Abstract
While the majority of DNA is enclosed within the nucleus, the mitochondria also contain their own, separate DNA, the mitochondrial DNA (mtDNA). Mutations in mtDNA are associated with various human diseases, demonstrating the importance of mtDNA. Intensive studies over the last 18 years have demonstrated the presence of two distinct classes of mtDNA replication intermediates in mammals. One involves leading-strand DNA synthesis in the absence of synchronous lagging-strand DNA synthesis. Currently there are competing models in which the lagging-strand template is either systematically hybridized to processed mitochondrial transcripts, or coated with protein, until the lagging-strand DNA synthesis takes place. The other class of mtDNA replication intermediates has many properties of conventional, coupled leading- and lagging-strand DNA synthesis. Additionally, the highly unusual arrangement of DNA in human heart mitochondria suggests a third mechanism of replication. These findings indicate that the mtDNA replication systems of humans and other mammals are far more complex than previously thought, and thereby will require further research to understand the full picture of mtDNA replication.
Keywords: bootlace model, coupled leading- and lagging-strand DNA synthesis, mitochondrial DNA replication, RITOLS, strand-displacement mechanism model
Mitochondria constitute a subcellular compartment in eukaryotic cells with many important functions. The etymology of mitochondrion is from the Greek words ‘mitos’ (thread) and ‘chondros’ (granule) (1, 2), and mitochondria are often referred to as ‘the powerhouses of the cell’, as they produce energy to fuel cellular activities. A remarkable feature of the organelle is that they contain their own multicopy genome, the mitochondrial DNA (mtDNA) (3). Human mtDNAs are double-stranded, circular molecules of 16,569 bp (4) and are arranged as protein-DNA complexes, called mitochondrial nucleoids, inside the mitochondrial matrix (5). In contrast to nuclear DNA, mtDNA is inherited maternally and typically exists in several hundreds to thousands of copies per cell (Fig. 1A). The two strands of mtDNA are distinguished by nucleotide composition and are named heavy (H)- and light (L)-strands. mtDNA encodes 13 subunits of the oxidative phosphorylation (OXPHOS) complexes, and 2 ribosomal RNAs (rRNAs) and 22 transfer RNAs (tRNAs) (4) that are necessary for the synthesis of the subunits in the mitochondria-specific translation system within mitochondria (Fig. 1B). Through the process of OXPHOS, mitochondria generate most of the cellular ATP and mitochondrial membrane potential (ΔΨm) that is requisite for various mitochondrial activities. The OXPHOS complexes are composed of approximately 90 subunits and four out of the five complexes (Complexes I, III, IV and V) require 13 mitochondrially encoded subunits [e.g. Figure 1 in (6)]. The rest of the OXPHOS complex subunits and all other proteins found within the mitochondria, including mitochondrial ribosomal proteins, metabolic enzymes and mtDNA replication factors [>1,000 proteins (7)] are encoded in the nuclear genome, which are then translated in the cytoplasm and imported into mitochondria. Gene organization of human mtDNA is compact, lacking intronic structure in protein-coding genes. Apart from the so-called ‘major noncoding region’ (NCR) of approximately 1 kb in length between tRNAPhe and tRNAPro genes, mtDNA encodes genes with minimal gaps between them (Fig. 1B). The NCR is also referred to as the control region. In some cases, the ends of reading frames of neighbouring genes overlap each other (e.g. ATP synthase subunits A8 and A6, and NADH-ubiquinone oxidoreductase subunits ND4L and ND4). Owing to the compactness of the mitochondrial genome structure, gene density in mtDNA is much higher than that in nuclear DNA. Despite the small size of mtDNA, mRNAs transcribed from mtDNA make up a surprisingly high proportion of cellular mRNA, nearly 30% of the total cellular mRNA content in the heart and 5–25% in other tissues (8). Transcription of human mtDNA is thought to initiate from three promoters, one L-strand promoter (LSP) and two H-strand promoters (HSP1 and HSP2) (Fig. 1B). Mitochondrial transcripts are produced as polycistronic precursors from both strands of mtDNA and are then processed into individual RNAs (9–11). The transcripts undergo RNA maturation, including the addition of poly A tails to mRNAs and post-transcriptional modification of tRNAs and rRNAs (12–14).
Fig. 1.
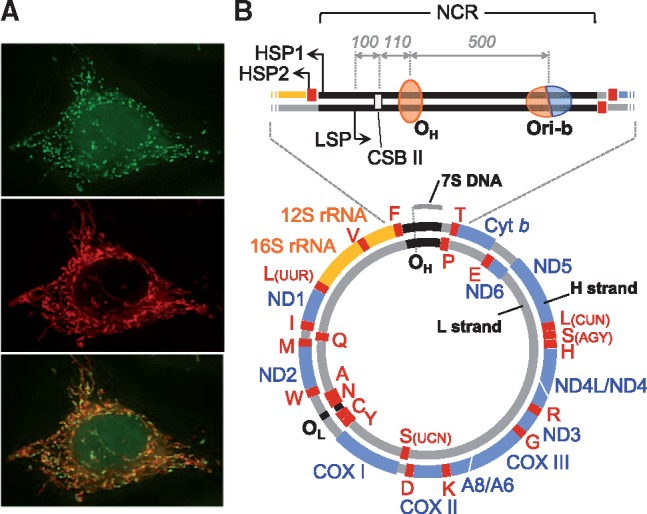
Staining image and genetic map of human mtDNA. (A) Fluorescent microscopic images of mtDNA (small green dots) (top) and mitochondria (red tubes) (middle) of culture human cells. The merged image is shown (bottom). (B) Schematic presentation of human mtDNA. Gene coding regions for proteins, ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) are shown as blue, orange and red thick lines, respectively. Encoded proteins are ND1, 2, 3, 4, 4L, 5 and 6 subunits of NADH-ubiquinone oxidoreductase (Complex I); cytochrome b (cyt b), a subunit of ubiquinone-cytochrome c oxidoreductase (Complex III); COXI, II and III, subunits of cytochrome c oxidase (Complex IV); A6 and 8, subunits of ATP synthase (Complex V). The 12S rRNA and 16S rRNA are mitochondrial rRNA genes. Mitochondrial tRNA genes are shown by their cognate amino acids with single letter notation. The position of the major noncoding region (NCR) is shown as black lines between tRNAPhe and tRNAPro genes. Transcription from the light (L)-strand promoter (LSP) is responsible for ND6 mRNA and 8 tRNAs. While transcription from the heavy (H)-strand promoter 1 (HSP1) generates 2 rRNAs and 2 tRNAs and terminates at the 3′ end of the 16S rRNA gene, transcription from HSP2 produces almost the entirety of the H-strand transcripts. OH and OL are the major replication initiation sites of H- and L-strands, respectively, under strand-asynchronous mtDNA replication models. The approximate position of OH on mtDNA is shown as an orange oval, representing the unidirectionality of this origin. Ori-b is an initiation region that appears to function as the second initiation site for unidirectional asynchronous synthesis of the H-strand and as a bidirectional initiation origin. The approximate position of Ori-b is shown as an orange/blue oval representing the uni- and bidirectionality of this origin. Conserved sequence block II (CSB II) is shown as a pale-grey rectangle. The position of 7S DNA is shown as a grey bar outside mtDNA. Approximate distances between LSP and CSB II, CSB II and OH and OH and Ori-b in human mtDNA are shown with grey italic numbers.
Because of the vital contributions of mtDNA gene products to the OXPHOS system, mtDNA maintenance is essential for cells and therefore for life. The importance of mtDNA maintenance can be recognized by the fact that mutations in mtDNA, such as base substitutions and deletions, and severe reductions in mtDNA copy number (mtDNA depletion) are associated with human disease [a compendium of mutations in human mtDNA can be found at MITOMAP (‘https://www.mitomap.org/MITOMAP’)]. Such abnormalities impair the OXPHOS system and result in diverse clinical symptoms that frequently affect the central nervous system, skeletal muscle and heart (6, 15, 16). It is obvious that proper maintenance and expression of the mitochondrial genome are dependent on the faithful copying of mtDNA molecules. Mitochondria have their own DNA replication system that involves mitochondria-specific proteins, such as DNA polymerase γ, Twinkle DNA helicase (c10orf2), mitochondrial single-stranded DNA binding protein (mtSSB) and mitochondrial RNA polymerase (POLRMT/mtRNAP) (17, 18). In addition to these proteins, RNase H1, DNA ligase III and topoisomerase 3α (Top3α), which are dually localized to the nucleus and mitochondria are likely to be key components in the mtDNA replication system (19–22). mtDNA defects are often caused by mutations in proteins that are involved in mtDNA replication and nucleotide biosynthesis (23). Unlike nuclear DNA, mtDNA replicates in differentiated cells, such as neuronal and cardiac cells, as well as in proliferating cells (24, 25). This indicates that mtDNA turns over in post-mitotic cells and that mtDNA replication is a continuous, indispensable event throughout the life of an organism. Therefore, the investigation of mtDNA replication mechanisms is a critical topic in mitochondrial biology and medicine. These investigations will also provide valuable information on the universality and diversity of DNA replication systems. A displacement loop model (strand-displacement model) was proposed for the replication of cultured mouse cell mtDNA in 1972 (26), and then it had been widely accepted to be the sole mechanism of mammalian mtDNA replication. This was up until the year 2000, when mtDNA replication intermediates (RIs) were detected that had properties of conventional, coupled leading- and lagging-strand DNA synthesis in human and mouse tissues and cultured human cells (27). In the years following, intensive research by Holt, Jacobs and colleagues and research performed by other groups have made considerable progress in our understanding of mtDNA replication (18, 28–31). This article will address proposed mechanisms of mammalian mtDNA replication; however, we will not go into the protein components of mtDNA replication because they have been discussed well in prior review articles (17, 31, 32).
Early studies and a strand-displacement model of replication
Initial studies of mammalian mtDNA replication were conducted by analysing replicating mtDNA molecules using electron microscopy (EM) long before the publication of human and mouse mtDNA sequences in 1981 (4, 33). In 1972, Vinograd and colleagues (26) proposed a strand-displacement mechanism (SDM), based on the arrangement of replicating mtDNA molecules observed under an electron microscope. The term ‘displacement’ came from the observation of double-forked circular molecules, in which one of the daughter segments was double-stranded and the other single-stranded. The model was then refined by Clayton and others [(34) and references therein]. According to the SDM model (34, 35), mtDNA replication initiates with the synthesis of the H-strand from a specific position in the NCR called OH (Fig. 1B). H-strand synthesis proceeds continuously and unidirectionally (clockwise in Fig. 1B) without synchronous synthesis of the opposite strand. The opposite strand, L-strand, begins to replicate from a physically and temporally distinct position called OL, which is located approximately 11 kb away from OH, and L-strand replication initiates only after H-strand synthesis passes over this point. L-strand synthesis is also continuous and unidirectional (counterclockwise in Fig. 1B). SDM is also referred to as the strand-asynchronous replication mechanism. Characteristic feature of this model is that leading-strand synthesis does not accompany synchronous, or coupled, synthesis of the lagging strand; therefore no Okazaki fragments are involved, unlike nuclear DNA replication. Thus, virtually all RIs will have a maximum of 11 kb single-stranded stretches of displaced parental H-strands, out of the 16.5 kb contour length, during the synthesis of nascent H-strands from OH to OL using parental L-strands as templates.
It should be noted that another line of microscopic examinations by Wolstenholme and colleagues in the late 1960s and early 1970s (36–38) are less recognized at present than the studies described above. The authors observed two types of replicating molecules of rat mtDNA, one in which both daughter segments were fully double-stranded and another where one of the daughter segments was totally or partially single-stranded. Measurements of the single-stranded portion lengths led the researchers to suggest a step-wise (discontinuous) replication model [summarized in (39)]. It appears, however, that this replication model was not examined further, and SDM was a prevailing model of mammalian mtDNA replication for more than 20 years.
Intriguing features of mammalian mtDNA replication
mtDNA replication intermediates, complex and sophisticated
Neutral/neutral two-dimensional agarose gel electrophoresis (2D-AGE) is a preeminent method to study DNA replication [(40, 41) and see references in (42)]. It was developed in the 1980s and has been successfully used to investigate replication in eukaryotes, prokaryotes and viruses. Although DNA images obtained using this method are not always straightforward or simple to interpret, they contain ample information to understand replication mechanisms from the analysed DNA. Two-dimensional gels fractionate branched DNA molecules based not only on their mass but also on their structure, enabling a fine characterization of RIs. For instance, when DNA is replicated via coupled leading- and lagging-strand DNA synthesis (e.g. nuclear DNA), characteristic electrophoresis patterns including Y arcs (fork arcs) and bubble arcs (initiation arcs) are observed (Fig. 2A–C) [for details on 2D-AGE, e.g. (42, 43)]. Importantly, the SDM would not produce standard Y or bubble arcs (see Fig. 2).
Fig. 2.
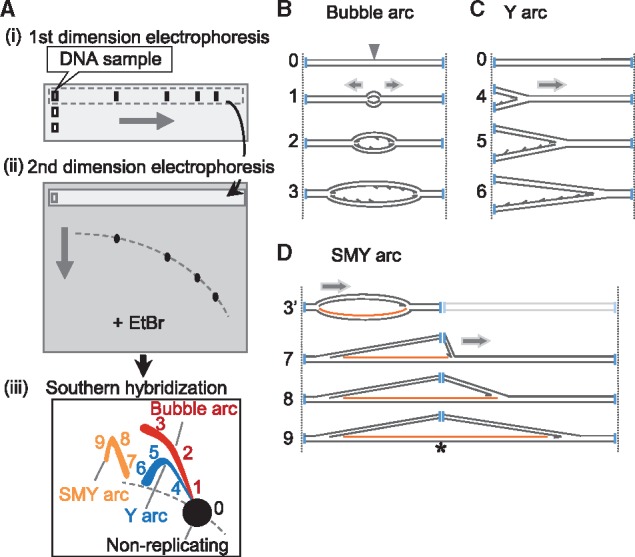
Principles of neutral/neutral two-dimensional agarose gel electrophoresis (2D-AGE). (A) Schematic diagram of 2D-AGE. (i) DNA is digested into appropriately sized fragments using restriction enzyme(s) and is run on a first-dimension gel. (ii) Then, the gel lane is excised and a second-dimension gel containing ethidium bromide (EtBr) is cast around the gel slice. (iii) After electrophoresis, the gel is blotted onto a solid support and DNA fragments of interest are visualized using Southern hybridization. (B–D) Schematic drawings of RIs. (B) Replication initiates at an origin (grey arrowhead) located in the middle of a DNA fragment (0) and progresses bidirectionally (1→3), forming a series of bubble structure molecules until the bubble ‘bursts’ as the replication forks exit the fragment. Restriction digest sites are indicated with vertical blue lines. A complete bubble arc (or initiation arc) is generated from the series of such molecules in 2D-AGE [1→3 in A-(iii)]. (C) Replication is initiated outside the fragment and the replication fork traverses from one end to the other, forming a series of ‘Y’ structure molecules (4→6) which generate a Y arc (or fork arc) in 2D-AGE [4→6 in A-(iii)]. (D) When replication is initiated and proceeds unidirectionally from an origin near the end of the fragment (3′), a bubble structure will be formed before the replisome reaches a restriction site. Then, the presence of ribonucleotides [the ribonucleotide-incorporated strand is represented as orange lines] at a restriction site blocks restriction enzyme digestion (indicated as an asterisk), giving rise to a series of molecules with one moving replication fork and one static fork. After the replisome goes beyond the restriction site, slow-moving Y-like (SMY) arcs will be formed (7→9). SMY arcs are generated [7→9 in A-(iii)] from the blockage of more than one restriction sites [see details in (45)].
At the beginning of the twenty-first century, a surprising study by Holt et al. (27) contained 2D-AGE images with clearly detectable Y and bubble arcs from human and mouse mtDNA. Their observations indicate that RIs with apparent properties of conventional coupled leading- and lagging-strand DNA synthesis are present in mammalian mitochondria. Further, such RIs were resistant to treatments with the single-stranded DNA cutting enzyme, S1 nuclease, which supported the idea that both daughter segments of the replicating mtDNA molecules are fully double-stranded. mtDNA RIs with these properties cannot be reconciled with SDM. Nevertheless, there were many other mtDNA RIs that were sensitive to S1 nuclease [Figure 2 in (27)], and therefore compatible with SDM. However, a follow-up study from the same group revised their initial interpretation of the S1 nuclease-sensitive species and revealed an unexpected characteristic of mtDNA replication (44) (see below).
The second study using 2D-AGE revealed novel Y-like replication arcs with unexpectedly large molecular masses, and notable by their absence were the widely dispersed S1 nuclease-sensitive species that were consistent with SDM replication (44). The crucial difference between the initial and subsequent studies was that the mitochondria of the later studies were of higher purity, suggesting that something was lost or damaged in the cruder preparations of mitochondria. In the initial publication, after the disruption of tissues or cells, differential centrifugation was performed to fractionate crude mitochondrial preparations that were used to study mtDNA RIs (27). In the subsequent publication (44), mtDNA was prepared from mitochondria that were further purified using ultracentrifugation with a sucrose density-gradient (SDG). With the aid of in vitro RNase H treatments, which selectively degrade the RNA in RNA/DNA hybrids, it was shown that the novel Y-like replication arcs (named SMY arcs, for slow-moving Y-like arcs) incorporate ribonucleotides extensively in nascent L-strands and that the RIs are essentially duplex (44). The large mass of the SMY arcs is thus due to ribonucleotides incorporated at restriction enzyme sites that are refractive to digestion, thereby producing SMY arcs that contain two (or more) contiguous restriction fragments (Fig. 2A-(iii) and D). Crucially, mild RNase H treatment of the more highly purified mtDNA converted the SMY arcs into molecular species similar to those seen in mtDNA obtained from crude mitochondrial preparations (44). These findings imply that ribonucleotides within the replicating mtDNA are prone to degradation by trace contaminations of RNase H, or other similar enzymes, in crude mitochondrial preparations and that SDG centrifugation minimizes such nuclease activities, allowing of preparations of mtDNA RIs without substantial loss of ribonucleotides. Partially single-stranded mtDNA molecules, which have been previously observed (27) and ascribed to SDM replication products, were therefore suggested to be molecules that lost ribonucleotides from nascent L-strands during the extraction process from crude mitochondrial preparations. Further characterization of ribonucleotide-rich RIs indicated that ribonucleotides are incorporated across virtually the entire lagging strand (L-strand) while leading-strand (H-strand) DNA synthesis proceeds (Fig. 3). Extraction of replicating molecules from 2D-AGE gels and re-fractionation after denaturation revealed RNAs of 200–600 nucleotides in length from the lagging strands. This led to the proposal that ribonucleotide (or RNA) incorporation occurred throughout the lagging strand, a process termed RITOLS (45). Additionally, it was shown that at a later stage of replication, L-strand RNA was replaced with DNA, suggesting a temporary nature of the RNA strands. It appeared that the OL site and an additional site at around nucleotide 12,800, within the ND5 gene, were the preferred initiation sites of L-strand DNA synthesis in mouse liver mtDNA (45). The presence of additional origin(s) for L-strand DNA synthesis was also suggested by EM observation (46). Further support for the RITOLS mechanism of mtDNA replication came with the demonstration that mtDNA RIs can be immunopurified using a monoclonal antibody specific for RNA/DNA hybrids (47). And in the same study, transmission EM detected fully duplex RIs in highly purified mtDNA samples from cells and tissues, whereas partially single-stranded RIs were detected after treatment with RNase H. Additionally, the single-stranded nascent strands that could be released systematically from SDM-predicted mtDNA RIs by strand separation (branch migration) were undetected in 2D-AGE. Instead, a prominent bubble arc, suggestive of the duplex nature of mtDNA RIs, was observed [Figure 6 in (47)]. These data indicate that mtDNA RIs are essentially duplex throughout their length and that SDM-predicted RIs are not present, or if present, it would account for a fairly small proportion of mtDNA RIs.
Fig. 3.
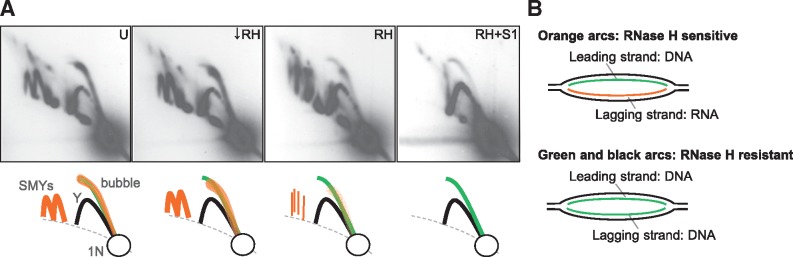
mtDNA RIs are composed of two classes of molecules with different sensitivity to nucleases. (A) 2D-AGE analysis of OH-containing fragments of mouse liver mtDNA digested with the restriction enzyme BclI and treated with nucleases as follows: left untreated (U) or treated with low levels of RNase H (↓RH), standard levels of RNase H (RH) or RH and S1 nuclease (S1) [Figure 5 in (45) for details]. These panels are reuse of those presented in Yasukawa et al. (45). Interpretations of arcs visualized using Southern hybridization are shown below each panel. Prominent bubble arcs (bubble) and SMY arcs (SMYs) were modified by RNase H and were degraded with the addition of S1 nuclease, suggesting that they are ribonucleotide-containing RIs. On the other hand, a fraction of the bubble arcs and Y arcs were resistant to nuclease treatments, indicating that such RIs have properties of those from conventional coupled leading- and lagging-strand DNA synthesis. Nuclease-resistant bubble arcs were also detected from non-NCR-containing fragments. 1N indicates non-replicating fragments. (B) Schematic drawings of proposed molecular structures of arcs observed in (A). ‘Orange arcs’ and ‘green and black arcs’ indicate arcs drawn in orange colour and arcs in green and black colours in (A), respectively.
One might wonder why previous EM studies observed replicating mtDNA molecules with single-stranded regions. The observation can be ascribed to the application of ethidium bromide–CsCl density-gradient centrifugation to mtDNA preparations (26, 46) because it was shown that ethidium bromide–CsCl density-gradient centrifugation can generate partially single-stranded forms from SDG-purified mtDNA RIs (47). Wolstenholme and colleagues also observed partially single-stranded forms along with fully duplex forms (37, 38). It appears that mitochondria were obtained without SDG centrifugation and that their mitochondria were so-called ‘crude preparation’ under the criteria described above. As a result, degradation of the RNA lagging strands may have occurred during mtDNA extraction.
The bootlace model: RNA hybridizes to the lagging-strand template during replication
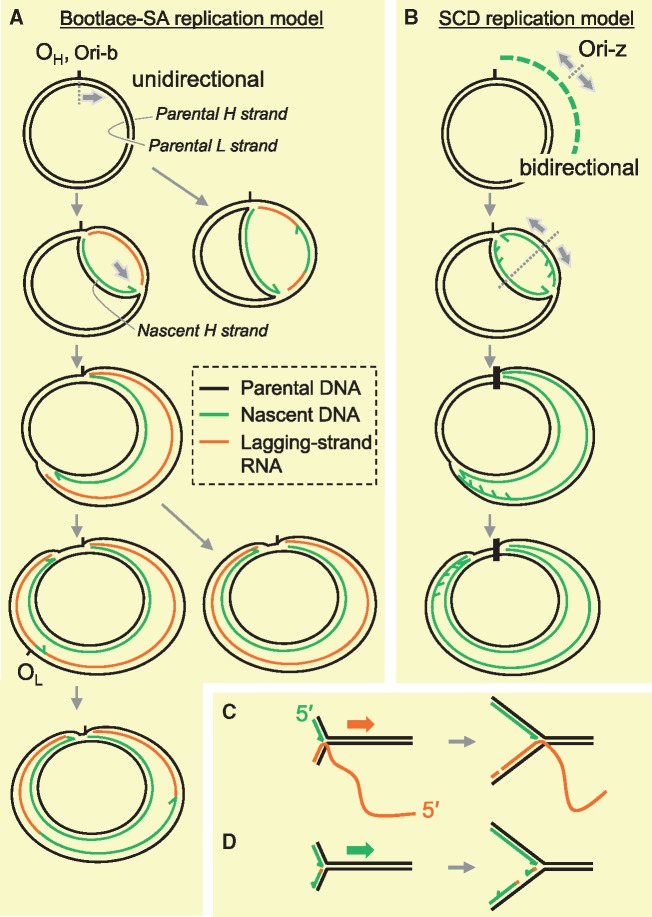
Further to the above investigations, more efforts were made to understand the RNA lagging strands after the proposal of RITOLS. Metabolic labelling of isolated mitochondria suggests that the lagging-strand RNA is not synthesized concurrently with the leading-strand DNA, but pre-existing RNA anneals to the parental (H) strands as replication (synthesis of nascent H-strand DNA) proceeds (48). Also, after the refinement of mtDNA preparation methods to preserve fragile mtDNA RIs, it was observed that the source of RNA incorporated in the RITOLS RIs was preformed L-strand transcripts (48). These findings led to the proposal of a ‘bootlace model’, in which processed transcripts are successively hybridized (or ‘threaded’) on to the lagging-strand template as the replication fork advances, and are replaced by lagging-strand DNA at a later stage of replication (30, 45, 48) (Fig. 4A and C). In short, it was proposed that RITOLS operates via the bootlace mechanism. To the best of our knowledge, the above-described involvement of RNA in DNA replication is unprecedented. This intriguing mechanism might have evolved to maintain the genome integrity in mammalian mitochondria. The replication speed of mammalian mtDNA is much slower than that of the eukaryotic nuclear and bacterial genomes (34); therefore, it would be vital to have a specific mechanism to prevent the long single strands from breaks during replication. Abundant mtDNA transcripts present in mitochondria that have complementary sequences to the H-strands are ideal substances to protect parental H-strands, by forming duplexes with the parental H-strands, while the synthesis of nascent H-strand DNA proceeds on parental L-strands. (Fig. 4A and C). Evolutionary and functional perspectives on the mechanism are discussed extensively in a recent publication (30). If one excludes the RNA involvement and only considers the asynchronous DNA synthesis of H- and L-strands in this replication mechanism, then replicating mtDNA structures are consistent with the SDM model that was proposed over four decades ago. Nevertheless, the series of studies discussed so far in this review strongly suggest that the configurations of mtDNA RIs are explained by the replication model using provisional RNA strands.
Fig. 4.
Proposed models of mtDNA replication. (A, C) Bootlace strand-asynchronous replication (Bootlace-SA replication). Replication of this mode initiates with the synthesis of an H-strand at one of two sites, OH or Ori-b. H-strand DNA synthesis (leading-strand synthesis) proceeds unidirectionally with concurrent incorporation of RNA into the lagging strand. The RNA lagging strand entails hybridization (‘threading’) of processed mitochondrial transcripts to the parental H-strand, as illustrated in (C). Delayed L-strand DNA synthesis is initiated frequently, but not exclusively at OL in mammals and proceeds unidirectionally. RNA lagging strands are removed in this process. (B, D) Strand-coupled DNA replication (SCD replication). Most of the replication within this mechanism initiates from a broad zone of several kilobases, including the gene-encoding region of mtDNA (Ori-z). In this replication mode, replication is bidirectional and the OH region appears to function as a replication fork barrier. Characterization of RIs from this process suggests that syntheses of the leading and lagging strands are synchronous (coupled) and both strands are essentially composed of DNA. The mechanism of lagging-strand synthesis remains to be elucidated. A proposed model is that lagging strands are formed with multiple short DNA fragments (Okazaki fragments) which are primed by an as yet unidentified mitochondrial primase as illustrated in (D), similar to nuclear DNA replication.
Although the RITOLS/bootlace model RIs were unambiguously observed using multiple methods, it is possible that during mtDNA preparation, mitochondrial RNAs might have hybridized adventitiously in vitro to displaced parental H-strands in RIs that were predicted by SDM. To address this possibility, intact cells, isolated mitochondria and tissue homogenates were subjected to nucleic acid cross-linking, and subsequently, the nucleic acids were extracted from the treated samples and the mtDNA RIs were analysed. While RNA-containing RIs (SMY arcs) were modified by in vitro RNase H treatment, they were markedly more resistant to the same treatment after the cross-linking [Figure 1 in (48)], indicating that RNA/DNA hybrid formation in mtDNA RIs is an in vivo event; in other words, the hybrids were not formed during or after mtDNA extraction but were already present before the cross-linking step (48). It should be noted that the SDM model suggests that mtSSB coats the displaced single-stranded parental H-strands (the lagging-strand template) until the synthesis of the second strand (L-strand) proceeds using the parental H-strands. This idea was supported by an early EM study (49); one daughter segment was observed to be bound by mtSSB in apparently replicating mtDNA that was extracted as a mtDNA-protein complex from rat mitochondria. To reconcile this observation with the RNA bootlace model, a possible explanation could be that RNA lagging strands were degraded during the lysis of mitochondria prepared by differential centrifugation (i.e. crude mitochondria) (49) and that the resulting single-stranded portions were occupied by mtSSB. A recent study of mtSSB-directed mtDNA immunoprecipitation suggests strong binding bias of mtSSB toward the major arc region (the ‘clockwise’ region from OH to OL in Fig. 1B) of the H-strand in cultured cells (50). The result appears to agree with the mtSSB-coating SDM model. However, it is not known whether the majority of mtDNA RIs were captured by the immunoprecipitation. It is possible that the data might have been generated with only a minor fraction of mtDNA RIs that were bound by many mtSSB molecules, owing to the accidental slow ‘lay-down’ of RNA lagging strands. This inference seems to be consistent with the result that the cross-linked, RNase H-treated material did not perfectly reproduce the 2D-AGE pattern of untreated mtDNA RI in the nucleic acid cross-linking experiments (48) because the result might reflect a situation where there are some (random) gaps in the hybridized RNA and these are occupied by mtSSB.
In summary, two classes of RIs are proposed to be present in mammalian mtDNA (27, 45, 51) (Figs 3 and 4). One class is derived from strand-asynchronous DNA synthesis involving the hybridization of processed mitochondrial transcripts as provisional lagging strands (Fig. 4A and C), discussed in detail. The other class has many properties of coupled leading- and lagging-strand DNA synthesis (Fig. 4B). For simplicity, we refer to the two replication mechanisms as bootlace strand-asynchronous replication (Bootlace-SA replication) and strand-coupled DNA synthesis (SCD replication), respectively, in this article. It remains to be determined whether the SCD replication is the same as conventional coupled leading- and lagging-strand DNA synthesis with regards to lagging-strand priming. Two possibilities are considered; [1] one is that lagging-strand DNA synthesis is primed by multiple short RNA primers provided by a primase, similar to nuclear DNA replication, which is operated via conventional coupled leading- and lagging-strand DNA synthesis (Fig. 4D). [2] The other is that an enzyme(s), such as RNase H1, rapidly processes hybridized transcripts to provide multiple short RNAs that can then serve as primers. However, it is difficult to answer why this process does not take place in the former replication mode. A mammalian mitochondrial primase, an essential enzyme required for that model [1], has not been identified. POLRMT, which is responsible for polycistronic transcription of mtDNA, is a potential candidate because it was shown to synthesize short RNAs on single-stranded DNA templates, independent of LSP sequence in vitro (50, 52, 53). However, it is not known whether the in vitro activity reflects the in vivo properties of POLRMT. Despite uncertainty about the existence of a mammalian mitochondrial primase, we speculate that mtDNA RIs from SCD replication in animal tissues and cells cultured under normal conditions are products of conventional coupled leading- and lagging-strand DNA synthesis. Our speculation is based on differences in the directionality of initiation, from which two classes of mtDNA RIs appear to be derived (see below).
Unidirectional and bidirectional initiation of mtDNA replication
Initiation of replication is a critical step in the replication process because it determines whether the DNA will be duplicated or not. Using 2D-AGE and ligation-mediated PCR, it was proposed that the two origins appear to be used for initiation of leading strand (H-strand) synthesis in Bootlace-SA replication (45). These two origins are OH (54, 55) and less frequently Ori-b, which was identified approximately 0.5 kb downstream from OH within the NCR (51) (Fig. 1B). They are not single nucleotide positions but consist of several tightly clustered sites. Replication following this mechanism proceeds unidirectionally and generates RNase H-sensitive bubble arcs from fragments containing OH and/or Ori-b at one end of the fragments (45) (Figs 2D-3’ and 3). It has been proposed that transcription from the LSP, which is located upstream from OH (Fig. 1B), provides a long RNA strand as a primer for the initiation of H-strand DNA synthesis from OH (56). A recent report (21) demonstrated the presence of RNA-DNA covalently attached molecules whose 5′ RNA ends are mapped to LSP, with RNA to DNA transition sites located at OH and Ori-b in cultured mouse cells in which endogenous RNase H1 was inducibly knocked out. The presence of such molecules assures the validity of the LSP-dependent priming model and involvement of RNase H1 in the removal of the RNA after priming. Further, it was suggested that suppression of mitochondrial transcription initiation complexes, through simultaneous depletion of two components of the initiation complexes, mitochondrial transcription factor B2 (TFB2M) and POLRMT (31, 57), lowered the intensities of SMY arc signals in 2D-AGE Southern hybridization in cultured human cells (58), supporting the idea that RNA from LSP primes H-strand DNA synthesis in Bootlace-SA replication. It remains to be clarified how the fate of a transcription event from LSP is determined to be used for replication primer formation or genome-length transcription. Recently, mitochondrial transcription elongation factor (TEFM) (59) was proposed to function in this determination of ‘replication-transcription switching’ based on an in vitro assay in which TEFM was able to prevent transcription termination at a CG-rich element called conserved sequence block II (CSB II) (60) (Fig. 1B). It should be noted that OH is located approximately 100 nucleotides away from CSB II in human mtDNA and that Ori-b is located about 500 nucleotides further downstream. The idea that CSB II is an RNA-DNA transition site for H-strand synthesis could be supported by a recent report that the depletion of the MGME1 nuclease in cultured cells results in the emergence of multiple 5′-DNA ends that were mapped to the region upstream from OH, including CSB II (61). At this time, however, it is difficult to reconcile the ‘switching at CSB II’ model with the detection of RNA-DNA covalently attached molecules whose (major) transition sites are OH and Ori-b, upon loss of RNase H1 activity (21). Further studies will be required to understand the regulation of replication and transcription.
Detection of bubble (initiation) arcs on 2D-AGE is a hallmark of the presence of replication origins in the analysed fragments (40, 62). An important finding came from examining various restriction-digested mtDNA fragments from human, rodent and chicken tissues, where bubble arcs were observed in fragments lacking OH and even in those lacking the entire NCR, indicating that mtDNA replication can commence from positions other than OH (63, 64). Careful inspection of 2D-AGE images suggested that these replication events are bidirectional and that most of them occur across a broad zone of several kilobases (Ori-z) (63, 64) (Fig. 4B). Furthermore, bubble arcs of this type are resistant to RNase H treatments, which is indicative of SCD replication (45), and the NCR appears to function as a replication termination region in these replication events (63, 64). Particularly significant in these findings are the following two points: it was demonstrated that OH is not the sole replication initiation position and that a bidirectional replication mechanism exists in mammalian and avian mitochondria. Outstanding issues in these bidirectional initiations are molecular processes that include the details of DNA unwinding at the initiation points and initial RNA primer synthesis on both the leading strands. Further research is required to identify the protein factors involved in these processes.
Variation and manipulation of mtDNA replication
When investigating 2D-AGE images of mtDNA RIs, one notices that the relative intensities vary between the two classes of RIs, Bootlace-SA RIs and SCD RIs. The Bootlace-SA RIs are abundant in mtDNA from rodent livers and cultured human 143B osteosarcoma (143B) and HeLa cells (45, 47, 48, 51). Metabolic labelling of isolated rat liver mitochondria showed more efficient labelling in Bootlace-SA RIs than SCD RIs, suggesting more rapid or frequent replication of the former in rat liver mitochondria (48). In contrast, SCD RIs appear to give stronger signals in thymidine kinase 1-deficient 143B cells [143B (TK−)] (20) and in a cultured cell line (Yasukawa, unpublished observation). It is currently unknown why variations of the ratio of the two classes of RIs exist. Taking into consideration that mtDNA replicates not only in proliferating cells but also in non-dividing cells, and that mitochondrial status differs substantially between tissues and cells, it would be reasonable to infer that the mtDNA replication system needs plasticity and fine regulation to maintain genomic stability and appropriate copy number under various conditions. In support of this idea, as well as adding more complexity to mammalian mtDNA replication, analysis of human heart mtDNA revealed that θ-type RIs (i.e. the RIs described so far) are absent, but instead a prominent arc was detected on 2D-AGE (65, 66), a so-called 2n spike or X arc, which arises from molecules containing four-way junctions (43). Also, molecules forming complex junctional networks were found to be abundantly present in human heart mtDNA preparations. These data imply that human heart mtDNA replication proceeds via a non-θ-type replication mechanism (66), the details of which remain unknown. Additionally, mtDNA multigenomic complexes found in the human heart were also observed in the human and mouse brains though to a lesser extent (66), and variations in the relative abundance of Bootlace-SA RIs, SCD RIs and 2n spikes were observed between six different tissues in mice (67).
Plasticity of mtDNA replication systems can also be recognized by substantial changes in mtDNA RI patterns in143B and HeLa cells, in response to transient depletion of mtDNA using reversible mtDNA replication inhibitors, ethidium bromide and 2′,3′-dideoxycytosine (27, 51). When cultured cells are treated with these chemicals within a certain range of concentrations, mtDNA replication is specifically inhibited without apparent toxicity to cells. Using this method, mtDNA copy numbers can be reduced to 10% of normal levels after several days of incubation, and upon withdrawal of the chemicals from the medium, cells replicate mtDNA to restore copy number. While mtDNA RIs in 143B and HeLa cells cultured in normal medium are a mixture of those produced by Bootlace-SA replication and SCD replication, RIs were shifted toward the latter class substantially during the mtDNA recovery stage (27, 51). Also, analyses of mtDNA from 143B cells showed that the shift was accompanied by a prominent bidirectional origin close to the tRNAPro gene end of the NCR, which was named Ori-b or cluster II (51) (Fig. 1B). We already discussed Ori-b as a second unidirectional origin of asynchronous H-strand synthesis (45), but it was initially identified as a bidirectional origin under mtDNA recovery (51). From these findings, two possibilities can be inferred. [1] Bootlace-SA replication and SCD replication are distinct modes of replication and Ori-b functions as initiation sites for both. SCD replication uses Ori-b frequently under the mtDNA recovery stage as a result of contraction of the initiation zone found in animal tissue mtDNA. [2] SCD-type RIs emerge under mtDNA recovery as a consequence of induced rapid maturation of the L-strand RNA in Bootlace-SA replication. During mtDNA restoration, the transient RNA lagging strands are processed rapidly enough to provide multiple RNA primers, and frequent short DNA fragments are synthesized. This can give rise to RIs that are similar in structure, but not necessarily in mechanism, to those generated by SCD replication in animal tissue or in cells cultured under normal conditions. Apart from the apparent bidirectional initiation from Ori-b, this possibility would nicely explain the phenomenon. Examination of possibilities [1] and [2] will deepen our understanding of mtDNA replication regulation.
Third strands present within the NCR
As discussed above, the NCR contains cis-elements for replication and transcription. Another notable feature of the NCR in mammals is the formation of a short triple-stranded region, or a displacement loop (D-loop) (68–70). The third strand, designated 7S DNA, has an H-strand sequence spanning from OH toward the tRNAPro gene end of the NCR (Fig. 1B) and is approximately 650 nucleotides long in humans. Frequencies of mtDNA molecules containing 7S DNA are rather variable [Table 1 in (70)]. Although the D-loop was discovered over four decades ago, the role(s) remains unclear. It was hypothesized that 7S DNA is an aborted product or a primer for H-strand DNA synthesis or that 7S DNA is involved in the control of transcription initiation (34). When TFB2M and POLRMT, components of the transcription initiation complexes, were acutely depleted using siRNA in HeLa cells, mitochondrial mRNA contents, 7S DNA levels and mtDNA replication were differentially influenced (58). This result might be an implication of regulatory associations between 7S DNA synthesis and replication and transcription of mtDNA. Also, 7S DNA, or the D-loop, was proposed to be the binding centre for proteins that regulate dynamics of mitochondrial nucleoids (71, 72). Additionally, a recent publication (73) showed that a fraction of mtDNA molecules has RNA strands that are complementary to 7S DNA and form an ‘R-loop’ in the NCR. This non-coding RNA was named L-strand, control region RNA (LC-RNA) (73), and the LC-RNA or the R-loop might directly or indirectly contribute to segregation of mtDNA molecules in vivo. We should also mention that Top3α was recently suggested to be essential for mtDNA segregation after replication (22). Investigation of functional relationships between these third strands (7S DNA and LC-RNA) and the protein factors proposed to be involved in mtDNA segregation may provide a deeper understanding of mtDNA maintenance.
Perspectives
It is now clear that replication of mammalian mtDNA is a much more complex and finely regulated system than previously considered, and there are still many questions to be explored. Given the variability of tissues and cells in the requirement of mitochondrial energy production and metabolic state, one can speculate that mitochondria have the capacity for modulating mtDNA replication dramatically and that such modulation is essential for mitochondrial function. mtDNA replication modulation is likely linked to the variation in patterns of mtDNA organization. While mtDNA frequently exists as a simple circle, many molecules of mtDNA are organized as complex networks containing abundant junctions in the human heart (66), which have some similarities to mtDNA organization in plants and malarial parasites (74, 75). Further studies of the mechanistic details of each replication mode, including identification of all protein factors involved, are certainly required. Why and how mitochondria operate and regulate multiple DNA replication mechanisms are imperative open questions. A deeper understanding of the mtDNA replication system will offer prospects of increasing our understanding of the molecular basis of human diseases with mtDNA abnormalities.
Acknowledgements
This work was supported in part by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science [JSPS KAKENHI grant numbers JP26840011 and JP17K07504 to T.Y. and JP17H01550 to D.K.].
Conflict of Interest
None declared.
Glossary
Abbreviations
- Bootlace-SA replication
bootlace strand-asynchronous replication
- CSB II
conserved sequence block II
- 2D-AGE
two-dimensional agarose gel electrophoresis
- D-loop
displacement loop
- EM
electron microscopy
- HSP
heavy-strand promoter
- H-strand
heavy-strand
- LC-RNA
L-strand, control region RNA
- LSP
light-strand promoter
- L-strand
light-strand
- mtDNA
mitochondrial DNA
- mtSSB
mitochondrial single-stranded DNA binding protein
- NCR
major noncoding region
- OXPHOS
oxidative phosphorylation
- POLRMT
mitochondrial RNA polymerase
- RIs
replication intermediates
- RITOLS
ribonucleotide incorporation throughout the lagging strand
- rRNA
ribosomal RNA
- SCD replication
strand-coupled DNA synthesis
- SDG
sucrose density-gradient
- SDM
strand-displacement mechanism
- SMY arcs
slow-moving Y-like arcs
- TEFM
mitochondrial transcription elongation factor
- TFB2M
mitochondrial transcription factor B2
- Top3α
topoisomerase 3α
- tRNA
transfer RNA
References
- 1. Benda C. (1898) Ueber die Spermatogenese der Vertebraten und höherer Evertebraten, II. Theil: Die Histiogenese der Spermien. Arch. Anat. Physiol. 73, 393–398 [Google Scholar]
- 2. Ernster L., Schatz G. (1981) Mitochondria: a historical review. J. Cell Biol. 91, 227s–255s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nass M.M., Nass S. (1963) Intramitochondrial fibers with DNA characteristics. I. Fixation and electron staining reactions. J. Cell Biol. 19, 593–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., Schreier P.H., Smith A.J., Staden R., Young I.G. (1981) Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 [DOI] [PubMed] [Google Scholar]
- 5. Spelbrink J.N. (2010) Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life 62, 19–32 [DOI] [PubMed] [Google Scholar]
- 6. Schon E.A., DiMauro S., Hirano M. (2012) Human mitochondrial DNA: roles of inherited and somatic mutations. Nat. Rev. Genet. 13, 878–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calvo S.E., Clauser K.R., Mootha V.K. (2016) MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–D1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mercer T.R., Neph S., Dinger M.E., Crawford J., Smith M.A., Shearwood A.M., Haugen E., Bracken C.P., Rackham O., Stamatoyannopoulos J.A., Filipovska A., Mattick J.S. (2011) The human mitochondrial transcriptome. Cell 146, 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Asin-Cayuela J., Gustafsson C.M. (2007) Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 32, 111–117 [DOI] [PubMed] [Google Scholar]
- 10. Scarpulla R.C. (2008) Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 11. Bestwick M.L., Shadel G.S. (2013) Accessorizing the human mitochondrial transcription machinery. Trends Biochem. Sci. 38, 283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nagaike T., Suzuki T., Ueda T. (2008) Polyadenylation in mammalian mitochondria: insights from recent studies. Biochim. Biophys. Acta 1779, 266–269 [DOI] [PubMed] [Google Scholar]
- 13. Suzuki T., Nagao A., Suzuki T. (2011) Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 45, 299–329 [DOI] [PubMed] [Google Scholar]
- 14. Bohnsack M.T., Sloan K.E. (2018) The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease. Cell. Mol. Life Sci. 75, 241–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greaves L.C., Reeve A.K., Taylor R.W., Turnbull D.M. (2012) Mitochondrial DNA and disease. J. Pathol. 226, 274–286 [DOI] [PubMed] [Google Scholar]
- 16. Ylikallio E., Suomalainen A. (2012) Mechanisms of mitochondrial diseases. Ann. Med. 44, 41–59 [DOI] [PubMed] [Google Scholar]
- 17. Wanrooij S., Falkenberg M. (2010) The human mitochondrial replication fork in health and disease. Biochim. Biophys. Acta 1797, 1378–1388 [DOI] [PubMed] [Google Scholar]
- 18. Holt I.J., Reyes A. (2012) Human mitochondrial DNA replication. Cold Spring Harb. Perspect. Biol. 4, a012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simsek D., Furda A., Gao Y., Artus J., Brunet E., Hadjantonakis A.K., Van Houten B., Shuman S., McKinnon P.J., Jasin M. (2011) Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature 471, 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruhanen H., Ushakov K., Yasukawa T. (2011) Involvement of DNA ligase III and ribonuclease H1 in mitochondrial DNA replication in cultured human cells. Biochim. Biophys. Acta 1813, 2000–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holmes J.B., Akman G., Wood S.R., Sakhuja K., Cerritelli S.M., Moss C., Bowmaker M.R., Jacobs H.T., Crouch R.J., Holt I.J. (2015) Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication. Proc. Natl. Acad. Sci. U S A 112, 9334–9339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nicholls T.J., Nadalutti C.A., Motori E., Sommerville E.W., Gorman G.S., Basu S., Hoberg E., Turnbull D.M., Chinnery P.F., Larsson N.G., Larsson E., Falkenberg M., Taylor R.W., Griffith J.D., Gustafsson C.M. (2018) Topoisomerase 3alpha is required for decatenation and segregation of human mtDNA. Mol. Cell 69, 9–23 e26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Young M.J., Copeland W.C. (2016) Human mitochondrial DNA replication machinery and disease. Curr. Opin. Genet. Dev. 38, 52–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Magnusson J., Orth M., Lestienne P., Taanman J.W. (2003) Replication of mitochondrial DNA occurs throughout the mitochondria of cultured human cells. Exp. Cell Res. 289, 133–142 [DOI] [PubMed] [Google Scholar]
- 25. Ylikallio E., Tyynismaa H., Tsutsui H., Ide T., Suomalainen A. (2010) High mitochondrial DNA copy number has detrimental effects in mice. Hum. Mol. Genet. 19, 2695–2705 [DOI] [PubMed] [Google Scholar]
- 26. Robberson D.L., Kasamatsu H., Vinograd J. (1972) Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc. Natl. Acad. Sci. U S A 69, 737–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holt I.J., Lorimer H.E., Jacobs H.T. (2000) Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell 100, 515–524 [DOI] [PubMed] [Google Scholar]
- 28. Yasukawa T., Poulton J. (2009) Mitochondrial DNA replication In Molecular Themes in DNA Replication (Cox L.S., ed.) pp. 316–345, Royal Society of Chemistry, London [Google Scholar]
- 29. McKinney E.A., Oliveira M.T. (2013) Replicating animal mitochondrial DNA. Genet. Mol. Biol. 36, 308–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holt I.J., Jacobs H.T. (2014) Unique features of DNA replication in mitochondria: a functional and evolutionary perspective. Bioessays 36, 1024–1031 [DOI] [PubMed] [Google Scholar]
- 31. Gustafsson C.M., Falkenberg M., Larsson N.G. (2016) Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 85, 133–160 [DOI] [PubMed] [Google Scholar]
- 32. Kasiviswanathan R., Collins T.R., Copeland W.C. (2012) The interface of transcription and DNA replication in the mitochondria. Biochim. Biophys. Acta 1819, 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bibb M.J., Van Etten R.A., Wright C.T., Walberg M.W., Clayton D.A. (1981) Sequence and gene organization of mouse mitochondrial DNA. Cell 26, 167–180 [DOI] [PubMed] [Google Scholar]
- 34. Clayton D.A. (1982) Replication of animal mitochondrial DNA. Cell 28, 693–705 [DOI] [PubMed] [Google Scholar]
- 35. Kasamatsu H., Grossman L.I., Robberson D.L., Watson R., Vinograd J. (1974) The replication and structure of mitochondrial DNA in animal cells. Cold Spring Harb. Symp. Quant. Biol. 38, 281–288 [DOI] [PubMed] [Google Scholar]
- 36. Kirschner R.H., Wolstenholme D.R., Gross N.J. (1968) Replicating molecules of circular mitochondrial DNA. Proc. Natl. Acad. Sci. U S A 60, 1466–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wolstenholme D.R., Koike K., Cochran-Fouts P. (1973) Single strand-containing replicating molecules of circular mitochondrial DNA. J. Cell Biol. 56, 230–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koike K., Wolstenholme D.R. (1974) Evidence for discontinuous replication of circular mitochondrial DNA molecules from Novikoff rat ascites hepatoma cells. J. Cell Biol. 61, 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wolstenholme D.R., Koike K., Cochran-Fouts P. (1974) Replication of mitochondrial DNA: replicative forms of molecules from rat tissues and evidence for discontinuous replication. Cold Spring Harb. Symp. Quant. Biol. 38, 267–280 [DOI] [PubMed] [Google Scholar]
- 40. Brewer B.J., Fangman W.L. (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51, 463–471 [DOI] [PubMed] [Google Scholar]
- 41. Brewer B.J., Fangman W.L. (1988) A replication fork barrier at the 3' end of yeast ribosomal RNA genes. Cell 55, 637–643 [DOI] [PubMed] [Google Scholar]
- 42. Reyes A., Yasukawa T., Holt I.J. (2007) Analysis of replicating mitochondrial DNA by two-dimensional agarose gel electrophoresis. Methods Mol. Biol. 372, 219–232 [DOI] [PubMed] [Google Scholar]
- 43. Friedman K.L., Brewer B.J. (1995) Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 262, 613–627 [DOI] [PubMed] [Google Scholar]
- 44. Yang M.Y., Bowmaker M., Reyes A., Vergani L., Angeli P., Gringeri E., Jacobs H.T., Holt I.J. (2002) Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell 111, 495–505 [DOI] [PubMed] [Google Scholar]
- 45. Yasukawa T., Reyes A., Cluett T.J., Yang M.Y., Bowmaker M., Jacobs H.T., Holt I.J. (2006) Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 25, 5358–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brown T.A., Cecconi C., Tkachuk A.N., Bustamante C., Clayton D.A. (2005) Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 19, 2466–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pohjoismäki J.L., Holmes J.B., Wood S.R., Yang M.Y., Yasukawa T., Reyes A., Bailey L.J., Cluett T.J., Goffart S., Willcox S., Rigby R.E., Jackson A.P., Spelbrink J.N., Griffith J.D., Crouch R.J., Jacobs H.T., Holt I.J. (2010) Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J. Mol. Biol. 397, 1144–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reyes A., Kazak L., Wood S.R., Yasukawa T., Jacobs H.T., Holt I.J. (2013) Mitochondrial DNA replication proceeds via a ′bootlace′ mechanism involving the incorporation of processed transcripts. Nucleic Acids Res. 41, 5837–5850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Van Tuyle G.C., Pavco P.A. (1985) The rat liver mitochondrial DNA-protein complex: displaced single strands of replicative intermediates are protein coated. J. Cell Biol. 100, 251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miralles Fuste J., Shi Y., Wanrooij S., Zhu X., Jemt E., Persson O., Sabouri N., Gustafsson C.M., Falkenberg M. (2014) In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet. 10, e1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yasukawa T., Yang M.Y., Jacobs H.T., Holt I.J. (2005) A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol. Cell 18, 651–662 [DOI] [PubMed] [Google Scholar]
- 52. Wanrooij S., Fuste J.M., Farge G., Shi Y., Gustafsson C.M., Falkenberg M. (2008) Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl. Acad. Sci. U S A 105, 11122–11127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fukuoh A., Ohgaki K., Hatae H., Kuraoka I., Aoki Y., Uchiumi T., Jacobs H.T., Kang D. (2009) DNA conformation-dependent activities of human mitochondrial RNA polymerase. Genes Cells 14, 1029–1042 [DOI] [PubMed] [Google Scholar]
- 54. Crews S., Ojala D., Posakony J., Nishiguchi J., Attardi G. (1979) Nucleotide sequence of a region of human mitochondrial DNA containing the precisely identified origin of replication. Nature 277, 192–198 [DOI] [PubMed] [Google Scholar]
- 55. Kang D., Miyako K., Kai Y., Irie T., Takeshige K. (1997) In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J. Biol. Chem. 272, 15275–15279 [DOI] [PubMed] [Google Scholar]
- 56. Shadel G.S., Clayton D.A. (1997) Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 66, 409–435 [DOI] [PubMed] [Google Scholar]
- 57. Hillen H.S., Morozov Y.I., Sarfallah A., Temiakov D., Cramer P. (2017) Structural basis of mitochondrial transcription initiation. Cell 171, 1072–1081.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Qu J., Yasukawa T., Kang D. (2016) Suppression of mitochondrial transcription initiation complexes changes the balance of replication intermediates of mitochondrial DNA and reduces 7S DNA in cultured human cells. J. Biochem. 160, 49–57 [DOI] [PubMed] [Google Scholar]
- 59. Minczuk M., He J., Duch A.M., Ettema T.J., Chlebowski A., Dzionek K., Nijtmans L.G., Huynen M.A., Holt I.J. (2011) TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 39, 4284–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Agaronyan K., Morozov Y.I., Anikin M., Temiakov D. (2015) Replication-transcription switch in human mitochondria. Science 347, 548–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nicholls T.J., Zsurka G., Peeva V., Scholer S., Szczesny R.J., Cysewski D., Reyes A., Kornblum C., Sciacco M., Moggio M., Dziembowski A., Kunz W.S., Minczuk M. (2014) Linear mtDNA fragments and unusual mtDNA rearrangements associated with pathological deficiency of MGME1 exonuclease. Hum. Mol. Genet. 23, 6147–6162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brewer B.J., Fangman W.L. (1991) Mapping replication origins in yeast chromosomes. Bioessays 13, 317–322 [DOI] [PubMed] [Google Scholar]
- 63. Bowmaker M., Yang M.Y., Yasukawa T., Reyes A., Jacobs H.T., Huberman J.A., Holt I.J. (2003) Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J. Biol. Chem. 278, 50961–50969 [DOI] [PubMed] [Google Scholar]
- 64. Reyes A., Yang M.Y., Bowmaker M., Holt I.J. (2005) Bidirectional replication initiates at sites throughout the mitochondrial genome of birds. J. Biol. Chem. 280, 3242–3250 [DOI] [PubMed] [Google Scholar]
- 65. Kajander O.A., Karhunen P.J., Holt I.J., Jacobs H.T. (2001) Prominent mitochondrial DNA recombination intermediates in human heart muscle. EMBO Rep. 2, 1007–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pohjoismäki J.L., Goffart S., Tyynismaa H., Willcox S., Ide T., Kang D., Suomalainen A., Karhunen P.J., Griffith J.D., Holt I.J., Jacobs H.T. (2009) Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J. Biol. Chem. 284, 21446–21457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Herbers E., Kekalainen N.J., Hangas A., Pohjoismäki J.L., Goffart S. (2018) Tissue specific differences in mitochondrial DNA maintenance and expression. Mitochondrion. 10.1016/j.mito.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 68. Arnberg A., van Bruggen E.F., Borst P. (1971) The presence of DNA molecules with a displacement loop in standard mitochondrial DNA preparations. Biochim. Biophys. Acta 246, 353–357 [DOI] [PubMed] [Google Scholar]
- 69. Kasamatsu H., Robberson D.L., Vinograd J. (1971) A novel closed-circular mitochondrial DNA with properties of a replicating intermediate. Proc. Natl. Acad. Sci. U S A 68, 2252–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nicholls T.J., Minczuk M. (2014) In D-loop: 40 years of mitochondrial 7S DNA. Exp. Gerontol. 56, 175–181 [DOI] [PubMed] [Google Scholar]
- 71. He J., Mao C.C., Reyes A., Sembongi H., Di Re M., Granycome C., Clippingdale A.B., Fearnley I.M., Harbour M., Robinson A.J., Reichelt S., Spelbrink J.N., Walker J.E., Holt I.J. (2007) The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 176, 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Di Re M., Sembongi H., He J., Reyes A., Yasukawa T., Martinsson P., Bailey L.J., Goffart S., Boyd-Kirkup J.D., Wong T.S., Fersht A.R., Spelbrink J.N., Holt I.J. (2009) The accessory subunit of mitochondrial DNA polymerase gamma determines the DNA content of mitochondrial nucleoids in human cultured cells. Nucleic Acids Res. 37, 5701–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Akman G., Desai R., Bailey L.J., Yasukawa T., Dalla Rosa I., Durigon R., Holmes J.B., Moss C.F., Mennuni M., Houlden H., Crouch R.J., Hanna M.G., Pitceathly R.D., Spinazzola A., Holt I.J. (2016) Pathological ribonuclease H1 causes R-loop depletion and aberrant DNA segregation in mitochondria. Proc. Natl. Acad. Sci. U S A 113, E4276–E4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Preiser P.R., Wilson R.J., Moore P.W., McCready S., Hajibagheri M.A., Blight K.J., Strath M., Williamson D.H. (1996) Recombination associated with replication of malarial mitochondrial DNA. EMBO J. 15, 684–693 [PMC free article] [PubMed] [Google Scholar]
- 75. Backert S., Borner T. (2000) Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr. Genet. 37, 304–314 [DOI] [PubMed] [Google Scholar]