Diffuse intrinsic pontine glioma (DIPG) is a lethal childhood brain cancer and patients face a grim prognosis with few treatment options [7]. Targeted therapies based on actionable genetic mutations may offer DIPG patients novel treatment regimens [9, 10]. Although whole exome sequencing (WES) of tumor tissue can fully characterize the somatic mutational profile, it requires a surgical procedure and is relatively costly and time consuming. Consequently, less invasive and more rapid diagnostic tests are needed to detect actionable brain cancer mutations.
Brain tumors and metastases to the brain shed circulating tumor DNA (ctDNA) into the cerebrospinal fluid (CSF), which can be leveraged for the detection of tumor-associated genetic mutations from minimally invasive lumbar punctures [16]. Droplet digital PCR (ddPCR) is an ultrasensitive PCR method that can detect low copy numbers of DNA, including ctDNA, in CSF [13]. It has proven adept for the detection of ctDNA mutations in CSF from patients with primary brain tumors [3, 5, 14] and central nervous system (CNS) metastases from other cancers [3, 8, 12, 14, 15, 17].
The majority of DIPGs possess a recurrent, potentially actionable mutation to histone 3 (either H3F3A or HIST1H3B) at lysine position 27 (K27M). H3K27M detection in CSF by a combination of nested PCR and Sanger sequencing in DIPG patients [6] as well as by ddPCR in older diffuse midline glioma patients has been reported [11]. Thus far, there have been no extensive studies using ddPCR to quantify ctDNA in the CSF of younger pediatric DIPG patients. Additionally, there are significant gaps in our knowledge, including whether ctDNA abundance depends on location of sample collection and whether ctDNA can quantify tumor growth and treatment response. To help answer these questions, we developed a novel ddPCR assay for the H3F3A K27M mutation and applied it to four pediatric patients with H3F3A K27M-mutant DIPG and non-brainstem GBM, including multi-focal sampling of one patient. Additionally, we generated an in vitro co-culture model of DIPG cells and astrocytes (NHA), evaluating their release of DNA into cell culture media as a means to simulate ctDNA release into the CSF.
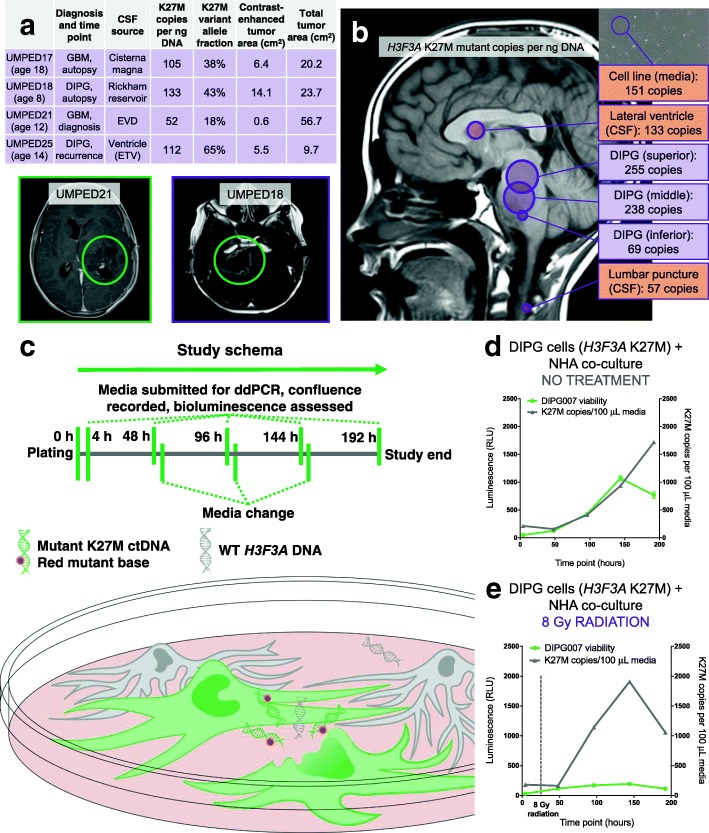
We employed ddPCR because it is a rapid, simple, and ultra-sensitive method of DNA detection capable of accurate quantification down to very low copy number [13]. We designed PCR probes specific to wild-type (WT) H3F3A and mutant K27M sequences (Additional file 1: supporting information, SI), which were validated for low copy detection and linearity by serial dilution of synthetic K27M mutant sequence oligonucleotide (Additional file 2: Figure S1), as well as in control CSF (no CNS tumor) with and without synthetic K27M oligonucleotide (Additional file 2: Figure S2). This fully validated ddPCR method was then used on experimental samples (Fig. 1a). In a prospective cohort of patients who were enrolled in the IRB-approved University of Michigan Brain Tumor CSF Registry, CSF ddPCR results were compared to contrast-enhancing and total tumor cross-sectional area on MRI.
Fig. 1.
a CSF ddPCR results from experimental samples correlated with contrast-enhancing and total tumor cross-sectional area on MRI. b ddPCR of multi-focal sampling shows K27M copy number varies between tumor (purple) and CSF (orange) regions c Co-culture scheme of bioluminescent human DIPG007 cells with NHAs. d DIPG007 cells release ctDNA in proportion to their proliferation. e 8 Gy radiation results in an increase in mutant ctDNA from DIPG007 cells
We found that ddPCR was able to detect the K27M mutation in patient CSF and that the closest relationship emerged between mutant K27M copies per ng of total DNA (hereafter K27M copies) and contrast-enhancing cross-sectional tumor area on MRI (Fig. 1a). We then used ddPCR for multi-focal sampling of an eight-year-old patient with DIPG at autopsy (UMPED18) and observed that K27M copies varied throughout the tumor (Fig. 1b). The number of K27M copies was two-fold higher in CSF from the lateral ventricle as compared to CSF from a lumbar puncture, in accordance with prior studies that have suggested that ctDNA release into the CSF may be reliant upon the location of the tumor adjacent to a CSF reservoir [16]. If this finding is confirmed in future cases with multi-focal sampling, lumbar samples may have reduced sensitivity for CSF ctDNA compared to ventricular samples.
To better understand changes in K27M copy number in response to both growth and treatment of DIPG cells, we created an experimental in vitro model of bioluminescent human DIPG007 cells co-cultured with NHAs (Fig. 1c). We found that DIPG007 cells released more ctDNA into culture media in proportion to their proliferation (Fig. 1d), even when the media was changed frequently to approximate the constant production and resorption of CSF. This suggests that, at least in part, ctDNA correlates with tumor cell proliferation. However, irradiation with 8 Gy resulted in a dramatic increase in mutant ctDNA approximately 72–120 h post radiotherapy (Fig. 1e) before tapering off. The results suggest ddPCR may be a viable method for monitoring response to therapy with an early release of ctDNA indicative of an effective treatment.
The dawn of precision medicine, and its potential benefit to patients, has spurred research into faster, simpler, and less invasive methods of detection of actionable tumor-associated mutations. Due to its great sensitivity and low limit of detection, ddPCR has been used to detect tumor mutations in CSF from a range of cancer patients [1–5, 8, 12, 14, 15, 17]. However, there has been little elaboration within the literature on whether tumor size may be related to the amount of ctDNA detected by ddPCR or its suitability to track response to treatment.
Our pilot study suggests that H3F3A K27M copies in the CSF of children with DIPG and high-grade glioma have a linear relationship with contrast-enhancing cross-sectional tumor area and confirms the importance of proximity of a sample to the tumor. The former observation was further supported by in vitro experiments showing that tumor cell proliferation results in increased ctDNA and that H3F3A K27M copies can be used to follow treatment response due to temporarily enhanced ctDNA release shortly after effective therapies. Our study lays the ground work for the inclusion of CSF analysis with surveillance MRIs in the treatment of this patient population.
Additional files
Supplemental Information. Detailed methods and H3F3A K27M assay design. (DOCX 28 kb)
Figure S1. Serial dilution of K27M mutant oligonucleotide in constant background of wild-type DNA demonstrates consistent detection down to at least 2% VAF under typical experimental conditions, with the possibility of detection at even lower VAF under ideal conditions. One such dilution series is shown above, with (a) showing number of droplets positive for mutant or wild-type H3F3A sequence and (b) showing the corresponding VAF values. Figure S2. Plot of droplets (blue – positive mutant H3F3A K27M, green – positive wildtype H3F3A, grey – negative droplets) from ddPCR performed on (a) non-tumor human CSF spiked with synthetic K27M mutant sequence oligonucleotide and (b) non-tumor human CSF alone. (DOCX 268 kb)
Acknowledgments
The authors thank the patients and their families for participation in this study.
Funding
CK is supported by NIH/NINDS Grant K08-NS099427–01, Michigan Medicine Department of Pediatrics Gorman Scholar Award, The University of Michigan Chad Carr Pediatric Brain Tumor Center, the Chad Tough Foundation, and Hyundai Hope on Wheels. RH is supported by NIH/NINDS RO1-NS093079. RM is a Hyundai Hope on Wheels Scholar.
Availability of data and materials
All somatic mutation information generated from this study has been uploaded to dbGaP (https://www.ncbi.nlm.nih.gov/gap) under accession number phs000673.v2.p1.
Abbreviations
- CNS
Central nervous system
- CSF
Cerebrospinal fluid
- ctDNA
Circulating tumor DNA
- ddPCR
Droplet digital PCR
- DIPG
Diffuse intrinsic pontine glioma
- GBM
Glioblastoma
- LOD
Limit of detection
- NHA
Normal human astrocyte
- WES
Whole exome sequencing
- WT
Wild-type
Authors’ contributions
All authors have contributed in meaningful ways and reviewed the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients have consented to participate.
Consent for publication
All authors consent to publication of this work.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chen WW, Balaj L, Liau LM, Samuels ML, Kotsopoulos SK, Maguire CA, Loguidice L, Soto H, Garrett M, Zhu LD, et al. BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol Ther Nucleic Acids. 2013;2:e109. doi: 10.1038/mtna.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connolly ID, Li Y, Pan W, Johnson E, You L, Vogel H, Ratliff J, Hayden Gephart M. A pilot study on the use of cerebrospinal fluid cell-free DNA in intramedullary spinal ependymoma. J Neuro-Oncol. 2017;135:29–36. doi: 10.1007/s11060-017-2557-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Mattos-Arruda L, Mayor R, Ng CK, Weigelt B, Martinez-Ricarte F, Torrejon D, Oliveira M, Arias A, Raventos C, Tang J, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiemcke-Jiwa LS, Minnema MC, Radersma-van Loon JH, Jiwa NM, de Boer M, Leguit RJ, de Weger RA, Huibers MMH. The use of droplet digital PCR in liquid biopsies: a highly sensitive technique for MYD88 p.(L265P) detection in cerebrospinal fluid. Hematol Oncol. 2018;36:429–435. doi: 10.1002/hon.2489. [DOI] [PubMed] [Google Scholar]
- 5.Hirano M, Ohka F, Maeda S, Chalise L, Yamamichi A, Aoki K, Kato A, Tanahashi K, Motomura K, Nishimura Y. et al (2018) A novel high-sensitivity assay to detect a small fraction of mutant IDH1 using droplet digital PCR. Brain Tumor Pathol. Doi: 10.1007/s10014-018-0310-7 [DOI] [PubMed]
- 6.Huang TY, Piunti A, Lulla RR, Qi J, Horbinski CM, Tomita T, James CD, Shilatifard A, Saratsis AM. Detection of histone H3 mutations in cerebrospinal fluid-derived tumor DNA from children with diffuse midline glioma. Acta neuropathologica communications. 2017;5:28. doi: 10.1186/s40478-017-0436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johung TB, Monje M. Diffuse intrinsic pontine glioma: new pathophysiological insights and emerging therapeutic targets. Curr Neuropharmacol. 2017;15:88–97. doi: 10.2174/1570159X14666160509123229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Pan W, Connolly ID, Reddy S, Nagpal S, Quake S, Gephart MH. Tumor DNA in cerebral spinal fluid reflects clinical course in a patient with melanoma leptomeningeal brain metastases. J Neuro-Oncol. 2016;128:93–100. doi: 10.1007/s11060-016-2081-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linzey JR, Marini BL, Pasternak A, Smith C, Miklja Z, Zhao L, Kumar-Sinha C, Paul A, Harris N, Robertson PL, et al. Development of the CNS TAP tool for the selection of precision medicine therapies in neuro-oncology. J Neuro-Oncol. 2018;137:155–169. doi: 10.1007/s11060-017-2708-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marini BL, Benitez LL, Zureick AH, Salloum R, Gauthier AC, Brown J, Wu YM, Robinson DR, Kumar C, Lonigro R, et al. Blood-brain barrier-adapted precision medicine therapy for pediatric brain tumors. Transl Res. 2017;188:27.e21–27.e14. doi: 10.1016/j.trsl.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Ricarte F, Mayor R, Martinez-Saez E, Rubio-Perez C, Pineda E, Cordero E, Cicuendez M, Poca MA, Lopez-Bigas N, Ramon YCS et al (2018) Molecular diagnosis of diffuse gliomas through sequencing of cell-free circulating tumour DNA from cerebrospinal fluid. Clin Cancer Res. Doi: 10.1158/1078-0432.ccr-17-3800 [DOI] [PubMed]
- 12.Momtaz P, Pentsova E, Abdel-Wahab O, Diamond E, Hyman D, Merghoub T, You D, Gasmi B, Viale A, Chapman PB. Quantification of tumor-derived cell free DNA(cfDNA) by digital PCR (DigPCR) in cerebrospinal fluid of patients with BRAFV600 mutated malignancies. Oncotarget. 2016;7:85430–85436. doi: 10.18632/oncotarget.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olmedillas-Lopez S, Garcia-Arranz M, Garcia-Olmo D. Current and emerging applications of droplet digital PCR in oncology. Mol Diagn Ther. 2017;21:493–510. doi: 10.1007/s40291-017-0278-8. [DOI] [PubMed] [Google Scholar]
- 14.Pan W, Gu W, Nagpal S, Gephart MH, Quake SR. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem. 2015;61:514–522. doi: 10.1373/clinchem.2014.235457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siravegna G, Geuna E, Mussolin B, Crisafulli G, Bartolini A, Galizia D, Casorzo L, Sarotto I, Scaltriti M, Sapino A, et al. Genotyping tumour DNA in cerebrospinal fluid and plasma of a HER2-positive breast cancer patient with brain metastases. ESMO Open. 2017;2:e000253. doi: 10.1136/esmoopen-2017-000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Springer S, Zhang M, McMahon KW, Kinde I, Dobbyn L, Ptak J, Brem H, Chaichana K, Gallia GL, et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci U S A. 2015;112:9704–9709. doi: 10.1073/pnas.1511694112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J, Ye X, Xu Y, Chen M, Zhong W, Sun Y, Yang Z, Zhu G, Gu Y, Wang M. EGFR mutation status of paired cerebrospinal fluid and plasma samples in EGFR mutant non-small cell lung cancer with leptomeningeal metastases. Cancer Chemother Pharmacol. 2016;78:1305–1310. doi: 10.1007/s00280-016-3155-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information. Detailed methods and H3F3A K27M assay design. (DOCX 28 kb)
Figure S1. Serial dilution of K27M mutant oligonucleotide in constant background of wild-type DNA demonstrates consistent detection down to at least 2% VAF under typical experimental conditions, with the possibility of detection at even lower VAF under ideal conditions. One such dilution series is shown above, with (a) showing number of droplets positive for mutant or wild-type H3F3A sequence and (b) showing the corresponding VAF values. Figure S2. Plot of droplets (blue – positive mutant H3F3A K27M, green – positive wildtype H3F3A, grey – negative droplets) from ddPCR performed on (a) non-tumor human CSF spiked with synthetic K27M mutant sequence oligonucleotide and (b) non-tumor human CSF alone. (DOCX 268 kb)
Data Availability Statement
All somatic mutation information generated from this study has been uploaded to dbGaP (https://www.ncbi.nlm.nih.gov/gap) under accession number phs000673.v2.p1.