Abstract
Extracellular vesicles are promising delivery vesicles for therapeutic RNAs. Small interfering RNA (siRNA) conjugation to cholesterol enables efficient and reproducible loading of extracellular vesicles with the therapeutic cargo. siRNAs are typically chemically modified to fit an application. However, siRNA chemical modification pattern has not been specifically optimized for extracellular vesicle-mediated delivery. Here we used cholesterol-conjugated, hydrophobically modified asymmetric siRNAs (hsiRNAs) to evaluate the effect of backbone, 5′-phosphate, and linker chemical modifications on productive hsiRNA loading onto extracellular vesicles. hsiRNAs with a combination of 5′-(E)-vinylphosphonate and alternating 2′-fluoro and 2′-O-methyl backbone modifications outperformed previously used partially modified siRNAs in extracellular vesicle-mediated Huntingtin silencing in neurons. Between two commercially available linkers (triethyl glycol [TEG] and 2-aminobutyl-1-3-propanediol [C7]) widely used to attach cholesterol to siRNAs, TEG is preferred compared to C7 for productive exosomal loading. Destabilization of the linker completely abolished silencing activity of loaded extracellular vesicles. The loading of cholesterol-conjugated siRNAs was saturated at ∼3,000 siRNA copies per extracellular vesicle. Overloading impaired the silencing activity of extracellular vesicles. The data reported here provide an optimization scheme for the successful use of hydrophobic modification as a strategy for productive loading of RNA cargo onto extracellular vesicles.
Keywords: extracellular vesicles, siRNA, oligonucleotides, nanovesicles, chemical modification, RNA therapy, exosomes
Graphical Abstract
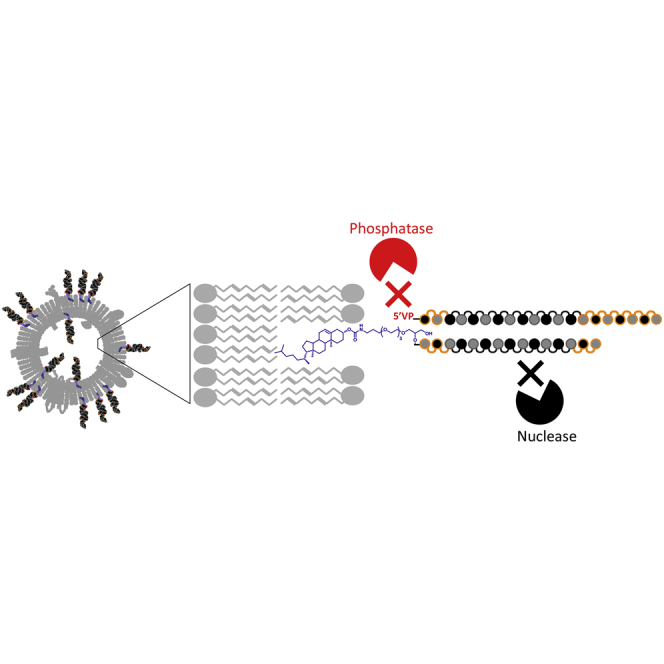
Haraszti et al. optimize hydrophobic modification as a strategy to productively load extracellular vesicles with therapeutic siRNA. The combination of chemical modifications on all riboses, 5′-(E)-vinylphosphonate, TEG linker between the cholesterol and the siRNA, and an siRNA-to-extracellular vesicle (EV) ratio of 3,000 outperforms previously used loading schemes.
Introduction
Extracellular vesicles are being explored for therapeutic RNA delivery due to (1) their small size (50–150 nm) allowing penetration through some biological barriers,1, 2 (2) their unique protein composition enabling target cell specificity,3, 4 and (3) their natural capacity to transfer RNA between cells.5, 6 Mesenchymal stem cell-derived extracellular vesicles can deliver therapeutic miRNAs to brain.7, 8, 9, 10, 11, 12 Small interfering RNAs (siRNAs), similar to therapeutic microRNAs (miRNAs), are capable of selective gene silencing.13 Thus, siRNAs offer a therapeutic option for genetically defined diseases, such as Huntington’s disease. However, delivery to target tissues remains the bottleneck for clinical application of therapeutic RNAs, including siRNAs. Extracellular vesicles represent a strategy to overcome the delivery challenge.14, 15
Cholesterol conjugation-mediated loading of siRNAs onto extracellular vesicles is among the most reproducible and scalable loading strategies,14, 16, 17 characterized by efficient transfer of the loaded cholesterol-siRNA to target cells. However, productive gene silencing induced by the transferred cholesterol-siRNA was variable. We speculate that these variations are due to differences in siRNA chemical modification patterns (45%–71% of riboses modified), cholesterol placement position (5′ or 3′ of the sense strand), siRNA-to-extracellular vesicle loading ratio (100s versus 1,000s of siRNAs per extracellular vesicle), and siRNA concentrations used in silencing studies (∼50–1,500 nM).14, 16, 17 All studies used a version of pyrimidine-modified siRNAs, which have been shown to provide stabilization against nucleases in vitro in serum.18, 19, 20 Cholesterol-conjugated, but not chemically modified, siRNAs have been shown to degrade in the presence of extracellular vesicles due to endogenous RNase activity.21
Advances in oligonucleotide chemistry have enabled the expansion of siRNA use from in vitro serum-rich environments to systemic delivery in vivo.18, 22, 23, 24, 25 In particular, siRNAs with modification of all riboses22, 23, 26, 27, 28, 29 (with 2′-fluoro and 2′-O-methyl) were 10,000-fold more active in vivo30 than partially modified siRNAs similar to siRNAs originally used for extracellular vesicle loading.14, 16, 17 A second type of modification, 5′-(E)-vinylphosphonate, also improved the activity of systemically administered conjugated siRNAs.31, 32, 33 However, full chemical modification did not affect the activity of non-conjugated siRNAs delivered in cationic liposomes.24, 34, 35 Extracellular vesicle-mediated delivery of hydrophobically modified asymmetric siRNAs (hsiRNAs) combines principles of siRNA conjugation and lipid nanoparticle technology. The impact of siRNA chemical modifications on efficacy of extracellular vesicle-mediated delivery is, therefore, difficult to predict and remains unknown.
Among many synthetic approaches on cholesterol attachment to the siRNA, TEG (triethyl glycol) and C7 (2-aminobutyl-1-3-propanediol) linkers are frequently used and commercially available. In the amino linker class, the C7 linker was optimal for siRNA passive uptake.36 Despite the common use of both linkers, no systematic comparison has been published to date.
Here we evaluated the impact of siRNA chemical modification patterns, cholesterol attachment via different linkers, and siRNA-to-extracellular vesicle loading ratio on functional extracellular vesicle-mediated delivery of siRNAs. We used siRNA concentrations ranging from 23 to 1,500 nM in all experiments. We used mesenchymal stem cells (derived from umbilical cord Wharton’s jelly) as extracellular vesicle producer cells, because they have been proven safe in numerous clinical trials,37, 38 provide higher extracellular vesicle yield compared to other origins of mesenchymal stem cells (R.A.H., unpublished data), and mesenchymal stem cells have been shown to be beneficial in Huntington’s disease,39 the disease model used in this study.
Results
Linker Chemistry Influences Efficiency of Cholesterol-Mediated Loading of siRNAs onto Extracellular Vesicles
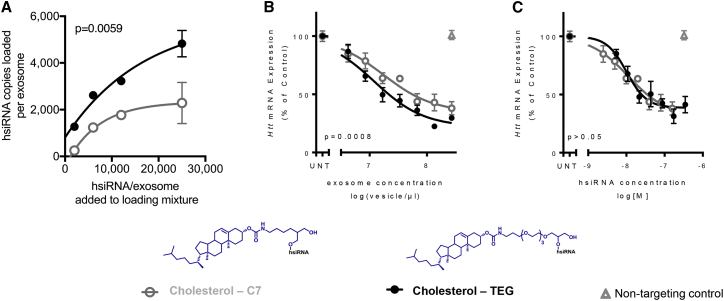
To compare two commercially available strategies to conjugate cholesterol to siRNAs (TEG and C7 linkers), we used a previously developed asymmetric siRNA scaffold,18, 40 characterized by a short duplex region (15 bp) and a fully phosphorothioated tail assisting membrane association18, 41, 42 (hsiRNAs). hsiRNAs are either partially modified with 2′-fluoro pyrimidines on the antisense strand and 2′-O-methyl pyrimidines on the sense strand, or they are fully modified using alternating 2′-O-methyl and 2′-fluoro pattern providing endonuclease stability and protection from innate immune response.23, 43, 44 We synthesized fully modified cholesterol-hsiRNAs targeting Huntingtin mRNA40 using either TEG or a C7 linkers (Figure 1). Cholesterol-hsiRNA variants were loaded onto extracellular vesicles at increasing hsiRNA-to-extracellular vesicle ratios (Figure 1A) Both variants showed efficient loading onto extracellular vesicles with saturation kinetics (Figure 1A). Cholesterol-TEG-hsiRNAs loaded more efficiently onto extracellular vesicles than cholesterol-C7-hsiRNAs at all ratios tested (Figure 1A; p = 0.0059). More efficient loading led to more potent Huntingtin mRNA silencing, when primary neurons were treated with extracellular vesicles loaded to saturation (cholesterol-TEG IC50 ∼8 × 106 extracellular vesicles, cholesterol-C7 IC50 ∼22 × 106 extracellular vesicles; p = 0.0008; Figure 1B). hsiRNA uptake into neurons was confirmed using peptide-nucleic acid (PNA) hybridization assay (Figure S3E). Normalization to hsiRNA content eliminated the observed differences (p > 0.05; Figure 1C) in silencing. Thus, silencing potency of the two hsiRNA variants was the same. Improved silencing activity upon extracellular vesicle-mediated delivery could be fully explained by better loading of cholesterol-TEG-hsiRNA onto extracellular vesicles. Therefore, cholesterol-TEG-hsiRNA was used for subsequent experiments.
Figure 1.
Triethyl Glycol Linker for Cholesterol Is Favorable
Fluorescent, fully modified hsiRNA was loaded onto extracellular vesicles, primary murine cortical neurons were treated for 1 week, and target Htt mRNA silencing was measured using QuantiGene (Affymetrix). (A) hsiRNA conjugated to cholesterol with either a TEG (triethyl glycerol) or a C7 (2-aminobutyl-1-3-propanediol) linker was loaded onto extracellular vesicles at varying hsiRNA-to-extracellular vesicle ratios. (B) Huntingtin mRNA silencing in primary neurons 1 week after treatment with varying concentrations of hsiRNA-loaded extracellular vesicles. mRNA levels were normalized to housekeeping gene Hprt and expressed as percentage of untreated control. UNT, untreated; n = 3; mean ± SEM. (C) Level of silencing from (B) was normalized to hsiRNA content of loaded extracellular vesicles.
Optimization of Cholesterol-hsiRNA-to-Extracellular Vesicle Ratio: 1,000–3,000 hsiRNAs per Extracellular Vesicle Is Preferred
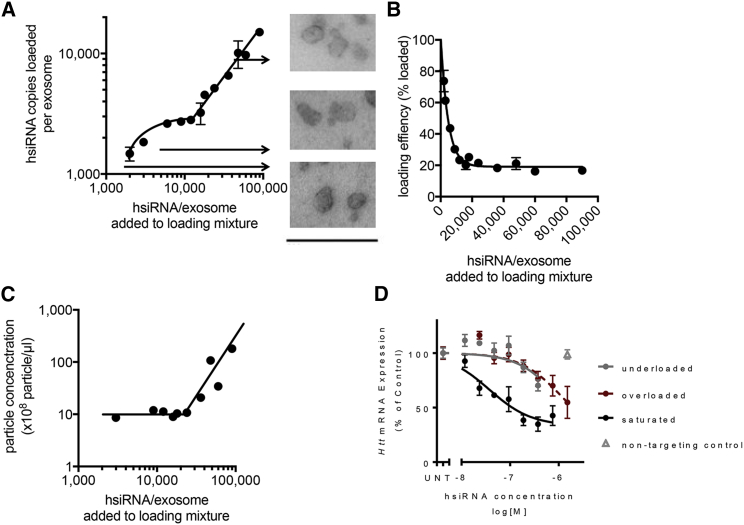
Contrary to conventional siRNA extracellular vesicle-loading approaches (i.e., electroporation or overexpression in parent cells), hydrophobic modifications of siRNA enable the association of a large number of RNA molecules per extracellular vesicle. To define an optimal hsiRNA-to-extracellular vesicle ratio, i.e., ratio supporting productive target mRNA silencing, we evaluated the effect of hsiRNA concentration during the loading process (1,000 to 100,000 hsiRNA copies per extracellular vesicle added to the loading mixture). The addition of 6,000 hsiRNAs per extracellular vesicle into the loading mixture resulted in ∼2,600 hsiRNAs associated per vesicle (Figure 2A), leading to 43% loading efficiency (Figure 2B). A further increase in the amount of hsiRNAs added to the loading mixture (9,000 and 12,000 per vesicle) did not support an increase in the amount of extracellular vesicle-associated hsiRNAs (Figure 2A), indicating a level of intermediate saturation at ∼2,500–3000 hsiRNAs per vesicle. As the amount of loaded hsiRNAs stayed constant, the estimated loading efficiency decreased from 43% to 23% (Figure 2B).
Figure 2.
Loading of hsiRNA to Extracellular Vesicles Is a Partially Saturatable Process
(A) Cholesterol-hsiRNA was loaded onto extracellular vesicles at different hsiRNA-to-extracellular vesicles ratios. The loading curve shows and initial saturation phase followed by a secondary linear phase. Transmission electron microscopy images correspond to hsiRNA-loaded extracellular vesicles at an hsiRNA-to-extracellular vesicle ratio of 3,000, 10,000, and 100,000. Scale bar represents 500 nm. (B) Loading efficiency at varying hsiRNA-to-extracellular vesicle ratios. (C) Particle concentration as assessed by nanoparticle-tracking analysis after loading at varying hsiRNA-to-extracellular vesicle ratios. (D) Huntingtin mRNA silencing in primary neurons 1 week after treatment with extracellular vesicles loaded with hsiRNAs at hsiRNA-to-extracellular vesicles ratios of 3,000, 10,000, and 100,000 (1,000, 3,000, and 18,000 RNA molecules per vesicle loaded). UNT, untreated; n = 3; mean ± SEM.
Following this initial saturation phase, we observed a linear increase in the amount of hsiRNAs loaded per extracellular vesicle, starting at approximately 20,000 hsiRNA per extracellular vesicle added to the loading mixture, representing a constant loading efficiency of 18% (Figure 2B). Transmission electron microscopy showed similar lipid bilayer-surrounded vesicles post-loading at hsiRNA-to-extracellular vesicle ratios below the initial saturation phase (3,000), at the initial saturation phase (10,000), and in the linear increase phase (100,000) (Figure 2A). Increasing hsiRNA-to-extracellular vesicle loading ratio beyond 20,000 resulted in an increased total particle number (Figure 2C). Loading hsiRNAs onto extracellular vesicles did not alter the protein content (Figures S2A–S2C) or size (Figure S2D) of vesicles. The same phenomenon of initial saturation phase followed by a linear increase phase was observed when loading cholesterol-C7-hsiRNA to extracellular vesicles (Figure S3B) or both cholesterol-TEG-hsiRNA (Figure S3C) and cholesterol-C7-hsiRNA (Figure S3D) to conventional liposomes. During saturation phase, more hsiRNAs could be loaded onto liposomes than to extracellular vesicles (Figures S3C and S3D), suggesting that the protein content of extracellular vesicle membrane may interfere with hsiRNA loading. The formation of extra particles in the presence of vesicles may be explained by hsiRNA aggregation.
Next, we evaluated how hsiRNA-to-extracellular vesicle ratio affected the ability of loaded extracellular vesicles to silence Huntingtin mRNA in primary neurons. 3,000, 10,000, and 100, 000 hsiRNAs (per vesicle) were added to the loading mixture, generating extracellular vesicles with 1,000, 3,000, and 18,000 RNA molecules loaded per vesicle, respectively. From three hsiRNA-to-extracellular vesicle ratios tested, extracellular vesicles containing 3,000 hsiRNAs per vesicle performed the best with an IC50 of 37 nM (Figure 2D). In contrast, extracellular vesicles underloaded (1,000 hsiRNAs per extracellular vesicle) or overloaded (18,000 hsiRNAs per extracellular vesicle) were less efficient in Huntingtin mRNA silencing (IC50 1,330 nM and 1,164 nM, respectively) (Figure 2D). As extracellular vesicles loaded with 3,000 hsiRNAs (saturation level) were 36-fold more potent than underloaded and 31-fold more potent than overloaded extracellular vesicles, extracellular vesicles loaded with 3,000 hsiRNAs were used for subsequent experiments. Extracellular vesicles were more efficient at delivering hsiRNA and inducing Huntingtin silencing than conventional liposomes or hsiRNA delivered carrier-free (p < 0.0001; Figure S3A).
Full Chemical Stabilization of RNA Cargo Improves Productive Loading onto Extracellular Vesicles
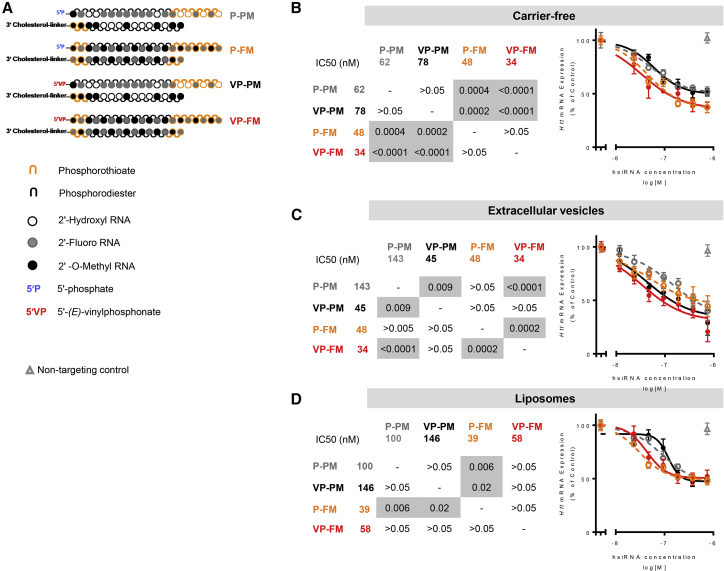
To evaluate the impact of chemical modifications on extracellular vesicle-mediated delivery of hsiRNAs, we synthesized four different hsiRNA variants: (1) partially modified (all pyrimidines modified, similar to commercially available siRNAs18, 19), (2) partially modified with 5′-(E)-vinylphosphonate,31, 32, 33, 45 (3) fully modified (100% of riboses modified22, 23, 26, 27, 28, 29), and (4) fully modified with 5′-(E)-vinylphosphonate (Figure 3A). In fully modified hsiRNA variants, four additional phosphorothioate modifications were introduced compared to partially modified variants, to provide additional stabilization from exonuclease activity (Figure 3A).
Figure 3.
Full Stabilization of hsiRNA Is Beneficial for Extracellular Vesicle-Mediated Delivery
(A) Scheme of chemically modified hsiRNAs. P-PM, partially modified backbone with 5′-phosphate on guide strand; P-FM, fully modified backbone with 5′-phosphate on guide strand; VP-PM, partially modified backbone with 5′-(E)-vinylphosphonate on guide strand; VP-FM, fully modified backbone with 5′-(E)-vinylphosphonate on guide strand. (B) Primary murine cortical neurons were incubated for 1 week with cholesterol-hsiRNA variants with different extents of 2′ ribose and 5′ end modifications either alone (carrier-free) or loaded onto extracellular vesicles or conventional liposomes, target Huntingtin mRNA silencing was measured, and silencing potency was calculated (IC50). n = 3, mean ± SEM. Pairwise comparison of curves was conducted using two-way ANOVA with Tukey’s post hoc test. Significance is depicted in gray. (C) Experiment from (B) conducted with extracellular vesicle-mediated delivery. (D) Experiment from (B) conducted with liposome-mediated delivery.
Surprisingly, only 5′-(E)-vinylphosphonate modification improved Huntingtin mRNA-silencing activity of hsiRNA-loaded extracellular vesicles (p = 0.009), whereas full modification of hsiRNA backbone alone had no effect (p > 0.05) (Figure 3C). The same effects were observed when using PPIB-targeting hsiRNAs (Figures S4E–S4G). However, when combined with 5′-(E)-vinylphosphonate, full hsiRNA backbone modification further improved silencing (p < 0.0001) compared to 5′-(E)-vinylphosphonate alone (p = 0.009) (Figure 3C). Thus, phosphatase resistance provided a larger benefit than nuclease resistance during extracellular vesicle-mediated delivery of hsiRNAs to neurons. An opposite effect was observed with carrier-free (i.e., no extracellular vesicles, liposomes, or transfection reagents used for delivery) hsiRNA uptake, where full modification of hsiRNA backbone improved silencing (p = 0.0004), whereas 5′-(E)-vinylphosphonate did not (p > 0.05) (Figure 3B). Indeed, previous data showed no effect of 5′-(E)-vinylphosphonate modification on silencing activity of siRNAs delivered carrier-free to HeLa cells.33 As another control, we used conventional neutral liposomes with a size range (Figure S1) and loading technique identical to that of extracellular vesicles (i.e., hsiRNAs residing predominantly on the surface of liposomes). When hsiRNAs were delivered in neutral liposomes, the effect of chemical modifications resembled carrier-free delivery, with full modification of hsiRNA backbone improving silencing activity the most (p = 0.006).
Thus, the biological origin and contents of extracellular vesicles represent a likely reason for increased relative importance of 5′-(E)-vinylphosphonate over chemical modification of siRNA backbone. Indeed, a variety of nucleases, including 5′-nucleotidase, was detected in extracellular vesicles purified from umbilical cord-derived mesenchymal stem cells using mass spectrometry (Table S2).
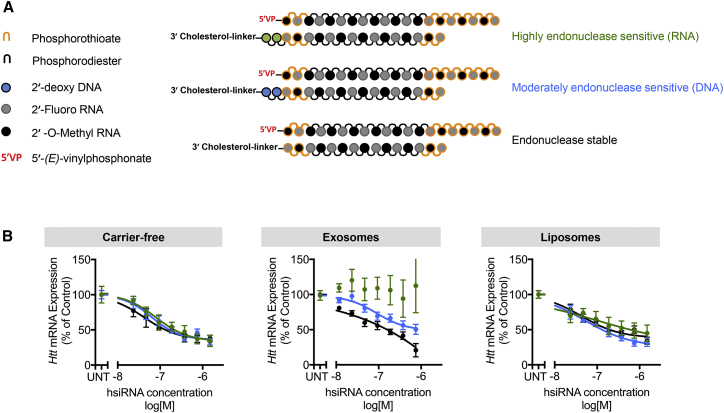
The Stability of the Cholesterol Linker Is Essential for Productive hsiRNA Loading onto Extracellular Vesicles
Loading of hsiRNAs onto extracellular vesicles is dependent on the presence of the hydrophobic cholesterol conjugate,14 which anchors the hsiRNA onto the membrane. Stable association with membranes may potentially trap siRNAs in endosomes and limit loading into cytoplasmic RNA-induced silencing complex (RISC), thus impairing silencing activity.46 Introduction of cleavable linkers has been used as a successful strategy to enhance silencing activity of conjugated siRNAs.47 To test whether the use of cleavable linkers is an advantage in extracellular vesicle-mediated delivery of cholesterol-hsiRNAs, we synthesized three hsiRNA variants with varying stability of the linker connecting cholesterol to the hsiRNA. We used a fully chemically modified hsiRNA variant containing 5′-(E)-vinylphosphonate for these studies. Incorporation of moderately (2′-deoxy-DNA) or highly (2′-hydroxyl-RNA) endonuclease-sensitive bases between the sense strand and the linker was used to modulate the rate of cholesterol cleavage (Figure 4A). Destabilization of the linker chemistry greatly impaired hsiRNA-silencing activity when delivered via extracellular vesicles. Huntingtin mRNA silencing was completely abolished (p < 0.0001) upon the incorporation of two 2′-hydroxyl RNA residues (highly endonuclease sensitive) and significantly reduced (p = 0.024) upon the incorporation of 2′-deoxy DNA residues (moderately endonuclease sensitive) (Figure 4B, middle panel). On the contrary, chemical stability of the linker had no effect on hsiRNA-silencing activity when delivered carrier-free (Figure 4B, right panel) or in neutral liposomes (Figure 4B, left panel). Thus, use of a stable linker is essential for productive loading of extracellular vesicles with the RNA cargo.
Figure 4.
Cleavable Cholesterol Impairs Activity of Cholesterol-hsiRNA-Loaded Extracellular Vesicles
(A) Incorporation of moderately (2′-deoxy-DNA, blue) or highly (2′-hydroxy-RNA, green) endonuclease-sensitive bases into the sense strand is a strategy to facilitate cleavage of cholesterol from cholesterol-conjugated hsiRNA. (B) Huntingtin mRNA silencing in primary neurons 1 week after treatment with hsiRNA variants alone (carrier-free) or loaded onto extracellular vesicles or liposomes at varying concentrations. UNT, untreated; n = 3; mean ± SEM.
Discussion
Extracellular vesicles are promising delivery vesicles for therapeutic RNAs. Cholesterol conjugation to siRNAs is a simple, scalable, and widely used method to load extracellular vesicles with RNA cargo that has proven useful in both in vitro16, 17 and in vivo14 experiments. Fluorescent cholesterol-hsiRNA loaded to extracellular vesicles has been shown to distribute and silence target mRNA bilaterally in brain following unilateral infusion.14 Furthermore, cholesterol-conjugated RNA aptamers have been able to direct extracellular vesicles to tumors in vivo.48 However, there is a lack of knowledge on the importance of typical siRNA chemical modifications18, 22, 23, 24, 25 on extracellular vesicle-mediated delivery.
The presence of nucleases in extracellular vesicle preparations bears consideration during the rational design of RNA cargo. This study found that previously used partial modification of siRNA14, 16, 17 is suboptimal for extracellular vesicle-mediated siRNA delivery. Instead, siRNAs with a combination of 5′-(E)-vinylphosphonate31, 32, 33 and alternating 2′-fluoro and 2′-O-methyl modifications22, 23, 26, 27, 28, 29 performed best at extracellular vesicle-mediated delivery and Huntingtin mRNA silencing in neurons. Furthermore, incorporation of nuclease-sensitive bases into the cholesterol linker impaired extracellular vesicle-mediated delivery of hsiRNAs. The benefit of chemical modifications may stem from the localization of hsiRNAs (i.e., on the surface of extracellular vesicles14), altered cellular internalization pathway,14 as well exosomal protein content, including a variety of nucleases (Figure S2)49 and nuclease activity.21
Here we showed that the cholesterol conjugation strategy to siRNA plays an important role in extracellular vesicle-mediated delivery and, therefore, should be included in optimized RNA cargo design. Between two commercially available strategies to conjugate cholesterol to the 3′ end of the siRNA sense strand, TEG proved to be favorable compared to C7 for productive exosomal loading of cholesterol-siRNA. Both length and hydrophilic character of the TEG linker might favor the lipid bilayer geometry and promote more efficient hsiRNA loading.
The mechanism of extracellular vesicle uptake is not fully understood. There are data supporting endocytosis-mediated uptake,50 membrane fusion,51 phagocytosis,52 and macropinocytosis.53 We speculate that extracellular vesicles may use multiple pathways for cellular uptake and delivery of hsiRNA. Confocal microscopy has shown intracellular punctate of fluorescent hsiRNA when delivered via extracellular vesicles14 and dominantly plasma membrane fluorescence when hsiRNA was delivered carrier-free.40 These previous observations suggest that the uptake mechanism of hsiRNA-extracellular vesicle complexes is distinct from the uptake mechanism of hsiRNA alone.
Our data suggest that hsiRNA loading onto extracellular vesicles saturates at ∼3,000 hsiRNAs molecules per extracellular vesicle. In addition, overloading (more than 5,000 hsiRNAs per extracellular vesicle) may induce aggregation via interfering with membrane curvature and vesicle integrity, and thereby it may adversely affect productive gene silencing. Therefore, we suggest ∼3,000 hsiRNAs per extracellular vesicle as optimal loading capacity. This number might be altered by the nature of the hydrophobic conjugate of hsiRNA (i.e., other than cholesterol) and extracellular vesicle membrane composition, which varies greatly depending on the parent cell.4, 54 Thus, the maximal hsiRNA copy number per extracellular vesicle will need to be defined for each hydrophobic conjugate of siRNA and extracellular vesicle cell source separately.
The data presented here provide a detailed framework for the successful use of hydrophobic modification as a strategy for productive loading of RNA cargo onto extracellular vesicles. A similar chemical optimization strategy is likely needed when taking advantage of extracellular vesicles for in vivo delivery of other oligonucleotide species, including CRISPR guide RNAs, artificial miRNAs, small mRNAs, RNA tethers, aptamers, or antisense oligonucleotides.
Materials and Methods
Oligonucleotides
Oligonucleotides were synthesized using standard phosphoramidite chemistry as described previously.28, 33, 40 siRNA sequences and chemical modification patterns used in this study are described in Table S1.
Cell Culture
Umbilical cord, Wharton’s jelly-derived mesenchymal stem cells (PCS-500-010, ATCC, Manassas, VA) were cultured in appropriate stem cell medium (PCS-500-030, ATCC, Manassas, VA) in the presence of 2% fetal bovine serum (FBS) and growth factors (PCS-500-040, ATCC, Manassas, VA) at 37°C and 5% CO2. Medium was changed every 3 days, and cells were expanded until passage 12 s, to reach a total of 3,000 cm2 surface in T500 triple flasks.
Isolation of Extracellular Vesicles
Medium on umbilical cord-derived mesenchymal stem cells was changed to extracellular vesicle-depleted medium (centrifuged at 100,000 × g for at least 17 hr) and incubated for 48 hr. Extracellular vesicles were then purified from this conditioned medium via differential ultracentrifugation as described previously.14 Briefly, cell debris was pelleted at 300 × g (10 min). Larger vesicles were pelleted at 10,000 × g (30 min), then supernatant was filtered through a 0.2-μm membrane (Nalgene aPES, Thermo Fisher Scientific, Waltham, MA) and extracellular vesicles were pelleted at 100,000 × g (90 min) using 70-mL polycarbonate bottles (Beckman Coulter, Brea, CA; 355622) and Type 45 Ti rotor (Beckman Coulter, Brea, CA; 339160). Extracellular vesicle pellet was then washed once in 1 mL sterile PBS and centrifuged again for 90 min at 100,000 × g in a tabletop ultracentrifuge using a TLA-110 rotor (Beckman Coulter, Brea, CA; 366730).
Characterization of Extracellular Vesicles
Nanoparticle Tracking Analysis (NanoSight NS300, Malvern, Malvern, UK) was used to measure concentration and size distribution of extracellular vesicles. Briefly, samples were diluted in PBS 1:1,000, manually injected into the instrument, and videos were acquired at ambient temperature at camera level 9 for 1 min per sample (n = 3) (Figure S1). Extracellular vesicles were then frozen at −80°C in 0.1 M sucrose and appropriate dilution of protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO; P8340) until further use. Transmission electron microscopy of extracellular vesicles was conducted at Mass General Hospital using a JEOL 1100 transmission electron microscope (JEOL, Peabody, MA) at 60 kV, as described previously.4 For western blotting, extracellular vesicle or cell pellets were suspended in radioimmunoprecipitation assay (RIPA) buffer (Pierce 899000, Thermo Fisher Scientific, Waltham, MA) containing PMSF (36978, Thermo Fisher Scientific, Waltham, MA) and protease inhibitor cocktail (cOmplete Mini, 11836153001, Roche, Indianapolis, IN), and samples were sonicated for 15 min. Insoluble material was pelleted by centrifugation for 15 min at 13,000 rpm at 4°C. Proteins (50 μg) were loaded and simultaneously analyzed on NuPAGE 4%–12% Bis-Tris gels (Invitrogen). After transfer to polyvinylidene fluoride (PVDF) (Bio-Rad, Hercules, CA) membranes, antibody incubation and development were performed using the Odyssey system (LI-COR Biosciences, Bad Homburg, Germany), according to the manufacturer’s instructions. Primary antibodies used were Calnexin (C5C9, Cell Signaling Technology, Danvers, MA), CD63 (H193, Santa Cruz Biotechnology, Dallas, TX), Tsg101 (4A10, Abcam, Cambridge, MA), and CD81 (B11, Santa Cruz Biotechnology, Dallas, TX). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) of extracellular vesicles was performed as described previously.4
Liposome Preparation
Dioleoyl-phosphatidylcholine (DOPC) (Avanti Polar Lipids, Alabaster, AL; 850375) and cholesterol (Avanti Polar Lipids, Alabaster, AL; 700000) were diluted in chloroform at a concentration of 50 mg/mL. 35 μL DOPC and 15 μL cholesterol was transferred into a glass vial, and chloroform was evaporated under argon flow. The resulting lipid film was rehydrated in 500 μL PBS (Dulbecco’s Phosphate Buffered Saline, Corning, Manassas, VA; 21-031-CV), sonicated for 15 min in water bath (Branson ultrasonic cleaner 40 kHz, Cleanosonic, Richmond VA; BB5510), and then extruded using Mini-Extruder (Avanti Polar Lipids, Alabaster, AL; 610000) through a 50-nm pore-sized polycarbonate membrane (Whatman Nucleopore, MilliporeSigma, St. Louis, MO; WHA800308).
Loading hsiRNAs onto Extracellular Vesicles and Liposomes
Known numbers of extracellular vesicles or liposomes were co-incubated with known amounts of hsiRNAs at 37°C for 1 hr in 500 μL PBS (i.e., loading mixture). Then the extracellular vesicle-hsiRNA mixture was centrifuged at 100,000 × g for 90 min, and the supernatant containing unloaded hsiRNA was removed (supernatant). Pellet was taken up in 500 μL PBS for fluorescence measurement or in 300 μL Neural Q medium for treatment of primary neurons.
To quantify loading of Cy3-labeled hsiRNA, a 200-μL aliquot was taken from resuspended extracellular vesicle pellet or from the supernatant. Fluorescence was assessed at 550-nm excitation and 570-nm emission on a TECAN instrument. Percentage of loaded hsiRNA was calculated as follows: pellet/(pellet + supernatant). To estimate hsiRNA copy number per extracellular vesicle, the following formula was used: (percent of loaded hsiRNA) × (amount of hsiRNA initially mixed in with extracellular vesicles [mol]) × (Avogadro number)/(number of extracellular vesicles initially mixed in).
To quantify loading of not fluorescently labeled hsiRNAs, a PNA hybridization based assay was used.33, 55 Briefly, a 10-μL aliquot of resuspended extracellular vesicle pellet loaded with hsiRNAs was diluted in 90 μL RIPA buffer (Pierce 899000, Thermo Fisher Scientific, Waltham, MA) and sonicated for 15 min. Then 30 μL 3M KCl was added to precipitate SDS from the lysate, and precipitated SDS was pelleted at 5,000 × g for 15 min. Supernatant was then removed and a Cy3-labeled PNA oligonucleotide (PNA Bio, Newbury Park, CA) fully complementary to hsiRNA guide strand was added to the extracellular vesicle lysate and annealed at 95°C for 15 min, followed by incubation at 50°C for 15 min and cooling to room temperature. Then PNA-annealed extracellular vesicle lysate was injected into high-performance liquid chromatography (HPLC) anion exchange column (Dionex DNAPac PA100, Thermo Fisher Scientific, Waltham, MA) using an autosampler (1260 Infinity system, Agilent, Santa Clara, CA). The mobile phase used for HPLC was 50% acetonitrile, 25 mM Tris-HCl (pH 8.5), and 1 mM EDTA in water, and 0–800 mM NaClO4 salt gradient was used to elute the hsiRNA-PNA hybrid. Quantification was performed using a calibration curve of known amounts of hsiRNAs.
Preparation of Primary Cortical Neurons
All animal procedures were approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee (IACUC, protocol number A-2411). Primary cortical neurons were isolated from embryonic day (E)15.5 mouse embryos of wild-type FVBNj mice. Pregnant females were anesthetized by either intraperitoneal injection of ketamine (100 mg/kg, KETASET, Zoetis, Kalamazoo, MI) and xylazine (10 mg/kg, AnaSed, AKORN, Laker Forest, IL; NDC59399-111-50) or isoflurane (Isoflurane, USP, Piramal Cricital Care, Betlehem, PA; NDC66794-013-010), and cervical dislocation followed. Embryos were removed and transferred to ice-cold DMEM/F12 medium (Invitrogen, Carlsbad, CA; 11320). Brains were removed from DMEM and meninges were carefully detached under a microscope. Cortices were isolated and transferred into pre-warmed (37°C) papain-DNase solution for 30 min at 37°C and 5% CO2 to dissolve the tissue. Papain (Worthington, Lakewood, NJ; 54N15251) was dissolved in 2 mL Hibernate E (Brainbits, Springfield, IL; HE) and supplemented with 0.25 mL 10 mg/mL DNase1 (Worthington, Lakewood, NJ; 54M15168) in Hibernate E. After 30-min incubation, the papain solution was removed and 1 mL NeuralQ (Sigma-Aldrich, St. Louis, MO; N3100) supplemented with 2.5% FBS was added to the tissue. Tissues were then dissociated by trituration through a fire-polished, glass Pasteur pipet. Neurons were counted using Neubauer chamber and diluted at 106 cells/mL. 105 neurons per well were plated on 96-well plates pre-coated with poly-L-lysine (BD BIOCOAT, Corning, NY; 356515). After overnight incubation at 37°C and 5% CO2, an equal volume of NeuralQ supplemented with anti-mitotics, 0.484 μL/mL 5′UtP (Sigma, St. Louis, MO; U6625) and 0.2402 μL/mL 5′FdU (Sigma, St. Louis, MO; F3503), was added to prevent the growth of non-neuronal cells. Half of the volume of media was replaced with fresh NeuralQ containing anti-mitotic every 48 hr until the experiments were performed.
Measurement of hsiRNA-Silencing Activity in Neurons
Neurons were treated with hsiRNA-loaded extracellular vesicles (resuspended in NeuralQ medium) and incubated for 7 days at 37°C and 5% CO2 post-treatment. Neurons were then lysed and mRNA quantification was performed using the QuantiGene 2.0 assay kit (Affymetrix, QS0011, Thermo Fisher Scientific, Waltham, MA), as described previously.56 Catalog numbers for probes used in QuantiGene 2.0 assay kit are as follows: mouse Htt (Affymetrix, SB-14150, Thermo Fisher Scientific, Waltham, MA), and mouse Hprt (Affymetrix, SB-15463, Thermo Fisher Scientific, Waltham, MA). Datasets were normalized to housekeeping gene Hprt and presented as percentage of untreated control. Each measurement was run in duplicates. hsiRNAPPIB was used as non-targeting control for Huntingtin silencing, and hsiRNAHTT was used as a non-targeting control for PPIB silencing.
Measurement of hsiRNA Uptake to Neurons
hsiRNA guide strands in neuron cell lysates were quantified using a PNA hybridization assay.28, 33, 55, 57 PNAs are oligonucleotides where the sugar-phosphate backbone is replaced with a polyamide backbone. PNAs have no charge and have a high hybridization energy to RNA. SDS from leftover neuron lysates after mRNA quantification was precipitated with 3 M KCl and pelleted at 4,000 × g for 15 min. hsiRNA guide strands in cleared supernatant were hybridized to fully complementary Cy3-labeled PNA strands (PNABio, Thousand Oaks, CA). hsiRNA-PNA duplexes were injected into HPLC DNAPac PA100 anion exchange column (Thermo Scientific, Carlsbad, CA), and Cy3 fluorescence was monitored and peaks integrated. The mobile phase for HPLC was 50% water 50% acetonitrile, 25 mM Tris-HCl (pH 8.5), and 1 mM EDTA and the salt gradient was 0–800 mM NaClO4. For the calibration curve, a known amount of hsiRNA duplex was spiked into cell lysis solution.
Statistical Analysis
Data were analyzed using GraphPad Prism 7, version 7.04 (GraphPad Software, La Jolla, CA). In in vitro-silencing experiments, the IC50 values were determined by fitting dose-response curve using log(inhibitor) vs. response – variable slope (four parameters) equation. Curves were compared using two-way ANOVA. Differences in all comparisons were considered significant at p values < 0.05.
Author Contributions
Conceptualization, R.A.H., M.-C.D., J.F.A., M.R.H., L.R., and A.K.; Methodology, R.A.H. and M.-C.D.; Investigation, R.A.H., R.M., and E.S.; Writing – Original Draft, R.A.H., N.A., and A.K.; Funding Acquisition, N.A. and A.K.; Resources, A.B., L.R., J.F.A., M.R.H., and D.E.; Supervision, M.D., N.A., and A.K.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
We thank Andrew Coles for assistance in primary neuron preparations and John Leszyk for assistance in proteomics (LC-MS/MS). This work was supported by NIH UH3 grant TR 000888 05 and UMass grant CCTS UL1 TR000161 to N.A. and A.K.; NIH grants RO1GM10880304, RO1NS10402201, and S10 OD020012 to A.K.; and the CHDI Foundation (Research Agreement A-6119, JSC A6367) to N.A. Marie-Cecile Didiot was supported by a Huntington’s Disease Society of America Postdoctoral Fellowship. This publication is part of the NIH Extracellular RNA Communication Consortium paper package and was supported by the NIH Common Fund’s exRNA Communication Program.
Footnotes
Supplemental Information includes four figures and two tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.05.024.
Contributor Information
Neil Aronin, Email: neil.aronin@umassmed.edu.
Anastasia Khvorova, Email: anastasia.khvorova@umassmed.edu.
Supplemental Information
References
- 1.Grapp M., Wrede A., Schweizer M., Hüwel S., Galla H.J., Snaidero N., Simons M., Bückers J., Low P.S., Urlaub H. Choroid plexus transcytosis and exosome shuttling deliver folate into brain parenchyma. Nat. Commun. 2013;4:2123. doi: 10.1038/ncomms3123. [DOI] [PubMed] [Google Scholar]
- 2.Yang T., Martin P., Fogarty B., Brown A., Schurman K., Phipps R., Yin V.P., Lockman P., Bai S. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm. Res. 2015;32:2003–2014. doi: 10.1007/s11095-014-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haraszti R.A., Didiot M.C., Sapp E., Leszyk J., Shaffer S.A., Rockwell H.E., Gao F., Narain N.R., DiFiglia M., Kiebish M.A. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles. 2016;5:32570. doi: 10.3402/jev.v5.32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 6.Zomer A., Maynard C., Verweij F.J., Kamermans A., Schäfer R., Beerling E., Schiffelers R.M., de Wit E., Berenguer J., Ellenbroek S.I.J. In Vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161:1046–1057. doi: 10.1016/j.cell.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xin H., Li Y., Buller B., Katakowski M., Zhang Y., Wang X., Shang X., Zhang Z.G., Chopp M. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells. 2012;30:1556–1564. doi: 10.1002/stem.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xin H., Li Y., Liu Z., Wang X., Shang X., Cui Y., Zhang Z.G., Chopp M. MiR-133b promotes neural plasticity and functional recovery after treatment of stroke with multipotent mesenchymal stromal cells in rats via transfer of exosome-enriched extracellular particles. Stem Cells. 2013;31:2737–2746. doi: 10.1002/stem.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H.K., Finniss S., Cazacu S., Xiang C., Brodie C. Mesenchymal stem cells deliver exogenous miRNAs to neural cells and induce their differentiation and glutamate transporter expression. Stem Cells Dev. 2014;23:2851–2861. doi: 10.1089/scd.2014.0146. [DOI] [PubMed] [Google Scholar]
- 10.Cui C., Ye X., Chopp M., Venkat P., Zacharek A., Yan T., Ning R., Yu P., Cui G., Chen J. miR-145 Regulates Diabetes-Bone Marrow Stromal Cell-Induced Neurorestorative Effects in Diabetes Stroke Rats. Stem Cells Transl. Med. 2016;5:1656–1667. doi: 10.5966/sctm.2015-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xin H., Wang F., Li Y., Lu Q.E., Cheung W.L., Zhang Y., Zhang Z.G., Chopp M. Secondary Release of Exosomes From Astrocytes Contributes to the Increase in Neural Plasticity and Improvement of Functional Recovery After Stroke in Rats Treated With Exosomes Harvested From MicroRNA 133b-Overexpressing Multipotent Mesenchymal Stromal Cells. Cell Transplant. 2017;26:243–257. doi: 10.3727/096368916X693031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Chopp M., Liu X.S., Katakowski M., Wang X., Tian X., Wu D., Zhang Z.G. Exosomes Derived from Mesenchymal Stromal Cells Promote Axonal Growth of Cortical Neurons. Mol. Neurobiol. 2017;54:2659–2673. doi: 10.1007/s12035-016-9851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 14.Didiot M.-C., Hall L.M., Coles A.H., Haraszti R.A., Godinho B.M.D.C., Chase K., Sapp E., Ly S., Alterman J.F., Hassler M.R. Exosome-mediated Delivery of Hydrophobically Modified siRNA for Huntingtin mRNA Silencing. Mol. Ther. 2016;24:1836–1847. doi: 10.1038/mt.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamerkar S., LeBleu V.S., Sugimoto H., Yang S., Ruivo C.F., Melo S.A., Lee J.J., Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Loughlin A.J., Mäger I., de Jong O.G., Varela M.A., Schiffelers R.M., El Andaloussi S., Wood M.J.A., Vader P. Functional Delivery of Lipid-Conjugated siRNA by Extracellular Vesicles. Mol. Ther. 2017;25:1580–1587. doi: 10.1016/j.ymthe.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stremersch S., Vandenbroucke R.E., Van Wonterghem E., Hendrix A., De Smedt S.C., Raemdonck K. Comparing exosome-like vesicles with liposomes for the functional cellular delivery of small RNAs. J. Control. Release. 2016;232:51–61. doi: 10.1016/j.jconrel.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Byrne M., Tzekov R., Wang Y., Rodgers A., Cardia J., Ford G., Holton K., Pandarinathan L., Lapierre J., Stanney W. Novel hydrophobically modified asymmetric RNAi compounds (sd-rxRNA) demonstrate robust efficacy in the eye. J. Ocul. Pharmacol. Ther. 2013;29:855–864. doi: 10.1089/jop.2013.0148. [DOI] [PubMed] [Google Scholar]
- 19.Leake, D., Reynolds, A., Khvorova, A., Marshall, W., and Scaringe, S. (2004). Stabilized polynucleotides for use in RNA interference. U.S. patent, WO/2004/090105.
- 20.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 21.Stremersch S., Brans T., Braeckmans K., De Smedt S., Raemdonck K. Nucleic acid loading and fluorescent labeling of isolated extracellular vesicles requires adequate purification. Int. J. Pharm. 2017 doi: 10.1016/j.ijpharm.2017.10.022. Published online October 12, 2017. [DOI] [PubMed] [Google Scholar]
- 22.Nair J.K., Willoughby J.L., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 23.Allerson C.R., Sioufi N., Jarres R., Prakash T.P., Naik N., Berdeja A., Wanders L., Griffey R.H., Swayze E.E., Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 24.Choung S., Kim Y.J., Kim S., Park H.O., Choi Y.C. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 25.Czauderna F., Fechtner M., Dames S., Aygün H., Klippel A., Pronk G.J., Giese K., Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coelho T., Adams D., Silva A., Lozeron P., Hawkins P.N., Mant T., Perez J., Chiesa J., Warrington S., Tranter E. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 27.Fitzgerald K., White S., Borodovsky A., Bettencourt B.R., Strahs A., Clausen V., Wijngaard P., Horton J.D., Taubel J., Brooks A. A Highly Durable RNAi Therapeutic Inhibitor of PCSK9. N. Engl. J. Med. 2017;376:41–51. doi: 10.1056/NEJMoa1609243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikan M., Osborn M.F., Coles A.H., Godinho B.M., Hall L.M., Haraszti R.A., Hassler M.R., Echeverria D., Aronin N., Khvorova A. Docosahexaenoic Acid Conjugation Enhances Distribution and Safety of siRNA upon Local Administration in Mouse Brain. Mol. Ther. Nucleic Acids. 2016;5:e344. doi: 10.1038/mtna.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alterman J.F., Hall L.M., Coles A.H., Hassler M.R., Didiot M.C., Chase K., Abraham J., Sottosanti E., Johnson E., Sapp E. Hydrophobically Modified siRNAs Silence Huntingtin mRNA in Primary Neurons and Mouse Brain. Mol. Ther. Nucleic Acids. 2015;4:e266. doi: 10.1038/mtna.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassler M.R., Turanov A.A., Alterman J.F., Haraszti R.A., Coles A.H., Osborn M.F., Echeverria D., Nikan M., Salomon W.E., Roux L. Comparison of partially and fully chemically-modified siRNA in conjugate-mediated delivery in vivo. Nucleic Acids Res. 2018;46:2185–2196. doi: 10.1093/nar/gky037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prakash T.P., Kinberger G.A., Murray H.M., Chappell A., Riney S., Graham M.J., Lima W.F., Swayze E.E., Seth P.P. Synergistic effect of phosphorothioate, 5′-vinylphosphonate and GalNAc modifications for enhancing activity of synthetic siRNA. Bioorg. Med. Chem. Lett. 2016;26:2817–2820. doi: 10.1016/j.bmcl.2016.04.063. [DOI] [PubMed] [Google Scholar]
- 32.Parmar R., Willoughby J.L., Liu J., Foster D.J., Brigham B., Theile C.S., Charisse K., Akinc A., Guidry E., Pei Y. 5′-(E)-Vinylphosphonate: A Stable Phosphate Mimic Can Improve the RNAi Activity of siRNA-GalNAc Conjugates. ChemBioChem. 2016;17:985–989. doi: 10.1002/cbic.201600130. [DOI] [PubMed] [Google Scholar]
- 33.Haraszti R.A., Roux L., Coles A.H., Turanov A.A., Alterman J.F., Echeverria D., Godinho B.M.D.C., Aronin N., Khvorova A. 5′-Vinylphosphonate improves tissue accumulation and efficacy of conjugated siRNAs in vivo. Nucleic Acids Res. 2017;45:7581–7592. doi: 10.1093/nar/gkx507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrissey D.V., Lockridge J.A., Shaw L., Blanchard K., Jensen K., Breen W., Hartsough K., Machemer L., Radka S., Jadhav V. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 35.Olearczyk J., Gao S., Eybye M., Yendluri S., Andrews L., Bartz S., Cully D., Tadin-Strapps M. Targeting of hepatic angiotensinogen using chemically modified siRNAs results in significant and sustained blood pressure lowering in a rat model of hypertension. Hypertens. Res. 2014;37:405–412. doi: 10.1038/hr.2013.155. [DOI] [PubMed] [Google Scholar]
- 36.Petrova N.S., Chernikov I.V., Meschaninova M.I., Dovydenko I.S., Venyaminova A.G., Zenkova M.A., Vlassov V.V., Chernolovskaya E.L. Carrier-free cellular uptake and the gene-silencing activity of the lipophilic siRNAs is strongly affected by the length of the linker between siRNA and lipophilic group. Nucleic Acids Res. 2012;40:2330–2344. doi: 10.1093/nar/gkr1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiner A.T., Witwer K.W., van Balkom B.W.M., de Beer J., Brodie C., Corteling R.L., Gabrielsson S., Gimona M., Ibrahim A.G., de Kleijn D. Concise Review: Developing Best-Practice Models for the Therapeutic Use of Extracellular Vesicles. Stem Cells Transl. Med. 2017;6:1730–1739. doi: 10.1002/sctm.17-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lener T., Gimona M., Aigner L., Börger V., Buzas E., Camussi G., Chaput N., Chatterjee D., Court F.A., Del Portillo H.A. Applying extracellular vesicles based therapeutics in clinical trials - an ISEV position paper. J. Extracell. Vesicles. 2015;4:30087. doi: 10.3402/jev.v4.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee M., Liu T., Im W., Kim M. Exosomes from adipose-derived stem cells ameliorate phenotype of Huntington’s disease in vitro model. Eur. J. Neurosci. 2016;44:2114–2119. doi: 10.1111/ejn.13275. [DOI] [PubMed] [Google Scholar]
- 40.Alterman J.F., Hall L.M., Coles A.H., Hassler M.R., Didiot M.C., Chase K., Abraham J., Sottosanti E., Johnson E., Sapp E. Hydrophobically Modified siRNAs Silence Huntingtin mRNA in Primary Neurons and Mouse Brain. Mol. Ther. Nucleic Acids. 2015;4:e266. doi: 10.1038/mtna.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ly S., Navaroli D.M., Didiot M.C., Cardia J., Pandarinathan L., Alterman J.F., Fogarty K., Standley C., Lifshitz L.M., Bellve K.D. Visualization of self-delivering hydrophobically modified siRNA cellular internalization. Nucleic Acids Res. 2017;45:15–25. doi: 10.1093/nar/gkw1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Nallagatla S.R., Bevilacqua P.C. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elkayam E., Parmar R., Brown C.R., Willoughby J.L., Theile C.S., Manoharan M., Joshua-Tor L. siRNA carrying an (E)-vinylphosphonate moiety at the 5′ end of the guide strand augments gene silencing by enhanced binding to human Argonaute-2. Nucleic Acids Res. 2017;45:3528–3536. doi: 10.1093/nar/gkw1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dovydenko I., Tarassov I., Venyaminova A., Entelis N. Method of carrier-free delivery of therapeutic RNA importable into human mitochondria: Lipophilic conjugates with cleavable bonds. Biomaterials. 2016;76:408–417. doi: 10.1016/j.biomaterials.2015.10.075. [DOI] [PubMed] [Google Scholar]
- 47.Chen Q., Butler D., Querbes W., Pandey R.K., Ge P., Maier M.A., Zhang L., Rajeev K.G., Nechev L., Kotelianski V. Lipophilic siRNAs mediate efficient gene silencing in oligodendrocytes with direct CNS delivery. J. Control. Release. 2010;144:227–232. doi: 10.1016/j.jconrel.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 48.Pi F., Binzel D.W., Lee T.J., Li Z., Sun M., Rychahou P., Li H., Haque F., Wang S., Croce C.M. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat. Nanotechnol. 2018;13:82–89. doi: 10.1038/s41565-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keerthikumar S., Chisanga D., Ariyaratne D., Al Saffar H., Anand S., Zhao K., Samuel M., Pathan M., Jois M., Chilamkurti N. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016;428:688–692. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heusermann W., Hean J., Trojer D., Steib E., von Bueren S., Graff-Meyer A., Genoud C., Martin K., Pizzato N., Voshol J. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J. Cell Biol. 2016;213:173–184. doi: 10.1083/jcb.201506084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montecalvo A., Larregina A.T., Shufesky W.J., Stolz D.B., Sullivan M.L., Karlsson J.M., Baty C.J., Gibson G.A., Erdos G., Wang Z. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuyama K., Sun H., Mitsutake S., Igarashi Y. Sphingolipid-modulated exosome secretion promotes clearance of amyloid-β by microglia. J. Biol. Chem. 2012;287:10977–10989. doi: 10.1074/jbc.M111.324616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sagar G., Sah R.P., Javeed N., Dutta S.K., Smyrk T.C., Lau J.S., Giorgadze N., Tchkonia T., Kirkland J.L., Chari S.T., Mukhopadhyay D. Pathogenesis of pancreatic cancer exosome-induced lipolysis in adipose tissue. Gut. 2016;65:1165–1174. doi: 10.1136/gutjnl-2014-308350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Griffiths S.G., Cormier M.T., Clayton A., Doucette A.A. Differential Proteome Analysis of Extracellular Vesicles from Breast Cancer Cell Lines by Chaperone Affinity Enrichment. Proteomes. 2017;5:E25. doi: 10.3390/proteomes5040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roehl, I., Schuster, M., and Seiffert, S. (2011).Oligonucleotide detection method. U.S. patent, 13,124,411.
- 56.Coles A.H., Osborn M.F., Alterman J.F., Turanov A.A., Godinho B.M., Kennington L., Chase K., Aronin N., Khvorova A. A High-Throughput Method for Direct Detection of Therapeutic Oligonucleotide-Induced Gene Silencing In Vivo. Nucleic Acid Ther. 2016;26:86–92. doi: 10.1089/nat.2015.0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nikan M., Osborn M.F., Coles A.H., Biscans A., Godinho B.M.D.C., Haraszti R.A., Sapp E., Echeverria D., DiFiglia M., Aronin N., Khvorova A. Synthesis and evaluation of parenchymal retention and efficacy of a metabolically stable O-Phosphocholine-N-docosahexaenoyl-l-serine siRNA conjugate in mouse brain. Bioconjug. Chem. 2017;28:1758–1766. doi: 10.1021/acs.bioconjchem.7b00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.