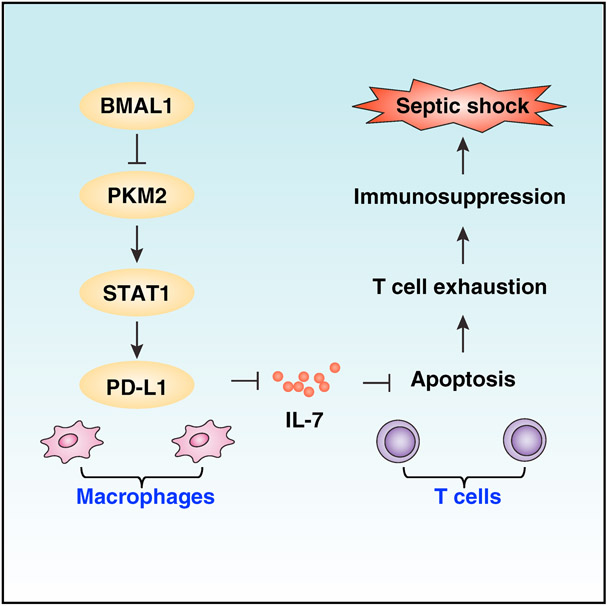
SUMMARY
Sepsis and septic shock are associated with life-threatening organ dysfunction caused by an impaired host response to infections. Although circadian clock disturbance impairs the early inflammatory response, its impact on post-septic immunosuppression remains poorly elucidated. Here, we show that Bmal1, a core circadian clock gene, plays a role in the regulation of host immune responses in experimental sepsis. Mechanistically, Bmal1 deficiency in macrophages increases PKM2 expression and lactate production, which is required for expression of the immune checkpoint protein PD-L1 in a STAT1-dependent manner. Consequently, targeted ablation of Pkm2 in myeloid cells or administration of anti-PD-L1-neutralizing antibody or supplementation with recombinant interleukin-7 (IL-7) facilitates microbial clearance, inhibits T cell apoptosis, reduces multiple organ dysfunction, and reduces septic death in Bmal1-deficient mice. Collectively, these findings suggest that the circadian clock controls the immune checkpoint pathway in macrophages and therefore represents a potential therapeutic target for lethal infection.
In Brief
Deng et al. demonstrate that BMAL1 plays a role in preventing the development of a sepsis phenotype during severe infection through counter-regulating PD-L1 expression and T cell exhaustion. These findings indicate that targeting the circadian clock and immunometabolism pathway has potential for treating infectious diseases that lead to lethal sepsis.
Graphical Abstract
INTRODUCTION
Despite decades of research and advances in clinical management, sepsis and septic shock remain the leading causes of death among intensive care unit patients. The complex interaction between the infecting pathogen and the host immune system in sepsis involves multiple distinct pathways. Among the most frequently isolated pathogens in sepsis patients are Gram-negative bacteria, such as Escherichia coli. Pattern-recognition receptors (e.g., toll-like receptors) are the central components of the innate immune system that recognize exogenous pathogen-associated molecular patterns (PAMPs) and endogenous damage-associated molecular patterns (DAMPs) and trigger both adaptive and maladaptive aspects of the immune response during severe infection (Hotchkiss et al., 2016). The maladaptive host response is generally characterized by two distinct stages: excessive inflammation at early stage and sustained immune suppression at late stage (van der Poll et al., 2017). In particular, immune suppression from T cell dysfunction or apoptosis is frequently associated with sepsis-related deaths in patients (Boomer et al., 2011; Spec et al., 2016). In fact, increasing T cell viability or function using interleukin (IL)-7 has been shown to protect against septic lethality in experimental and clinical settings (Unsinger et al., 2010; Venet et al., 2017).
Circadian rhythms are driven by the circadian clock, which consists of interlocked transcriptional-translational feedback loops. During the daytime, the transcription factors circadian locomotor output cycles kaput (CLOCK) and brain and muscle aryl hydrocarbon receptor nuclear translocator-like 1 (BMAL1) heterodimerize and bind to E-box promoter elements to drive the gene expression of Period (Per1-3) and Cryptochrome (Cry1/2). PER/CRY protein complexes reside in the cytoplasm throughout the day and later relocate into the nucleus, where they inhibit the activity of the CLOCK-BMAL1 complex. Disruption of the circadian clock has been linked to various disorders, including metabolic, inflammatory, and immunological diseases (Asher and Sassone-Corsi, 2015; Curtis et al., 2014).
In this study, we provide evidence that BMAL1 plays a role in preventing the development of a sepsis phenotype during severe infection through counter-regulating programmed cell death 1 ligand 1 (PD-L1), an immune checkpoint protein. Pyruvate kinase M2 (PKM2) upregulation was required for lactate production and PD-L1 expression in Bmal1-deficient macrophages. Consequently, conditional deletion of Bmal1 in myeloid cells accelerated death in polymicrobial sepsis, a phenotype that was reversed with anti-PD-L1-neutralizing antibody or recombinant IL-7 protein or deletion of Pkm2. These findings reveal a direct link between the circadian clock, metabolism, and immune suppression in the pathophysiology of sepsis.
RESULTS
Altered Circadian and Sepsis Phenotype in Bmal1Mye−/− Mice
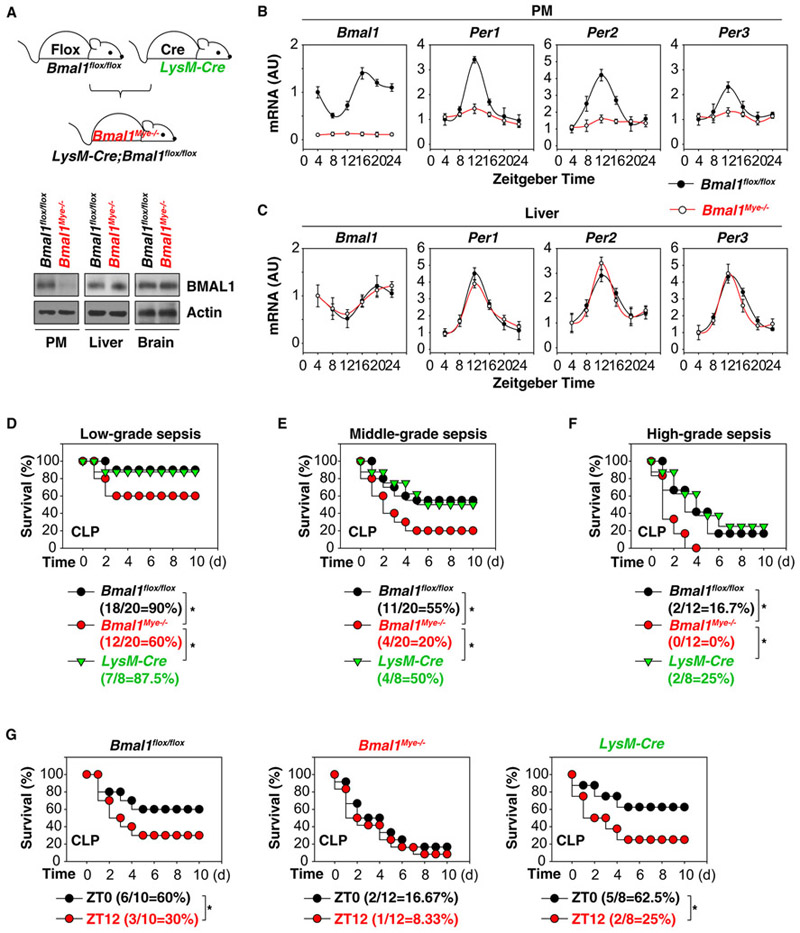
Myeloid cells, including monocytes and macrophages, are key effectors and regulators of inflammation and the innate immune response. To evaluate the role of the core clock gene Bmal1 in vivo, we generated myeloid cell-specific Bmal1-knockout mice (termed Bmal1Mye−/− mice) by crossing Bmal1 flox/flox and LysM-Cre. As expected, the protein expression of BMAL1 was diminished in peritoneal macrophages (PMs) from Bmal1Mye−/− mice, but not from control Bmal1flox/flox mice (Figure 1A). BMAL1 protein levels in other tissues, such as the brain and liver, remained unchanged, confirming the myeloid-cell-specific ablation of Bmal1 in mice (Figure 1A).
Figure 1. Altered Circadian and Sepsis Phenotype in Bmal1Mye−/− Mice.
(A) Western blot analysis of BMAL1 expression in PMs, livers, and brains from Bmal1 flox/flox and Bmal1Mye−/− mice.
(B) Ablation of Bmal1 in the myeloid cells results in disrupted circadian expression of clock genes Bmal1, Per1, Per2, and Per3 in PMs (n = 3 mice/group; data expressed as means ± SD).
(C) Ablation of Bmal1 in myeloid cells does not disrupt circadian expression of clock genes Bmal1, Per1, Per2, and Per3 in liver (n = 3 mice/group; data expressed as means ± SD)
(D−F) Indicated mice were subjected to CLP during ZT0–4 with syringe needles with gauges ranging from 27G (D; ‘low-grade sepsis’), 22G (E; ‘middle-grade sepsis’), to 17G (F; ‘high-grade sepsis’). Animal survival was assayed (n = 12–20 mice/group; *p < 0.05; Kaplan-Meier survival analysis).
(G) Indicated mice were subjected to CLP-induced middle-grade sepsis at ZT0 or ZT12. Animal survival was assayed (n = 8–12 mice/group; *p < 0.05; Kaplan-Meier survival analysis).
As BMAL1 is a core component of the circadian clock, we sought to determine whether targeted ablation of Bmal1 in myeloid cells affects the circadian oscillator in macrophages. Although mRNA expression of Bmal1, Per1, Per2, and Per3 in PMs was abolished in Bmal1Mye−/− mice (Figure 1B), their circadian expression in the liver tissue remained unaffected (Figure 1C). Thus, the myeloid-cell-specific ablation of Bmal1 affects circadian gene expression only within macrophages but does not influence the function of other tissues (Curtis et al., 2015; Gibbs et al., 2012).
A clinically relevant experimental model of sepsis is induced by a surgical procedure termed cecal ligation and puncture (CLP), where the cecal appendage is ligated and then perforated with needles to cause fecal leakage as typically seen in gastroin-testinal perforations in humans (Buras et al., 2005). Zeitgeber time is a measure of time (hours) after the lights go on in the mouse facility. ZT0 corresponds to zeitgeber time 0 (namely, ‘lights on’), whereas ZT12 corresponds to zeitgeber time 12 (namely, ‘lights off’). Mice with myeloid-cell-specific ablation of Bmal1 as well as control animals were subjected to CLP during ZT0–3 (namely, ‘morning time’) using needles that varied in gauge from 17G to 27G. Increasing needle thickness decreased percent survival from 90% (using a 27G needle; ‘low-grade sepsis’), to 60% (using a 22G needle; ‘middle-grade sepsis’), to 16.7% (using a 17G needle; “high-grade sepsis’) in control mice (Bmal1 flox/flox or LysM-Cre mice; Figures 1D–1F). In contrast, Bmal1Mye−/− mice had a significant increase in CLP-induced 10-day death rate across all three levels of sepsis severity (Figures 1D–1F). Consistent with a previous finding using a lethal endotoxemia model (Curtis et al., 2015; Nguyen et al., 2013), control mice (Bmal1 flox/flox or LysM-Cre mice) were less susceptible to the CLP-induced middle-grade sepsis at ZT0 versus ZT12 (Figure 1G) compared to Bmal1Mye−/− mice (Figure 1G). These findings highlight the importance of an intact circadian clock gene Bmal1 in suppressing a lethal response to severe infection.
The Circadian Clock Blocks PD-L1 Expression in Activated Macrophages and Monocytes
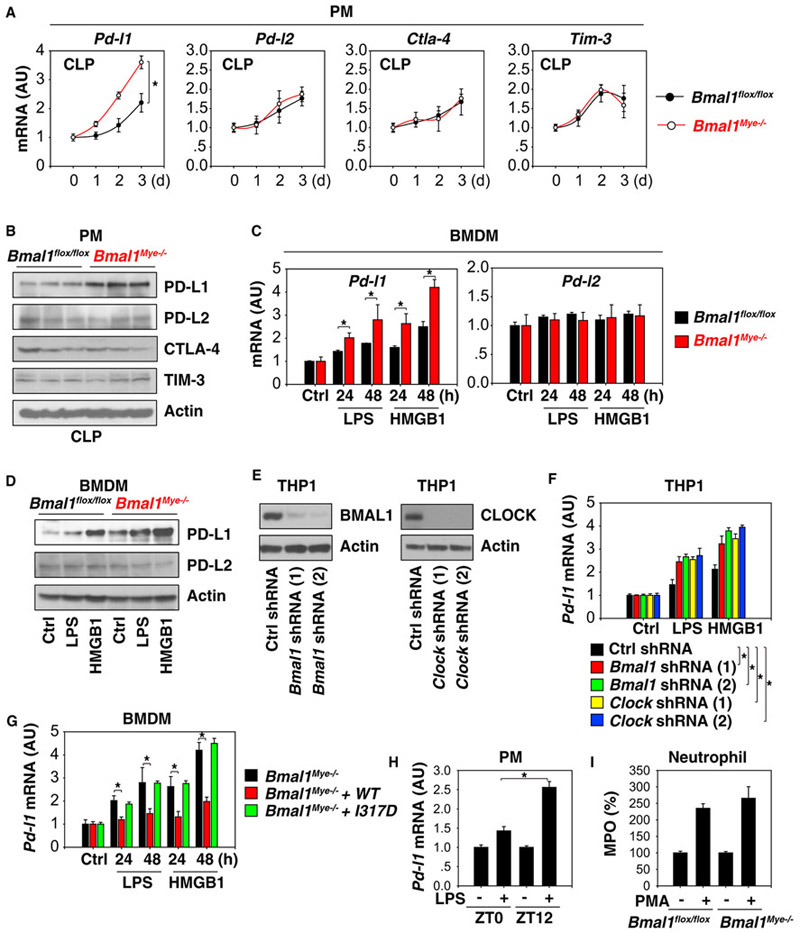
Immune checkpoints are negative regulators of the immune system and emerge as a modulator of inflammation-associated immune suppression during sepsis (Delano and Ward, 2016). To establish whether the altered septic phenotype in Bmal1Mye−/− mice may be due to abnormal immune responses, we determined the expression of several immune checkpoint genes in macrophages. Pd-l1 was significantly upregulated in PMs (Figure 2A) from septic Bmal1Mye−/− mice. Other immune checkpoints, such as Pd-l2, cytotoxic T-lymphocyte associated protein 4 (Ctla-4), and T cell immunoglobulin mucin family member 3 (Tim-3), were not increased in Bmal1-deficient macrophages (Figure 2A). Consistently, the protein levels of PD-L1 (but not PD-L2, CTLA-4, and TIM-3) were upregulated in PMs from septic Bmal1Mye−/− mice (Figure 2B). We also analyzed the effects of Bmal1 deletion on PD-L1 expression in bone-marrow-derived macrophages (BMDMs) following stimulation with a prototypic PAMP (e.g., lipopolysaccharide [LPS]) or DAMP (e.g., high mobility group box 1 [HMGB1]; Wang et al., 1999). The loss of Bmal1 increased LPS- or HMGB1-induced Pd-l1 mRNA (Figure 2C) and protein (Figure 2D) expression in BMDMs. Loss of Bmal1 failed to affect the expression of PD-L2 (Figures 2C and 2D). Similarly, the knockdown of Bmal1 or Clock by two different short hairpin RNAs (shRNAs) increased LPS- or HMGB1-induced Pd-l1 mRNA expression in a human leukemia monocytic THP1 cell line (Figures 2E and 2F). A Bmal1 I317D mutant exhibits a loss of affinity for CLOCK in the circadian clock (Huang et al., 2012). We observed that Bmal1 I317D mutant similarly failed to inhibit the LPS- or HMGB1-induced Pd-l1 mRNA expression in Bmal1Mye−/− BMDMs (Figure 2G). These findings indicate that circadian clock genes selectively suppress PD-L1 expression in activated macrophages and monocytes.
Figure 2. The Circadian Clock Limits PD-L1 Expression in Activated Macrophages and Monocytes.
(A) qPCR analysis of expression of indicated genes in PMs from middle-grade septic Bmal1 flox/flox and Bmal1Mye−/− mice (n = 3 mice/group; data expressed as means ± SD; *p < 0.05; ANOVA LSD test).
(B) Western blot analysis of expression of indicated proteins in PMs at day 3 from middle-grade septic Bmal1 flox/flox and Bmal1Mye−/− mice (n = 3 mice/group).
(C) qPCR analysis of Pd-l1 and Pd-l2 mRNA expression in indicated BMDMs treated with LPS (100 ng/mL) or HMGB1 (500 ng/mL) for 24–48 hr (n = 3; data expressed as means ± SD; *p < 0.05; t test).
(D) Western blot analysis of PD-L1 and PD-L2 protein expression in indicated BMDMs treated with LPS (100 ng/mL) or HMGB1 (500 ng/mL) for 48 hr.
(E) Western blot analysis of BMAL or CLOCK protein expression in indicated knockdown THP1 cells.
(F) Knockdown of Bmal1 or Clock increased LPS (100 ng/mL)- or HMGB1 (500 ng/mL)-induced Pd-l1 mRNA expression in THP1 cells (n = 3; data expressed as means ± SD; *p < 0.05; t test).
(G) Bmal1 wild-type, but not I317D mutant, restored the inhibition of LPS (100 ng/mL)- or HMGB1 (500 ng/mL)-induced Pd-l1 mRNA expression in Bmal1Mye−/− BMDMs (n = 3; data expressed as means ± SD; *p < 0.05; t test).
(H) Wild-type PMs were harvested at ZT0 and ZT12 and treated immediately with LPS (100 ng/mL) for 24 hr and analyzed for Pd-l1 mRNA expression (n = 3; data expressed as means ± SD; *p < 0.05; t test).
(I) Indicated neutrophils harvested from middle-grade septic Bmal1 flox/flox and Bmal1Mye−/− mice were exposed to phorbol 12-myristate 13-acetate (PMA; 0.08 μM) for 30 min and analyzed for myeloperoxidase (MPO) release (n = 3; data expressed as means ± SD).
Coincident with the increased Pd-l1 expression in activated Bmal1Mye−/− macrophages (Figure 2C), the LPS-induced Pd-l1 mRNA expression was greater in wild-type PMs isolated at ZT12 versus at ZT0 (Figure 2H), supporting that the circadian clock controls PD-L1 expression during lethal infection. Neutrophil activation by phorbol 12-myristate 13-acetate was not different between isolated circulating neutrophils from septic Bmal1 flox/flox and Bmal1Mye−/− mice (Figure 2I). It suggests that conditional knockout of Bmal1 in macrophages controls the activation of macrophages, but not neutrophils, during sepsis.
Lactate Metabolism Contributes to PD-L1 Expression
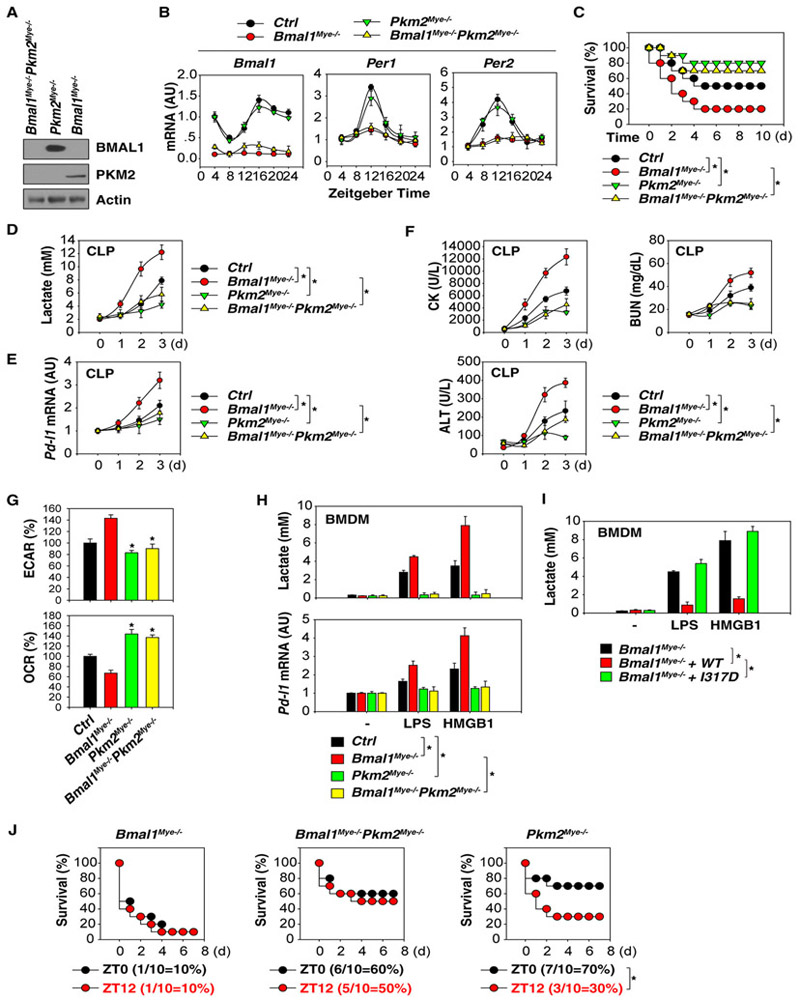
Sepsis and septic shock are associated with lactic acidosis (Thomas-Rueddel et al., 2015), supporting a metabolic role of lactate in the regulation of immune response in sepsis. We and others have demonstrated that PKM2-mediated lactate metabolism in macrophages contributes to the inflammatory response, organ injury, and septic death (Huang et al., 2017; Palsson-McDermott et al., 2015; Xie et al., 2016; Yang et al., 2014; Zhang et al., 2016). To investigate whether PKM2 is responsible for the increased risk of sepsis lethality in Bmal1Mye−/− mice, we generated myeloid-cell-specific Bmal1 and Pkm2 double knockout mice (Bmal1Mye−/−;Pkm2Mye−/− mice). As expected, the protein expression of both BMAL1 and PKM2 were absent in PMs from Bmal1Mye−/−;Pkm2Mye−/− mice (Figure 3A). The loss of Pkm2 did not alter the expression of Bmal1, Per1, and Per2 mRNA in PMs isolated from Bmal1Mye−/−;Pkm2Mye−/− mice compared to Bmal1Mye−/− mice (Figure 3B). However, the absence of Pkm2 increased survival (Figure 3C), reduced serum lactate levels (Figure 3D), and blocked Pd-l1 mRNA expression in PMs (Figure 3E) in septic Bmal1Mye−/−;Pkm2Mye−/− mice. Biochemical measurement of circulating levels of tissue injury enzymes also revealed protective effects of Pkm2 depletion against dysfunction or injury of the heart (creatine kinase [CK]), kidney (blood urea nitrogen [BUN]), and liver (alanine aminotransferase [ALT]) in septic Bmal1Mye−/−;Pkm2Mye−/− mice (Figure 3F). The absence of Pkm2 significantly inhibited glycolysis (as measured by extracellular acidification rate [ECAR]) and increased oxidative phosphorylation (as measured by oxygen consumption rate [OCR]) in PMs from septic Pkm2Mye−/− or Bmal1Mye−/−;Pkm2Mye−/− mice compared to Bmal1Mye−/− mice (Figure 3G). These findings support the notion that overcoming PKM2-dependent aerobic glycolysis (the Warburg effect) in macrophages may lead to better outcomes in experimental sepsis (Huang et al., 2017; Xie et al., 2016; Yang et al., 2014; Zhang et al., 2016).
Figure 3. BMAL1 Limits PD-L1 Expression in a Lactate-Metabolism-Dependent Manner.
(A) Western blot analysis of BMAL1 and PKM2 protein expression in indicated PMs.
(B) qPCR analysis of Bmal1, Per1, and Per2 mRNAs in indicated PMs (n = 3 mice/group; data expressed as means ± SD).
(C) Indicated mice were subjected to CLP with 22G syringe needles (middle-grade sepsis), and animal survival was assayed (n = 20 mice/group; *p < 0.05; Kaplan-Meier survival analysis).
(D–F) Analysis of serum lactate (D), Pd-l1 mRNA in PMs (E), and serum tissue function enzyme (F) in indicated middle-grade septic mice (n = 3 mice/group; data expressed as means ± SD).
(G) Basal ECAR (indicative of glycolysis) and OCR (indicative of oxidative phosphorylation) were monitored at isolated PMs from indicated CLP (72 hr)-induced septic mice (n = 3 mice/group; *p < 0.05 versus Bmal1Mye−/− group; data expressed as means ± SD).
(H) Analysis of lactate production and Pd-l1 mRNA in indicated BMDMs following treatment with LPS (100 ng/mL) or HMGB1 (500 ng/mL) for 48 hr (n = 3; data expressed as means ± SD; *p < 0.05; ANOVA LSD test).
(I) Bmal1 wild-type, but not I317D mutant, restored the inhibition of LPS (100 ng/mL)- or HMGB1 (500 ng/mL)-induced lactate production in Bmal1Mye−/− BMDMs (n = 3; data expressed as means ± SD; *p < 0.05; ANOVA LSD test).
(J) Indicated mice were subjected to LPS (15 mg/kg)-induced endotoxemia at ZT0 or ZT12. Animal survival was assayed (n = 10 mice/group; *p < 0.05; Kaplan-Meier survival analysis).
*p < 0.05.
To further test the hypothesis that BMAL1 limits PD-L1 expression in a PKM2-dependent manner, we examined the effects of BMAL1 and PKM2 on the LPS- and HMGB1-induced glycolysis and PD-L1 expression in BMDMs. The levels of lactate production and Pd-l1 mRNA expression were increased in Bmal1Mye−/−, but not in Pkm2Mye−/− or Bmal1Mye−/−;Pkm2Mye−/− BMDMs in response to LPS or HMGB1 (Figure 3H). LPS- or HMGB1-induced lactate production was also not inhibited in macrophages expressing the BMAL I317D mutant (Figure 3I) in Bmal1Mye−/− BMDMs, indicating that CLOCK-BMAL1 transcriptional activator complex is required for the control of lactate production. Pkm2Mye−/− mice were less susceptible to the LPS-induced mortality at ZT0 versus ZT12 compared to Bmal1Mye−/− or Bmal1Mye−/−;Pkm2Mye−/− mice (Figure 3J). These findings further support the idea that BMAL1 plays an important role in gating LPS-induced mortality under circadian control.
PKM2 Is Required for STAT1-Dependent PD-L1 Upregulation
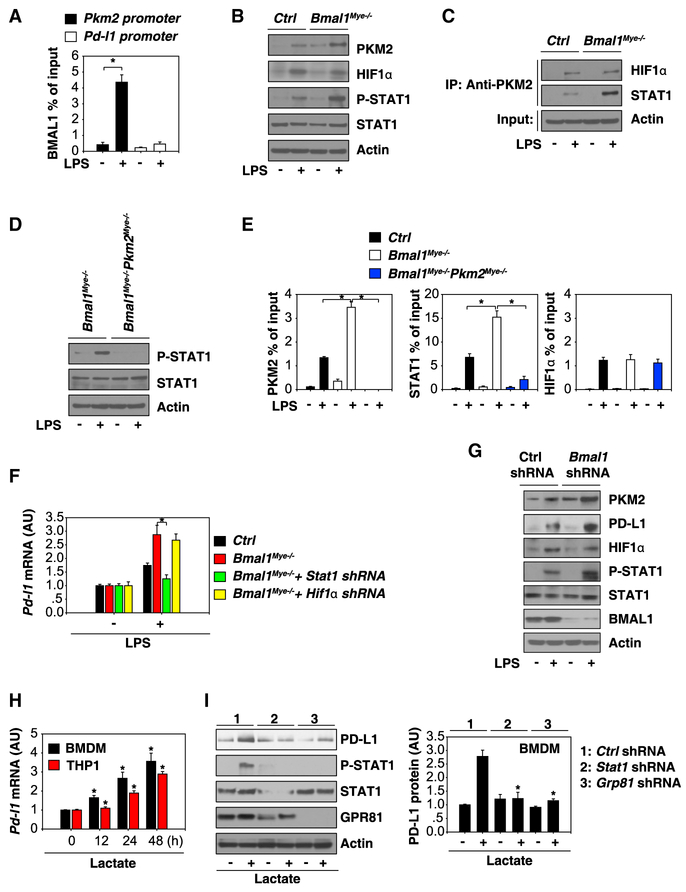
Like the BMAL1/CLOCK target genes that usually carry an E-box DNA motif on their promoters, bioinformatics analysis also revealed the presence of a classical E-box DNA motif (‘CAGCTG’) in the promoter region of Pkm2, but not Pd-l1 gene (Table S1). Chromatin immunoprecipitation (ChIP) analysis confirmed that BMAL1 could bind directly to the promoter of Pkm2, but not Pd-l1 gene, in LPS-stimulated BMDMs (Figure 4A). Moreover, the depletion of Bmal1 increased LPS-induced PKM2 expression in BMDMs (Figure 4B), suggesting that BMAL1 inhibits glycolysis by directly repressing the expression of PKM2 in activated BMDMs.
Figure 4. BMAL1-Mediated PKM2 Expression Contributes to STAT1-Dependent PD-L1 Upregulation.
(A) Binding of BMAL1 to Pd-l1 or Pkm2 promoter was analyzed by ChIP-qPCR in BMDMs following treatment with LPS (100 ng/mL) for 48 hr (n = 3; data expressed as means ± SD; *p < 0.05; t test).
(B) Western blot analysis of indicated proteins in BMDMs following treatment with LPS (100 ng/mL) for 48 hr.
(C) Immunoprecipitation analysis of indicated protein complexes in BMDMs following treatment with LPS (100 ng/mL) for 48 hr.
(D) Western blot analysis of indicated proteins in BMDMs following treatment with LPS (100 ng/mL) for 48 hr.
(E) Binding of PKM2, STAT1, or HIF1α to Pd-l1 promoter was analyzed using ChIP-qPCR in indicated BMDMs following treatment with LPS (100 ng/mL) for 48 hr (n = 3; data expressed as means ± SD; *p < 0.05; t test).
(F) qPCR analysis of Pd-l1 mRNA in indicated BMDMs following treatment with LPS (100 ng/mL) for 48 hr (n = 3; data expressed as means ± SD; *p < 0.05; t test).
(G) Western blot analysis of expression of indicated proteins in BMDMs following treatment with LPS (100 ng/mL) for 48 hr.
(H) qPCR analysis of Pd-l1 mRNA in BMDMs and THP1 cells following treatment with lactate (10 mM) for 12–48 hr (n = 3; data expressed as means ± SD; *p < 0.05 versus untreated group; t test).
(I) Knockdown of STAT1 and GPR81 inhibited lactate (10 mM; 24 hr)-induced PD-L1 protein expression in BMDMs (n = 3; data expressed as means ± SD; *p < 0.05 versus control shRNA group; t test).
See also Table S1.
Although PKM2 might be required for the LPS-induced PD-L1 expression in macrophages through a hypoxia-inducible factor 1α (HIF1α)-dependent mechanism (Palsson-McDermott et al., 2017), we did not observe a significant change in the LPS-induced HIF1α protein expression (Figure 4B) or its interaction with PKM2 (Figure 4C) in Bmal1Mye−/− BMDMs compared to control cells. In contrast, the depletion of Bmal1 led to an elevation of the LPS-induced phosphorylation of STAT1 (a core transactional factor for Pd-l1 expression; Figure 4B) and PKM2-STAT1 interaction (Figure 4C). Consistently, the LPS-induced STAT1 phosphorylation was impaired in Bmal1Mye−/−;Pkm2Mye−/− BMDMs (Figure 4D). The binding of PKM2 and STAT1 (but not HIF1α) to Pd-l1 promoter were increased in Bmal1Mye−/− BMDMs, but not in Bmal1Mye−/−;Pkm2Mye−/− BMDMs (Figure 4E). Similar to the depletion of Pkm2 (Figure 3H), the knockdown of Stat1 (but not Hif1α) reduced LPS-induced Pd-l1 expression in Bmal1-deficient BMDMs (Figure 4F). Like Bmal1Mye−/− BMDMs, knockdown of Bmal1 by shRNA exhibited similar effects on LPS-induced PKM2, PD-L1, and p-STAT1 expression in BMDMs (Figure 4G). These findings indicate that PKM2 is a transcriptional coactivator of STAT1 responsible for promoting Pd-l1 expression in Bmal1-deficient BMDMs.
To further define the role of lactate in the regulation of PD-L1 expression, we directly stimulated macrophage and monocyte cell cultures with lactate. Exogenously supplemented lactate induced Pd-l1 mRNA expression in BMDM and THP1 cells (Figure 4H). Furthermore, the knockdown of G-protein-coupled receptor 81 (Gpr81, a lactate receptor) or Stat1 by shRNA inhibited the lactate-induced STAT1 phosphorylation and PD-L1 protein expression in BMDMs (Figure 4I), supporting that the STAT1 pathway is required for lactate-induced PD-L1 expression in macrophages.
PD-L1 Pathway Contributes to Septic Death in Bmal1Mye−/− Mice
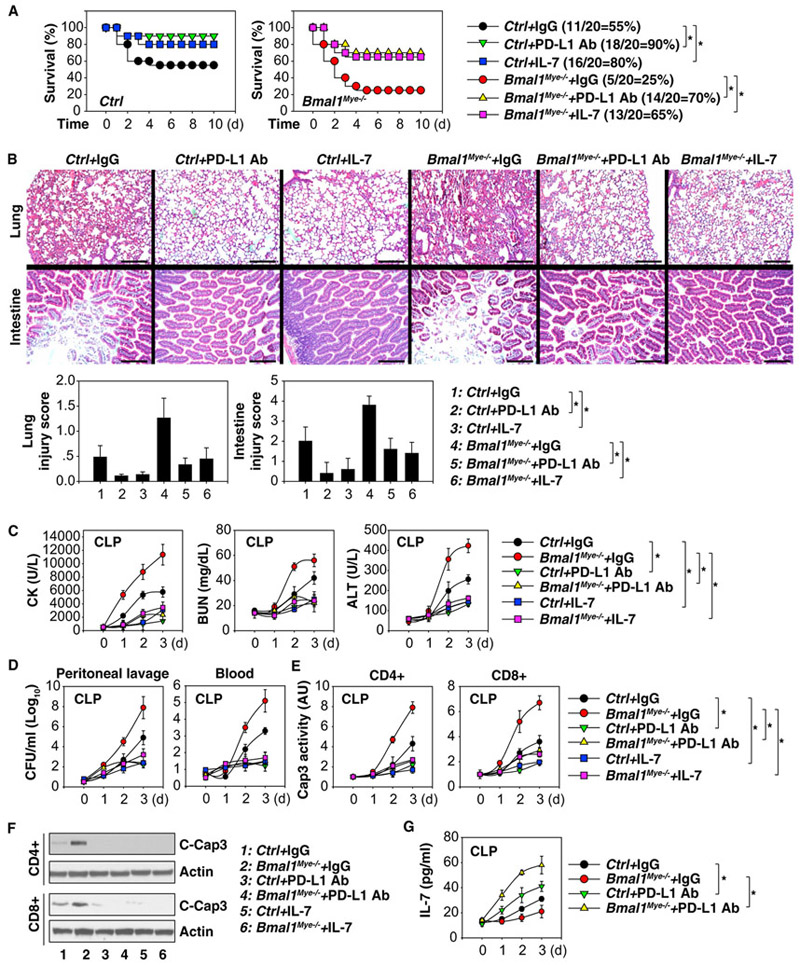
To determine whether the PD-L1 pathway is responsible for the increase in sepsis lethality in Bmal1Mye−/− mice, we treated mice with anti-PD-L1 neutralizing antibody. Compared to the administration of control immunoglobulin G (IgG), anti-PD-L1 neutralizing antibody prolonged animal survival in CLP-induced sepsis in Bmal1Mye−/− mice (Figure 5A). This was also associated with reduced injury in the lung and small intestine (Figure 5B). Lung from septic mice exhibited hemorrhage, leukocyte infiltration, alveolar septal thickening, and edema, whereas intestines displayed markers of injury characterized by the loss of mucosal villi (Figure 5B). These CLP-induced histological changes were attenuated by anti-PD-L1 neutralizing antibody administration in septic Bmal1Mye−/− mice (Figure 5B). Biochemical measurement of circulating tissue enzymes (CK, BUN, and ALT) also confirmed the protective effects of anti-PD-L1 neutralizing antibody against organ dysfunction and injury (Figure 5C).
Figure 5. The PD-L1 Pathway Contributes to Septic Death in Bmal1Mye−/− Mice.
(A) Repeated administration of anti-PD-L1 antibody (20 mg/kg) or IL-7 (5 mg/mice) at 3, 24, 48, and 72 hr after CLP (22G needle) significantly increased survival in indicated mice (n = 20 mice/group; *p < 0.05; Kaplan-Meier survival analysis).
(B–G) In parallel, tissue H&E staining and injury score (day 3; scale bars, 200 αM; B), serum enzyme activity (C), bacterial loading (D), and caspase 3 activity in T cells (E) were assayed (n = 3–5 mice/group; data expressed as means ± SD; *p < 0.05; ANOVA LSD test).
(F) Western blot analysis of cleaved-caspase 3 in indicated T cells.
(G) Serum levels of IL-7 in indicated septic mice (n = 3 mice/group; data expressed as means ± SD; *p < 0.05; ANOVA LSD test).
We next investigated the impact of PD-L1 blockage on bacterial loads and T cell apoptosis. The administration of anti-PD-L1 neutralizing antibody resulted in a reduction of bacterial loads in the blood and peritoneal lavage fluid (Figure 5D) and an inhibition of caspase 3 activity (an indicator of apoptosis) in splenic CD4+ and CD8+T cells (Figure 5E). Consistently, increased cleaved-caspase 3 protein in splenic CD4+ and CD8+T cells from septic Bmal1Mye−/− mice were blocked by anti-PD-L1 neutralizing antibody (Figure 5F), supporting a role of PD-L1 in the regulation of lymphocyte survival. In contrast, serum levels of IL-7, a potent anti-apoptotic cytokine for lymphocyte proliferation, were increased in septic Bmal1Mye−/− mice following anti-PD-L1 neutralizing antibody treatment (Figure 5G). The administration of recombinant IL-7 also increased animal survival (Figure 5A) and reversed disease phenotype (Figures 5C–5F) in septic Bmal1Mye−/− mice. Thus, inhibition of PD-L1 or administration of IL-7 protects mice from sepsis-induced lethality and displays reduced organ damage, improved bacterial clearance, and increased T cell survival.
DISCUSSION
Sepsis is the leading cause of death in intensive care units and recognized as a global health priority by the current World Health Organization (Reinhart et al., 2017). In the present study, we demonstrate that circadian clock controls the immune checkpoint pathway in sepsis. In macrophages, BMAL1-mediated circadian oscillations are required for lactate metabolism, which controls PD-L1 expression to affect T cell survival in sepsis. Genetic depletion of Bmal1 increased the risk of septic death, whereas anti-PD-L1 antibody rendered mice more resistant to polymicrobial sepsis. These findings not only provide insights into the molecular function of the circadian clock gene in immunity but also reinforce the notion that targeting immune checkpoints have potential for treating infectious diseases that lead to lethal sepsis (Hutchins et al., 2014).
Abnormal circadian rhythms have been associated with various pathological conditions, inducing lethal inflammation and immune dysfunction. Disruption of the molecular clock in mice via genetic ablation of the myeloid Baml1 gene or artificially mimicking jet lag increases the acute inflammatory response during endotoxemia (Castanon-Cervantes et al., 2010; Curtis et al., 2015; García et al., 2015; Gibbs et al., 2012). Accordingly, proinflammatory cytokines (e.g., IL-6, IL-12, and tumor necrosis factor) and chemokines (e.g., C-X-C motif chemokine ligand 1, C-C motif chemokine ligand 5, and C-C motif chemokine ligand 2 [CCL2]) were excessively induced by endotoxemia in animals with abnormal circadian rhythms (Castanon-Cervantes et al., 2010; Curtis et al., 2015; García et al., 2015; Gibbs et al., 2012; Hashiramoto et al., 2010; Keller et al., 2009; Nakazato et al., 2017; Nguyen et al., 2013). Mechanistically, circadian control of the innate inflammatory response involves the regulation of Toll-like receptor expression (Silver et al., 2012), nuclear factor κB (NF-κB) activation (Spengler et al., 2012), miR-155 induction (Curtis et al., 2015), and epigenomic states of enhancers (Oishi et al., 2017). Depletion of the clock component Nr1d1/Rev-erbα (a transcriptional repressor of BMAL1) in mice exhibits a loss of a gated inflammatory response (especially IL-6 production) to LPS (Gibbs et al., 2012). Moreover, Nr1d1/Rev-erbaα is a direct transcriptional repressor of the Ccl2 in activated macrophages (Sato et al., 2014), supporting that the circadian clock plays a crucial role in the regulation of the inflammation response.
Our present study has suggested the involvement of BMAL1 in the control of immune checkpoint protein PD-L1 expression during polymicrobial sepsis. We observed an increase in CLP-induced lethality in Bmal1Mye−/− mice, which was associated with elevated lactate production and PD-L1 expression, highlighting a potential role of lactate as an immunosuppressor that contributes to sepsis development and severity. Indeed, lactic acidosis has been suggested as an independent predictor of sepsis mortality (Thomas-Rueddel et al., 2015). We also observed that LPS and HMGB1 promote lactate production in macrophages through PKM2-dependent glycolysis. Consequently, conditional depletion of Pkm2 in myeloid cells prevented the enhanced sepsis phenotype and reduced animal survival observed in Bmal1Mye−/− mice, supporting the pathologic role of metabolic reprogramming in sepsis. The impact of the circadian clock on the regulation of glycolysis may depend on tissue type. For example, Bmal1−/− skeletal muscle exhibits impaired glycolysis due to reduced HIF1α expression under hypoxia (Peek et al., 2017). Our current data indicate that LPS-induced HIF1α expression in macrophages is not affected by BMAL1. The loss of Cry1/2 also does not affect hypoxia-induced HIF1α expression (Peek et al., 2017). Thus, these circadian clock genes may not share a common mechanism in the regulation of HIF1α expression in different stresses or cells.
Glycolysis metabolites, including lactate, have emerged as critical regulators of the inflammation and the immune response (O’Neill et al., 2016). Besides its role in the regulation of inflammasome activation (Xie et al., 2016) and T cell function (Haas et al., 2015), we observed that lactate induced PD-L1 expression in macrophages in a GPR81-dependent manner. GPR81 acts as a cell-surface receptor for lactate and promotes the expression of PD-L1 by activation of STAT1, a transcription factor in interferon-dependent responses. Our data suggested that PKM2, but not PD-L1, is a transcriptional target of BMAL1 in macrophages. PKM2 upregulation by Bmal1 deficiency is required for lactate production and STAT1-mediated PD-L1 expression in macrophages. Our findings are broadly consistent with recent findings that PKM2 regulates PD-L1 expression in tumor cells and immune cells (Palsson-McDermott et al., 2017). Our study also suggests a specific role for PKM2-mediated transcription and upregulation of PD-L1 in Bmal1−/− macrophages following LPS treatment. Of note, bone marrow cells from Bmal1−/− mice exhibit downregulation of PD-L1 expression during Mycobacterium tuberculosis infection (Sutton et al., 2017). The context-dependent activity and cofactor of BMAL1 in the regulation of PD-L1 expression remains to be further explored.
Sepsis and cancer share many immunosuppressive mechanisms, including the increased expression of PD-1 and PD-L1 with T cell exhaustion (Hotchkiss and Moldawer, 2014). PD-1 and its ligand, PD-L1, are increasingly recognized as powerful targets to enhance tumor-directed cytotoxic T cell function (Chen and Han, 2015). We observed that the loss of Bmal1 increased PD-L1 expression in macrophages. Importantly, anti-PD-L1 neutralizing antibodies improved septic survival, reduced lymphocyte apoptosis, and improved bacterial clearance in Bmal1Mye−/− mice. IL-7 is a promising immunotherapeutic agent that reverses immunosuppression and improves survival in septic animals and patients through enhancing T cell viability and function (Unsinger et al., 2010; Venet et al., 2017). Our study indicates that supplementation with IL-7 or administration of anti-PD-L1 neutralizing antibodies may ameliorate the sepsis-induced loss of immune effector CD4 and CD8 T cells in the spleen as well as organ dysfunction and injury in Bmal1Mye−/− mice. We would anticipate that IL-7 and immune checkpoint inhibition have differing functions in reversing sepsis-induced immunosuppression (Shindo et al., 2015). Therefore, the recent successes of several immunomodulatory agents in cancer could provide hope for and insight into potential immunostimulatory therapies in sepsis as well (Patera et al., 2016; Shindo et al., 2017).
In summary, we have uncovered an essential role of the core clock gene Bmal1 in protecting against lethal sepsis through regulating macrophage-metabolism-associated PD-L1 expression and T cell apoptotic cell death. Further studies will be aimed at defining the molecular and cellular mechanisms within immune cells implicated in circadian responses, possibly offering opportunities to design immunotherapeutic strategies for sepsis.
EXPERIMENTAL PROCEDURES
Reagents
The antibodies to BMAL1 (no. 14020), CLOCK (no. 5157), TIM-3 (no. 83882), PKM2 (no. 4053), cleaved caspase-3 (no. 9661), STAT1 (no. 14994), phospho-STAT1 (Tyr701; no. 9167), and actin (no. 3700) were obtained from Cell Signaling Technology. The antibody to GPR81 (no. orb335576) was obtained from Biorbyt. The antibodies to HIF-1α (no. ab2185) and BMAL1 (no. ab3350) were obtained from Abcam. The antibody to PD-L1 (no. MAB90781), PD-L2 (no. AF1022), and CTLA-4 (no. AF476) were obtained from R&D Systems. LPS (Escherichia coli LPS 0111:B4; no. L4391) and lactate (no. L6661) were obtained from Sigma. Recombinant mouse IL-7 (no. 577808), recombinant mouse HMGB1 (no. 764006), recombinant human HMGB1 (no. 557804), and anti-mouse PD-L1 neutralizing antibody (no. 124329) were obtained from BioLegend.
Cell Culture
THP1 cells (no. TIB-202) were obtained from American Type Culture Collection. BMDMs were obtained using L929-cell-conditioned medium as a source of granulocyte or macrophage colony stimulating factor. Mouse PMs (peritoneal exudate cells) were isolated from mice as previously described (Andújar et al., 2012). Mouse neutrophil was isolated from circulating blood using the Neutrophil Isolation Kit (no. 130-097-658; Miltenyi Biotec). These cells were cultured in DMEM (supplemented with 10% heat-inactivated fetal bovine serum and 100 units of penicillin and 100 mg/mL streptomycin) at 37°C, 95% humidity, and 5% CO2. All cells were mycoplasma free and authenticated by Short Tandem Repeat DNA Profiling Analysis.
Animal Model of Sepsis
We conducted all animal care and experiments in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care guidelines (https://www.aaalac.org) and with approval from our Institutional Animal Care and Use Committee.
LysM-Cre (no. 004781), Bmal1 flox/flox (no. 007668), and Pkm2 flox/flox (no. 024048) mice were purchased from The Jackson Laboratory. These mice were used to generate myeloid-cell-specific Bmal1-knockout, Pkm2-knockout, or double-knockout mice. Mice were housed under a regular 12 hr light and dark cycle (7:00–19:00 light period) at a constant temperature of 22°C ± 1°C and relative humidity of 40%–60%. During this period, food and water were available ad libitum. All animal experiments, including surgical procedures and sample harvests, were carried out under pathogen-free conditions.
Polymicrobial sepsis was induced in C57BL/6J mice (8–10 weeks old; 22–26 g; half male:halffemale) at ZT0–3 (except for Figure 1G) using a surgical procedure as previously described (Kang et al., 2016). Briefly, anesthesia was induced with ketamine and xylazine. A small midline abdominal incision was made and the cecum was exteriorized and ligated with 4–0 silk immediately distal to the ileocecal valve without causing intestinal obstruction. The cecum was then punctured twice with a 17G–27G needle. The abdomen was closed in two layers, and mice were injected subcutaneously with 1 mL Ringer’s solution including analgesia (0.05 mg/kg buprenorphine).
Endotoxemia model: LPS (Escherichia coli 0111:B4; no. L4391; Sigma) was dissolved in PBS. C57BL/6J mice (8–10 weeks old; 22–26 g; half male:half female) received LPS at 15 mg/kg intraperitoneally (i.p.).
Biochemical Assay
Lactate in culture medium or serum was measured with the L-Lactate Assay Kit (Abcam; no. ab65331). Activation of caspase-3 in cell lysates was measured with the caspase-3 Assay Kit (Abcam; no. ab39401). Neutrophil activity was measured with the Myeloperoxidase Activity Assay Kit (Abcam; no. ab105136). Activation of tissue enzymes (CK, BUN, and ALT) in serum was performed using the IDEXX Catalyst Dx Chemistry Analyzer (IDEXX Laboratories).
RNAi and Gene Transfection
Human Bmal1-shRNA-1 (sequence: 5’-CCGGCTTCTAGGCACATCGTGTTATCTCGAGATAACACGATGTGCCTAGAAGTTTTTG-3’); human Bmal1-shRNA-2 (sequence: 5’-CCGGGCAGAATGTCATAGGCAAGTTCTCGAGAACTTGCCTATGACATTCTGCTTTTT-3’), human Clock-shRNA-1 (sequence: 5’-CCGGCGCACACATAGGCCATCTTATCTCGAGATAAGATGGCCTATGTGTGCGTTTTT-3’); human Clock-shRNA-2 (sequence: 5’-CCGGCGACGAGAACTTGGCATTGAACTCGAGTTCAATGCCAAGTTCTCGTCGTTTTT-3’), mouse Bmal1-shRNA (sequence:5’-CCGGACATAGGCATCGATATGATAGCTCGAGCTATCATATCGATGCCTATGTTTTTTG-3’), mouse Grp81-shRNA (sequence: 5’-CCGGCCTGGAAGTCAAGCACTATTTCTCGAGAAATAGTGCTTGACTTCCAGGTTTTT-3’), and mouse Stat1-shRNA (sequence: 5’-CCGGGCTGTTACTTTCCCAGATATTCTCGAGAATATCTGGGAAAGTAACAGCTTTTTG-3’) were obtained from Sigma. Bmal1-wild-type and Bmal1-I317D mutant were a gift from Joseph Takahashi. Transfection was performed using lentiviral vector or Lipofectamine 3000 (no. L3000–015; Invitrogen). Puromycin (no. ant-pr-1; InvivoGen) was used to generate stable knockdown cell lines.
Western Blot Analysis
Western blot was used to analyze protein expression as described previously (Sun et al., 2016; Tang et al., 2010; Xie et al., 2017). In brief, after extraction, proteins in cell lysates were first resolved by 4%–12% Criterion XT Bis-Tris gels (Bio-Rad; no. 3450124) in XT MES running buffer (Bio-Rad; no. 1610789) and then transferred to polyvinylidene difluoride membrane (pore size 0.22 μM; Bio-Rad; no. 1620233). The membranes were blocked with Tris-buffered saline Tween 20 containing 5% skim milk for 1 hr at room temperature and then incubated with the indicated primary antibodies (1:500–1:2,000) overnight at 4°C. After incubation with peroxidase-conjugated secondary antibodies (1:2,000–1:4,000), the signals were visualized by enhanced chemiluminescence (no. 32106; Pierce).
qPCR
Total RNA was extracted using QIAGEN RNeasy Plus Kit. First-strand cDNA was synthesized from 1 μg of RNA using the iScript cDNA Synthesis kit (no. 1708890; Bio-Rad). Briefly, 20 μL reactions were prepared by combining 4 μL iScript Select reaction mix, 2 μL gene-specific enhancer solution, 1 mL reverse transcriptase, 1 μL gene-specific assay pool (20×; 2 μM), and 12 μL RNA diluted in RNase-free water. Real-time qPCR was carried out using synthesized cDNA, primers, and SsoFast EvaGreen Supermix (no. 172–5204; BioRad). The expression of target genes was calculated using the ddCt method relative to the expression of a house-keeping gene, β-actin (Song et al., 2018; Zhu et al., 2017). Data shown are the relative quantity (RQ), with RQ of the control cells set to one. The specific primers were listed as below: mouse Bmal1: 5’-ACCTCGCAGAATGTCACAGGCA-3’ and 5’- CTGAACCATCGACTTCGTAGCG-3’; mouse Per1: 5’- GAAACCTCTGGCTGTTCCTACC-3’ and 5’- AGGCTGAAGAGGCAGTGTAGGA-3’; mouse Per2: 5’- CTGCTTGTTCCAGGCTGTGGAT-3’ and 5’- CTTCTTGTGGATGGCGAGCATC-3’; mouse Per3: 5’- CACAGACATCGAAGGAGGTGCT-3’ and 5’-CTTACACGCCACGGCAACACTT-3’; human Pd-l1: 5’- TGCCGACTACAAGCGAATTACTG-3’ and 5’- CTGCTTGTCCAGATGACTTCGG-3’; mouse Pd-l1: 5’- TGCGGACTACAAGCGAATCACG-3’ and 5’- CTCAGCTTCTGGATAACCCTCG-3’; mouse Pd-l2: 5’- CTGGGACTACAAGTACCTGACG-3’ and 5’- CTCTAGCCTGGCAGGTAAGCTG-3’; mouse Ctla-4: 5’- GTACCTCTGCAAGGTGGAACTC-3’ and 5’- CCAAAGGAGGAAGTCAGAATCCG-3’; mouse Tim-3: 5’-ACAGACACTGGTGACCCTCCAT-3’ and 5’- CAGCAGAGACTCCCACTCCAAT-3’; mouse β-actin: 5’-CTGTCCCTGTATGCCTCTG-3’ and 5’-ATGTCACGCACGATTTCC-3’; and human β-actin: 5’-AGCGAG CATCCCCCAAAGTT-3’ and 5’-AGGGCA CGAAGGCTCATCATT-3’.
Oxidative Phosphorylation and Glycolysis Assay
Cellular OXPHOS and glycolysis were monitored using the Seahorse Bioscience Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA) by measuring basal OCR (indicative of respiration) and ECAR (indicative of glycolysis) as previously described (Yang et al., 2014).
ChIP
ChIP assay was performed using the Pierce ChIP Kit (no. 26156; Thermo Scientific). One-twentieth of the immunoprecipitated DNA was used in qPCR. Results were shown as percentage of input. BMAL1, STAT1, PKM2, and HIF1α antibodies used for ChIP were acquired from Cell Signaling Technology or Abcam. The following primers were used for qPCR of DNA quantification: Pd-l1 promoter: 5’-CACTGGCTCCTGAGTACTGG-3’ and 5’-CTGTCTGTGAAACCGAAGCC-3’; Pkm2 promoter: 5’-TGGTCTACGATGTCCTTCCG-3’ and 5’-TGAGAGAAGCTCAGGGTAGG-3’.
Histologic Examination
After fixation in 4% phosphate-buffered formaldehyde, tissues were embedded in optimum cutting temperature cryomedium (Sakura Finetek) and cut into 4-μm sections. H&E (modified Harris Hematoxylin, Thermo Scientific; Eosin-Y, Thermo Scientific) staining was performed for each section to examine histomorphologic features (Kang et al., 2017). All histologic examinations were performed by a pathologist who specializes in the pathophysiology of sepsis. Lung and intestine injury was evaluated and scored according to previous criterions (Matute-Bello et al., 2011; Oktar et al., 2002).
CD4+ and CD8+ T Cell Isolation
A single-cell suspension from mouse spleen was prepared using gentleMACS Dissociator (no. 130-093-235; Miltenyi Biotec). CD4+ and CD8+ T cells were then isolated from this single-cell suspension using the magnetic CD4+ T Cell Isolation Kit (no. 130-104-454; Miltenyi Biotec) or CD8+ T Cell Isolation Kit (no. 130-104-075; Miltenyi Biotec) according to the manufacturer’s instructions.
Measurement of Bacterial Counts
The peritoneal cavity was washed with 1 mL PBS, and the peritoneal lavage was collected under sterile conditions. Samples of blood and peritoneal lavage fluid were serially 10-fold diluted in sterile saline and cultured on tryptic soy agar pour plates (Becton Dickinson; no. BA-256665.02). Plates were incubated (37°C) for 24–48 hr, and colony counts were performed (Kang et al., 2016).
Statistical Analysis
Unpaired Student’s t tests were used to compare the means of two groups. One-way ANOVA was used for comparison among the different groups. When ANOVA was significant, post hoc testing of differences between groups was performed using the least significant difference (LSD) test. The Kaplan-Meier method was used to compare differences in mortality rates between groups. A p value < 0.05 was considered statistically significant. In all experiments, animals were randomized to different treatment groups without blinding. Sample size in experiments was specified in each figure legend. We did not exclude samples or animals. For every figure, statistical tests are justified as appropriate. All data meet the assumptions of the tests. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those generally employed in the field. SigmaPlot 11.0 was used to analyze the data.
Supplementary Material
Highlights.
Bmal1 deficiency in myeloid cells increases polymicrobial infection in mice
The circadian clock blocks PD-L1 expression in activated macrophages and monocytes
PKM2 is required for STAT1-dependent PD-L1 upregulation
PD-L1 pathway contributes to septic death in BmalMye−/− mice
ACKNOWLEDGMENTS
We thank Christine Heiner (Department of Surgery, University of Pittsburgh) for her critical reading of the manuscript. This work was supported by grants from the NIH (R01GM115366, R01CA160417, R01AT005076, R01GM063075, R01GM44100, and R01CA211070), the Natural Science Foundation of Guangdong Province (2016A030308011), the American Cancer Society (Research Scholar Grant RSG-16-014-01-CDD), the National Natural Science Foundation of China (31671435, 81400132, 81772508, and 81570154), the Natural Science Foundation of Hunan Province (2016JJ3171), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2017), Lin He’s Academician Workstation of New Medicine and Clinical Translation (2017), and International Scientific and Technology Cooperation Program of China (2015DFA31490).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes one table and can be found with this article online at https://doi.org/10.10167j.celrep.2018.06.026.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Andújar I, Ríos JL, Giner RM, Miguel Cerdá J, and Recio, Mdel. C (2012). Beneficial effect of shikonin on experimental colitis induced by dextran sulfate sodium in BALB/c mice. Evid. Based Complement. Alternat. Med 2012, 271606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, and Sassone-Corsi P (2015). Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92. [DOI] [PubMed] [Google Scholar]
- Boomer JS, To K, Chang KC, Takasu O, Osborne DF, Walton AH, Bricker TL, Jarman SD 2nd, Kreisel D, Krupnick AS, et al. (2011). Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA 306, 2594–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buras JA, Holzmann B, and Sitkovsky M (2005). Animal models of sepsis: setting the stage. Nat. Rev. Drug Discov 4, 854–865. [DOI] [PubMed] [Google Scholar]
- Castanon-Cervantes O, Wu M, Ehlen JC, Paul K, Gamble KL, Johnson RL, Besing RC, Menaker M, Gewirtz AT, and Davidson AJ (2010). Dysregulation of inflammatory responses by chronic circadian disruption. J. Immunol 185, 5796–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, and Han X (2015). Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J. Clin. Invest 125, 3384–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis AM, Bellet MM, Sassone-Corsi P, and O’Neill LA (2014). Circadian clock proteins and immunity. Immunity 40, 178–186. [DOI] [PubMed] [Google Scholar]
- Curtis AM, Fagundes CT, Yang G, Palsson-McDermott EM, Wochal P, McGettrick AF, Foley NH, Early JO, Chen L, Zhang H, et al. (2015). Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc. Natl. Acad. Sci. USA 112, 7231–7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano MJ, and Ward PA (2016). Sepsis-induced immune dysfunction: can immune therapies reduce mortality? J. Clin. Invest 126, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García JA, Volt H, Venegas C, Doerrier C, Escames G, Loópez LC, and Acuña-Castroviejo D (2015). Disruption of the NF-κB/NLRP3 connection by melatonin requires retinoid-related orphan receptor-α and blocks the septic response in mice. FASEB J 29, 3863–3875. [DOI] [PubMed] [Google Scholar]
- Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, Farrow SN, Else KJ, Singh D, Ray DW, and Loudon AS (2012). The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl. Acad. Sci. USA 109, 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas R, Smith J, Rocher-Ros V, Nadkarni S, Montero-Melendez T, D’Acquisto F, Bland EJ, Bombardieri M, Pitzalis C, Perretti M, et al. (2015). Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. PLoS Biol 13, e1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiramoto A, Yamane T, Tsumiyama K, Yoshida K, Komai K, Yamada H, Yamazaki F, Doi M, Okamura H, and Shiozawa S (2010). Mammalian clock gene Cryptochrome regulates arthritis via proinflammatory cytokine TNF-alpha. J. Immunol 184, 1560–1565. [DOI] [PubMed] [Google Scholar]
- Hotchkiss RS, and Moldawer LL (2014). Parallels between cancer and infectious disease. N. Engl. J. Med 371, 380–383. [DOI] [PubMed] [Google Scholar]
- Hotchkiss RS, Moldawer LL, Opal SM, Reinhart K, Turnbull IR, and Vincent JL (2016). Sepsis and septic shock. Nat. Rev. Dis. Primers 2, 16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, Green CB, Zhang H, and Takahashi JS (2012). Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science 337, 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu K, Zhu S, Xie M, Kang R, Cao L, and Tang D (2017). AMPK regulates immunometabolism in sepsis. Brain Behav. Immun Published online November 3, 2017 10.1016/j.bbi.2017.11.003. [DOI] [PubMed] [Google Scholar]
- Hutchins NA, Unsinger J, Hotchkiss RS, and Ayala A (2014). The new normal: immunomodulatory agents against sepsis immune suppression. Trends Mol. Med 20, 224–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Zeng L, Xie Y, Yan Z, Zhou B, Cao L, Klionsky DJ, Tracey KJ, Li J, Wang H, et al. (2016). A novel PINK1- and PARK2-dependent protective neuroimmune pathway in lethal sepsis. Autophagy 12, 2374–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Xie Y, Zhang Q, Hou W, Jiang Q, Zhu S, Liu J, Zeng D, Wang H, Bartlett DL, et al. (2017). Intracellular HMGB1 as a novel tumor suppressor of pancreatic cancer. Cell Res 27, 916–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk HD, Kramer A, and Maier B (2009). A circadian clock in macrophages controls inflammatory immune responses. Proc. Natl. Acad. Sci. USA 106, 21407–21412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute-Bello G, Downey G, Moore BB, Groshong SD, Matthay MA, Slutsky AS, and Kuebler WM; Acute Lung Injury in Animals Study Group (2011). An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am. J. Respir. Cell Mol. Biol 44, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazato R, Hotta S, Yamada D, Kou M, Nakamura S, Takahata Y, Tei H, Numano R, Hida A, Shimba S, et al. (2017). The intrinsic microglial clock system regulates interleukin-6 expression. Glia 65, 198–208. [DOI] [PubMed] [Google Scholar]
- Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, and Chawla A (2013). Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science 341, 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LA, Kishton RJ, and Rathmell J (2016). A guide to immunometabolism for immunologists. Nat. Rev. Immunol 16, 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi Y, Hayashi S, Isagawa T, Oshima M, Iwama A, Shimba S, Okamura H, and Manabe I (2017). Bmal1 regulates inflammatory responses in macrophages by modulating enhancer RNA transcription. Sci. Rep 7, 7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktar BK, Gülpinar MA, Bozkurt A, Ghandour S, Cetinel S, Moini H, Yeğen BC, Bilsel S, Granger DN, and Kurtel H (2002). Endothelin receptor blockers reduce I/R-induced intestinal mucosal injury: role of blood flow. Am. J. Physiol. Gastrointest. Liver Physiol 282, G647–G655. [DOI] [PubMed] [Google Scholar]
- Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MA, Sheedy FJ, Gleeson LE, van den Bosch MW, Quinn SR, Domingo-Fernandez R, Johnston DG, et al. (2015). Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab 21, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson-McDermott EM, Dyck L, Zasłona Z, Menon D, McGettrick AF, Mills KHG, and O’Neill LA (2017). Pyruvate kinase M2 is required for the expression of the immune checkpoint PD-L1 in immune cells and tumors. Front. Immunol 8, 1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patera AC, Drewry AM, Chang K, Beiter ER, Osborne D, and Hotchkiss RS (2016). Frontline Science: Defects in immune function in patients with sepsis are associated with PD-1 or PD-L1 expression and can be restored by antibodies targeting PD-1 or PD-L1. J. Leukoc. Biol 100, 1239–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek CB, Levine DC, Cedernaes J, Taguchi A, Kobayashi Y, Tsai SJ, Bonar NA, McNulty MR, Ramsey KM, and Bass J (2017). Circadian clock interaction with HIF1α mediates oxygenic metabolism and anaerobic glycolysis in skeletal muscle. Cell Metab 25, 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart K, Daniels R, Kissoon N, Machado FR, Schachter RD, and Finfer S. (2017). Recognizing sepsis as a global health priority - a WHO resolution. N. Engl. J. Med 377, 414–417. [DOI] [PubMed] [Google Scholar]
- Sato S, Sakurai T, Ogasawara J, Takahashi M, Izawa T, Imaizumi K, Taniguchi N, Ohno H, and Kizaki T (2014). A circadian clock gene, Rev-erbα, modulates the inflammatory function of macrophages through the negative regulation of Ccl2 expression. J. Immunol 192, 407–417. [DOI] [PubMed] [Google Scholar]
- Shindo Y, Unsinger J, Burnham CA, Green JM, and Hotchkiss RS (2015). Interleukin-7 and anti-programmed cell death 1 antibody have differing effects to reverse sepsis-induced immunosuppression. Shock 43, 334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo Y, McDonough JS, Chang KC, Ramachandra M, Sasikumar PG, and Hotchkiss RS (2017). Anti-PD-L1 peptide improves survival in sepsis. J. Surg. Res 208, 33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver AC, Arjona A, Walker WE, and Fikrig E (2012). The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity 36, 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zhu S, Xie Y, Liu J, Sun L, Zeng D, Wang P, Ma X, Kroemer G, Bartlett DL, et al. (2018). JTC801 induces pH-dependent death JTC801 induces pH-dependent death specifically in cancer cells and slows growth of tumors in mice. Gastroenterology 154, 1480–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spec A, Shindo Y, Burnham CA, Wilson S, Ablordeppey EA, Beiter ER, Chang K, Drewry AM, and Hotchkiss RS (2016). T cells from patients with Candida sepsis display a suppressive immunophenotype. Crit. Care 20, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler ML, Kuropatwinski KK, Comas M, Gasparian AV, Fedtsova N, Gleiberman AS, Gitlin II, Artemicheva NM, Deluca KA, Gudkov AV, et al. (2012). Core circadian protein CLOCK is a positive regulator of NF-κB-mediated transcription. Proc. Natl. Acad. Sci. USA 109, E2457–E2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R, and Tang D. (2016). Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 63, 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton CE, Finlay CM, Raverdeau M, Early JO, DeCourcey J, Zaslona Z, O’Neill LAJ, Mills KHG, and Curtis AM (2017). Loss of the molecular clock in myeloid cells exacerbates T cell-mediated CNS autoimmune disease. Nat. Commun 8, 1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ 3rd, and Lotze MT (2010). Endogenous HMGB1 regulates autophagy. J. Cell Biol 190, 881–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Rueddel DO, Poidinger B, Weiss M, Bach F, Dey K, Häberle H, Kaisers U, Rüddel H, Schädler D, Scheer C, et al. (2015). Hyperlactatemia is an independent predictor of mortality and denotes distinct subtypes of severe sepsis and septic shock. J. Crit. Care 30, 439.e1–439.e6. [DOI] [PubMed] [Google Scholar]
- Unsinger J, McGlynn M, Kasten KR, Hoekzema AS, Watanabe E, Muenzer JT, McDonough JS, Tschoep J, Ferguson TA, McDunn, JE, et al. (2010). IL-7 promotes T cell viability, trafficking, and functionality and improves survival in sepsis. J. Immunol 184, 3768–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Poll T, van de Veerdonk FL, Scicluna BP, and Netea MG (2017). The immunopathology of sepsis and potential therapeutic targets. Nat. Rev. Immunol 17, 407–420. [DOI] [PubMed] [Google Scholar]
- Venet F, Demaret J, Blaise BJ, Rouget C, Girardot T, Idealisoa E, Rimmelé T, Mallet F, Lepape A, Textoris J, and Monneret G (2017). IL-7 restores T lymphocyte immunometabolic failure in septic shock patients through mTOR activation. J. Immunol 199, 1606–1615. [DOI] [PubMed] [Google Scholar]
- Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, Frazier A, Yang H, Ivanova S, Borovikova L, et al. (1999). HMG-1 as a late mediator of endotoxin lethality in mice. Science 285, 248–251. [DOI] [PubMed] [Google Scholar]
- Xie M, Yu Y, Kang R, Zhu S, Yang L, Zeng L, Sun X, Yang M, Billiar TR, Wang H, et al. (2016). PKM2-dependent glycolysis promotes NLRP3 and AIM2 inflammasome activation. Nat. Commun 7, 13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Zhu S, Song X, Sun X, Fan Y, Liu J, Zhong M, Yuan H, Zhang L, Billiar TR, et al. (2017). The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep 20, 1692–1704. [DOI] [PubMed] [Google Scholar]
- Yang L, Xie M, Yang M, Yu Y, Zhu S, Hou W, Kang R, Lotze MT, Billiar TR, Wang H, et al. (2014). PKM2 regulates the Warburg effect and promotes HMGB1 release in sepsis. Nat. Commun 5, 4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Deng W, Kang R, Xie M, Billiar T, Wang H, Cao L, and Tang D (2016). Plumbagin protects mice from lethal sepsis by modulating immuno-metabolism upstream of PKM2. Mol. Med 2016, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Zhang Q, Sun X, Zeh HJ 3rd, Lotze MT, Kang R, and Tang D (2017). HSPA5 regulates ferroptotic cell death in cancer cells. Cancer Res 77, 2064–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.