Abstract
Huntington’s disease (HD) is a dominantly inherited neurodegenerative disease caused by expansion of a polyglutamine tract in the huntingtin protein. HD symptoms include severe motor, cognitive, and psychiatric impairments that result from dysfunction and later degeneration of medium-sized spiny neurons (MSNs) in the striatum. A key early pathogenic mechanism is dysregulated synaptic transmission due to enhanced surface expression of juvenile NMDA-type glutamate receptors containing GluN3A subunits, which trigger the aberrant pruning of synapses formed by cortical afferents onto MSNs. Here, we tested the therapeutic potential of silencing GluN3A expression in YAC128 mice, a well-established HD model. Recombinant adeno-associated viruses encoding a short-hairpin RNA against GluN3A (rAAV-shGluN3A) were generated, and the ability of different serotypes to transduce MSNs was compared. A single injection of rAAV9-shGluN3A into the striatum of 1-month-old mice drove potent (>90%) and long-lasting reductions of GluN3A expression in MSNs, prevented dendritic spine loss and improved motor performance in YAC128 mice. Later delivery, when spine pathology is already apparent, was also effective. Our data provide proof-of-concept for GluN3A silencing as a beneficial strategy to prevent or reverse corticostriatal disconnectivity and motor impairment in HD and support the use of RNAi-based or small-molecule approaches for harnessing this therapeutic potential.
Keywords: GluN3A, synapse pruning, Huntington, NMDA receptor, RNAi, rAAV, dendritic spine, motor impairment, YAC128
Marco et al. assess the potential of viral delivery of small interfering RNAs (siRNAs) to correct aberrant expression of the GluN3A subunit of NMDA receptors, which play a causative early stage role in the pathogenesis of Huntington’s disease (HD). A recombinant AAV9 vector effectively delivered siRNAs that fully rescued dendritic spine loss and motor function in the YAC128 model of HD when injected into the striatum. Therapy prevented spine loss and motor symptoms when initiated at an early stage and reversed key aspects of the pathology when initiated later on.
Introduction
Huntington’s disease (HD) is a fatal neurodegenerative disorder with motor, cognitive, and psychiatric disturbances. It is caused by abnormal expansion of a polyglutamine repeat in the huntingtin (HTT) protein that triggers its misfolding and aggregation and alters interactions of HTT with multiple binding partners. The neuropathology involves early synapse failure and loss followed by cell death, especially in the striatum where medium-sized spiny neurons (MSNs) that make 90%–95% of all neurons are selectively vulnerable.1 Currently, there is no cure or treatment to slow disease progression, and therapies barely manage the symptoms.
The search for disease-modifying therapies has focused on lowering the amount of mutant HTT with antisense oligonucleotides or RNAi.2, 3, 4 Doing so while preserving wild-type (WT) protein levels has proven challenging, but ongoing clinical trials based on allele-specific or non-selective targeting of both WT and mutant HTT are beginning to yield promising results. Other therapeutic options include modulating autophagy, modifying HTT aggregation, or supplementing growth factors.5, 6, 7 An alternative that has been less explored, due to insufficient knowledge of the underlying molecular mechanisms, is to arrest the pathogenic process by counteracting key initiating events. Most prominent in HD is the progressive disconnection between the cortex and the striatum, which results from dysfunction and loss of excitatory glutamatergic synapses8, 9, 10 impinging on dendritic spines of MSNs. Supporting its relevance for disease pathogenesis, longitudinal imaging and clinical studies have demonstrated that degeneration of MSN spines and striatal atrophy occur in premanifest HD11, 12, 13 and are robust predictors of clinical onset and progression.14, 15
We recently identified aberrant expression of juvenile NMDA receptors containing GluN3A subunits (GluN3A-NMDARs)16 as a major driver of the HD synaptic pathology.17 Sequestration of the GluN3A-selective endocytic adaptor PACSIN1 by mutant HTT1, 17 causes accumulation of GluN3A-NMDARs at MSN synapses, altering MSN excitability and reactivating pruning mechanisms normally restricted to postnatal developmental stages.18, 19 Importantly, dysregulated GluN3A expression can be detected from very early disease stages and is a feature shared by HD brains and a variety of mouse models (expressing N-terminal or full-length HTT, and knockin mice). Suppressing GluN3A expression by crossing YAC128 mice, a well-established mouse model, with GluN3A null mice prevented all aspects of the synapse pathology, as well as cognitive and motor dysfunction and reduced MSN loss.17, 18 While this prevention was encouraging, genetic suppression is neither an amenable therapeutic strategy nor definitive proof of principle given known compensatory effects in knockout mice. Further validation was needed, including defining a therapeutic window and developing feasible therapeutic approaches, if possible exclusively targeted to vulnerable cell types to minimize adverse effects.
Here, we validate GluN3A as a therapeutic target by using recombinant adeno-associated vectors (rAAVs) to deliver RNAi triggers that correct the aberrant GluN3A expression. rAAVs offer many advantages for gene transfer in neurodegenerative disease, and different serotypes exhibit different tropisms allowing to target a variety of cell types.20, 21 Of several serotypes tested, rAAV9-driven GluN3A silencing was most efficient for stably and selectively transducing MSNs and fully prevented the synaptic pathology and motor deficits in YAC128 mice. Protection was achieved when rAAVs were delivered at the onset of pathology but also at later disease stages. Together, our results support the development of RNAi-based therapies or alternative approaches targeting GluN3A to treat HD.
Results
Intrastriatal rAAV9-shGluN3A Drives Sustained GluN3A Silencing in MSNs
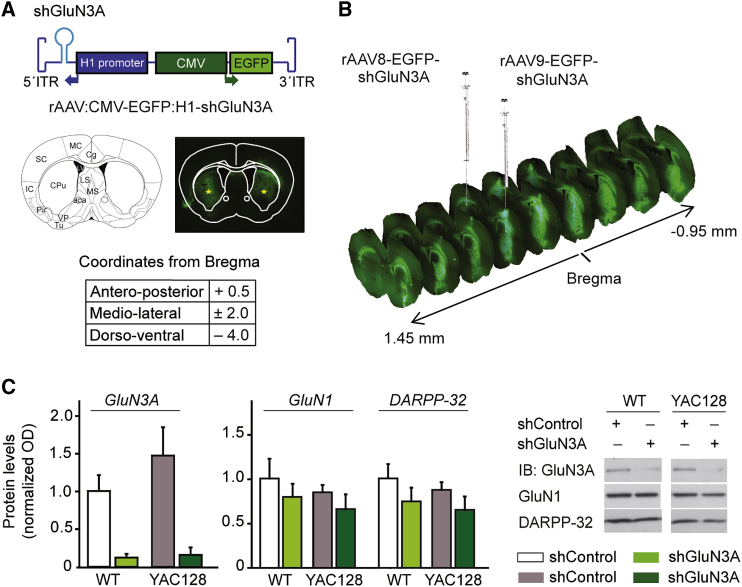
We generated rAAVs expressing a previously validated short hairpin RNA (shRNA) directed to GluN3A22 (shGluN3A) under the regulation of the H1 non-coding RNA promoter. A GFP reporter gene driven by a cytomegalovirus (CMV) promoter was included in the vector to aid detection of transduced cells (Figure 1A). In a first series of experiments, we compared the ability of different rAAV serotypes to spread throughout the mouse striatum and efficiently transduce MSNs, the vulnerable cell population. Serotypes 8, 9, and 10 were chosen based on previous reports.23, 24 Viral particles were delivered into the striatum using coordinates determined with the Paxinos mouse brain atlas25 (Figure 1A) at the doses indicated in Figure S1. GFP expression was visualized 2 weeks after surgery by fluorescence microscopy analysis of series of mosaic images spanning the entire striatum. All serotypes yielded detectable GFP expression, but a quantitative assessment demonstrated that the spread capacity of rAAV9 was higher and more consistent across individuals (Figure S1); a single injection of rAAV9 resulted in widespread transduction that extended 2.4 mm across the anteroposterior axis of the striatum (Figure 1B).
Figure 1.
Silencing GluN3A in MSNs by Intrastriatal Injection of rAAV9-Driven shRNAs
(A) Top, cartoon of rAAV construct encoding shRNAs against the NMDAR subunit GluN3A flanked by inverted terminal repeat (ITR) sequences. shRNA expression is driven by the H1 promoter, and expression of EGFP by the CMV promoter. Middle, schematic of the brain section where injections were placed, along with example of an injected mouse brain. Stars show the tip of the needle. CPu, caudate putamen. Bottom, stereotaxic coordinates relative to bregma (in mm). (B) Widespread viral transduction upon single rAAV injections into the striatum. Injection sites and antero-posterior coordinates relative to bregma (in mm) are shown. (C) Immunoblot analysis of in vivo silencing efficiency of rAAV9-shGluN3A. Quantification and representative immunoblots showing levels of the indicated proteins in extracts from 3-month-old WT and YAC128 mice striata receiving rAAV9-shGluN3A at 1 month of age, compared to contralateral striata injected with control rAAV9. Data are mean ± SEM normalized to WT striatum injected with rAAV9-shControl (n = 5 animals per group). Two-way ANOVA showed no interaction between genotype and shRNA treatment (F(1,14) = 0.74, p = 0.403) but revealed a strong effect of rAAV9shGluN3A on GluN3A protein levels relative to shControl (F(1,14) = 19, p = 0.0007).
We next evaluated the in vivo silencing efficacy of rAAV9-shGluN3A. rAAV9s expressing shGluN3A or control shRNA were delivered into the striatum of 1-month-old WT and YAC128 mice (injection volume, 2 μL; 6.4–8.8 × 1010 viral genomes), and GluN3A expression was quantified 2 months after injection. Immunoblot analysis demonstrated that GluN3A protein levels were dramatically reduced in the rAAV9-shGluN3A-injected striatum relative to contralateral striata receiving rAAV9-shControl (Figure 1C; p = 0.0007). Silencing was persistent and lasted for at least 11 months after injection, which was the latest time point analyzed; the reduction in GluN3A protein levels observed in rAAV9-shGluN3A-injected striata of old (12-month-old) WT and YAC128 mice was of similar magnitude as it was in younger animals (to 10%–15% of shControl-injected striata; p < 0.0001). The effects of shGluN3A were target specific, as neither levels of the obligatory NMDAR subunit GluN1 nor the MSN marker DARPP-32 were altered (Figure 1C).
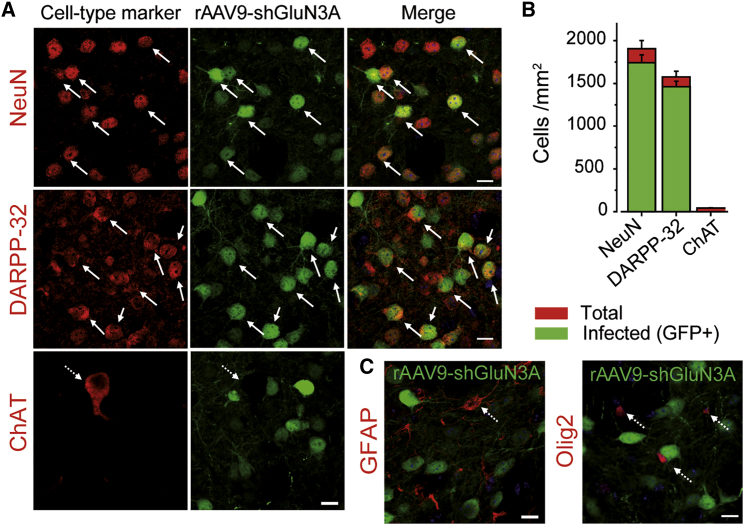
We finally characterized the transduction profile of rAAV9-shGluN3A by quantifying the co-localization of GFP with specific cell type markers. rAAV9-shGluN3A transduced a majority of striatal neurons and exhibited strong tropism for DARPP-32-labeled MSNs (92.6% ± 4% of MSNs were GFP positive), with virtually no cholinergic interneurons transduced (Figures 2A and 2B). Transduction of GFAP-positive astrocytes was very low and restricted to the site of injection (Figure 2C). GFP expression was also absent from striatal oligodendrocytes labeled with Olig2 (Figure 2C). Similarly, high efficiency and analogous neuronal distribution of infection by rAAV9 were observed throughout the duration of the experiment (Figure S2), in line with the sustained reductions in GluN3A protein measured by western blot. Thus, rAAV9-shGluN3A shows a specific neuronal tropism in the striatum and preferentially targets MSNs, the vulnerable population in HD, without modifying GluN3A levels in other neuronal or non-neuronal populations.
Figure 2.
In Vivo Transduction Efficiency in Immunohistochemically Identified Cell Types
(A and C) Single confocal images of striatal sections from WT mice receiving rAAV9-shGluN3A at 1 month of age (green) and stained by immunohistochemistry for specific cell type markers upon sacrifice, 2 weeks after delivery (red) (A: NeuN, all neurons; DARPP-32, MSNs; ChAT, cholinergic neurons; C: GFAP, astrocytes; Olig2, oligodendrocytes). Solid arrows, examples of colocalization; dashed arrows, no colocalization. Scale bars, 10 μm. (B) Quantification of striatal cells positive for a given marker transduced by rAAV9-shGluN3A. Data are mean ± SEM (n = 4 animals per group, five striatal fields at different antero-posterior levels were analyzed per mouse).
Intrastriatal rAAV9-shGluN3A Prevents and Reverses Spine Loss in MSNs
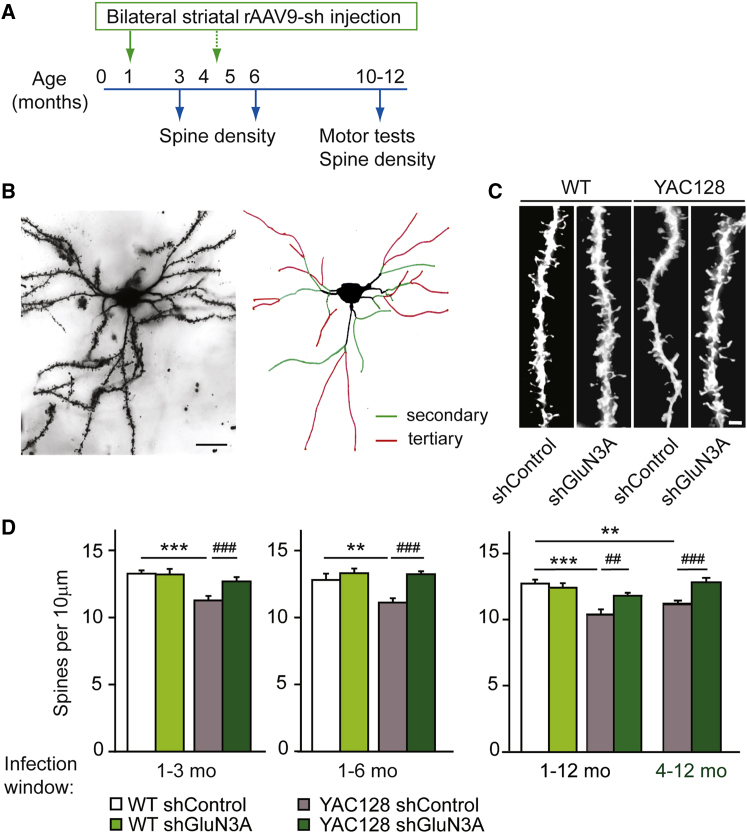
To evaluate the beneficial effects of suppressing GluN3A expression in MSNs, we performed single bilateral injections of rAAV9-shGluN3A or rAAV9-shControl into the striatum of WT and YAC128 mice (Figure 3A). YAC128 mice express full-length HTT with 128 glutamine repeats under the control of the human promoter and recapitulate many characteristics of HD, including early dysfunction and loss of MSN synapses prior to motor and cognitive deficits, later followed by cell death.26 A first set of injections was timed to match the onset of synapse dysfunction, which can be detected by 1 month of age in YAC128 mice.17 The status of synaptic connectivity was monitored over the course of disease progression (3, 6, and 12 months) by measuring spine densities on MSNs using the Golgi impregnation technique and applying quantitative criteria previously described (Figures 3A and 3B). MSNs from YAC128 mice injected with rAAV9-shControl showed significantly decreased spine densities at all ages examined, consistent with previous work (Figures 3C and 3D, white versus gray bars). Spine loss was fully prevented by rAAV9-shGluN3A; the effect was persistent as it was observed 2, 5, and even 11 months after surgery (Figure 3D, dark green versus gray bars). By contrast, rAAV9-shGluN3A did not affect spine density in WT MSNs at any age examined (Figure 3D, white versus bright green bars) as could be expected given the low levels of juvenile GluN3A subunits expressed by adult MSNs. This latter finding confirmed that GluN3A suppression specifically targets the pathological trait, i.e., enhanced GluN3A-mediated pruning in MSNs, without affecting physiological mechanisms used by MSNs for spine maintenance.
Figure 3.
GluN3A Silencing Prevents and Reverses Spine Loss
(A) Experimental design followed in this study. Times of injection are indicated in green. (B) Photomicrograph of a Golgi-impregnated MSN from a 12-month-old YAC128 mouse injected with rAAV9-shGluN3A, along with schematic showing criteria adopted for defining secondary (green) or tertiary (red) dendrites to be used in quantification. Scale bar, 20 μm. (C) Representative dendritic segments of tertiary dendrites from MSNs of 6-month-old WT and YAC128 mice injected with rAAV9-shControl/shGluN3A at 1 month of age. Scale bar, 2 μm. (D) Quantification of spine densities in MSNs from mice receiving rAAV9-shControl or shGluN3A over the indicated time windows. Two-way ANOVA 1–3 months: genotype × shRNA interaction, F(1,91) = 9.24, p = 0.0031; shRNA, F(1,91) = 4.76, p = 0.0317; genotype, F(1,91) = 12.6, p = 0.0006; 1–6 months: genotype × shRNA, F(1,70) = 5.78, p = 0.0189; shRNA, F(1,70) = 15, p = 0.0002; genotype, F(1,70) = 6.90, p = 0.016; 1–12 months: genotype × shRNA, F(1, 102) = 8.85, p = 0.0037; shRNA, F(1,102) = 2.44, p = 0.1214; genotype, F(1,102) = 20.4, p < 0.0001; 4–12 months: genotype × shRNA, F(1,109) = 11.6, p = 0.0009; shRNA, F(1,109) = 3.88, p = 0.0515; genotype, F(1,109) = 0.538, p = 0.465. Data are mean ± SEM (n = 14–35 neurons from 4–6 animals per group; **p < 0.01, ***p < 0.001, ##p < 0.01, ###p < 0.001, post-hoc Bonferroni multiple comparison test).
In order to define a potential therapeutic window, we injected rAAV9-shGluN3A or shControl into the striatum of 4- to 5-month-old mice, an age by which spine loss is well established in the YAC128 model. End-study analysis of spine density was conducted at 12 months of age. A significant reversal of spine loss was observed in YAC128 mice receiving rAAV9-shGluN3A relative to mice injected with control shRNA (Figure 3D, far right gray versus dark green bars; p < 0.001, two-way ANOVA followed by Bonferroni post-hoc test). Cumulatively, the results show that suppressing GluN3A in MSNs prevents and rescues the synaptic pathology even when treatment is started after overt spine loss has taken place.
Intrastriatal rAAV9-shGluN3A Improves Motor Performance
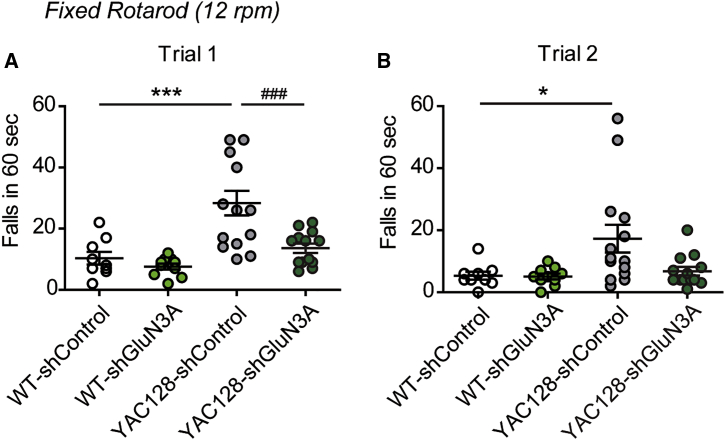
We finally investigated whether the synaptic rescue was associated to improvements in motor performance. Motor coordination and balance were assayed on 10- to 12-month-old mice bilaterally injected with rAAV9-shGluN3A or shControl. By this age, YAC128 mice displayed impairments in motor coordination in a fixed rotarod task when compared to WT (Figure 4A, white versus gray circles; p < 0.001, two-way ANOVA followed by Bonferroni post-hoc test). rAAV9-shGluN3A significantly decreased the number of falls from the fixed rotarod relative to mice receiving control shRNA (Figure 4A; p < 0.001, dark green versus gray circles; p < 0.001, two-way ANOVA followed by Bonferroni post-hoc test) and fully restored the performance of YAC128 mice upon two trials (Figure 4B).
Figure 4.
rAAV9-shGluN3A Improves Motor Coordination in YAC128 Mice
(A and B) Number of falls from a fixed rotarod set at 12 rpm for 10- to 12-month-old mice of the indicated genotypes that received bilateral intrastriatal injections of rAAV9-shControl or shGluN3A at 1 month of age. Data are represented for each of two trials conducted (trial 1, A; trial 2, B). Two-way ANOVA trial 1: genotype × shRNA interaction, F(1,42) = 4.53, p = 0.0392; shRNA, F(1,42) = 9.59, p = 0.0035; genotype, F(1,91) = 18.2, p = 0.0001; trial 2: genotype × shRNA, F(1,42) = 3.03, p = 0.0893; shRNA, F(1,42) = 3.31, p = 0.0762; genotype, F(1,42) = 5.31, p = 0.0262. Data are mean ± SEM (n = 9–14 mice per group; *p < 0.05, ***p < 0.001, ###p < 0.001, post-hoc Bonferroni multiple comparison test).
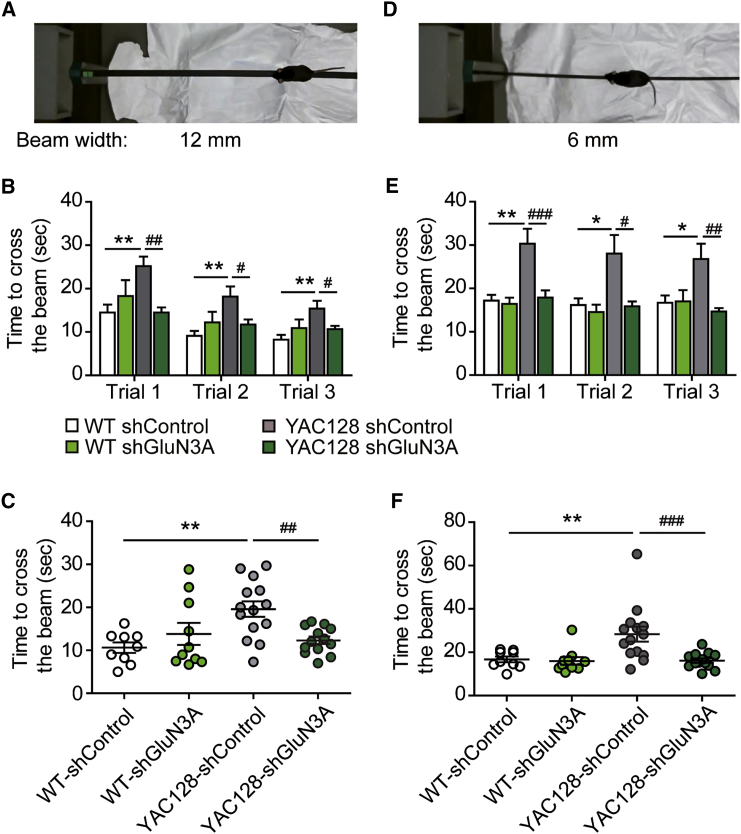
Similar results were observed in the balance beam test, where fine motor coordination and balance can be tested by measuring the time it takes for the mouse to traverse an elevated narrow beam to reach a safe platform. Here, two different beam widths, 12 and 6 mm, were used (Figures 5A and 5D). YAC128 mice took longer to cross a 12-mm beam than WT (albeit their performance improved across trials; Figures 5B and 5C); motor deficits were more prominent on a 6-mm beam, and practice was not sufficient to overcome the defect (Figures 5E and 5F). By contrast, YAC128 mice injected with rAAV9-shGluN3A were indistinguishable from WT animals in their ability to traverse both beams (Figures 5B, 5C, 5E, and 5F). Injection of rAAV9-shGluN3A in WT mice did not affect motor performance in either task, as noted in the rotarod task. Amelioration of the motor phenotype was not due to changes in body weight or motor strength because neither parameter was affected by rAAV9-shGluN3A in YAC128 mice (Figure S3).
Figure 5.
Rescue of Balance Deficits by rAAV9-shGluN3A in YAC128 Mice
(A and D) Photographs of the balance beam task showing two different beam widths (12 mm, A; 6 mm, D) and escape box to the left. (B, C, E, and F) Time to cross a 12 (B and C) or 6 (E and F)-mm-wide beam for 10- to 12-month-old mice of the indicated genotypes and treatments. Data in (B) and (E) are per trial. Two-way ANOVA genotype × shRNA interaction: 12-mm beam trial 1, F(1,42) = 9.62, p = 0.0034; trial 2, F(1,42) = 5.88, p = 0.0197; trial 3, F(1,42) = 5.851, p = 0.020; 6-mm beam: trial 1, F(1,42) = 5.45, p = 0.0245; trial 2, F(1,42) = 3.30, p = 0.0765; trial 3, F(1,42) = 5.633, p = 0.0223. Data in (C) and (F) are individual averages across different trials. Two-way ANOVA genotype × shRNA interaction: 12-mm beam, F(1,42) = 9.13, p = 0.043; 6-mm beam, F(1,42) = 5.59, p = 0.0228. Data are mean ± SEM (n = 9–14 mice per group; *p < 0.05, **p < 0.01, #p < 0.05, ##p < 0.01, ###p < 0.001, post-hoc Bonferroni multiple comparison test).
Discussion
Here, we provide validation of GluN3A as a target to treat early pathogenic mechanisms in HD and develop and test RNAi-based tools to harness this therapeutic potential. Our previous findings that (1) impaired glutamatergic synaptic transmission and excessive synapse pruning resulting from aberrant GluN3A expression are among the earliest events in HD pathophysiology and major drivers of the pathogenic process, and (2) genetic GluN3A suppression prevents early- to late-disease phenotypes,17, 18 led us to explore viable therapeutic strategies. We show that upon a single intrastriatal injection, rAAV9s encoding shRNAs against GluN3A drove highly efficient, long-lasting GluN3A silencing that selectively targeted MSNs. Silencing expression in MSNs was sufficient for halting the synaptic pathology and restoring motor performance.
The present work corroborates and extends the earlier results and has major translational implications. First, silencing GluN3A exclusively in MSNs (that normally express low GluN3A levels in adult stages) while sparing other cell types in the striatum (such as cholinergic interneurons that retain high expression into adulthood)17 permits to block specifically the pathological event, i.e., age-inappropriate reactivation of GluN3A in vulnerable populations, while sparing normal function. Such specificity could provide clinical benefit while minimizing adverse effects. The observation that injecting rAAV9-shGluN3A does not affect spine density in WT MSNs, nor motor performance, supports this rationale. Second, rAAV9-driven GluN3A silencing was effective when timed with the onset of the synaptic pathology but also when initiated later in the disease course, establishing a window of opportunity over which preexisting phenotypes can be reversed. Third, the approach targets one of the earliest disease mechanisms when intervention is more likely to be efficacious and before a point of no return has been reached.27 Electrophysiological and morphological evidence of early dysfunction and loss of MSN synapses is extensive in HD mouse models9, 18 and humans.11, 12 Likewise, longitudinal imaging and functional studies in humans report significant striatal atrophy years prior to diagnosable HD,28 which is strongly correlated with time to disease onset, performance, and clinical progression.14, 15 And, GluN3A has been shown to be required for the multivariate dysfunction of synaptic transmission onto MSNs that precedes morphological signs (chosen as readout in this study), including early enhanced synaptic currents mediated by AMPA and NMDA-type glutamate receptors and NMDA “spikes” or “upstates.”17, 18
Within the experimental time frame of the present study, loss of MSNs is not yet detectable in YAC128 mice and we could not determine whether GluN3A knockdown would reduce the neurodegeneration that is seen at later stages. In our previous report, genetic cross of YAC128 mice with GluN3A null mice conferred partial protection from cell death,17 suggesting that preventing synaptic damage preserves pro-survival signaling pathways driven by afferent synaptic activity.29 However, multiple other factors such as deficient neurotrophic signaling, transcriptional dysregulation, or altered proteostasis have been linked to cell death in preclinical HD studies, and establishing whether these are related or independent of the synaptic pathology needs to be carefully addressed.
While HTT-lowering drugs seem the therapy of choice, developing allele-specific strategies has proven to be difficult and will likely require targeting single-nucleotide polymorphisms residing only in the mutant allele or alternative approaches such as genome editing.30, 31 Clinical trials testing the safety of allele-specific antisense oligonucleotides that silence the mutant HTT gene (Wave Precision-HDs) are currently active, but the polymorphisms targeted are present only in a subset of people with the HD gene and in some cases do not discriminate between mutant and normal HTT.32 Antisense oligonucleotides against both WT and mutant HTT (IONIS-HTTRx) have recently yielded promising dose-response reductions in protein levels along with appropriate safety and tolerability profiles in a completed phase 1 clinical trial. However, the efficacy of these approaches (as well as their tolerability over the extended periods of time likely required to treat chronic neurodegeneration) remains to be tested through larger trials. Our study provides the necessary proof-of-principle for exploring therapies targeting aberrant GluN3A expression in HD that could be used alone or in combination with HTT-lowering treatments. Efforts to be undertaken include further evaluation of the current RNA-based approach or investing in screenings for small molecules blocking GluN3A expression, function, or downstream signaling pathways. In the current RNA-based approach, delivery continues to be a challenge, as it would require direct intraparenchymal administration into the striatum. Nevertheless, our data show that rAAV9 yields remarkably long-lasting knockdown of GluN3A protein levels associated to motor improvement upon a single injection, which would limit the invasiveness of the procedure.
Materials and Methods
RNAi Constructs
A 59-bp fragment encoding a 19-bp-long shRNA specific for rat and mouse GluN3A (sh-GluN3A target sequence: 5′-CTACAGCTGAGTTTAGAAA-3′)22 or a control shRNA were expressed under control of the H1 promoter. For serotype comparison experiments in Figure S1, rAAV expression vectors carried only EGFP driven by a CMV promoter and were produced at the Vector Core Facility of the University of Barcelona (Spain). rAAV9-shControl and rAAV9-shGluN3A were manufactured by Vector Biolabs (Malvern, PA). Titers of rAAV9-shControl or shGluN3A used ranged between 3.2 and 4.4 × 1013 vg/mL.
Animals
WT and YAC128 (line 55 homozygotes) male mice in a FVB/N background crossed for at least four generations into a C57Bl6J background were used. Animals were housed four to six per cage with ad libitum access to food and water and maintained in a temperature-controlled environment on a 12-hr dark/light cycle. All procedures were conducted in accordance with the European and Spanish regulations (86/609/EEC; RD1201/2005) and were approved by the Ethical Committee of the Government of Navarra.
Stereotaxic Injections
Mice were anesthetized with ketamine/xylazine (80/10 mg/kg, intraperitoneally [i.p.]) and placed in a stereotaxic frame. The scalp was shaved, a longitudinal incision was made to expose the skull surface, and two burr holes were drilled above the infusion sites. A 2-μL virus suspension was stereotaxically injected into the striatum of anesthetized mice according to the Paxinos and Franklin mouse brain atlas.25 A 5 μL Hamilton syringe was used for the infusion (Hamilton, Reno, NV). The infusion rate was 200 nL/ min, and the needle remained in place for 5 min after infusion for vector absorption before removal of the syringe. Finally, the site was stitched closed.
Immunohistochemistry
Animals were deeply anesthetized and transcardially perfused with 4% paraformaldehyde in 0.1 M phosphate buffer (PB) (pH 7.4). Brains were removed, post-fixed overnight, and stored in 30% sucrose in PB at 4°C. Thirty-micrometer coronal sections were cut with a freezing sliding microtome and stored at −20°C in cryoprotectant solution (30% ethylene glycol, 30% glycerol, 20 mM PB) until processing. Free-floating brain sections were washed in PBS, blocked for 30 min in 1% BSA, 0.1% Triton X-100, and 4% normal goat or donkey serum (NS) in PBS, and incubated with primary antibody in 1% BSA, 0.1% Triton X-100, and 1% NS in PBS overnight at 4°C. The following primary antibodies were used: mouse anti-NeuN (1:500, Millipore, MAB377), rabbit anti-DARPP-32 (1:500, Cell Signaling, 19A3), goat anti-choline acetyltransferase (ChAT) (1:100, Chemicon AB144P), rabbit anti-GFAP (1:500, Dako, 20334), and rabbit anti-Olig2 (1:1000, Millipore AB9610). After washing with PBS, sections were incubated with appropriate Cy3-conjugated secondary antibody (1:1000, Jackson ImmunoResearch) in 1% BSA, 0.1% Triton X-100 in PBS for 1 hr at room temperature. After several washes in PBS, sections were mounted onto Superfrost Plus slides (Thermo Fisher Scientific), air-dried, and coverslipped with Mowiol-DABCO solution.
Image Acquisition and Analysis
To quantify transduction efficiency, mosaic pictures of sequential rAAV-injected brain sections (at the level of the striatum and spaced 240 μm apart) were captured with a 2.5× objective using a Nikon Eclipse E600 epifluorescence microscope. Positive EGFP signal was defined as three times the value of fluorescent intensity in the green channel in an adjacent but non-injected region of the brain. Regions with GFP signal within the striatum were automatically detected with a set threshold of 3-fold over matched non-injected regions, and their areas were summed to calculate the total infected area. For cell tropism, colocalization of GFP fluorescence with neuronal/glial markers was analyzed with Fiji software on images acquired on a Zeiss LSM800 confocal microscope (Carl Zeiss) using 10× or 40× objectives.
Western Blot Analysis
Striata were dissected and homogenized into 10 volumes of cold homogenization buffer (0.32 M sucrose, 10 mM HEPES [pH 7.4], 2 mM EDTA) supplemented with protease inhibitors (Roche). Protein concentrations were determined using the BCA protein assay (Pierce). Twenty to fifty micrograms of total protein were resolved by SDS-PAGE and transferred onto PVDF membranes (GE Healthcare). Antibodies to mouse anti-GluN1 (1:2,000, Millipore MAB363), mouse anti-GluN3A (1:100, kind gift from J.S. Trimmer), and mouse anti-DARPP-32 (1:500; BD Biosciences, 611520) were used for immunoblotting, followed by incubation with horseradish peroxidase-conjugated secondary antibodies (1:10,000, GE Healthcare). Blots were developed using enhanced chemiluminescence (ECL) plus substrate (Pierce) and exposed to film (GE Healthcare), and band intensities were quantified on a densitometer (Bio-Rad).
Spine Measurements
Fresh brains were processed following the Golgi-Cox method.17 The slides were randomly coded, and the experimenter was blind to genotype during image acquisition and analysis. Bright-field images of Golgi-impregnated MSNs were captured with a 100× oil objective. Only fully impregnated MSNs with their soma and at least four orders of dendrites entirely within the thickness of the section were included in the analysis. Image z stacks were taken every 0.75 μm and analyzed with Fiji software. A total of 2.536 dendritic segments (25–124-μm long; average, 50.67 μm) were traced through different layers of the stack and spines counted. Spine density per neurons was calculated in as many tertiary dendrites of length >25 μm as could be unequivocally identified per neuron (range, 7–15 dendrites).
Behavioral Tests
Two independent male mice cohorts of 10–12 months of age were used. For motor coordination assessment, naive mice were placed on a rotarod with a fixed speed of 12 rpm (two trials/day, spaced 2 hr) and the number of falls was recorded until the sum of latencies to fall reached a total of 60 s per trial. A balance beam test was used to evaluate fine motor coordination and balance.33 Mice were placed at one end of a 12-mm-wide, 90-cm-long beam, and the time to reach an escape box containing nesting material and located in the opposite end was recorded. Mice were allowed to rest for 15 s before next trial with a total of three consecutive trials. After 2 hr, the test was repeated using a 6-mm beam width. Muscular strength was measured as described previously.34 In brief, mice were placed on top of a standard wire cage lid (surrounded by tape to prevent mice walking off the edge), and after light shaking so mice gripped the wires, the lid was turned upside down. Latency to the first fall was recorded.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5. Sample sizes for each experiment were determined based on previous studies from our laboratory. One- or two-way ANOVA followed by Bonferroni post-hoc tests was used to assess differences between groups. In all figures, data are presented as means ± SEM.
Author Contributions
S.M. performed surgeries and behavioral experiments; S.M. and A.M. performed jointly all other experiments and data analysis. I.P.-O. and S.M. conceived the study, designed experiments, analyzed data, and wrote the paper.
Conflicts of Interest
The authors have no conflict of interest.
Acknowledgments
The authors thank Luis García-Rabaneda and John Wesseling for advice with the behavioral studies and manuscript writing and Miguel Pérez-Otaño for help with graphic design. Work was funded by the Spanish Ministry of Science (grants CSD2008-00005 and SAF2016-80895-R to I.P.-O., SAF2013-48983-R to I.P.-O. and J.F.W., SEV-2013-0317 to the Instituto de Neurociencias, and fellowship BES-2014-069359 to A.M.), UTE project CIMA, the Marató TV3 Foundation, Beca Josefina Garre (to I.P.-O.), and the Fundación Caja Navarra (to S.M.).
Footnotes
Supplemental Information includes three figures and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.05.013.
Supplemental Information
References
- 1.Wesseling J.F., Pérez-Otaño I. Modulation of GluN3A expression in Huntington disease: a new n-methyl-D-aspartate receptor-based therapeutic approach? JAMA Neurol. 2015;72:468–473. doi: 10.1001/jamaneurol.2014.3953. [DOI] [PubMed] [Google Scholar]
- 2.Boudreau R.L., McBride J.L., Martins I., Shen S., Xing Y., Carter B.J., Davidson B.L. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kordasiewicz H.B., Stanek L.M., Wancewicz E.V., Mazur C., McAlonis M.M., Pytel K.A., Artates J.W., Weiss A., Cheng S.H., Shihabuddin L.S. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drouet V., Perrin V., Hassig R., Dufour N., Auregan G., Alves S., Bonvento G., Brouillet E., Luthi-Carter R., Hantraye P., Déglon N. Sustained effects of nonallele-specific Huntingtin silencing. Ann. Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 5.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O’Kane C.J., Rubinsztein D.C. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 6.Bates G.P., Dorsey R., Gusella J.F., Hayden M.R., Kay C., Leavitt B.R., Nance M., Ross C.A., Scahill R.I., Wetzel R. Huntington disease. Nat. Rev. Dis. Primers. 2015;1:15005. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 7.Jimenez-Sanchez M., Lam W., Hannus M., Sönnichsen B., Imarisio S., Fleming A., Tarditi A., Menzies F., Dami T.E., Xu C. siRNA screen identifies QPCT as a druggable target for Huntington’s disease. Nat. Chem. Biol. 2015;11:347–354. doi: 10.1038/nchembio.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross C.A., Tabrizi S.J. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 9.Milnerwood A.J., Raymond L.A. Early synaptic pathophysiology in neurodegeneration: insights from Huntington’s disease. Trends Neurosci. 2010;33:513–523. doi: 10.1016/j.tins.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Veldman M.B., Yang X.W. Molecular insights into cortico-striatal miscommunications in Huntington’s disease. Curr. Opin. Neurobiol. 2018;48:79–89. doi: 10.1016/j.conb.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graveland G.A., Williams R.S., DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington’s disease. Science. 1985;227:770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- 12.Ferrante R.J., Kowall N.W., Richardson E.P., Jr. Proliferative and degenerative changes in striatal spiny neurons in Huntington’s disease: a combined study using the section-Golgi method and calbindin D28k immunocytochemistry. J. Neurosci. 1991;11:3877–3887. doi: 10.1523/JNEUROSCI.11-12-03877.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabrizi S.J., Scahill R.I., Durr A., Roos R.A., Leavitt B.R., Jones R., Landwehrmeyer G.B., Fox N.C., Johnson H., Hicks S.L., TRACK-HD Investigators Biological and clinical changes in premanifest and early stage Huntington’s disease in the TRACK-HD study: the 12-month longitudinal analysis. Lancet Neurol. 2011;10:31–42. doi: 10.1016/S1474-4422(10)70276-3. [DOI] [PubMed] [Google Scholar]
- 14.Tabrizi S.J., Scahill R.I., Owen G., Durr A., Leavitt B.R., Roos R.A., Borowsky B., Landwehrmeyer B., Frost C., Johnson H., TRACK-HD Investigators Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington’s disease in the TRACK-HD study: analysis of 36-month observational data. Lancet Neurol. 2013;12:637–649. doi: 10.1016/S1474-4422(13)70088-7. [DOI] [PubMed] [Google Scholar]
- 15.Harrington D.L., Liu D., Smith M.M., Mills J.A., Long J.D., Aylward E.H., Paulsen J.S. Neuroanatomical correlates of cognitive functioning in prodromal Huntington disease. Brain Behav. 2014;4:29–40. doi: 10.1002/brb3.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez-Otaño I., Larsen R.S., Wesseling J.F. Emerging roles of GluN3-containing NMDA receptors in the CNS. Nat. Rev. Neurosci. 2016;17:623–635. doi: 10.1038/nrn.2016.92. [DOI] [PubMed] [Google Scholar]
- 17.Marco S., Giralt A., Petrovic M.M., Pouladi M.A., Martínez-Turrillas R., Martínez-Hernández J., Kaltenbach L.S., Torres-Peraza J., Graham R.K., Watanabe M. Suppressing aberrant GluN3A expression rescues synaptic and behavioral impairments in Huntington’s disease models. Nat. Med. 2013;19:1030–1038. doi: 10.1038/nm.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahfooz K., Marco S., Martínez-Turrillas R., Raja M.K., Pérez-Otaño I., Wesseling J.F. GluN3A promotes NMDA spiking by enhancing synaptic transmission in Huntington’s disease models. Neurobiol. Dis. 2016;93:47–56. doi: 10.1016/j.nbd.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Kehoe L.A., Bellone C., De Roo M., Zandueta A., Dey P.N., Pérez-Otaño I., Muller D. GluN3A promotes dendritic spine pruning and destabilization during postnatal development. J. Neurosci. 2014;34:9213–9221. doi: 10.1523/JNEUROSCI.5183-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borel F., Kay M.A., Mueller C. Recombinant AAV as a platform for translating the therapeutic potential of RNA interference. Mol. Ther. 2014;22:692–701. doi: 10.1038/mt.2013.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson B.L., McCray P.B., Jr. Current prospects for RNA interference-based therapies. Nat. Rev. Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan T., Mameli M., O’Connor E.C., Dey P.N., Verpelli C., Sala C., Perez-Otano I., Lüscher C., Bellone C. Expression of cocaine-evoked synaptic plasticity by GluN3A-containing NMDA receptors. Neuron. 2013;80:1025–1038. doi: 10.1016/j.neuron.2013.07.050. [DOI] [PubMed] [Google Scholar]
- 23.Aschauer D.F., Kreuz S., Rumpel S. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS ONE. 2013;8:e76310. doi: 10.1371/journal.pone.0076310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cearley C.N., Vandenberghe L.H., Parente M.K., Carnish E.R., Wilson J.M., Wolfe J.H. Expanded repertoire of AAV vector serotypes mediate unique patterns of transduction in mouse brain. Mol. Ther. 2008;16:1710–1718. doi: 10.1038/mt.2008.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paxinos G., Franklin K. Elsevier Academic Press; San Diego, CA: 2001. The Mouse Brain in Stereotaxic Coordinates. [Google Scholar]
- 26.Slow E.J., van Raamsdonk J., Rogers D., Coleman S.H., Graham R.K., Deng Y., Oh R., Bissada N., Hossain S.M., Yang Y.Z. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 27.Rubinsztein D.C., Orr H.T. Diminishing return for mechanistic therapeutics with neurodegenerative disease duration?: There may be a point in the course of a neurodegenerative condition where therapeutics targeting disease-causing mechanisms are futile. BioEssays. 2016;38:977–980. doi: 10.1002/bies.201600048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aylward E.H., Sparks B.F., Field K.M., Yallapragada V., Shpritz B.D., Rosenblatt A., Brandt J., Gourley L.M., Liang K., Zhou H. Onset and rate of striatal atrophy in preclinical Huntington disease. Neurology. 2004;63:66–72. doi: 10.1212/01.wnl.0000132965.14653.d1. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto S., Pouladi M.A., Talantova M., Yao D., Xia P., Ehrnhoefer D.E., Zaidi R., Clemente A., Kaul M., Graham R.K. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat. Med. 2009;15:1407–1413. doi: 10.1038/nm.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keiser M.S., Kordasiewicz H.B., McBride J.L. Gene suppression strategies for dominantly inherited neurodegenerative diseases: lessons from Huntington’s disease and spinocerebellar ataxia. Hum. Mol. Genet. 2016;25(R1):R53–R64. doi: 10.1093/hmg/ddv442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monteys A.M., Ebanks S.A., Keiser M.S., Davidson B.L. CRISPR/Cas9 editing of the mutant Huntingtin allele in vitro and in vivo. Mol. Ther. 2017;25:12–23. doi: 10.1016/j.ymthe.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kay C., Skotte N.H., Southwell A.L., Hayden M.R. Personalized gene silencing therapeutics for Huntington disease. Clin. Genet. 2014;86:29–36. doi: 10.1111/cge.12385. [DOI] [PubMed] [Google Scholar]
- 33.Luong T.N., Carlisle H.J., Southwell A., Patterson P.H. Assessment of motor balance and coordination in mice using the balance beam. J. Vis. Exp. 2011 doi: 10.3791/2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canals J.M., Pineda J.R., Torres-Peraza J.F., Bosch M., Martín-Ibañez R., Muñoz M.T., Mengod G., Ernfors P., Alberch J. Brain-derived neurotrophic factor regulates the onset and severity of motor dysfunction associated with enkephalinergic neuronal degeneration in Huntington’s disease. J. Neurosci. 2004;24:7727–7739. doi: 10.1523/JNEUROSCI.1197-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.