Abstract
Background & objectives:
Hepatitis A virus (HAV) infection is a major cause of childhood hepatitis, prevalent worldwide. HAV is classified into seven genotypes I-VII; genotypes III and I are the most common among humans. The present work was carried out to identify the genotypes prevalent in children suspected to have acute viral hepatitis (AVH), hospitalized at a tertiary care centre in northwest India.
Methods:
A total of 1269 blood samples from children (0-15 yr of age) clinically suspected of viral hepatitis were screened for anti-HAV IgM. Acute phase serum was processed for RNA extraction and amplified by nested polymerase chain reaction (PCR) followed by sequencing of representative samples.
Results:
Among the 1269 samples tested, 642 (50.59%) were positive for anti-HAV IgM; among the positive samples, 171 patients having a history of less than seven days were tested by PCR, of whom 141 (82.45%) were found to be PCR positive. Nucleotide sequencing of a representative 44 samples showed high homology; all the samples were found to be of genotype IIIA.
Interpretation & conclusions:
Hepatitis A was prevalent during July to September and in predominantly children less than five years age. Only genotype IIIA was detected in all the samples.
Keywords: Genotyping, HAV, hepatitis A, phylogenetic analysis, polymerase chain reaction, serology
Hepatitis is a major health problem in both developing and developed countries. An estimated 1.4 million clinical cases of hepatitis A occur worldwide every year1. Hepatitis A occurs sporadically and also as outbreaks. Outbreaks of hepatitis A virus (HAV) have been reported from different States of India with varying positivity of 37.7-100 per cent (HAV- IgM) from Kerala (37.7-87%), Himachal Pradesh (63%) and Tamil Nadu (100%)2,3,4.
Ninety per cent of children are exposed to HAV by the age of five years, and almost all by adolescence1, majority of hepatitis A patients recover completely and fatality rate is low. The estimated mortality rate is 0.1 per cent for children less than 15 yr, 0.3 per cent for adults aged 15-39 yr and 2.1 per cent for adults aged 40 and above5. HAV infection in patients with pre-existing chronic liver disease is associated with high mortality5.
The HAV virion is a non-enveloped, spherical particle with a diameter of 27-32 nm. It contains 7.5-kb single-stranded RNA genome, which displays a high degree of antigenic (amino acid) and genetic (nucleotide) conservation. Based on its nucleotide sequence analysis of 5’UTR region, it has been classified into seven different genotypes [human (I-III & VII) and simian (IV-VI) genotypes]. Genotypes I and III are each divided into sub-genotypes A and B5,6. The 5’UTR region of its genome contains a complicated stem and loop structure with two pyrimidine-rich tracts, indicating the presence of a complex secondary structure, with enough genetic diversity, which is useful in identification of its genotypes and sub-genotypes. Majority of human HAV genotypes belong to the genotypes I and III; different genotypes (IA, IB and IIIA) with a predominance of genotype IIIA are known to be circulating in India2,3,4,7,8.
The present study was planned to do molecular characterization of HAV IgM positivity in children hospitalized at a tertiary care centre at Jaipur, Rajasthan, India, to find out the genotypes prevalent in this part of the country.
Material & Methods
A total of 1269 blood samples were collected from the patients between 0 and 15 yr of age, hospitalized at J.K. Lone Hospital, a paediatric hospital attached to SMS Medical College, Jaipur, having signs and symptoms suggestive of acute viral hepatitis (AVH). Duration of the study was from January 2012 to December 2014. Questionnaire specifying patient's name, age, sex, address, duration of illness, sign and symptoms of hepatitis was filled, and informed written consent was taken from parent/guardian. Peripheral blood samples (5 ml) were collected in a plain vial. Serum was separated and aliquoted in two vials; one was used for serological analysis and other was stored at −80°C until further use. Serum biochemistry [bilirubin, serum aspartate aminotranferase (AST), serum alanine aminotransferase (ALT), etc.] values were recorded from reports of investigations. Institutional Ethics Committee approval was also taken for the study. Sample collection, serological testing and molecular characterization were carried out at the Virology Laboratory, Advanced Basic Sciences and Clinical Research Laboratory, SMS Medical College, Jaipur.
The minimum sample size required for the study was calculated as 625 (based on the prevalence rate of 6.7%, at 95% confidence interval, 2% absolute error and 0.8 power) employing the previously described formula9. As only 1269 samples were included, power of the study was 0.9 at 99 per cent confidence interval.
Inclusion & exclusion criteria: All paediatric patients hospitalized with signs and symptoms of AVH (0-15 yr age group) from January 2012 to December 2014 were included in the study. Outdoor patients and patients not having signs or symptoms of hepatitis were excluded from the study.
Enzyme-linked immunosorbent assay (ELISA): ELISA was performed for detection of anti-HAV IgM antibody using commercial kit (DiaSorin, Italy) as per the manufacturer's instructions. The reading was taken at 450 nm on Infinite N200 Pro NanoQuant Spectrophotometer (Tecan, Switzerland). Samples were also tested for hepatitis B surface antigen, hepatitis C virus total antibody and hepatitis E virus IgM antibody at Virus Diagnostic Research Laboratory (ICMR-VDRL).
Nucleic acid extraction and nested polymerase chain reaction (PCR): Viral RNA was extracted from (171 ELISA-positive samples with less than seven days history of illness) frozen aliquots of serum sample with easyMAG extractor (bioMerieux, France) and RNA quantity was checked using Infinite N200 Pro NanoQuant Spectrophotometer (Tecan, Switzerland) by calculating the ratio of absorbance at 260 and 280 nm. Sample containing an amount of 1-2 μg total RNA was used for complementary DNA (cDNA) synthesis by High Capacity cDNA synthesis kit (Applied Biosystems, USA). Nested PCR was done to determine virus genotype, using two sets of primers [5’NCR (outer) forward primer (GGCTACGGGTGAAACCTCTT) and 5’NCR (outer) reverse primer (CCAATTTTGCAACTTCATG)] and [5’NCR (inner) forward primer (TAACAGCGGCGGATATTGGTG) and 5’NCR (inner) reverse primer (GGTCAAGGCCACTCCCAAC)]2. The amplicons were analyzed on 2 per cent agarose/Tris-borate-EDTA (TBE) gel stained with ethidium bromide and visualized on gel documentation system (Bio-Rad, USA) and inner amplified products were excised from gel for further purification.
Sequencing: Representative PCR-positive products (44) were processed for sequencing. Briefly; inner amplified PCR products were purified using PureLink Quick Gel Extraction and PCR Purification Combo Kit (Invitrogen, USA). These were further checked on 2 per cent agarose/Tris borate EDTA (TBE) gel for the purity and integrity of the band. Both the forward and reverse strands of the amplicons were sequenced using BigDye Terminator Cycle Sequencing Ready Reaction Kit version 3.0 (Applied Biosystems, USA) on 3500 DX Genetic analyzer (ABI, USA).
Phylogenetic analysis: The sequences generated from the sequencer were confirmed by BLAST analysis (www.ncbi.nlm.nih.gov/blast) and aligned by NJ algorithm at 1000 bootstrap replications using MEGA6.010 (Mega Software Solution Inc., USA). The phylogenetic tree generated was viewed using Tree View.
Statistical analysis: Comparison between data sets was performed using one-way analysis of variance followed by Student's t test and Chi-square test. All statistical analyses were performed using GraphPad Prism 5.0 (Graph Pad Software Inc., San Diego, CA, USA).
Results
Of the 1269 patients enrolled in the study, 839 (66.11%) were males and 430 (33.88%) were females and 42.31 per cent patients (n=537) were less than five years of age. A total of 642 (50.6%) patients were positive for anti-HAV IgM. Total serum bilirubin level was found to be <5 mg/dl in 779 patients (61.38%), 6-10 mg/dl in 283 patients (22.30%) and >11 mg/dl in 207 patients (16.31%). The levels of AST and ALT were found to be raised with mean AST level of 1315.76±1085.42 in HAV-positive samples versus 1015.42±921.23 in HAV-negative samples. Mean ALT in HAV-positive samples was 1349.67±1039.18 versus 979.63±791.67 in HAV-negative samples.
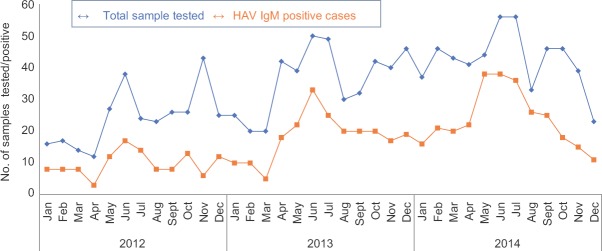
Difference in age-wise positivity (P=0.014) was significant (Table). Of the 642 (50.59%) samples found positive for anti-HAV IgM antibody, 435 (34.27%) were from male patients and 207 (16.33%) female patients. Among them, number of children belonging to age group 0-10 yr were higher (n=567, 55.64%) than other age groups (n=75, 29.88%). Peak positivity of HAV (50 to 86.6%) was seen during June to September, and least positivity (25 to 38%) was seen during December to February (Fig. 1). Of the 171 samples tested by PCR, 141 (82.45%) were found to be positive. On sequencing of representative 44 HAV PCR-positive samples, all were found to be of genotype IIIA (Gene bank Accession number: KT891942-KT891985) as shown in phylogenetic tree (Fig. 2). All the samples showed similarity with other Indian samples from Chandigarh (JX481901-JX481910; JX390729-JX390733)11, Shimla (FJ227129-FJ227134)3, Kottayam (DQ004690-DQ004693)2 and samples from Norway (AJ299464)12 and Japan (AB279734)13.
Table.
Age-wise distribution of hepatitis A virus (HAV) ELISA-positive cases
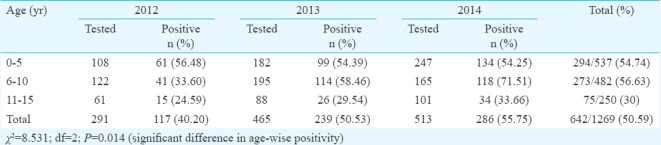
Fig. 1.
Seasonal trends of hepatitis A virus (HAV) from January 2012 to December 2014.
Fig. 2.
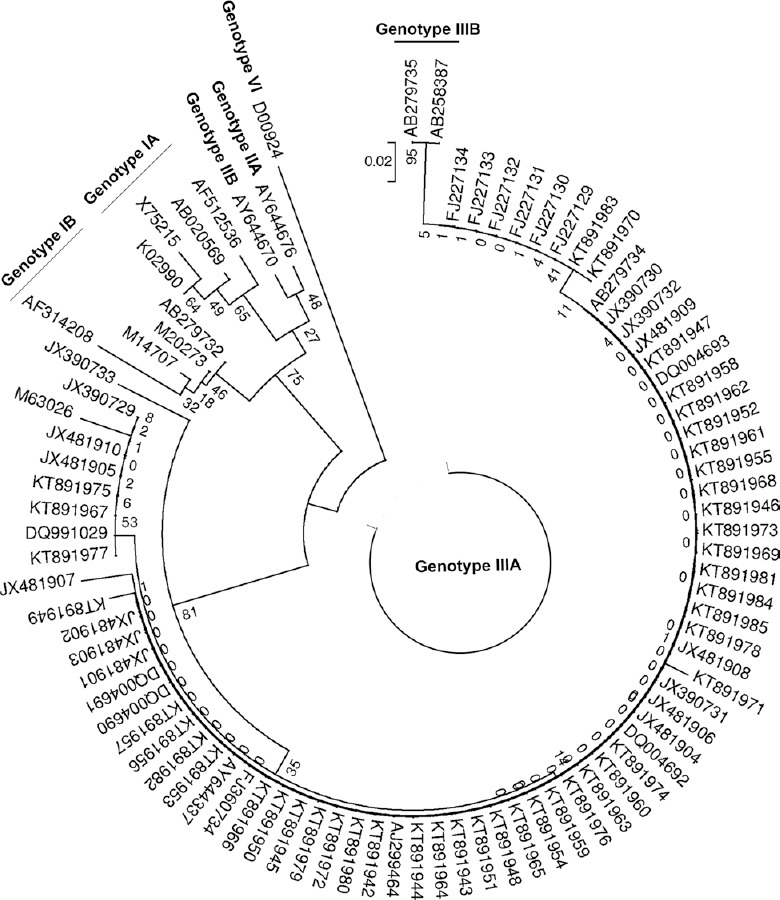
Phylogenetic analysis of 44 PCR positive representative hepatitis A virus samples based on 5’ UTR region.
Substitutions between nucleotides 500 and 700 of the 5’UTR region were observed in seven samples. Substitution of G to A at nucleotide 578 was detected in three samples (KT891967, KT891975 and KT891977) and A to G at nucleotide 629 in one sample (KT891949). Substitution of A to T at nucleotide 598 was detected in one sample (KT891971), while C to T substitution at nucleotide 605 was detected in two samples (KT891970, KT891983) when compared with wild-type HAV GBM/WT RNA (X75215).
Discussion
AVH due to HAV occurs worldwide and is endemic in Asia, Africa, Latin America and the Middle East14. In India, it remains a major public health problem despite improving sanitation, health awareness and socio-economic conditions. The present study was done on children with AVH; 50.59 per cent samples were found positive for anti-HAV IgM antibody. A wide variation in positivity (27.2-74.5%) has been reported from hospitalized patients in India: from Lucknow (27.2%)15, Madras (now Chennai) (38.6%)16 and Hyderabad (74.5%)17. Even wider variation in positivity has been reported among non-hospitalized paediatric patients from 10.3 to 92.85 per cent from various parts of India3,4,7,18.
In the present study, higher HAV IgM positivity of 56.64 per cent was observed in children of age group 6-10 yr; similar finding has been reported from Delhi (59.5%)19. However, other authors have grouped together 0-10 yr age and reported 80.95 per cent positivity from Shimla3 and 60 per cent from Chennai16.
Genotyping of samples provides insight into type of transmission whether the infection is due to indigenous strain or imported strain. In our study on phylogenetic analysis, all the samples tested belonged to genotype IIIA. Studies from various parts of India, such as, Jabalpur7, Himachal Pradesh3, Maharashtra20, Tamil Nadu4, Chandigarh11, also reported genotype IIIA as the only genotype found, circulating in the various parts of India. Circulation of other genotypes have also been reported from different parts of India. Genotype IA was reported in 21 per cent patients from Guwahati along with genotype IIIA in 79 per cent patients21. In a study from Delhi also, genotype III was reported as the predominant genotype (70%) followed by genotype IA (30%)8. Moreover, co-circulation of genotypes IIIA (74.2%) and IB (9.7%) and co-infection (16.1%) with both IIIA and IB genotypes have been reported from Pune22.
Similar to our study, genotype IIIA have been reported to be prevalent in the United States, Central Asian countries, Denmark, Korea, etc6,23,24. Both genotype IA and IIIA have also been found prevalent in Korea23,24 and Spain25. In China, both genotype IA and IB were found to be prevalent26. Genotype IA was prevalent in Mexico, Canada and Argentina, while both genotype IA and IB are known to be prevalent in Jordan, Iraq, Japan, Egypt, Turkey, Brazil, Italy, North Africa, South Africa, Norway, Greece, Thailand and Tunisia27,28,29,30, whereas genotype II has been reported from Netherland, France and Sierra Leone6.
On analyzing genetic diversity by 5’UTR region sequencing, 98-100 per cent nucleotide identity was observed in our samples taking wild type HAV GBM/WT RNA (X75215) as control. Our results were in concordance with various other studies reported from India which showed approximately 98-99.9 per cent nucleotide identity11,31. However, base substitution was observed in seven samples between nucleotides 500 and 700 of the 5’UTR region. Singh et al11 also reported base substitution at 200-500 region of 5’NTR while comparing their strains with wild type HAV GBM/WT RNA (X75215). They observed G to A substitution at nucleotide 324 position in all their samples (which is responsible for stable structure of its genome but has no correlation with disease severity) as was also reported by Fujiwara et al30. Other substitutions such as A161G (33.3%), C207T (53.3%), A559G (20%), C105T (100%), T148C (40%), C187T (100%), G213A (100%), G266A (100%), domain IV substitution (G490A, T517C) and insertion at 431 position have also been reported by Singh et al11; however, no correlation to disease severity has been reported in these substitutions. No correlation could be ascertained between biochemical and clinical profiles of patients with genotypes as only one genotype was detected in our studied patients. However, Yoon et al24 from Korea reported that genotype IIIA was more virulent than genotype IA, while Fujiwara et al30 from Japan reported no apparent associations between disease severity and sequences of the genotype-determining region.
Our study had some limitations. One major limitation was that all our samples were from hospitalized patients, with more severe disease, and higher than normal ALT and AST levels. It is important to study non-hospitalized patients as well as also samples from outbreak cases. Another limitation of our study was that the patients were not followed up, so the information about the clinical course of the disease was not available.
In conclusion, HAV was found to be positive in about 50 per cent of patients, predominantly from June to September. HAV IgM positivity was detected mostly in children below 10 yr of age. Only genotype IIIA was found in our patients and as a result, no correlation with disease severity could be ascertained.
Footnotes
Financial support & sponsorship: Authors acknowledge the financial support from the Indian Council of Medical Research, New Delhi, to the first author (BM) for establishment of ICMR Grade-I Virology Laboratory and Senior Research Fellowship to the second author (AK).
Conflicts of Interest: None.
References
- 1.World Health Organization. [accessed on April 30, 2018];WHO/CDS/CSR/EDC/2014. Hepatitis A. Department of Communicable Disease Surveillance and Response. Available at: http://www.who.int7csr/disease/hepatitis/HepatitisA-whocdscsredc2014-pdf . [Google Scholar]
- 2.Arankalle VA, Sarada Devi KL, Lole KS, Shenoy KT, Verma V, Haneephabi M. Molecular characterization of hepatitis A virus from a large outbreak from Kerala, India. Indian J Med Res. 2006;123:760–9. [PubMed] [Google Scholar]
- 3.Chobe LP, Arankalle VA. Investigation of a hepatitis A outbreak from Shimla Himachal Pradesh. Indian J Med Res. 2009;130:179–84. [PubMed] [Google Scholar]
- 4.Raju S, Rajendran P, Gunasekaran P, Skeriff AK, Mukesh Kumar DJ, Ashok G. Molecular characterization of hepatitis a virus from sporadic and epidemic cases of jaundice in Tamil Nadu, India. Pak J Biotechnol. 2011;8:45–53. [Google Scholar]
- 5.Hollinger FB, Ticehurst JR. Hepatitis A virus. In: Fields virology. Knipe DM, Howley PM, editors. Vol. 1. Philadelphia: Lippincott Williams & Wilkins; 1996. pp. 735–82. [Google Scholar]
- 6.Nainan OV, Xia G, Vaughan G, Margolis HS. Diagnosis of hepatitis A virus infection: a molecular approach. Clin Microbiol Rev. 2006;19:63–79. doi: 10.1128/CMR.19.1.63-79.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barde PV, Shukla MK, Pathak R, Kori BK, Bharti PK. Circulation of hepatitis A genotype IIIA virus in paediatric patients in central India. Indian J Med Res. 2014;139:940–4. [PMC free article] [PubMed] [Google Scholar]
- 8.Hussain Z, Husain SA, Almajhdi FN, Kar P. Immunological and molecular epidemiological characteristics of acute and fulminant viral hepatitis A. Virol J. 2011;8:254. doi: 10.1186/1743-422X-8-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniel WW. Biostatistics: A foundation for analysis in the health sciences. 7th ed. New York: John Wiley & Sons; 1999. [Google Scholar]
- 10.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh MP, Majumdar M, Thapa BR, Gupta PK, Khurana J, Budhathoki B, et al. Molecular characterization of hepatitis A virus strains in a tertiary care health set up in North western India. Indian J Med Res. 2015;141:213–20. doi: 10.4103/0971-5916.155577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stene JK, Skaug K, Blystad H. Surveillance of hepatitis A by molecular epidemiologic studies. Tidsskr Nor Laegeforen. 1999;119:3725–9. [PubMed] [Google Scholar]
- 13.Endo K, Takahashi M, Masuko K, Inoue K, Akahane Y, Okamoto H. Full-length sequences of subgenotype IIIA and IIIB hepatitis A virus isolates: characterization of genotype III HAV genomes. Virus Res. 2007;126:116–27. doi: 10.1016/j.virusres.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Franco E, Meleleo C, Serino L, Sorbara D, Zaratti L. Hepatitis A: Epidemiology and prevention in developing countries. World J Hepatol. 2012;4:68–73. doi: 10.4254/wjh.v4.i3.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain P, Prakash S, Gupta S, Singh KP, Shrivastava S, Singh DD, et al. Prevalence of hepatitis A virus, hepatitis B virus, hepatitis C virus, hepatitis D virus and hepatitis E virus as causes of acute viral hepatitis in North India: A hospital based study. Indian J Med Microbiol. 2013;31:261–5. doi: 10.4103/0255-0857.115631. [DOI] [PubMed] [Google Scholar]
- 16.Malathi S, Mohanavalli B, Menon T, Srilatha P, Sankaranarayanan VS, Raju BB, et al. Clinical and viral marker pattern of acute sporadic hepatitis in children in Madras, South India. J Trop Pediatr. 1998;44:275–8. doi: 10.1093/tropej/44.5.275. [DOI] [PubMed] [Google Scholar]
- 17.Syed R, Mohammed AH, Sindiri PK, Nathani AA, Rao VVR, Satti VP. Sero-epidemiology of hepatitis A virus in Hyderabad, South India. J Med Allied Sci. 2012;2:58–61. [Google Scholar]
- 18.Joshi MS, Bhalla S, Kalrao VR, Dhongade RK, Chitambar SD. Exploring the concurrent presence of hepatitis A virus genome in serum, stool, saliva, and urine samples of hepatitis A patients. Diagn Microbiol Infect Dis. 2014;78:379–82. doi: 10.1016/j.diagmicrobio.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 19.Kotwal A, Singh H, Verma AK, Gupta RM, Jain S, Sinha S, et al. A study of Hepatitis A and E virus seropositivity profile amongst young healthy adults in India. Med J Armed Forces India. 2014;70:225–9. doi: 10.1016/j.mjafi.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chadha MS, Lole KS, Bora MH, Arankalle VA. Outbreaks of hepatitis A among children in Western India. Trans R Soc Trop Med Hyg. 2009;103:911–6. doi: 10.1016/j.trstmh.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 21.Bose M, Bose S, Saikia A, Medhi S, Deka M. Molecular epidemiology of hepatitis A virus infection in Northeast India. J Med Virol. 2015;87:1218–24. doi: 10.1002/jmv.24168. [DOI] [PubMed] [Google Scholar]
- 22.Chitambar S, Joshi M, Lole K, Walimbe A, Vaidya S. Cocirculation of and coinfections with hepatitis A virus subgenotypes IIIA and IB in patients from Pune, Western India. Hepatol Res. 2007;37:85–93. doi: 10.1111/j.1872-034X.2007.00025.x. [DOI] [PubMed] [Google Scholar]
- 23.Lee KO, Jeong SJ, Seong HS, Kim KT, Hwang YS, Kim GY, et al. Genetic analysis of hepatitis A virus isolated from Korea. J Bacteriol Virol. 2009;39:165–71. [Google Scholar]
- 24.Yoon YK, Yeon JE, Kim JH, Sim HS, Kim JY, Park DW, et al. Comparative analysis of disease severity between genotypes IA and IIIA of hepatitis A virus. J Med Virol. 2011;83:1308–14. doi: 10.1002/jmv.22139. [DOI] [PubMed] [Google Scholar]
- 25.Pina S, Buti M, Jardí R, Clemente-Casares P, Jofre J, Girones R. Genetic analysis of hepatitis A virus strains recovered from the environment and from patients with acute hepatitis. J Gen Virol. 2001;82(Pt 12):2955–63. doi: 10.1099/0022-1317-82-12-2955. [DOI] [PubMed] [Google Scholar]
- 26.Cao J, Wang Y, Song H, Meng Q, Sheng L, Bian T, et al. Hepatitis A outbreaks in China during 2006: Application of molecular epidemiology. Hepatol Int. 2009;3:356–63. doi: 10.1007/s12072-008-9116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barameechai K, Sa-Nguanmoo P, Suwannakarn K, Thongmee C, Payungporn S, Chongsrisawat V, et al. Molecular characterisation of the hepatitis A virus circulating in the 2001-2005 outbreaks in Thailand. Ann Trop Med Parasitol. 2008;102:247–57. doi: 10.1179/136485908X278775. [DOI] [PubMed] [Google Scholar]
- 28.Poovorawan K, Chattakul P, Chattakul S, Thongmee T, Theamboonlers A, Komolmit P, et al. The important role of early diagnosis and preventive management during a large-scale outbreak of hepatitis A in Thailand. Pathog Glob Health. 2013;107:367–72. doi: 10.1179/2047773213Y.0000000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kokkinos P, Ziros P, Filippidou S, Mpampounakis I, Vantarakis A. Molecular characterization of hepatitis A virus isolates from environmental and clinical samples in Greece. Virol J. 2010;7:235. doi: 10.1186/1743-422X-7-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujiwara K, Yokosuka O, Imazeki F, Saisho H, Saotome N, Suzuki K, et al. Analysis of the genotype-determining region of hepatitis A viral RNA in relation to disease severities. Hepatol Res. 2003;25:124–34. doi: 10.1016/s1386-6346(02)00245-0. [DOI] [PubMed] [Google Scholar]
- 31.Kulkarni MA, Walimbe AM, Cherian S, Arankalle VA. Full length genomes of genotype IIIA hepatitis A virus strains (1995-2008) from India and estimates of the evolutionary rates and ages. Infect Genet Evol. 2009;9:1287–94. doi: 10.1016/j.meegid.2009.08.009. [DOI] [PubMed] [Google Scholar]