ABSTRACT
Nitric oxide (NO·) produced by mammalian cells exerts antimicrobial actions that result primarily from the modification of protein thiols (S-nitrosylation) and metal centers. A comprehensive approach was used to identify novel targets of NO· in Salmonella enterica serovar Typhimurium (S. Typhimurium). Newly identified targets include zinc metalloproteins required for DNA replication and repair (DnaG, PriA, and TopA), protein synthesis (AlaS and RpmE), and various metabolic activities (ClpX, GloB, MetE, PepA, and QueC). The cytotoxic actions of free zinc are mitigated by the ZntA and ZitB zinc efflux transporters, which are required for S. Typhimurium resistance to zinc overload and nitrosative stress in vitro. Zinc efflux also ameliorates NO·-dependent zinc mobilization following internalization by activated macrophages and is required for virulence in NO·-producing mice, demonstrating that host-derived NO· causes zinc stress in intracellular bacteria.
KEYWORDS: Salmonella, nitric oxide, pathogenesis, transporters, zinc homeostasis
IMPORTANCE
Nitric oxide (NO·) is produced by macrophages in response to inflammatory stimuli and restricts the growth of intracellular bacteria. Mechanisms of NO·-dependent antimicrobial actions are incompletely understood. Here, we show that zinc metalloproteins are important targets of NO· in Salmonella, including the DNA replication proteins DnaG and PriA, which were hypothesized to be NO· targets in earlier studies. Like iron, zinc is a cofactor for several essential proteins but is toxic at elevated concentrations. This study demonstrates that NO· mobilizes free zinc in Salmonella and that specific efflux transporters ameliorate the cytotoxic effects of free zinc during infection.
INTRODUCTION
Nitric oxide (NO·) is generated by the inducible nitric oxide synthase (iNOS) of phagocytic cells during infection. Sustained production of NO. restricts the replication of intracellular bacteria, and iNOS deficiency increases host susceptibility to infection (1, 46). NO· and related reactive nitrogen species (RNS) covalently modify thiols, tyrosine residues, metal centers, nucleotides, and lipids to impair bacterial growth and modulate bacterial virulence (2, 3, 7, 48–50). S-Nitrosylation is a reversible thiol modification that can disrupt protein function, sometimes by interfering with disulfide bonding or mobilizing metal cofactors. Although some examples of S-nitrosylation have been demonstrated, the full extent of bacterial proteins subject to S-nitrosylation and the protein targets responsible for the antimicrobial actions of NO· are incompletely understood.
Zinc metalloproteins typically comprise 3 to 5% of the bacterial proteome (4, 51). A previous study has shown that intracellular zinc can be mobilized during treatment of bacterial cells with an NO· donor (5). Zinc is typically coordinated by either histidine or cysteine residues, the latter representing potential targets of S-nitrosylation (6, 7). In Salmonella enterica serovar Typhimurium, intracellular zinc is tightly restricted by two zinc-sensing transcriptional regulators and multiple uptake and efflux transport systems. S. Typhimurium acquires zinc from the environment via the high-affinity ZnuABC system and the lower-affinity ZupT transporter (8, 52). Expression of znuABC is regulated by Zur, a member of the Fur family of bacterial regulators (8, 53–55). Under zinc-replete conditions, Zur binds to zinc in the cytoplasm and represses znuABC transcription. Under low-zinc conditions, apo-Zur is incapable of DNA binding, and the repression of znuABC transcription is relieved. The regulation of zupT expression is uncharacterized and may be constitutive (9). Cells typically maintain intracellular free zinc at picomolar to femtomolar levels (10, 11). As free zinc levels rise, zinc is bound by the transcriptional activator ZntR, which activates transcription of the high-affinity efflux transporter ZntA (12, 56, 57). Additional efflux transporters, called ZntB, ZitB, and YiiP, have been described in Escherichia coli and S. Typhimurium (13, 21, 22, 58). Recent work has suggested that ZitB acts in conjunction with ZntA to protect S. Typhimurium from zinc overload, but the contribution of these transport systems in maintaining zinc homeostasis during infection in vivo and the mechanisms by which their expression is regulated have yet to be fully elucidated (14).
The ZnuABC and ZupT zinc uptake systems are required for full virulence of S. Typhimurium (15, 59–61). However, zinc efflux has only recently been implicated in Salmonella virulence (14). We hypothesized that zinc mobilized from metalloproteins by phagocyte-derived NO· might be exported by zinc efflux systems to mitigate the consequences of zinc stress during infection. Here, we show that Salmonella zinc metalloproteins are targets of S-nitrosylation, and zinc efflux is required for Salmonella resistance to nitrosative stress both in vitro and in a murine model of infection.
RESULTS
The S-nitrosoproteome of S. Typhimurium includes proteins involved in essential cellular functions.
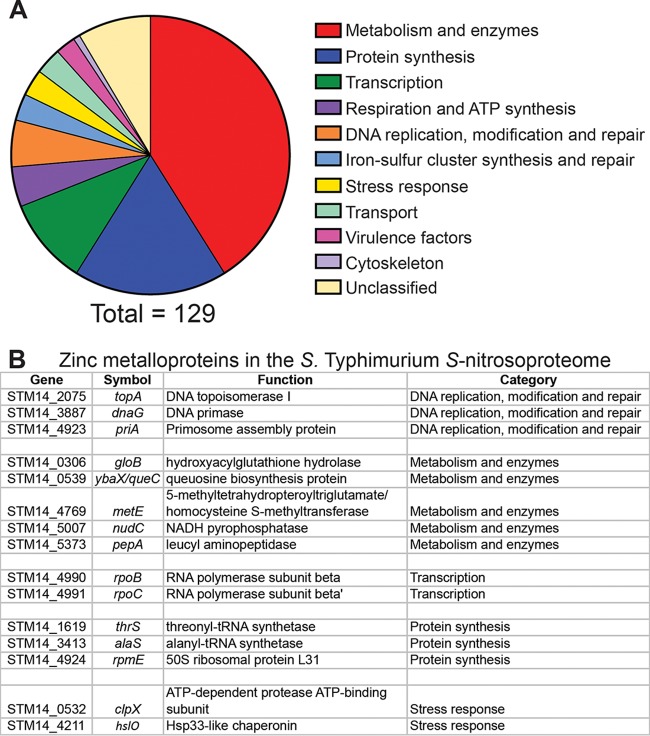
To identify S. Typhimurium proteins susceptible to S-nitrosylation, cell lysates were treated with the NO· donor diethylamine (DEA) NONOate (DEANO; half-life, 2 min), which generates a rapid bolus of NO·, for 10 min. S-nitrosylated proteins were selectively enriched by binding to a phenyl mercury column. Following on-column digestion with trypsin, 144 peptides from 129 proteins were identified by mass spectrometry (MS). The full list of S-nitrosylated peptides can be found in Table S1 in the supplemental material. The identified proteins are representative of a wide variety of functional categories, including transcription, respiration, iron-sulfur cluster metabolism, stress responses, and DNA replication or repair, with the majority consisting of enzymes or proteins involved in protein synthesis (Fig. 1A). Thirty-three of the targets are categorized as essential proteins in either S. Typhimurium or E. coli (16, 62, 63). Previously identified targets of S-nitrosylation, such as OxyR, LpdA, and FabB, were identified, along with many novel targets (17, 64, 65). To verify one of these novel targets, the enzymatic activity of GlyA (serine hydroxymethyltransferase [SHMT]) was assayed in cell lysates treated with either DEANO or DEA as a control. In vitro GlyA activity was monitored by measuring optical density at 420 nm (OD420) to determine the concentration of formaldehyde consumed. Lysates treated with DEANO displayed a greater than 50% reduction in GlyA activity compared to DEA-treated lysates, confirming that GlyA is a novel target of nitric oxide inhibition (Fig. S1).
FIG 1 .
Classification of proteins in the S. Typhimurium S-nitrosoproteome. (A) Functional classification of proteins with cysteine residues modified by NO· treatment. A total of 141 modified proteins were identified, and classification is shown as a percentage of the total. (B) Of the proteins modified by NO·, 15 (~10%) were found to be zinc metalloproteins. The metalloproteins are sorted by functional category and listed in numerical order by gene identifier in S. Typhimurium strain 14028s.
GlyA enzymatic activity is inhibited by NO·. S. Typhimurium expressing GlyA from a plasmid was treated with 10 mM diethylamine (DEA) as a mock treatment or 10 mM diethylamine NONOate (DEANO). Cell lysates were assayed for serine hydroxymethyltransferase activity by monitoring the glycine-dependent reduction of formaldehyde using a spectrophotometer at 420 nm. The specific activities of the DEANO-treated lysates were normalized to the activities of the corresponding mock DEA-treated lysate. Data are presented as percent specific activity compared to mock treatment. Error bars represent standard deviation from the mean for three biological replicates. Each biological replicate was analyzed in triplicate. The asterisk indicates statistical significance of P = 0.01 by paired two-tailed t test. Download FIG S1, TIF file, 0.2 MB (247.5KB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Peptides identified in the analysis of the S-nitrosoproteome of S. Typhimurium. Download TABLE S1, XLS file, 0.1 MB (102.5KB, xls) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
NO· targets zinc metalloproteins and disrupts zinc homeostasis.
A significant number of zinc metalloproteins were shown to be S-nitrosylated by NO· (Fig. 1B). These include DnaG and PriA, which were previously suggested to be targets of NO· inhibition but not experimentally verified (5). As the modification of zinc-coordinating cysteine ligands would be anticipated to disrupt metal binding and release free zinc, we examined the effects of NO· on the expression of zinc transport systems (Fig. 2A). The transcriptional regulators ZntR and Zur sense and respond to altered levels of intracellular free zinc to regulate expression of zntA and znuABC, respectively (11, 18, 55, 66).
FIG 2 .
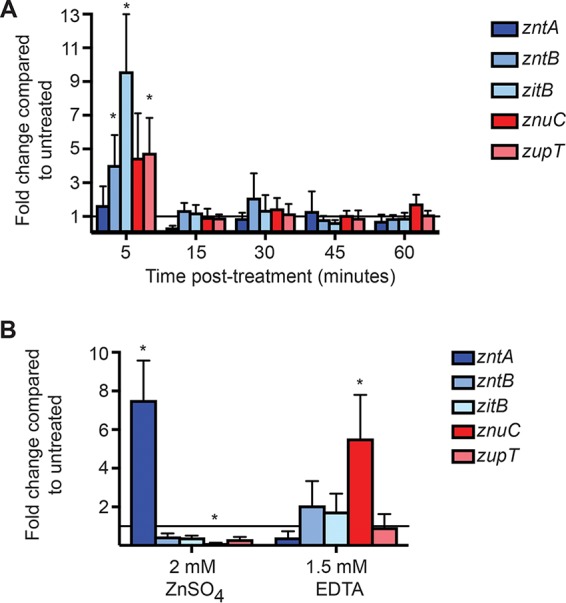
Expression of zinc transport systems in S. Typhimurium. qPCR data are presented as a positive fold change of treated compared to untreated cells with zinc efflux systems shown in blue and zinc acquisition systems shown in red. The solid line indicates a fold change of 1 to delineate between upregulation (>1) and downregulation (<1). (A) At 5 min after treatment with the NO· donor diethylamine NONOate (DEANO), expression of all zinc transport systems except zntA (dark blue bar) was modestly upregulated. The expression changes for zntB, zitB, and zupT achieved statistical significance with P values of 0.05, 0.004, and 0.03, respectively. At later time points, there was no significant difference in zinc transporter expression between treated and untreated cells. (B) In the presence of excess zinc (2 mM ZnSO4), expression of the high-affinity zinc efflux system zntA (dark blue bar) was significantly upregulated (P = 0.04), while expression of the high-affinity acquisition system znuABC (represented by znuC, red bar) was downregulated (P = 0.005). When cultures were treated with 1.5 mM EDTA to chelate zinc in the medium, expression of the znuABC acquisition system increased (P < 0.001). *, statistical significance was determined by one-sample t test compared to theoretical means of 2 for upregulated genes and 0.5 for downregulated genes. Data are the means from 3 (ZnSO4) and 8 (EDTA) replicates. Error bars represent standard deviations.
Expression of genes encoding three zinc efflux systems, zntA, zntB, and zitB (Fig. 2A, blue), and two zinc acquisition systems, znuABC and zupT (Fig. 2A, red), was monitored by quantitative PCR (qPCR) following treatment with 2 mM DEANO for 1 h. At 5 min posttreatment, transcript levels of both zinc uptake and export genes, with the exception of zntA, were elevated (Fig. 2A). By 15 min posttreatment, expression returned to a level that was not significantly different from untreated cells and subsequently remained constant for 60 min. Given the increased expression of all transport systems except ZntA, and the transient nature of expression in response to NO·, it is possible that qPCR was insufficiently sensitive to detect zntA expression. To investigate this possibility, the promoter regions of the S. Typhimurium hmp and zntA genes were fused to green fluorescent protein (GFP), and fluorescence was monitored by flow cytometry. Expression of Hmp (flavohemoglobin), an NO·-detoxifying enzyme regulated by the NO·-sensing transcriptional repressor NsrR, was monitored as a control (19). A significant increase in mean GFP intensity in response to NO· treatment was observed at all time points from both the hmp and zntA promoters (Fig. S2). Expression of the zinc transport systems in response to zinc supplementation or the metal chelator EDTA was as expected (Fig. 2B), with zntA expression (dark blue bars) increased in the presence of zinc and reduced in the presence of EDTA, whereas znuC expression (dark red bars) exhibited the opposite pattern. Expression of zntB, zitB, or zupT did not change significantly under either condition. Although the regulation of zntB expression has not been characterized, these results are consistent with expression patterns observed for zitB and zupT in E. coli (9, 20).
Mean fluorescence of GFP expressed from the S. Typhimurium hmp and zntA promoters increases in response to NO·. S. Typhimurium containing a plasmid expressing GFP from either the hmp (A) or zntA (B) promoter was treated with 2 mM DEANO and compared to untreated cells by flow cytometry at 5, 15, 30, 45, and 60 min. Mean fluorescence intensity increased in response to NO· for both promoter constructs at all times posttreatment, indicating that GFP expression occurred. Data are the means from three biological replicates, and error bars represent standard deviations. (A) An asterisk indicates statistical significance of P = 0.002 (5 min) or P < 0.001 (15, 30, 45, and 60 min) by unpaired two-tailed t test. (B) An asterisk indicates statistical significance of P = 0.04 (5 min), P = 0.009 (15 min), and P < 0.001 (30, 45, and 60 min) by unpaired two-tailed t test. Download FIG S2, TIF file, 0.8 MB (803.7KB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
After observing the simultaneous expression of zinc uptake and efflux systems in response to NO· treatment, the effect of NO· on total cellular zinc levels was determined by inductively coupled plasma mass spectrometry (ICP-MS). Within 5 min after treatment with the NO· donor DEANO, total cellular zinc levels dropped by about 20% compared to untreated cells (Fig. 3A). The level of total cellular zinc subsequently recovered, exceeding initial levels at 45 min posttreatment before returning to pretreatment levels by approximately 60 min. This brief accumulation of excess zinc prior to restoration of homeostasis may be due to the transient expression of zinc uptake systems also observed in response to NO· (Fig. 2A). To correlate these changes in cellular zinc levels with NO· levels, expression of the NO·-induced S. Typhimurium hmp gene, a reporter for nitrosative stress, was monitored in parallel (Fig. 3B) (19). Maximum hmp expression was observed 5 min posttreatment, suggesting that maximal NO· exposure also occurred within 5 min of DEANO treatment. The subsequent reduction in hmp expression at later time points indicates that NO· levels rapidly declined below the threshold required for restoration of NsrR-mediated repression. Total zinc levels began to recover at the same time that hmp expression declined. Together, these results suggest that NO· rapidly mobilizes zinc from metalloproteins and that zinc homeostasis is preserved by the excretion of this free zinc in response to nitrosative stress. Once NO· has been detoxified, zinc is reacquired from the environment until baseline levels are restored.
FIG 3 .
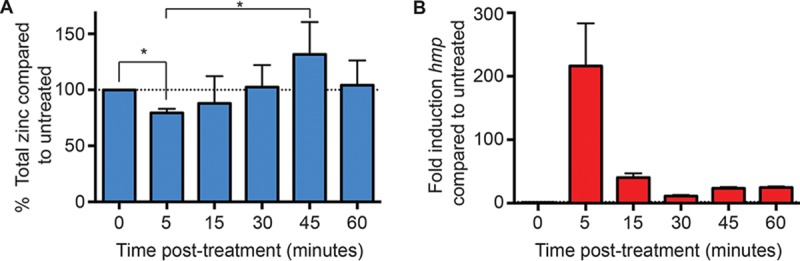
ICP-MS analysis of total cellular zinc content in S. Typhimurium following NO· treatment and transcriptional monitoring of NO· sensed by cells. (A) S. Typhimurium cells at an OD600 of ≈1 were treated with 2 mM diethylamine NONOate (DEANO), and total cellular zinc was measured at various times posttreatment. By 5 min posttreatment, total cellular zinc had fallen significantly compared to untreated cells, suggesting that zinc is effluxed from the cell following NO· treatment. Zinc levels gradually recovered to baseline levels over the course of 60 min. Statistical significance was determined by unpaired two-tailed t test; * indicates P values of <0.001 and 0.036, respectively. Error bars represent standard deviations. (B) A portion of each culture at each time point was also used to prepare RNA and cDNA for transcriptional analysis. Data are presented as the mean fold change in transcript level compared to untreated cells at each time point. The error bars represent standard deviations. Expression of hmp is regulated by the NO·-sensing NsrR regulator. The high level of expression at 5 min posttreatment indicates that the highest levels of NO· were present during this time period. At later time points, the level of hmp transcript declined significantly, indicating that the amount of NO· declined by 15 min posttreatment and remained low thereafter.
ZntA and ZitB are the primary zinc efflux systems in S. Typhimurium.
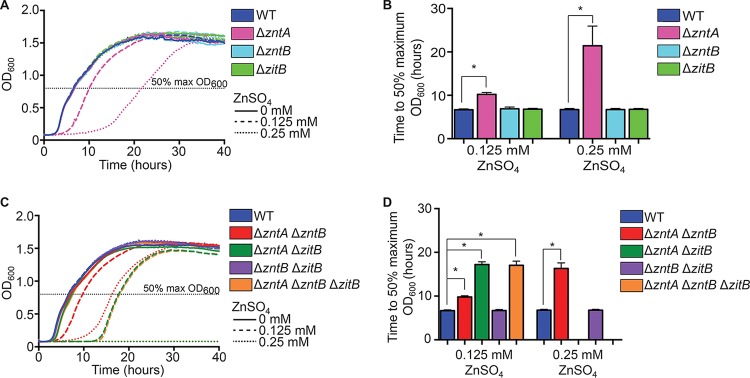
S. Typhimurium has 3 predicted zinc efflux systems (ZntA, ZntB, and ZitB) as well as a putative fourth system (YiiP), but the relative importance of these systems has not been established. To determine the relative roles played by these systems, mutations in the genes encoding zinc exporters were constructed singly or in combination and assessed for their effects on S. Typhimurium tolerance to various zinc concentrations (Fig. 4). Of the single mutants, only the ΔzntA mutant (Fig. 4A and B, pink) showed delayed growth in the presence of excess zinc. A ΔzntA ΔzntB mutant (Fig. 4C and D, red) was no more sensitive than a ΔzntA mutant alone. However, a ΔzntA ΔzitB double mutant was more sensitive to zinc than a ΔzntA mutant alone in medium supplemented with 0.125 mM zinc (Fig. 4C and D, green) and was unable to grow at higher concentrations. This indicates that these two efflux systems work cooperatively to protect cells from zinc overload, with ZntA able to compensate for the absence of ZitB but ZitB only partially capable of compensating for the absence of ZntA. A ΔzntA ΔzntB ΔzitB triple mutant (Fig. 4C and D, orange) exhibited susceptibility equivalent to that of a ΔzntA ΔzitB double mutant. To confirm that the observed mutant phenotypes resulted from the loss of the corresponding zinc efflux transporter, each mutant was complemented by expression in trans from its native promoter on a plasmid (Fig. S3). Plasmid-borne ZntA expression was able to fully complement ΔzntA or ΔzntA ΔzntB mutations (Fig. S3A, purple, and B, orange), but zntB expression failed to complement a ΔzntA ΔzntB mutant (Fig. S3B, green). Both the ΔzntA ΔzitB and ΔzntA ΔzntB ΔzitB mutant strains exhibited the full restoration of zinc tolerance when complemented by expression of either ZntA or ZitB on a plasmid (Fig. S3C, pink and gold, and D, violet and light green). The ability of the ZitB plasmid to fully compensate for the loss of both ZitB and ZntA, whereas chromosomal zitB only partially compensates for loss of zntA, is likely due to differences in gene dosage.
FIG 4 .
ZntA and ZitB are the primary zinc efflux transporters in S. Typhimurium. (A) A ΔzntA mutant (pink) was impaired for growth, represented by a delayed exit from lag phase, in both 0.125 mM ZnSO4 and 0.25 mM ZnSO4, while a ΔzntB mutant (aqua) and a ΔzitB mutant (light green) exhibited growth comparable to wild type (blue) under these conditions. (B) Significance of the growth defect for the ΔzntA mutant in panel A was determined by calculating the mean time required to reach 50% of the maximum final OD600 for each strain. (C) A ΔzntB ΔzitB double mutant (purple) exhibited growth comparable to wild type (blue) when exposed to elevated zinc concentrations. A ΔzntA ΔzntB mutant (red) behaved similarly to a ΔzntA mutant, whereas a ΔzntA ΔzitB mutant (green) displayed a more severe growth delay at 0.125 mM ZnSO4 and was unable to grow at 0.25 mM ZnSO4. A ΔzntA ΔzntB ΔzitB triple mutant (orange) exhibited growth characteristics identical to those of a ΔzntA ΔzitB double mutant. (D) Significance of the growth defects for the mutants in panel C was determined by calculating the mean time required to reach 50% of the maximum final OD600 for each strain. Statistical significance of the growth defects was determined by unpaired two-tailed t test, and an asterisk indicates P values of <0.001 for ΔzntA, ΔzntA ΔzntB, ΔzntA ΔzitB, and ΔzntA ΔzntB ΔzitB mutants compared to wild type at all concentrations. Error bars represent standard deviations.
Expression of either ZntA or ZitB, but not ZntB, is capable of correcting mutant growth defects in excess zinc. (A and B) The impaired growth of both a ΔzntA mutant (pink) and a ΔzntA ΔzntB mutant (red) can be fully complemented by expression of ZntA from its native promoter on a low-copy-number plasmid (purple [A] or orange [B]), but the growth defect of a ΔzntA ΔzntB mutant cannot be complemented by expression of ZntB (green [B]). (C and D) Both a ΔzntA ΔzitB mutant (green [C]) and a ΔzntA ΔzntB ΔzitB mutant (orange [D]) can be fully complemented by heterologous expression of either ZntA (pink [C] and violet [D]) or ZitB (gold [C] and light green [D]). Data are the means from 9 biological replicates from three trials. Statistical significance was determined by unpaired two-tailed t test. Download FIG S3, TIF file, 1 MB (1,004.9KB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
YiiP was initially identified as a putative iron efflux system but later characterized as a transporter of zinc and cadmium (21, 58). A ΔyiiP mutant was no more zinc-sensitive than wild-type (WT) S. Typhimurium, and ΔyiiP ΔzntA ΔzitB and ΔyiiP ΔzntA ΔzntB ΔzitB mutants were no more sensitive than the corresponding double and triple mutant strains, respectively (Fig. S4A and B). These results are consistent with previous observations with ΔyiiP mutant derivatives of E. coli and S. Typhimurium and suggest that YiiP does not play a role in zinc resistance under these conditions (14, 22).
YiiP does not contribute to zinc homeostasis in S. Typhimurium. (A) A ΔyiiP mutant (pink) is no more sensitive to zinc than wild type (wild-type), and both strains grew equally well in the presence or absence of excess zinc. (B) A ΔzntA ΔzitB ΔyiiP strain (purple) is no more sensitive to excess zinc than a ΔzntA ΔzitB strain (green). A ΔzntA ΔzntB ΔzitB ΔyiiP strain (gold) is no more zinc-sensitive than a ΔzntA ΔzntB ΔzitB strain (orange). Together, these results indicate that YiiP does not contribute to zinc homeostasis in S. Typhimurium under these experimental conditions, even in the absence of all other known zinc efflux systems. Data are the means from 12 biological replicates from four trials. Statistical significance was determined by unpaired two-tailed t test. Download FIG S4, TIF file, 0.7 MB (766.5KB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ZntA and ZitB are required for S. Typhimurium resistance to nitrosative stress.
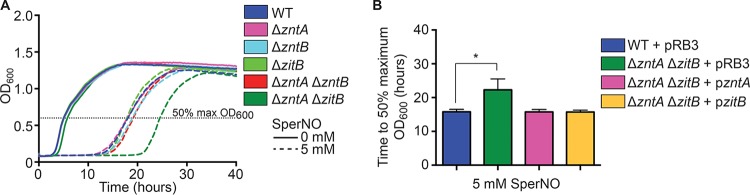
NO· rapidly mobilizes intracellular zinc in S. Typhimurium, which is followed by a reduction in total cellular zinc content. We therefore hypothesized that zinc efflux might play an important role in ameliorating the consequences of nitrosative stress. To test this hypothesis, mutant strains lacking zinc efflux transporters singly or in combination were cultured in the presence of the NO· donor spermine NONOate (SperNO; half-life, 39 min), which provides a sustained release of NO· for observation of growth phenotypes. Although none of the single efflux mutants was more sensitive to SperNO than the wild type, a ΔzntA ΔzitB double mutant exhibited a delayed exit from lag phase, indicative of enhanced sensitivity to NO· (Fig. 5A, dark green dashed line). The reduction in total cellular zinc displayed by wild-type S. Typhimurium in response to NO· was not observed in the ΔzntA ΔzitB double mutant, in which zinc levels remain steady or slightly elevated following NO· treatment (Fig. S5). Together these results indicate that ability to efflux free zinc is an important component of the response to nitrosative stress. The growth defect in the presence of SperNO was complemented by expression of either ZntA or ZitB from its native promoter on a low-copy-number vector (Fig. 5B). The sensitivity of the ΔzntA ΔzitB double mutant but not a ΔzntA single mutant suggests that the amount of zinc mobilized by NO· can be efficiently removed from the cell by ZitB in the absence of ZntA and that the level of zinc stress resulting from 5 mM SperNO was not as great as the zinc stress from the addition of 0.125 mM ZnSO4, which impaired the growth of a ΔzntA single mutant (Fig. 4A).
FIG 5 .
Zinc efflux by ZntA and ZitB is required for S. Typhimurium resistance to nitrosative stress in vitro. (A) Single zinc efflux mutants (pink, aqua, and light green) and a ΔzntA ΔzntB double mutant (red) were no more sensitive to NO· generated by the donor spermine NONOate (SperNO) than wild-type cells (blue). However, a ΔzntA ΔzitB double mutant (green) exhibited significantly delayed growth, indicating enhanced NO· sensitivity (*, P < 0.0001). (B) The growth defect of the ΔzntA ΔzitB mutant was complemented by expression of either ZntA or ZitB from its native promoter in trans. Statistical significance was determined by comparing the time required to reach 50% of the maximal OD600 (dashed line) by unpaired two-tailed t test.
A ΔzntA ΔzitB mutant does not show a decrease in total cellular zinc following NO· treatment. ΔzntA ΔzitB S. Typhimurium at an OD600 of ≈1 was treated with 2 mM diethlyamine NONOate (DEANO), and total cellular zinc was measured at various times posttreatment by ICP-MS. Total intracellular zinc was not significantly different from baseline (T0, untreated) at any of the times examined. Download FIG S5, TIF file, 0.8 MB (827.8KB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Macrophage-derived NO· induces free zinc accumulation in ΔzntA ΔzitB mutant Salmonella.
A genetically encoded zinc biosensor was used to determine whether macrophage-derived NO· mobilizes zinc in intracellular Salmonella and to measure the influence of zinc efflux transporters on zinc accumulation. The ZapCV5 zinc sensor contains the first two zinc fingers of the Zap1 transcription factor from Saccharomyces cerevisiae, coupled to an enhanced cyan fluorescent protein (CFP) fluorescence resonance energy transfer (FRET) donor and a circularly permuted Venus (cp173Venus) FRET acceptor protein (23). Each of the cysteine ligands in the zinc finger region was mutated to histidine in this construct, which only modestly reduces zinc sensitivity and renders the sensor insensitive to S-nitrosylation by NO·, which could otherwise confound interpretation of the results. Sensor function was confirmed by adding ZnSO4 to the growth medium to drive accumulation of free intracellular zinc and by measuring changes in the FRET ratio. In a ΔzntA ΔzitB mutant strain, the FRET ratio increased significantly following exposure to increasing ZnSO4 concentrations, indicating that absence of these efflux systems results in increased levels of free intracellular zinc, whereas wild-type cells were able to maintain their free zinc pool at a steady level (Fig. S6).
A fluorescence resonance energy transfer (FRET)-based biosensor for zinc shows accumulation of free intracellular zinc in a ΔzntA ΔzitB mutant but not in wild-type S. Typhimurium. A genetically encoded zinc biosensor, ZapCV5, was expressed constitutively from a plasmid in S. Typhimurium. Due to the constant presence of the sensor, a portion of sensor proteins are metallated during normal cell growth. The amount of FRET signal from the sensor was greater in a ΔzntA ΔzitB mutant than in wild type even when no exogenous ZnSO4 (0 mM) was added to the growth medium. At increased concentrations of ZnSO4, FRET signal increased in the ΔzntA ΔzitB mutant but not in the wild-type strain. None of the wild-type FRET ratios were significantly different from one another, whereas the ΔzntA ΔzitB FRET ratio was significantly greater than the wild-type ratio at each concentration (P < 0.001). The FRET ratios for the ΔzntA ΔzitB mutant at 0.25 mM and 0.5 mM ZnSO4 were also significantly elevated compared to the FRET ratio in the absence of exogenous zinc (P < 0.001). Statistical significance was analyzed by one-way analysis of variance. Download FIG S6, TIF file, 0.9 MB (897.8KB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
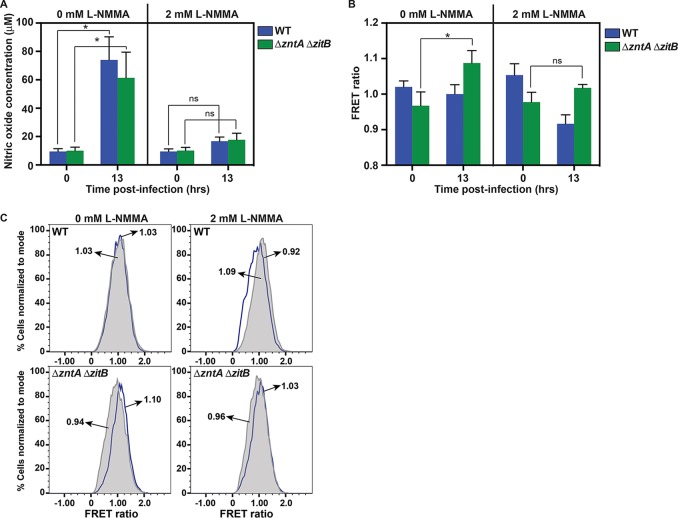
The zinc biosensor was next used to examine whether NO· production by infected murine macrophages mobilizes free zinc in S. Typhimurium. Production of NO· by iNOS peaks hours after infection (46), and a significant increase in nitric oxide was detected after 13 h (Fig. 6A, 0 mM NG-monomethyl-l-arginine [l-NMMA]). NO· production was inhibited by the addition of 2 mM NG-monomethyl-l-arginine monoacetate to the cell culture medium (Fig. 6A, 2 mM l-NMMA). In NO·-producing macrophages, an increase in the FRET ratio was observed in the ΔzntA ΔzitB strain 13 h postinfection but not in wild-type bacteria, in which all zinc efflux systems were functional (Fig. 6B, 0 mM l-NMMA, and C). This FRET ratio increase was abrogated by treatment with the NOS inhibitor l-NMMA. Together, these observations indicate that free intracellular zinc is mobilized from zinc metalloproteins in S. Typhimurium by macrophage-derived NO· and is subsequently exported from the cell by ZntA and ZitB. In the absence of ZntA and ZitB, mobilized free zinc remains available to bind to the sensor, leading to an increase in FRET. When NO· production is inhibited by l-NMMA, intracellular free zinc is reduced in both wild-type and mutant cells.
FIG 6 .
Free intracellular zinc levels increase in a ΔzntA ΔzitB S. Typhimurium mutant during macrophage infection in response to NO· production. (A) Changes in NO· production are shown at the time of infection (0 h) and 13 h postinfection in the presence or absence of the NOS inhibitor l-NMMA. IFN-γ-primed murine macrophages infected with either wild-type (blue) or ΔzntA ΔzitB (green) S. Typhimurium produced significant levels of NO· after 13 h in the absence of the NOS inhibitor l-NMMA (P < 0.001) but not in the presence of 2 mM l-NMMA (ns). (B) Changes in FRET ratio are shown immediately following infection (0 h) and 13 h postinfection. The FRET ratio of the ZapCV5 biosensor increased in ΔzntA ΔzitB mutants (green) isolated from murine macrophages after 13 h (P < 0.001), but not in wild-type S. Typhimurium, indicating that intracellular free zinc levels rise in the efflux-deficient mutant during infection. The increase in FRET was not observed when macrophages were treated with 2 mM l-NMMA to inhibit NO· production (ns). (C) Flow cytometry histograms from one representative experiment. The mean value of each histogram (T0, gray shape, and T13, blue line) is indicated. Data in panels A and B are presented as the means with error bars representing standard deviations. Statistical significance (*) was determined by one-way analysis of variance.
Zinc efflux is required for Salmonella virulence in NO·-producing mice.
As mutant Salmonella strains deficient in zinc efflux are more sensitive to nitrosative stress in vitro and accumulate intracellular free zinc following internalization by NO·-producing macrophages, we investigated whether zinc efflux is required for S. Typhimurium virulence in vivo in a murine infection model. Wild-type and ΔzntA ΔzitB S. Typhimurium strains in a 1:1 ratio were used to infect NO·-producing C3H/HeOuJ mice by intraperitoneal (i.p.) inoculation. Five days postinfection, the output ratios and competitive indexes (CI) were determined for bacteria recovered from the liver and spleen. A ΔzntA ΔzitB mutant was significantly outcompeted by isogenic wild-type S. Typhimurium in both the liver and the spleen (Fig. 7A), indicative of reduced virulence. To determine whether this defect was attributable to the mobilization of zinc by NO·, the infection was repeated in mice treated with the iNOS inhibitor l-N6-(1-iminoethyl)lysine dihydrochloride (l-NIL) (24). A ΔzntA ΔzitB mutant no longer exhibited a competitive disadvantage compared to wild-type S. Typhimurium in mice treated with l-NIL, indicating that zinc efflux is required for Salmonella virulence only in mice capable of NO· production (Fig. 7B).
FIG 7 .
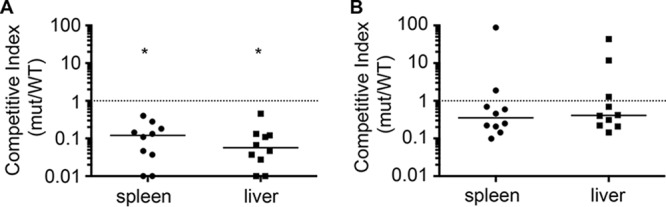
Virulence of ΔzntA ΔzitB mutant S. Typhimurium is attenuated in NO·-producing mice. Solid lines represent the median competitive index (CI) for each organ. The dotted line represents the expected CI if neither strain has a competitive advantage. (A) In wild-type C3H/HeOuJ mice, a ΔzntA ΔzitB mutant has a significant competitive disadvantage compared to wild type (P = 0.002 by Wilcoxon signed-rank test to a hypothetical median of 1 for both spleen and liver). (B) In C3H/HeOuJ mice that cannot produce NO· due to treatment with 500 µg ml−1 l-N6-(1-iminoethyl)lysine dihydrochloride (l-NIL), the mutant no longer has a statistically significant disadvantage compared to wild type, and the CIs are significantly different (*, P = 0.007 for spleen and P < 0.001 for liver by Mann-Whitney test) from the CI in untreated mice. A total of 10 animals were tested for each condition. Data points at a CI of 0.01 were at the limit of detection for the assay.
DISCUSSION
Nitric oxide (NO·) is an important mediator of the mammalian innate immune response to infection. Modification of protein thiols and metal centers by NO· produces pleiotropic effects on microbial physiology and confers NO· with broad-spectrum antimicrobial activity. Although some direct targets of NO· have been previously identified, many molecular targets of NO· are unknown. Our characterization of the Salmonella Typhimurium S-nitrosoproteome (see Table S1 in the supplemental material) confirmed several previously identified targets of NO· and identified many novel targets, including proteins with essential functions. Cysteines are known to play structural, metal-coordinating, catalytic, and regulatory roles within proteins; therefore, it is not surprising that a wide variety of targets from different functional categories were identified (25). S-Nitrosylation targets surface-exposed cysteines, often flanked by charged residues (26). Although some integral membrane proteins were detected, this category of proteins, along with secreted proteins and those present at low abundance under experimental conditions, is likely to be underrepresented or absent from the data set. Nevertheless, the S-nitrosoproteome of S. Typhimurium identified in this study provides a foundation for future studies and an opportunity to directly validate new targets of NO·-related antimicrobial activity.
The NO· targets identified in this study include a number of zinc metalloproteins (Fig. 1B), two of which were previously implicated in the inhibition of DNA replication by NO· (5). Many zinc metalloproteins bind metals via cysteine residues, which are no longer able to retain zinc following S-nitrosylation. The loss of a zinc cofactor would directly impair protein function, and the release of free zinc into the cytoplasm might also exert broad toxic effects on the cell if the zinc is not promptly removed or bound. Although the precise mechanistic basis of zinc toxicity is not known, it is thought to result, at least in part, from mismetallation of metalloproteins that ordinarily bind other divalent metals (27). Zinc is capable of forming more stable complexes with proteins than metals further down the Irving-Williams series, such as iron and manganese, and might thereby disrupt the function of proteins requiring these cofactors (28). Zinc has also been shown to target and destroy exposed 4Fe-4S clusters of dehydratase enzymes (29).
S-Nitrosylation of zinc metalloproteins was predicted to lead to release of free intracellular zinc, expression of the zntA zinc efflux system regulated by ZntR, and repression of the znuABC zinc acquisition system regulated by Zur (Fig. 8). However, following NO· treatment, a transient increase in expression of all zinc transporters was observed (Fig. 2A and S2). It is possible that zinc-binding residues within Zur were directly modified by NO·, altering its responsiveness, but these modifications may not have been detected in the S-nitrosoproteome assay due to the limited sensitivity of the assay for low-abundance proteins. It is presently uncertain whether NO· leads to dysregulation of transporter expression by direct or indirect actions. Nevertheless, despite the initial dysregulation of transporter expression, total cellular zinc levels were observed to fall immediately following NO· treatment before recovering to baseline levels as NO· levels decreased (Fig. 3).
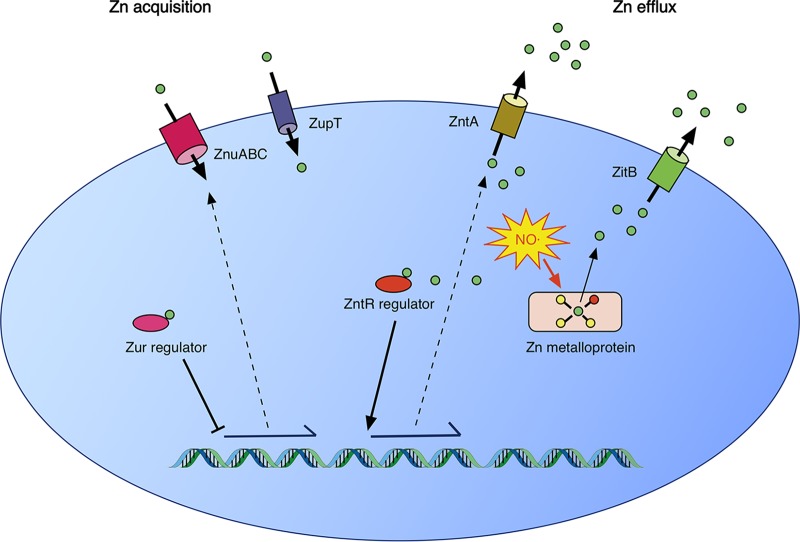
FIG 8 .
A model of zinc homeostasis in Salmonella Typhimurium. Under conditions of zinc deficiency, zinc is not available to bind to the Zur repressor, leading to expression of the ZnuABC zinc acquisition system. ZupT, whose regulation is uncharacterized, has also been shown to contribute to zinc acquisition. When zinc is abundant, Zur bound to zinc represses ZnuABC expression. In addition, free cytoplasmic zinc binds to the transcriptional activator ZntR to induce expression of the ZntA zinc efflux system. Zinc efflux in S. Typhimurium is also mediated by ZitB. Under conditions of nitrosative stress, S-nitrosylation of cysteine ligands in zinc metalloproteins leads to mobilization of free intracellular zinc. The zinc efflux activities of ZntA and ZitB are required for the resistance of S. Typhimurium to nitrosative stress.
Of the four putative zinc efflux systems in S. Typhimurium, this study demonstrates that ZntA and ZitB are required for resistance to zinc stress, corroborating other recent observations (14). The inducible high-affinity transporter ZntA appears to be the most important of these zinc efflux systems, as an S. Typhimurium ΔzntA mutant exhibits a growth defect at zinc concentrations that are not inhibitory for a ΔzitB mutant strain (Fig. 4). ZntA and ZitB are also required to efflux zinc mobilized by NO· and appear to play complementary roles, as either transporter alone is able to prevent NO· hypersusceptibility (Fig. 5). The mobilization of zinc by NO· and its subsequent efflux by ZntA and ZitB as observed in vitro also occur in bacteria following internalization by NO·-producing macrophages (Fig. 6) and during infection of NO·-producing mice, in which a ΔzntA ΔzitB mutant exhibits a competitive disadvantage in comparison to wild-type Salmonella (Fig. 7).
It has been suggested that macrophages may employ zinc as an antimicrobial mediator, which is opposed by the Salmonella pathogenicity island 1 (SPI-1) type III secretion system (30, 67). Our observations do not directly address this possibility, but it is nevertheless evident that zinc efflux is required for Salmonella virulence and may confer resistance to exogenous zinc as well as to NO·.
Our observations provide new insights into bacterial zinc homeostasis within the host environment. Although most previous studies have focused on the need for bacteria to acquire zinc within the host by competing with the activity of eukaryotic zinc transporters and the chelator calprotectin (31, 59, 68, 69), we have demonstrated that zinc sequestration and removal are also required for pathogenic bacteria to achieve a balance between acquiring zinc for essential cellular functions while minimizing the toxicity of free intracellular zinc mobilized by the actions of NO· on metalloproteins.
MATERIALS AND METHODS
Growth conditions.
Salmonella enterica serovar Typhimurium strain ATCC 14028s was used as the wild-type strain for all experiments. S. Typhimurium was grown aerobically in Luria-Bertani (LB; Difco) medium at 37°C with shaking at 250 rpm. Antibiotics were used at the following concentrations: 100 µg ml−1 ampicillin (Amp), 50 µg ml−1 kanamycin (Kan), 20 µg ml−1 chloramphenicol (Cm), and 20 µg ml−1 tetracycline (Tet).
Strain and plasmid construction.
All strains are listed in Table S2 in the supplemental material, and primers are listed in Table S3. The ΔzntA and ΔzitB S. Typhimurium mutant strains were generated using the method of Datsenko and Wanner with the pKD3 Cm or pKD4 Kan cassettes as the templates, and the ΔyiiP mutant was generated using the pKD3 template (32). The ΔzntB mutant was generated by lambda-red replacement with TetRA insertion (33). Mutations were transduced into a clean ATCC 14028s background using P22 bacteriophage to generate strains EF487 (ΔzntA), EF511 (ΔzntB), EF512 (ΔzitB), and EF561 (ΔyiiP). P22 bacteriophage transduction was also used to generate strains EF527 (ΔzntA ΔzntB), EF528 (ΔzntA ΔzitB), EF529 (ΔzntB ΔzitB), and EF530 (ΔzntA ΔzntB ΔzitB). The antibiotic resistance cassettes of these strains were recombinationally excised using pCP20 to generate strains EF531, EF532, EF533, and EF534 (32). Phenotypes of the excised and unexcised strains were identical. To generate strains EF562 and EF563, the ΔyiiP mutation was added by P22 bacteriophage transduction into EF532 and EF534. Strain JK377 was derived from strains TH6727 and BC1459 by P22 bacteriophage transduction into JK237 (34).
Strains and plasmids used in this study. Download TABLE S2, DOCX file, 0.1 MB (108.8KB, docx) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequences of primers used in this study. Download TABLE S3, DOCX file, 0.1 MB (101.6KB, docx) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
E. coli strain DH10B was used as the host strain for all cloning, and confirmed plasmids were subsequently electroporated into S. Typhimurium. The glyA gene was cloned into the NheI and HindIII sites of the pBAD18-Cm multiple cloning site (MCS) to generate plasmid pJK715 (35). All zinc mutant-complementing plasmids were generated using the stable pRB3-273C (Ampr) plasmid backbone (36). The complementing gene sequences for zntA, zntB, and zitB, including several hundred upstream bases encompassing the native promoter, were cloned into the BamHI and HindIII sites of the pRB3-273C MCS to generate plasmids pJK719, pJK720, and pJK721. To generate pAS3, the ZapCV5 sequence was subcloned from plasmid pcDNA3.1-zapCV5 and inserted into a modified pBAD vector at the BamHI and EcoRI sites (23). Control plasmids pAS4 and pAS5 were created by amplifying and cloning the CFP sequence and cp173Venus sequence into the same modified pBAD vector (23, 37). Plasmids pAS15, pAS16, and pAS17 were generated by amplifying the ZapCV5, CFP, and cp173Venus sequences from plasmids pAS3, pAS4, and pAS5 and cloning into the XbaI and HindIII sites of pHR103 (38). Plasmids pAS20 and pAS22 containing green fluorescent protein (GFP) fused to the promoter regions of the S. Typhimurium hmp and zntA genes, respectively, were generated from source plasmids pJK682 and pRU001. To generate pJK682, the gfp gene, without the C-terminal LVA tag, was amplified from plasmid pJBA111 and inserted into the NheI and AatII sites of pBR322 (39, 70). Plasmid pRU001 was created by digesting pJK682 with AvaI, digesting pFPV-mCherry with HindIII, filling cohesive ends using T4 polynucleotide kinase, and digesting both fragments with BamHI before ligating (40). The promoter regions of hmp and zntA were amplified by PCR and then digested and inserted into the EcoRI site of pRU001 to generate pAS20 and pAS22. Correct promoter orientation was validated by PCR and confirmed by sequencing.
S-Nitrosoproteome analysis.
S. Typhimurium was grown overnight in 5 ml LB and then diluted 1:100 into 500 ml fresh LB medium and grown to an optical density at 600 nm (OD600) of ~0.5. The cells were pelleted by centrifugation (5,000 × g, 5 min, 4°C), washed once in phosphate-buffered saline (PBS), and then resuspended in 3 ml HDN (250 mM HEPES [pH 7.7], 1 mM diethylenetriamine penta-acetic acid [DTPA], 0.1 mM neocuproine), and 1% Triton X-100. The cells were lysed with a French press at 20,000 lb/in2 twice. After the first pressing, 150 µl of 25× protease inhibitor cocktail mix (Roche Complete without EDTA) was added. The mixture was clarified by centrifugation at 10,000 × g for 15 min at 4°C. The supernatant was passed over a Bio-Gel P-6DG Econo-Pac column (Bio-Rad) equilibrated in 20 ml HDN and 1% Triton X-100 per the manufacturer’s protocol to remove low-molecular-weight thiols. Protein concentration in the eluate was determined using the Coomassie blue protein assay (Thermo Scientific), and 1-ml aliquots were stored at −80°C.
Lysates were diluted to 0.8 mg ml−1 in HDN, 1% Triton X-100. Ten milliliters of diluted lysate (~8 mg of total protein) was transferred to 50-ml dark conical tubes (Litesafe II) and treated with either 150 µM diethylamine NONOate (DEANO) or diethylamine (DEA) and incubated at 37°C for 10 min. Thirty milliliters of cold 100% acetone was added and incubated at −20°C for 20 min. The mixture was centrifuged at 3,500 × g for 5 min at 4°C, and the pellet was washed in 25 ml cold 75% acetone and centrifuged again. The pellet was resuspended in 10 ml blocking buffer (HDN, 2.5% SDS) and N-ethylmaleimide (Sigma-Aldrich) added to 50 mM and incubated at 50°C for 60 min, with vortexing every 5 min to block nonmodified cysteine residues. The proteins were precipitated with acetone as described above and resuspended in 4 ml loading buffer (250 mM 2-(N-morpholino)ethanesulfonic acid [MES], pH 6.0, 1 mM DTPA, 1% SDS). A sample was taken to monitor the level of S-nitrosylation using the biotin switch method (41).
The lysates were then passed over 4 ml of activated organic mercury resin and washed, and tryptically digested peptides were eluted and analyzed by mass spectrometry essentially as described previously (26).
Data for the DEANO treatment condition were compared to the DEA-only data. Peptides with equal total ion current (TIC) values in the two data sets (TIC DEA/NO − TIC DEA = 0) were discarded from the final analysis. Peptide annotation was performed by using Sorcerer Sequest to search against a UniProt database for Salmonella that included methionine dioxidation (+32 Da), cysteine trioxidation (+48 Da), and cysteine N-ethylamide alkylation (+125) as variable modifications. Cellular location was predicted using PSORTb (42). Classification as iron or zinc binding was determined by a search of available literature and crystal structures.
Assay of SHMT activity.
S. Typhimurium strains JK1284 and JK1285 containing pBAD18-Cm or pJK715 were grown overnight in LB-Cm and then subcultured 1:100 into 100 ml fresh LB-Cm with 0.2% arabinose and grown to an OD600 of 0.8 to 0.9. Cells were pelleted, washed twice in cold 0.85% NaCl, and then resuspended in two 50-ml aliquots. Each cell suspension was sonicated on ice three times for 15 s each at power level 2 with a Misonix Microson Ultrasonic Cell Disrupter XL. Lysates were centrifuged at 4°C to clear cell debris, and supernatants were pooled. Cleared lysates were purified over a Bio-Gel P-6DG Econo-Pac column (Bio-Rad), using the standard protocol to remove low-molecular-weight thiols, and eluted in 2.7 ml 0.85% saline to yield protein concentrations of approximately 4 mg ml−1. Two hundred fifty microliters of purified lysate was treated with a 10 mM concentration of either DEA or DEANO for 10 min at 37°C. Serine hydroxymethyltransferase (SHMT) activity assays were carried out according to previously described protocols (43, 44). Briefly, formaldehyde, ditetrahydrofolate, and Nash B reagent were made fresh each time. A 500× pyridoxal 5′-phosphate solution was made once, and aliquots were stored in the dark. Formaldehyde, ditetrahydrofolate, phosphate buffer (pH 7.5), pyridoxal 5′-phosphate, and lysate were combined with or without glycine, and the volume was brought to 1,200 µl with water. The reaction was allowed to proceed for 8 min at 37°C and then stopped by the addition of 300 µl 15% trichloroacetic acid. Samples were centrifuged to remove precipitated protein, and then 500 µl of the supernatant were incubated with 2 ml Nash B reagent for 45 min at 37°C with agitation. SHMT activity was monitored by reading the OD at 420 nm and comparing values to a standard curve generated with known concentrations of formaldehyde. All data were normalized to protein concentration and compared to control reaction mixtures in which glycine was absent. Data are presented as the mean with error bars representing standard deviations. Statistical significance was determined by paired two-tailed t test.
ICP-MS.
An overnight culture of S. Typhimurium strain JK377 lacking flagella (for efficient cell pelleting) was subcultured 1:100 into 100 ml fresh LB and grown to an OD600 of 1.0. The culture was then divided into 10 6-ml aliquots in glass tubes, and 5 were treated with 2 mM DEANO. At 0, 5, 15, 30, 45, and 60 min posttreatment, 4.5 ml of treated and untreated cultures was pelleted by centrifugation, washed once with 3 ml ultrapure water, and then resuspended in 500 µl analytic-grade nitric acid and incubated in an 85°C water bath for 30 min. The nitric acid solution was diluted 1:10 into MilliQ purified water before inductively coupled plasma-mass spectrometry (ICP-MS) analysis was performed by the Environmental Health Laboratory and Trace Organics Analysis Center at the University of Washington using an Agilent 7500 CE instrument.
RNA isolation, cDNA synthesis, and gene expression analysis.
Primers for quantitative PCR (qPCR) analysis were published previously (19). For induction by 2 mM DEANO, 1 ml of cells from the same cultures analyzed by ICP-MS was pelleted and resuspended in Trizol reagent. RNA and cDNA were prepared according to previously described protocols (45). qPCR was performed using SYBR green master mix on a Bio-Rad CFX96 real-time system. The rpoD gene was amplified for use as an internal control. Statistical significance was determined by one-sample t test comparing fold change (expression treated/expression untreated) to theoretical means of 2 for upregulated genes and 0.5 for downregulated genes.
Flow cytometry-based detection of GFP expression.
Overnight cultures of JK237, AS212, and AS214 were subcultured 1:100 into 25 ml fresh LB and grown to an OD600 of 1.0. Cultures were divided into 3-ml aliquots and either treated with DEANO or left untreated. At 5, 15, 30, 45, and 60 min posttreatment, 0.5 ml of culture was pelleted by centrifugation and fixed in 1 ml 2.5% paraformaldehyde for 30 min at 37°C followed by resuspension in 1 ml phosphate-buffered saline (PBS). Fixed bacterial cells were analyzed using an LSR II flow cytometer (Becton, Dickinson). Emission was collected using a 530/30 filter following excitation at 488 nm. Cells were gated according to forward and side scatter (FSC/SSC), and photomultiplier tube voltages were adjusted using wild-type S. Typhimurium strain JK237. A total of 10,000 events were collected, and mean fluorescence intensity of GFP was calculated and plotted using FlowJo v10.3 software (TreeStar, Inc.). Data were collected as three biological replicates analyzed on the same day. Statistical significance was determined by unpaired two-tailed t test.
Zinc sensitivity assays.
S. Typhimurium wild-type (WT) (JK237) and mutant strains lacking specific zinc efflux pumps (EF487, EF511, EF512, EF531, EF532, EF533, and EF534) were grown overnight in 5 ml LB and then diluted 1:1,000 into fresh LB with or without 0.125 mM or 0.25 mM ZnSO4 to a final volume of 300 µl in microtiter plate wells. Cultures were grown aerobically with shaking at 37°C in a Labsystems Bioscreen C machine (Growth Curves USA). Growth was monitored by measuring OD600 every 15 min. Differences between cultures were determined by calculating the time to reach 50% maximum OD600, and statistical significance was determined by unpaired two-tailed t tests. Complementation experiments were conducted as described above using strains JK895, EF535, EF536, EF539, EF540, EF553, EF543, EF544, EF545, EF548, EF549, and EF550.
Nitric oxide sensitivity assays.
S. Typhimurium wild type (JK237) and isogenic zinc efflux-deficient mutant derivatives (EF487, EF511, EF512, EF527, EF528, EF529, and EF530) were grown overnight in 5 ml LB and then diluted 1:1,000 into fresh LB with or without 5 mM SperNO to a final volume of 300 µl in microtiter plate wells. Cultures were grown aerobically with shaking at 37°C in a Labsystems Bioscreen C machine (Growth Curves USA). Growth was monitored by measuring OD600 every 15 min. Differences between cultures were determined by calculating the time to reach 50% maximum OD600, and statistical significance was determined by unpaired two-tailed t tests. Complementation experiments were performed as described above using strains JK895, EF543, EF544, and EF545.
Macrophage infections.
The murine macrophage-like cell line RAW 264.7 (TIB-71; ATCC) was maintained in Dulbecco’s modified Eagle medium, 4.5 g liter−1 d-glucose, 4 mM l-glutamine, 110 mg liter−1 sodium pyruvate (DMEM) (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Fisher Scientific), penicillin, and streptomycin. One day prior to infection, cells were seeded in 24-well plates (5 × 105 cells per well) with DMEM (minus phenol red) supplemented with 200 U ml−1 gamma interferon (IFN-γ) (Millipore) and 100 ng ml−1 S. Typhimurium lipopolysaccharide (Sigma). Nitric oxide production by iNOS was inhibited by addition of 2 mM NG-monomethyl-l-arginine monoacetate (l-NMMA; AG Scientific). Overnight AS168 and AS172 cultures were harvested, and 0.1 ml of cells was opsonized in mouse serum. Macrophages were infected with opsonized bacteria at a multiplicity of infection (MOI) of 10:1. The plate was spun at 1,000 rpm for 5 min to synchronize infection followed by 20 min of incubation in a CO2 incubator. Wells were washed three times with PBS followed by addition of fresh medium containing IFN-γ, l-NMMA, and 20 µg ml−1 gentamicin. Three wells each were immediately processed for T0 samples by lysing macrophages with 1% Triton X-100. After 13 h, the growth medium was collected for analysis of nitric oxide production by the Griess reaction (46). Wells were washed with PBS, and macrophages were lysed with 1% Triton X-100 to collect surviving bacteria. Bacteria from three wells were pooled and fixed for analysis by flow cytometry.
Detection of free zinc by FRET.
Bacterial cells were fixed in 1 ml 2.5% paraformaldehyde for 30 min at 37°C followed by resuspension in 1 ml phosphate-buffered saline (PBS). Fixed bacterial cells were analyzed using an LSR II flow cytometer (Becton, Dickinson). CFP and fluorescence resonance energy transfer (FRET) signals were detected after excitation at 405 nm, and emission was collected with a 450/50-nm filter for the CFP channel and a 530/30-nm filter for the FRET channel. The cp173Venus protein was excited at 488 nm, and emission was collected with a 530/30-nm filter. Cells were gated according to forward and side scatter (FSC/SSC), and photomultiplier tube voltages were adjusted using the signal from cells expressing only CFP (AS169) or only cp173Venus (AS170) (see Fig. S7 in the supplemental material). Data were processed with FlowJo v10.3 software (TreeStar, Inc.), and FRET ratios for each event were calculated in the double-positive cell population according to the following equation: FRET ratio = [FRET intensity − a × (CFP intensity) − b × (cp173Venus intensity)]/CFP intensity, where a is mean FRET intensity/CFP intensity in cells expressing CFP only and b is mean FRET intensity/cp173Venus intensity in cells expressing cp173Venus only. A total of 10,000 events were collected for each experiment.
Flow cytometry strategy for calculating FRET ratio. (A) Donor (CFP) bleed-through in the FRET channel was calculated as the mean value of FRET intensity/CFP intensity in CFP-expressing cells. Direct excitation of the acceptor (cp173Venus) was calculated as the mean FRET intensity/cp173Venus intensity in cp173Venus-expressing cells. These values were used to calculate the FRET ratio in cells expressing the full biosensor according to the formula listed in Materials and Methods. The images show histograms of Ratio_CFP and Ratio_cp173Venus along with their gating strategies. Live bacteria were gated from forward versus side scatter plots (FSC versus SSC). Live bacteria were further gated based on CFP and cp173Venus intensities. (B) The histograms and gating strategies of control strains used for infecting RAW 264.7 macrophages (AS169, AS170, JK237, and AS172). The FRET ratio was calculated for each event according to the formula presented in Materials and Methods. Histograms of each strain are shown from the cells present in the double-positive gate (CFP+, cp173Venus+). The double-positive gate was created on a CFP-versus-cp173Venus scatter plot. Download FIG S7, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To validate the response of the zinc FRET biosensor, strains AS168 and AS172 were grown overnight in LB-Amp and diluted to an OD600 of 0.2 in 5 ml fresh medium supplemented with ZnSO4. Following 30 min of growth, 1 ml of culture was harvested by centrifugation, and cells were fixed for analysis.
Competitive infections.
All mouse work was approved by the University of Washington Institutional Animal Care and Use Committee (IACUC) and performed according to protocol 3373-01. Ten-week-old female C3H/HeOuJ mice were obtained from The Jackson Laboratory. Three days prior to infection, one group was switched to drinking water containing 500 µg ml−1 l-N6-(1-iminoethyl)lysine dihydrochloride (l-NIL). Bacteria were grown overnight in 5 ml LB and then diluted in PBS. Wild-type and mutant bacteria (JK237 and EF528) were combined to form a 1:1 ratio of 2 × 103 total CFU ml−1, and mice were injected intraperitoneally with 500 µl of cells. Quantitative plating followed by selective patching of 100 colonies onto selective plates was used to determine the input CFU and input ratios. Five days postinfection, the mice were euthanized. Livers and spleens were homogenized in PBS, diluted, and plated for enumeration of CFU on LB agar. Colonies were then patched onto appropriate selective medium to determine the output ratio of wild-type to mutant organisms. The output ratio was compared to the input ratio to determine the competitive index (CI) (47). CI is the ratio of (mutant/wild type)output to (mutant/wild type)input. A CI of 1 indicates that wild-type and mutant cells have equal fitness during infection, while a CI of <1 indicates that the mutant has a competitive disadvantage. Statistical analysis was performed using the Wilcoxon rank sum test to determine the significance of each CI in GraphPad Prism (version 6).
ACKNOWLEDGMENTS
We thank Rodolfo Urbano for construction of the pRU001 plasmid. We thank Mary Stewart from the Cookson Lab at the University of Washington for construction of the BC1459 strain. We thank Carol Fierke from the Department of Biological Chemistry at the University of Michigan Medical School and Richard Thompson from the Department of Biochemistry and Molecular Biology at the University of Maryland School of Medicine for technical advice and gifts of reagents. We thank Amy Palmer from the Department of Chemistry and Biochemistry at the University of Colorado Boulder for the pcDNA3.1-ZapCV5 construct.
Funding for this work was supplied by NIH grant AI118962 to Ferric C. Fang, NIH fellowship AI112101 to Elaine R. Frawley, Rhodes College startup funding to Elaine R. Frawley, and grant HL54926 to Harry Ischiropoulos. The funding agencies had no role in the experimental design, data collection, data interpretation, or decision to publish this work.
E.R.F., J.E.K., S.J.L., A.S., P.-T.D., H.I., and F.C.F. designed experiments and analyzed data. J.E.K. prepared the samples for proteomic analysis based on methodology designed by P.-T.D. and H.I. P.-T.D. and H.I. carried out the experimental analysis and analyzed the data. E.R.F., J.E.K., and S.J.L. conducted the experiments related to zinc homeostasis. A.S. conducted the experiments involving FRET analysis. E.R.F. and F.C.F. wrote the paper with significant editorial input from J.E.K., S.J.L., A.S., P.-T.D., and H.I.
Footnotes
Citation Frawley ER, Karlinsey JE, Singhal A, Libby SJ, Doulias P-T, Ischiropoulos H, Fang FC. 2018. Nitric oxide disrupts zinc homeostasis in Salmonella enterica serovar Typhimurium. mBio 9:e01040-18. https://doi.org/10.1128/mBio.01040-18.
REFERENCES
- 1.MacMicking JD, Nathan C, Hom G, Chartrain N, Fletcher DS, Trumbauer M, Stevens K, Xie QW, Sokol K, Hutchinson N. 1995. Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell 81:641–650. doi: 10.1016/0092-8674(95)90085-3. [DOI] [PubMed] [Google Scholar]
- 2.Ischiropoulos H, Zhu L, Chen J, Tsai M, Martin JC, Smith CD, Beckman JS. 1992. Peroxynitrite-mediated tyrosine nitration catalyzed by superoxide dismutase. Arch Biochem Biophys 298:431–437. doi: 10.1016/0003-9861(92)90431-U. [DOI] [PubMed] [Google Scholar]
- 3.Urbano R, Karlinsey JE, Libby SJ, Doulias PT, Ischiropoulos H, Warheit-Niemi HI, Liggitt DH, Horswill AR, Fang FC. 2018. Host nitric oxide disrupts microbial cell-to-cell communication to inhibit staphylococcal virulence. Cell Host Microbe 23:594–606.e7. doi: 10.1016/j.chom.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreini C, Banci L, Bertini I, Rosato A. 2006. Zinc through the three domains of life. J Proteome Res 5:3173–3178. doi: 10.1021/pr0603699. [DOI] [PubMed] [Google Scholar]
- 5.Schapiro JM, Libby SJ, Fang FC. 2003. Inhibition of bacterial DNA replication by zinc mobilization during nitrosative stress. Proc Natl Acad Sci U S A 100:8496–8501. doi: 10.1073/pnas.1033133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binet MR, Cruz-Ramos H, Laver J, Hughes MN, Poole RK. 2002. Nitric oxide releases intracellular zinc from prokaryotic metallothionein in Escherichia coli. FEMS Microbiol Lett 213:121–126. doi: 10.1111/j.1574-6968.2002.tb11295.x. [DOI] [PubMed] [Google Scholar]
- 7.Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J. 1992. S-Nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Natl Acad Sci U S A 89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patzer SI, Hantke K. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol 28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 9.Grass G, Franke S, Taudte N, Nies DH, Kucharski LM, Maguire ME, Rensing C. 2005. The metal permease ZupT from Escherichia coli is a transporter with a broad substrate spectrum. J Bacteriol 187:1604–1611. doi: 10.1128/JB.187.5.1604-1611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Outten CE, O’Halloran TV. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 11.Osman D, Foster AW, Chen J, Svedaite K, Steed JW, Lurie-Luke E, Huggins TG, Robinson NJ. 2017. Fine control of metal concentrations is necessary for cells to discern zinc from cobalt. Nat Commun 8:1884. doi: 10.1038/s41467-017-02085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beard SJ, Hashim R, Membrillo-Hernández J, Hughes MN, Poole RK. 1997. Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol Microbiol 25:883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x. [DOI] [PubMed] [Google Scholar]
- 13.Worlock AJ, Smith RL. 2002. ZntB is a novel Zn2+ transporter in Salmonella enterica serovar Typhimurium. J Bacteriol 184:4369–4373. doi: 10.1128/JB.184.16.4369-4373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang K, Wang D, Frederiksen RF, Rensing C, Olsen JE, Fresno AH. 2017. Investigation of the role of genes encoding zinc exporters zntA, zitB, and fieF during Salmonella Typhimurium infection. Front Microbiol 8:2656. doi: 10.3389/fmicb.2017.02656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campoy S, Jara M, Busquets N, Pérez De Rozas AM, Badiola I, Barbé J. 2002. Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar Typhimurium virulence. Infect Immun 70:4721–4725. doi: 10.1128/IAI.70.8.4721-4725.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto M, Ichimura T, Mizoguchi H, Tanaka K, Fujimitsu K, Keyamura K, Ote T, Yamakawa T, Yamazaki Y, Mori H, Katayama T, Kato J. 2005. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol Microbiol 55:137–149. doi: 10.1111/j.1365-2958.2004.04386.x. [DOI] [PubMed] [Google Scholar]
- 17.Hausladen A, Privalle CT, Keng T, DeAngelo J, Stamler JS. 1996. Nitrosative stress: activation of the transcription factor OxyR. Cell 86:719–729. doi: 10.1016/S0092-8674(00)80147-6. [DOI] [PubMed] [Google Scholar]
- 18.Khan S, Brocklehurst KR, Jones GW, Morby AP. 2002. The functional analysis of directed amino-acid alterations in ZntR from Escherichia coli. Biochem Biophys Res Commun 299:438–445. doi: 10.1016/S0006-291X(02)02660-8. [DOI] [PubMed] [Google Scholar]
- 19.Karlinsey JE, Bang IS, Becker LA, Frawley ER, Porwollik S, Robbins HF, Thomas VC, Urbano R, McClelland M, Fang FC. 2012. The NsrR regulon in nitrosative stress resistance of Salmonella enterica serovar Typhimurium. Mol Microbiol 85:1179–1193. doi: 10.1111/j.1365-2958.2012.08167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D, Hosteen O, Fierke CA. 2012. ZntR-mediated transcription of zntA responds to nanomolar intracellular free zinc. J Inorg Biochem 111:173–181. doi: 10.1016/j.jinorgbio.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei Y, Fu D. 2005. Selective metal binding to a membrane-embedded aspartate in the Escherichia coli metal transporter YiiP (FieF). J Biol Chem 280:33716–33724. doi: 10.1074/jbc.M506107200. [DOI] [PubMed] [Google Scholar]
- 22.Grass G, Fan B, Rosen BP, Franke S, Nies DH, Rensing C. 2001. ZitB (YbgR), a member of the cation diffusion facilitator family, is an additional zinc transporter in Escherichia coli. J Bacteriol 183:4664–4667. doi: 10.1128/JB.183.15.4664-4667.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiedler BL, Van Buskirk S, Carter KP, Qin Y, Carpenter MC, Palmer AE, Jimenez R. 2017. Droplet microfluidic flow cytometer for sorting on transient cellular responses of genetically encoded sensors. Anal Chem 89:711–719. doi: 10.1021/acs.analchem.6b03235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore WM, Webber RK, Jerome GM, Tjoeng FS, Misko TP, Currie MG. 1994. L-N6-(1-iminoethyl)lysine: a selective inhibitor of inducible nitric oxide synthase. J Med Chem 37:3886–3888. doi: 10.1021/jm00049a007. [DOI] [PubMed] [Google Scholar]
- 25.Marino SM, Gladyshev VN. 2012. Analysis and functional prediction of reactive cysteine residues. J Biol Chem 287:4419–4425. doi: 10.1074/jbc.R111.275578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. 2010. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci U S A 107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imlay JA. 2014. The mismetallation of enzymes during oxidative stress. J Biol Chem 289:28121–28128. doi: 10.1074/jbc.R114.588814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irving H, Williams RJP. 1948. Order of stability of metal complexes. Nature 162:746–747. doi: 10.1038/162746a0. [DOI] [Google Scholar]
- 29.Xu FF, Imlay JA. 2012. Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl Environ Microbiol 78:3614–3621. doi: 10.1128/AEM.07368-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, Brandli I, Wang C, Tailleux L, Tilleul S, Charrière GM, Waddell SJ, Foti M, Lugo-Villarino G, Gao Q, Maridonneau-Parini I, Butcher PD, Castagnoli PR, Gicquel B, de Chastellier C, Neyrolles O. 2011. Mycobacterial p(1)-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe 10:248–259. doi: 10.1016/j.chom.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irazoki O, Aranda J, Zimmermann T, Campoy S, Barbé J. 2016. Molecular interaction and cellular location of RecA and CheW proteins in Salmonella enterica during SOS response and their implication in swarming. Front Microbiol 7:1560. doi: 10.3389/fmicb.2016.01560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlinsey JE. 2007. Lambda-Red genetic engineering in Salmonella enterica serovar Typhimurium. Methods Enzymol 421:199–209. doi: 10.1016/S0076-6879(06)21016-4. [DOI] [PubMed] [Google Scholar]
- 34.Aldridge PD, Wu C, Gnerer J, Karlinsey JE, Hughes KT, Sachs MS. 2006. Regulatory protein that inhibits both synthesis and use of the target protein controls flagellar phase variation in Salmonella enterica. Proc Natl Acad Sci U S A 103:11340–11345. doi: 10.1073/pnas.0602127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berggren RE, Wunderlich A, Ziegler E, Schleicher M, Duke RC, Looney D, Fang FC. 1995. HIV gp120-specific cell-mediated immune responses in mice after oral immunization with recombinant Salmonella. J Acquir Immune Defic Syndr Hum Retrovirol 10:489–495. doi: 10.1097/00042560-199510050-00001. [DOI] [PubMed] [Google Scholar]
- 37.Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. 2004. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci U S A 101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frawley ER, Crouch ML, Bingham-Ramos LK, Robbins HF, Wang W, Wright GD, Fang FC. 2013. Iron and citrate export by a major facilitator superfamily pump regulates metabolism and stress resistance in Salmonella Typhimurium. Proc Natl Acad Sci U S A 110:12054–12059. doi: 10.1073/pnas.1218274110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64:2240–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drecktrah D, Levine-Wilkinson S, Dam T, Winfree S, Knodler LA, Schroer TA, Steele-Mortimer O. 2008. Dynamic behavior of Salmonella-induced membrane tubules in epithelial cells. Traffic 9:2117–2129. doi: 10.1111/j.1600-0854.2008.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaffrey SR, Snyder SH. 2001. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 42.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stauffer GV, Brenchley JE. 1974. Evidence for the involvement of serine transhydroxymethylase in serine and glycine interconversions in Salmonella typhimurium. Genetics 77:185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scrimgeour KG, Huennekens FM. 1962. Serine hydroxymethylase. Methods Enzymol 3:838–843. [Google Scholar]
- 45.Will WR, Bale DH, Reid PJ, Libby SJ, Fang FC. 2014. Evolutionary expansion of a regulatory network by counter-silencing. Nat Commun 5:5270. doi: 10.1038/ncomms6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vazquez-Torres A, Jones-Carson J, Mastroeni P, Ischiropoulos H, Fang FC. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J Exp Med 192:227–236. doi: 10.1084/jem.192.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beuzón CR, Holden DW. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect 3:1345–1352. doi: 10.1016/S1286-4579(01)01496-4. [DOI] [PubMed] [Google Scholar]
- 48.Lancaster JR, Hibbs JB. 1990. EPR demonstration of iron-nitrosyl complex formation by cytotoxic activated macrophages. Proc Natl Acad Sci U S A 87:1223–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richardson AR, Soliven KC, Castor ME, Barnes PD, Libby SJ, Fang FC. 2009. The Base Excision Repair system of Salmonella enterica serovar Typhimurium counteracts DNA damage by host nitric oxide. PLoS Pathog 5:e1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freeman BA, Baker PR, Schopfer FJ, Woodcock SR, Napolitano A, d’Ischia M. 2008. Nitro-fatty acid formation and signaling. J Biol Chem 283:15515–15519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katayama A, Tsujii A, Wada A, Nishino T, Ishihama A. 2002. Systematic search for zinc-binding proteins in Escherichia coli. Eur J Biochem 269:2403–2413. [DOI] [PubMed] [Google Scholar]
- 52.Grass G, Wong MD, Rosen BP, Smith RL, Rensing C. 2002. ZupT is a Zn(II) uptake system in Escherichia coli. J Bacteriol 184:864–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patzer SI, Hantke K. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J Biol Chem 275:24321–24332. [DOI] [PubMed] [Google Scholar]
- 54.Outten CE, Tobin DA, Penner-Hahn JE, O'Halloran TV. 2001. Characterization of the metal receptor sites in Escherichia coli Zur, an ultrasensitive zinc(II) metalloregulatory protein. Biochemistry 40:10417–10423. [DOI] [PubMed] [Google Scholar]
- 55.Gilston BA, Wang S, Marcus MD, Canalizo-Hernandez MA, Swindell EP, Xue Y, Mondragon A, O'Halloran TV. 2014. Structural and mechanistic basis of zinc regulation across the E. coli Zur regulon. PLoS Biol 12:e1001987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rensing C, Mitra B, Rosen BP. 1997. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci U S A 94:14326–14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brocklehurst KR, Hobman JL, Lawley B, Blank L, Marshall SJ, Brown NL, Morby AP. 1999. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol Microbiol 31:893–902. [DOI] [PubMed] [Google Scholar]
- 58.Lu M, Fu D. 2007. Structure of the zinc transporter YiiP. Science 317:1746–1748. [DOI] [PubMed] [Google Scholar]
- 59.Ammendola S, Pasquali P, Pistoia C, Petrucci P, Petrarca P, Rotilio G, Battistoni A. 2007. High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect Immun 75:5867–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cerasi M, Liu JZ, Ammendola S, Poe AJ, Petrarca P, Pesciaroli M, Pasquali P, Raffatellu M, Battistoni A. 2014. The ZupT transporter plays an important role in zinc homeostasis and contributes to Salmonella enterica virulence. Metallomics 6:845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y, Zhang Q, Hu M, Yu K, Fu J, Zhou F, Liu X. 2015. Proteomic analyses of intracellular Salmonella enterica serovar Typhimurium reveal extensive bacterial adaptations to infected host epithelial cells. Infect Immun 83:2897–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kato J, Hashimoto M. 2007. Construction of consecutive deletions of the Escherichia coli chromosome. Mol Syst Biol 3:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knuth K, Niesalla H, Hueck CJ, Fuchs TM. 2004. Large-scale identification of essential Salmonella genes by trapping lethal insertions. Mol Microbiol 51:1729–1744. [DOI] [PubMed] [Google Scholar]
- 64.Richardson AR, Payne EC, Younger N, Karlinsey JE, Thomas VC, Becker LA, Navarre WW, Castor ME, Libby SJ, Fang FC. 2011. Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar Typhimurium. Cell Host Microbe 10:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brandes N, Rinck A, Leichert LI, Jakob U. 2007. Nitrosative stress treatment of E. coli targets distinct set of thiol-containing proteins. Mol Microbiol 66:901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ibáñez MM, Checa SK, Soncini FC. 2015. A single serine residue determines selectivity to monovalent metal ions in metalloregulators of the MerR family. J Bacteriol 197:1606–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kapetanovic R, Bokil NJ, Achard ME, Ong CY, Peters KM, Stocks CJ, Phan MD, Monteleone M, Schroder K, Irvine KM, Saunders BM, Walker MJ, Stacey KJ, McEwan AG, Schembri MA, Sweet MJ. 2016. Salmonella employs multiple mechanisms to subvert the TLR-inducible zinc-mediated antimicrobial response of human macrophages. FASEB J 30:1901–1912. [DOI] [PubMed] [Google Scholar]
- 68.Kehl-Fie TE, Skaar EP. 2010. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol 14:218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, Pasquali P, Lane TE, Chazin WJ, Vogl T, Roth J, Skaar EP, Raffatellu M. 2012. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11:227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, Crosa JH, Falkow S. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95–113. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GlyA enzymatic activity is inhibited by NO·. S. Typhimurium expressing GlyA from a plasmid was treated with 10 mM diethylamine (DEA) as a mock treatment or 10 mM diethylamine NONOate (DEANO). Cell lysates were assayed for serine hydroxymethyltransferase activity by monitoring the glycine-dependent reduction of formaldehyde using a spectrophotometer at 420 nm. The specific activities of the DEANO-treated lysates were normalized to the activities of the corresponding mock DEA-treated lysate. Data are presented as percent specific activity compared to mock treatment. Error bars represent standard deviation from the mean for three biological replicates. Each biological replicate was analyzed in triplicate. The asterisk indicates statistical significance of P = 0.01 by paired two-tailed t test. Download FIG S1, TIF file, 0.2 MB (247.5KB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Peptides identified in the analysis of the S-nitrosoproteome of S. Typhimurium. Download TABLE S1, XLS file, 0.1 MB (102.5KB, xls) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mean fluorescence of GFP expressed from the S. Typhimurium hmp and zntA promoters increases in response to NO·. S. Typhimurium containing a plasmid expressing GFP from either the hmp (A) or zntA (B) promoter was treated with 2 mM DEANO and compared to untreated cells by flow cytometry at 5, 15, 30, 45, and 60 min. Mean fluorescence intensity increased in response to NO· for both promoter constructs at all times posttreatment, indicating that GFP expression occurred. Data are the means from three biological replicates, and error bars represent standard deviations. (A) An asterisk indicates statistical significance of P = 0.002 (5 min) or P < 0.001 (15, 30, 45, and 60 min) by unpaired two-tailed t test. (B) An asterisk indicates statistical significance of P = 0.04 (5 min), P = 0.009 (15 min), and P < 0.001 (30, 45, and 60 min) by unpaired two-tailed t test. Download FIG S2, TIF file, 0.8 MB (803.7KB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of either ZntA or ZitB, but not ZntB, is capable of correcting mutant growth defects in excess zinc. (A and B) The impaired growth of both a ΔzntA mutant (pink) and a ΔzntA ΔzntB mutant (red) can be fully complemented by expression of ZntA from its native promoter on a low-copy-number plasmid (purple [A] or orange [B]), but the growth defect of a ΔzntA ΔzntB mutant cannot be complemented by expression of ZntB (green [B]). (C and D) Both a ΔzntA ΔzitB mutant (green [C]) and a ΔzntA ΔzntB ΔzitB mutant (orange [D]) can be fully complemented by heterologous expression of either ZntA (pink [C] and violet [D]) or ZitB (gold [C] and light green [D]). Data are the means from 9 biological replicates from three trials. Statistical significance was determined by unpaired two-tailed t test. Download FIG S3, TIF file, 1 MB (1,004.9KB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
YiiP does not contribute to zinc homeostasis in S. Typhimurium. (A) A ΔyiiP mutant (pink) is no more sensitive to zinc than wild type (wild-type), and both strains grew equally well in the presence or absence of excess zinc. (B) A ΔzntA ΔzitB ΔyiiP strain (purple) is no more sensitive to excess zinc than a ΔzntA ΔzitB strain (green). A ΔzntA ΔzntB ΔzitB ΔyiiP strain (gold) is no more zinc-sensitive than a ΔzntA ΔzntB ΔzitB strain (orange). Together, these results indicate that YiiP does not contribute to zinc homeostasis in S. Typhimurium under these experimental conditions, even in the absence of all other known zinc efflux systems. Data are the means from 12 biological replicates from four trials. Statistical significance was determined by unpaired two-tailed t test. Download FIG S4, TIF file, 0.7 MB (766.5KB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A ΔzntA ΔzitB mutant does not show a decrease in total cellular zinc following NO· treatment. ΔzntA ΔzitB S. Typhimurium at an OD600 of ≈1 was treated with 2 mM diethlyamine NONOate (DEANO), and total cellular zinc was measured at various times posttreatment by ICP-MS. Total intracellular zinc was not significantly different from baseline (T0, untreated) at any of the times examined. Download FIG S5, TIF file, 0.8 MB (827.8KB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A fluorescence resonance energy transfer (FRET)-based biosensor for zinc shows accumulation of free intracellular zinc in a ΔzntA ΔzitB mutant but not in wild-type S. Typhimurium. A genetically encoded zinc biosensor, ZapCV5, was expressed constitutively from a plasmid in S. Typhimurium. Due to the constant presence of the sensor, a portion of sensor proteins are metallated during normal cell growth. The amount of FRET signal from the sensor was greater in a ΔzntA ΔzitB mutant than in wild type even when no exogenous ZnSO4 (0 mM) was added to the growth medium. At increased concentrations of ZnSO4, FRET signal increased in the ΔzntA ΔzitB mutant but not in the wild-type strain. None of the wild-type FRET ratios were significantly different from one another, whereas the ΔzntA ΔzitB FRET ratio was significantly greater than the wild-type ratio at each concentration (P < 0.001). The FRET ratios for the ΔzntA ΔzitB mutant at 0.25 mM and 0.5 mM ZnSO4 were also significantly elevated compared to the FRET ratio in the absence of exogenous zinc (P < 0.001). Statistical significance was analyzed by one-way analysis of variance. Download FIG S6, TIF file, 0.9 MB (897.8KB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains and plasmids used in this study. Download TABLE S2, DOCX file, 0.1 MB (108.8KB, docx) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequences of primers used in this study. Download TABLE S3, DOCX file, 0.1 MB (101.6KB, docx) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Flow cytometry strategy for calculating FRET ratio. (A) Donor (CFP) bleed-through in the FRET channel was calculated as the mean value of FRET intensity/CFP intensity in CFP-expressing cells. Direct excitation of the acceptor (cp173Venus) was calculated as the mean FRET intensity/cp173Venus intensity in cp173Venus-expressing cells. These values were used to calculate the FRET ratio in cells expressing the full biosensor according to the formula listed in Materials and Methods. The images show histograms of Ratio_CFP and Ratio_cp173Venus along with their gating strategies. Live bacteria were gated from forward versus side scatter plots (FSC versus SSC). Live bacteria were further gated based on CFP and cp173Venus intensities. (B) The histograms and gating strategies of control strains used for infecting RAW 264.7 macrophages (AS169, AS170, JK237, and AS172). The FRET ratio was calculated for each event according to the formula presented in Materials and Methods. Histograms of each strain are shown from the cells present in the double-positive gate (CFP+, cp173Venus+). The double-positive gate was created on a CFP-versus-cp173Venus scatter plot. Download FIG S7, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2018 Frawley et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.