulation of fibronectin 1 encoding gene (FN1) was predicted to be mediated by the inactivation of JUN and SOX4. JUN was also predicted to be involved in the downregulation of other ECM modulators, namely LTBP1, MMP2, and SDC1. Moreover, FOXA2 and JUN were involved in the upregulation of genes associated with synaptic machinery and dendritic spine morphogenesis, such as ABCC8, CDH1, SST, HOMER1, and CD247. CEBPA and FOXA2 were also predicted to activate the expression of the neuronal migration regulator CXCR4.
Figure 5.
Upstream Regulator Analysis of Neurospheroids and 2D Differentiation
(A) Venn diagram of differentially expressed genes associated with extracellular space and plasma membrane in neurospheroids and 2D differentiation (Srikanth et al., 2015).
(B) Top five upstream regulators predicted in IPA to be responsible for the differential gene expression of exclusively modulated genes (extracellular space and plasma membrane genes only) in 3D (665 genes) and 2D (755 genes). The activation scores and p values are graphically represented by different colors and sphere sizes, respectively.
(C) Regulatory networks derived for 3D and 2D data based on IPA predictions for the top five upstream regulators.
See also Figure S5.
The regulatory network derived for the 2D cultures revealed that four out of the top five transcription regulators had a relevant role on the upregulation of laminin- and collagen-encoding genes (LAMA3, COL1A1, COL1A2, COL6A3, and COL16A1), namely the predicted activation of SMAD3, FOS, and MTPN and inactivation of SMAD7. The upregulation of ANXA4, LGALS3BP, predicted to be under the control of FOS, was also in agreement with our proteomics data for 2D differentiation described above.
Discussion
Neural microenvironment homeostasis plays an essential role in the maintenance of normal cellular function in CNS. Here, we addressed the cellular and extracellular remodeling occurring in vitro during 3D differentiation of hiPSC-NPCs, in a system devoid of exogenous ECM addition (e.g., Matrigel): neurospheroids differentiated in perfusion stirred-tank bioreactors. Changes in the expression patterns were analyzed via a two-level integrative approach, generating a large-scale quantitative transcriptome and proteome dataset. Such approaches can, for instance, highlight the contribution of non-genetic regulation mechanisms (Low et al., 2013, Sacco et al., 2016). Post-transcriptional regulation plays a critical role during neural development, namely in neuronal morphogenesis and connectivity (DeBoer et al., 2013, Frese et al., 2017). Our datasets reflect the relevance of post-transcriptional control mechanisms during neural differentiation compared with the neural progenitor cell state, with lower correlation coefficients being observed between mRNA and protein levels once neurospheroid differentiation started.
A global overview of the transcriptional changes occurring during neurospheroid differentiation revealed that these were mostly related to the modulation of cAMP, RhoGDI, and Ephrin signaling pathways. These pathways are well-known for their role during neurogenesis, regulating neuronal morphogenesis, survival, and migration (Hall and Lalli, 2010, Klein, 2004, Lonze and Ginty, 2002). The main transcriptional regulation predicted to be driving these modulations was the repression of REST, which is consistent with its function as a negative neurogenesis regulator (Palm et al., 1998). The observed upregulation of SRRM4 was identified as one possible contributor to REST repression, by promoting the expression of a less active truncated REST isoform via alternative splicing (Raj et al., 2011). The decreased REST repressive activity results in increased expression of ASCL1, NEUROG2, and TLX3, which are essential players in the development and specification of neuronal lineages (Borromeo et al., 2014, Casarosa et al., 1999, Parras et al., 2002). These transcriptional changes were followed by a gradual arrestment of the cell cycle, suggesting a progression toward mature post-mitotic neural cell populations. The neuronal cell-cycle suppressor CDK5 and its activator CDK5R1, which are typically expressed in post-mitotic neurons (Frese et al., 2017, Zhang and Herrup, 2011), were found to be upregulated during neurospheroid differentiation. Correlation with human brain tissue data revealed that the generated neurospheroids displayed higher similarity with cortical structure expression, despite the expression of fore-, mid-, and hindbrain developmental genes. To further elucidate the impact of regional identity on the expression patterns of ECM-associated genes, we adapted region-directed differentiation protocols (Ashton et al., 2015, Niclis et al., 2017, Yuan et al., 2016) for neurospheroid differentiation.
Our data show that neurospheroid differentiation induces a significant modulation of ECM components and CAMs, diverging greatly from the profile resulting from 2D differentiated cultures. Here, a distinct feature between both differentiation processes was the enrichment in neurospheroids of several PNN components, including proteoglycans (BCAN and VCAN), and link protein TNC, as well as increased gene expression of TNR and hyaluronan synthesis/binding proteins (HAS2, HAPLN2, and HAPLN4) (Yamaguchi, 2000). In addition to structural ECM components, the expression profile of cell surface proteins also diverged considerably between neurospheroids and monolayers. NRCAM, NFASC, L1CAM, and CHL1, key modulators of axon guidance (Maness and Schachner, 2007, Sakurai, 2012), were enriched in neurospheroids relative to 2D cultures. Several synaptic machinery proteins were also significantly enriched in neurospheroids, such as CAMs, which have been described as essential for synapse morphogenesis and organization. These include NCAM1, whose role in both early synaptogenesis and subsequent synaptic maturation is well documented; the presynaptic proteins NECTIN1 and NRXN2; the post-synaptic proteins NLGN2, SHANK2, HOMER1, and GPHN; and the four trans-synaptic CADM proteins CADM1, CADM2, CADM3, and CADM4 (Dityatev, 2004, Fogel et al., 2007, Togashi et al., 2009).
In contrast, 2D differentiation resulted in a collagen-rich ECM, which may result from signaling cues provided by the laminin-coated surfaces and by different stiffness, mechanical properties, or cell-cell interactions (e.g., neuron-astrocyte), relative to 3D cultures. This collagen-enriched profile suggests that 2D cultures offer a more distant representation of the healthy neural tissue microenvironment as compared with the profile obtained in the neurospheroids. Collagen deposition in neural tissue is often associated with pathological alterations, promoted for instance by gangliomas that induce the expression of many collagen-interacting proteins that were enriched in the 2D differentiated cultures (e.g., PXDN, LGAL1, LGALS3BP, A2M, ANXA4, and ITGA5) (Autelitano et al., 2014, Liu et al., 2010, Le Mercier et al., 2010).
To minimize any possible bias in the analysis and to increase the significance of our findings, we further compared our datasets with publicly available data derived from different protocols for 3D and 2D neural differentiation and fetal cortex tissue. The current limitations on standardized methods for proteome quantification and data availability hampered comparison studies at protein level between different in vitro protocols and/or tissue samples but allowed comparisons at transcriptome levels. This comparative analysis further supported our previous observations on the microenvironment differences between 3D and 2D systems. ECM organization-related genes were among the most significant genes contributing to the separation of 2D and 3D biological samples in the PCA. Similarly to what we observed for neurospheroids, cortical organoids and fetal cortex displayed lower expression levels of collagen- and laminin-related genes and increased the expression of neural proteoglycan-encoding genes, relative to 2D cultures. Between the two 3D systems, neurospheroids presented increased expression of genes related to synaptic machinery and ion channels. The higher intra- and inter-spheroid homogeneity (Simão et al., 2015) compared with cortical organoids (Lancaster and Knoblich, 2014, Lancaster et al., 2013) can contribute to these differences.
As in the neurospheroid differentiation process, REST repression together with TLX3 and ASCL1 activation were among the most relevant transcriptional regulators predicted to be driving the neural differentiation in 2D cultures. Still, the analysis of upstream regulators modulating the expression of ECM and plasma membrane related genes in neurospheroids and 2D cultures revealed clear differences between both systems. In the neurospheroids JUN and ATF4 were predicted to be repressed, inducing the downregulation of molecules such as LTBP1, MMP2, SDC1, and NID2 that regulate collagen biosynthesis, processing, and binding (Hori et al., 1998, Ishikawa and Kramer, 2010, Kohfeldt et al., 1998, Visse, 2003). The activation of CEBPA and FOXA2 regulated the upregulation of several synaptic proteins and the chemokine receptor CXCR4, key player during neuronal differentiation and migration (Pitcher et al., 2010). For 2D differentiation, all the identified transcription factors are involved in modulating the expression of collagens and collagen processing/organization proteins, including LUM, ASPN, LOX, SPARC, and DCN (Chakravarti et al., 1998, Lau et al., 2006, Nakajima et al., 2007).
Human 3D in vitro models, such as the neurospheroids described herein and cortical organoids, can be valuable tools to enable better understanding of the dynamics of ECM and soluble factor secretion/degradation. The two 3D culture methods present specific advantages and drawbacks, making them more suited for different applications. As demonstrated by several authors, cortical organoids, employing the methodology first published by Lancaster et al. (2013), are well suited to address questions regarding the human neurodevelopmental process (Akkerman and Defize, 2017). However, these structures typically present high batch-to-batch variability, rendering it less adequate for drug or toxicological screening. Neurospheroids present a simpler structure and are devoid of exogenous ECM addition. Thus, the increased homogeneity between individual structures and different batches makes it a more suitable system for preclinical applications and with potential to address disease-associated modulation of neuron-ECM interactions. Large-scale quantitative approaches, such as the one presented in this work, can be applied to address the link between changes in neural microenvironment and neurological disorders. The amount of evidence suggesting this link is increasing, including for instance the ECM remodeling following brain injury induced by reactive astrocytes (e.g., traumatic brain injury, stroke) (Kim et al., 2016, Zamanian et al., 2012).
Overall, we have demonstrated that the 3D differentiation of hNPCs as neurospheroids induces the expression of important cellular and extracellular proteins typically observed in the neural tissue. By combining a comprehensive transcriptomic and proteomic characterization, we demonstrate that neurospheroid cultures can better mimic the protein composition of the neural microenvironment as compared with the traditional monolayer-based cell models. The extension of this culture system to other cell sources (e.g., patient-derived iPSCs) and/or chemical and physical insults is therefore likely to contribute to a more accurate assessment of the role and link between ECM changes and neurological disorders.
Experimental Procedures
Generation of Neural Progenitors from Human Induced Pluripotent Stem Cells and Differentiation
Human iPSCs (Royan iPSC clone 1 [R1-hiPSC1] and clone 4 [R1-hiPSC4], also known as RIi001-A and RIi007-A, respectively) were derived from dermal fibroblasts as described previously (Totonchi et al., 2010). Neural progenitor cells generation, differentiation, and immunostaining were performed as described in Supplemental Experimental Procedures.
Quantitative Transcriptomics and Proteomics
All samples were processed for RNA-seq and SWATH-MS as described in Supplemental Experimental Procedures. Computational and statistical analysis of the datasets was performed using Perseus software environment (Tyanova et al., 2016), DAVID 6.8 (Huang et al., 2009), and IPA, as described in Supplemental Experimental Procedures.
Author Contributions
Conceptualization, D.S. and C.B.; Methodology, D.S., P.G.-A., N.R., and C.B.; Formal Analysis, D.S., P.G.-A., and N.R.; Investigation, D.S., M.M.S., A.P.T., and F.A.; Writing – Original Draft, D.S. and C.B.; Writing – Review & Editing, D.S., P.G.-A, N.R., and C.B.; Visualization, D.S.; Resources, M.F.Q.S., N.Z.M., T.S., N.R., P.G.-A., and P.M.A.; Funding Acquisition, N.R., P.M.A., and C.B.; Supervision, P.M.A. and C.B.
Acknowledgments
iNOVA4Health – UID/Multi/04462/2013, a program financially supported by Fundação para a Ciência e Tecnologia/Ministério da Educação e Ciência, through national funds and co-funded by FEDER under the PT2020 Partnership Agreement, is acknowledged. PD/BD/52473/2014, PD/BD/52481/2014, and PD/BD/128371/2017 PhD fellowships funded by FCT, Portugal. N.R. is supported by the European Research Council Starting Grant 337327. T.S. is supported by a grant provided by the Deutsche Forschungsgemeinschaft (DFG) (grant number SA 1382/7-1). The authors gratefully thank João Clemente for support on the bioreactor operation and Dr. Inês Isidro for fruitful discussions on data analysis. MS data were obtained by UniMS – Mass Spectrometry Unit, ITQB/iBET, Oeiras, Portugal.
Published: July 26, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.06.020.
Accession Numbers
Data have been deposited at the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) and Proteome Xchange Consortium via PRIDE (Vizcaíno et al., 2016). GEO: GSE102139 (RNA-seq); Proteome Xchange: PXD007130 (SWATH-MS).
Supplemental Information
References
- Akkerman N., Defize L.H.K. Dawn of the organoid era: 3D tissue and organ cultures revolutionize the study of development, disease, and regeneration. BioEssays. 2017;39 doi: 10.1002/bies.201600244. [DOI] [PubMed] [Google Scholar]
- Ashton R., Lippmann E., Estevez-Silva M., Ashton R. Chemically defined differentiation of human pluripotent stem cells to hindbrain and spinal cord neural stem cells with defined regional identities. Protocol Exch. 2015 [Google Scholar]
- Autelitano F., Loyaux D., Roudières S., Déon C., Guette F., Fabre P., Ping Q., Wang S., Auvergne R., Badarinarayana V. Identification of novel tumor-associated cell surface sialoglycoproteins in human glioblastoma tumors using quantitative proteomics. PLoS One. 2014;9:e110316. doi: 10.1371/journal.pone.0110316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandtlow C.E., Zimmermann D.R. Proteoglycans in the developing brain: new conceptual insights for old proteins. Physiol. Rev. 2000;80:1267–1290. doi: 10.1152/physrev.2000.80.4.1267. [DOI] [PubMed] [Google Scholar]
- Bikbaev A., Frischknecht R., Heine M. Brain extracellular matrix retains connectivity in neuronal networks. Sci. Rep. 2015;5:14527. doi: 10.1038/srep14527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borromeo M.D., Meredith D.M., Castro D.S., Chang J.C., Tung K.-C., Guillemot F., Johnson J.E. A transcription factor network specifying inhibitory versus excitatory neurons in the dorsal spinal cord. Development. 2014;2812:2803–2812. doi: 10.1242/dev.105866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa S., Fode C., Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- Chakravarti S., Magnuson T., Lass J.H., Jepsen K.J., LaMantia C., Carroll H. Lumican regulates collagen fibril assembly: Skin fragility and corneal opacity in the absence of lumican. J. Cell Biol. 1998;141:1277–1286. doi: 10.1083/jcb.141.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chu C., Kong X., Huang T., Cai Y.D. Discovery of new candidate genes related to brain development using protein interaction information. PLoS One. 2015;10:1–11. doi: 10.1371/journal.pone.0118003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassah M., Almeida D., Hahn R., Bonaldo P., Worgall S., Hajjar K.A. Annexin A2 mediates secretion of collagen VI, pulmonary elasticity and apoptosis of bronchial epithelial cells. J. Cell Sci. 2014;127:828–844. doi: 10.1242/jcs.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoer E.M., Kraushar M.L., Hart R.P., Rasin M.R. Post-transcriptional regulatory elements and spatiotemporal specification of neocortical stem cells and projection neurons. Neuroscience. 2013;248:499–528. doi: 10.1016/j.neuroscience.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dityatev A. Polysialylated neural cell adhesion molecule promotes remodeling and formation of hippocampal synapses. J. Neurosci. 2004;24:9372–9382. doi: 10.1523/JNEUROSCI.1702-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson R., Broglie J.J., Adcock A.F., Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel A.I., Akins M.R., Krupp A.J., Stagi M., Stein V., Biederer T. SynCAMs organize synapses through heterophilic adhesion. J. Neurosci. 2007;27:12516–12530. doi: 10.1523/JNEUROSCI.2739-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese C.K., Mikhaylova M., Stucchi R., Gautier V., Liu Q., Mohammed S., Heck A.J.R., Altelaar A.F.M., Hoogenraad C.C. Quantitative map of proteome dynamics during neuronal differentiation. Cell Rep. 2017;18:1527–1542. doi: 10.1016/j.celrep.2017.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Lalli G. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb. Perspect. Biol. 2010;2:1–18. doi: 10.1101/cshperspect.a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori Y., Katoh T., Hirakata M., Kaname S., Fukagawa M., Okuda T., Ohashi H., Fujita T., Miyazono K., Kurokawa K. Anti-latent TGF-β binding protein-1 antibody or synthetic oligopeptides inhibit extracellular matrix expression induced by stretch in cultured rat mesangial cells. Kidney Int. 1998;53:1616–1625. doi: 10.1046/j.1523-1755.1998.00908.x. [DOI] [PubMed] [Google Scholar]
- Howell M.D., Gottschall P.E. Lectican proteoglycans, their cleaving metalloproteinases, and plasticity in the central nervous system extracellular microenvironment. Neuroscience. 2012;217:6–18. doi: 10.1016/j.neuroscience.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Lempicki R.A., Sherman B.T. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Humpel C. Organotypic brain slice cultures: a review. Neuroscience. 2015;305:86–98. doi: 10.1016/j.neuroscience.2015.07.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Kramer R.H. Sdc1 negatively modulates carcinoma cell motility and invasion. Exp. Cell Res. 2010;316:951–965. doi: 10.1016/j.yexcr.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Porter B.E., Friedman A., Kaufer D. A potential role for glia-derived extracellular matrix remodeling in postinjury epilepsy. J. Neurosci. Res. 2016;94:794–803. doi: 10.1002/jnr.23758. [DOI] [PubMed] [Google Scholar]
- Klein R. Eph/ephrin signaling in morphogenesis, neural development and plasticity. Curr. Opin. Cell Biol. 2004;16:580–589. doi: 10.1016/j.ceb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Kohfeldt E., Sasaki T., Göhring W., Timpl R. Nidogen-2: a new basement membrane protein with diverse binding properties. J. Mol. Biol. 1998;282:99–109. doi: 10.1006/jmbi.1998.2004. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau Y.K.I., Gobin A.M., West J.L. Overexpression of lysyl oxidase to increase matrix crosslinking and improve tissue strength in dermal wound healing. Ann. Biomed. Eng. 2006;34:1239–1246. doi: 10.1007/s10439-006-9130-8. [DOI] [PubMed] [Google Scholar]
- Liu Y., Carson-Walter E.B., Cooper A., Winans B.N., Johnson M.D., Walter K.A. Vascular gene expression patterns are conserved in primary and metastatic brain tumors. J. Neurooncol. 2010;99:13–24. doi: 10.1007/s11060-009-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze B.E., Ginty D.D. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Low T.Y., vanHeesch S., vandenToorn H., Giansanti P., Cristobal A., Toonen P., Schafer S., Hübner N., vanBreukelen B., Mohammed S. Quantitative and qualitative proteome characteristics extracted from in-depth integrated genomics and proteomics analysis. Cell Rep. 2013;5:1469–1478. doi: 10.1016/j.celrep.2013.10.041. [DOI] [PubMed] [Google Scholar]
- Luo C., Lancaster M.A., Castanon R., Nery J.R., Knoblich J.A., Ecker J.R. Cerebral organoids recapitulate epigenomic signatures of the human fetal brain. Cell Rep. 2016;17:3369–3384. doi: 10.1016/j.celrep.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maness P.F., Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat. Neurosci. 2007;10:19–26. doi: 10.1038/nn1827. [DOI] [PubMed] [Google Scholar]
- Le Mercier M., Fortin S., Mathieu V., Kiss R., Lefranc F. Galectins and gliomas. Brain Pathol. 2010;20:17–27. doi: 10.1111/j.1750-3639.2009.00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M., Kizawa H., Saitoh M., Kou I., Miyazono K., Ikegawa S. Mechanisms for asporin function and regulation in articular cartilage. J. Biol. Chem. 2007;282:32185–32192. doi: 10.1074/jbc.M700522200. [DOI] [PubMed] [Google Scholar]
- Niclis J.C., Gantner C.W., Alsanie W.F., McDougall S.J., Bye C.R., Elefanty A.G., Stanley E.G., Haynes J.M., Pouton C.W., Thompson L.H. Efficiently specified ventral midbrain dopamine neurons from human pluripotent stem cells under xeno-free conditions restore motor deficits in parkinsonian rodents. Stem Cells Transl. Med. 2017;6:937–948. doi: 10.5966/sctm.2016-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm K., Belluardo N., Metsis M., Timmusk T. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J. Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras C.M., Schuurmans C., Scardigli R., Kim J., Anderson D.J., Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 2002;16:324–338. doi: 10.1101/gad.940902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher J., Shimizu S., Burbassi S., Meucci O. Disruption of neuronal CXCR4 function by opioids: preliminary evidence of ferritin heavy chain as a potential etiological agent in neuroAIDS. J. Neuroimmunol. 2010;224:66–71. doi: 10.1016/j.jneuroim.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B., O’Hanlon D., Vessey J.P., Pan Q., Ray D., Buckley N.J., Miller F.D., Blencowe B.J. Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Mol. Cell. 2011;43:843–850. doi: 10.1016/j.molcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Rutka J.T., Apodaca G., Stern R., Rosenblum M. The extracellular matrix of the central and peripheral nervous systems: structure and function. J. Neurosurg. 1988;69:155–170. doi: 10.3171/jns.1988.69.2.0155. [DOI] [PubMed] [Google Scholar]
- Sacco F., Silvestri A., Posca D., Pirrò S., Gherardini P.F., Castagnoli L., Mann M., Cesareni G. Deep proteomics of breast cancer cells reveals that metformin rewires signaling networks away from a pro-growth state. Cell Syst. 2016;2:159–171. doi: 10.1016/j.cels.2016.02.005. [DOI] [PubMed] [Google Scholar]
- Sakurai T. The role of NrCAM in neural development and disorders-Beyond a simple glue in the brain. Mol. Cell. Neurosci. 2012;49:351–363. doi: 10.1016/j.mcn.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Simão D., Pinto C., Piersanti S., Weston A., Peddie C.J., Bastos A.E.P.P., Licursi V., Schwarz S.C., Collinson L.M., Salinas S. Modeling human neural functionality in vitro: three-dimensional culture for dopaminergic differentiation. Tissue Eng. Part A. 2015;21:654–668. doi: 10.1089/ten.TEA.2014.0079. [DOI] [PubMed] [Google Scholar]
- Simão D., Terrasso A.P., Teixeira A.P., Brito C., Sonnewald U., Alves P.M. Functional metabolic interactions of human neuron-astrocyte 3D in vitro networks. Sci. Rep. 2016;6:33285. doi: 10.1038/srep33285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão D., Arez F., Terasso A.P., Pinto C., Sousa M.F.Q., Brito C., Alves P.M. Perfusion stirred-tank bioreactors for 3D differentiation of human neural stem cells. In: Turksen K., editor. Bioreactors in Stem Cell Biology. Springer; 2016. pp. 129–142. [DOI] [PubMed] [Google Scholar]
- Snow A.D., Sekiguchi R.T., Nochlin D., Kalaria R.N., Kimata K. Heparan sulfate proteoglycan in diffuse plaques of hippocampus but not of cerebellum in Alzheimer’s disease brain. Am. J. Pathol. 1994;144:337–347. [PMC free article] [PubMed] [Google Scholar]
- Soleman S., Filippov M.A., Dityatev A., Fawcett J.W. Targeting the neural extracellular matrix in neurological disorders. Neuroscience. 2013;253:194–213. doi: 10.1016/j.neuroscience.2013.08.050. [DOI] [PubMed] [Google Scholar]
- Srikanth P., Han K., Dana G., Kosik K.S., Dennis J., Young-pearse T.L., Srikanth P., Han K., Callahan D.G., Makovkina E. Genomic DISC1 disruption in hiPSCs alters Wnt signaling and neural cell fate article genomic DISC1 disruption in hiPSCs alters Wnt signaling and neural cell fate. Cell Rep. 2015;12:1414–1429. doi: 10.1016/j.celrep.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi H., Sakisaka T., Takai Y. Cell adhesion molecules in the central nervous system. Cell Adh. Migr. 2009;3:29–35. doi: 10.4161/cam.3.1.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totonchi M., Taei A., Seifinejad A., Tabebordbar M., Rassouli H., Farrokhi A., Gourabi H., Aghdami N., Hosseini-Salekdeh G., Baharvand H. Feeder- and serum-free establishment and expansion of human induced pluripotent stem cells. Int. J. Dev. Biol. 2010;54:877–886. doi: 10.1387/ijdb.092903mt. [DOI] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Visse R. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ. Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- Vizcaíno J.A., Csordas A., Del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fawcett J. The perineuronal net and the control of CNS plasticity. Cell Tissue Res. 2012;349:147–160. doi: 10.1007/s00441-012-1375-y. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y. Lecticans: organizers of the brain extracellular matrix. Cell. Mol. Life Sci. 2000;57:276–289. doi: 10.1007/PL00000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F., Fang K.-H., Cao S.-Y., Qu Z.-Y., Li Q., Krencik R., Xu M., Bhattacharyya A., Su Y.-W., Zhu D.-Y. Efficient generation of region-specific forebrain neurons from human pluripotent stem cells under highly defined condition. Sci. Rep. 2016;5:18550. doi: 10.1038/srep18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian J.L., Xu L., Foo L.C., Nouri N., Zhou L., Giffard R.G., Barres B.A. Genomic analysis of reactive astrogliosis. J. Neurosci. 2012;32:6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Herrup K. Nucleocytoplasmic Cdk5 is involved in neuronal cell cycle and death in post-mitotic neurons. Cell Cycle. 2011;10:1208–1214. doi: 10.4161/cc.10.8.15328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.