Abstract
Background
Genodermatoses represent genetic anomalies of skin tissues including hair follicles, sebaceous glands, eccrine glands, nails, and teeth. Ten consanguineous families segregating various genodermatosis phenotypes were investigated in the present study. Methods Homozygosity mapping, exome, and Sanger sequencing were employed to search for the disease-causing variants in the 10 families.
Results
Exome sequencing identified seven homozygous sequence variants in different families, including: c.27delT in FERMT1; c.836delA in ABHD5; c.2453C>T in ERCC5; c.5314C>T in COL7A1; c.1630C>T in ALOXE3; c.502C>T in PPOX; and c.10G>T in ALDH3A2. Sanger sequencing revealed three homozygous variants: c.1718 + 2A>G in FERMT1; c.10459A>T in FLG; and c.92delT in the KRT14 genes as the underlying genetic cause of skin phenotypes.
Conclusion
This study supports the use of exome sequencing as a powerful, efficient tool for identifying genes that underlie rare monogenic skin disorders.
Introduction
Skin, the largest organ that covers the human body and its appendages, has key homeostatic properties of both barrier protection and controlled regeneration. Genetic disruption of this homeostatic system results in a large number of genodermatoses, which may include both cutaneous and extracutaneous manifestations.1,2 Genodermatoses comprise a large group of Mendelian disorders, and almost half of them are inherited in an autosomal recessive manner.3 As genetic defects are involved in hereditary disorders, identification of gene variants plays an important role in research on and diagnosis of these diseases.
In the present study, we investigated 10 consanguineous Pakistani families with various forms of genodermatoses at clinical and molecular level. In seven families, variously affected with ichthyosis, porphyria, and xeroderma pigmentosum, exome sequencing was used to find out the diseases causing sequence variants. Moreover, homozygosity mapping and Sanger sequencing was performed for genetic analysis in one family with epidermolysis bullosa and two families with ichthyosis. As a final point, pathogenic sequence variants were identified in 10 consanguineous families in the genes: FERMT1, ABHD5,FLG, ERCC5, COL7A1, ALOXE3, PPOX, KRT14, and ALDH3A2.
Materials and methods
Human subjects
Approval of the study was obtained from the Institutional Review Boards (IRBs) of Quaid-i-Azam University, Islamabad, Pakistan; Baylor College of Medicine, Houston, TX, USA; University of Colorado, Denver, CO, USA; and Center for Medical Genetics, Ghent University Hospital, Ghent, Belgium. Written informed consent was obtained from the participating family members. Ten families (ED112, ED113, ED129, ED156, ED172, ED178, ED188, ED212, ED236, and ED238) were enrolled from different rural areas of Pakistan. Pedigrees of all families showed an apparent autosomal recessive mode of phenotypic inheritance. Affected members in the families were examined at local government hospitals. Blood samples were collected from affected individuals, their parents, and normal siblings.
Genotyping
Six families (ED112, ED113, ED129, ED172, ED178, and ED212) were genotyped using the Infinium® HumanCoreExome Array (Illumina, USA), which interrogates >500,000 SNP markers. Homozygosity mapping and linkage analysis were performed using HomozygosityMapper and MERLIN, respectively.4,5 Multipoint linkage analysis was performed using as parameters disease allele frequency of 0.001 and an autosomal recessive mode of inheritance with complete penetrance. Two families, ED156 and ED236, were genotyped using highly polymorphic microsatellite markers flanking candidate genes involved in related phenotypes.
Exome and Sanger sequencing
Exome sequencing was performed using a DNA sample from at least one affected individual from each of six families (Fig. 1a, c–f, h). In family ED238, exome sequencing was carried out in five family members, including three affected siblings and their unaffected parents. Sequence capture was performed in a solution using the Roche NimbleGen SeqCap EZ Human Exome Library v2.0 to target approximately 36.6 Mb of coding regions, and sequencing was carried out using the Illumina HiSeq or NextSeq 500 platforms. FASTQ files were aligned to human reference sequence (hg19) using the Burrows-Wheeler Aligner. The Genome Analysis Toolkit (GATK) was used for realignment of regions containing indels, recalibration of base qualities, and variant detection and calling,6 and variants were annotated using SeattleSeq137. Apparent novel homozygous variants were checked in public databases including dbSNP, 1000 Genomes, and Exome Variant Server (EVS), and in sequences of 400 normal individuals from the same ethnic groups. The FERMT1, FLG, and KRT14 genes were Sanger-sequenced as candidates in affected members of ED113, ED156, and ED236, respectively. Cosegregation of apparent pathogenic variants was tested by Sanger sequencing using DNA samples from the rest of the family members.
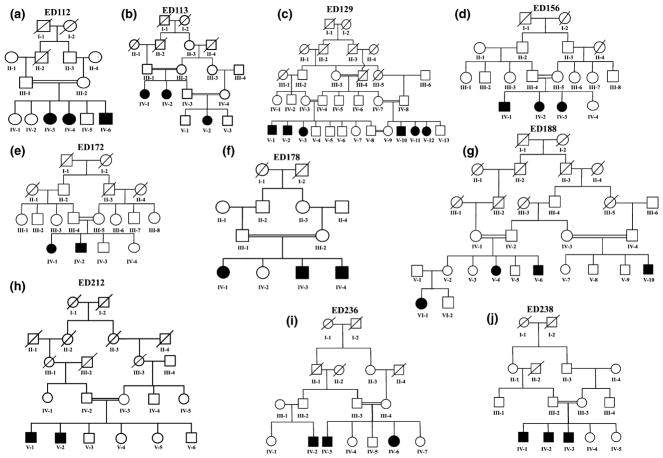
Figure 1.
(a–j) Pedigrees of 10 consanguineous families segregating various types of skin disorders. Open symbols represent unaffected individuals, and filled symbols represent affected individuals. A crossline on the symbol indicates a deceased individual
Results
Family structures and phenotypes
Family ED112 and ED113
Six affected individuals in these two families (Fig. 1a, b) showed features of mild skin blistering mostly over joints, and cutaneous atrophy, photosensitivity, random hypopigmentation and hyperpigmentation, and telangiectasia throughout the body (Fig. 2a–e; Table 1). These features are consistent with Kindler syndrome. Disease severity varied among affected individuals in the same family. The irregular pigmentation, cutaneous atrophy, and telangiectasia were reported to be progressive in both families. Family members reported decreased numbers of blisters with age.
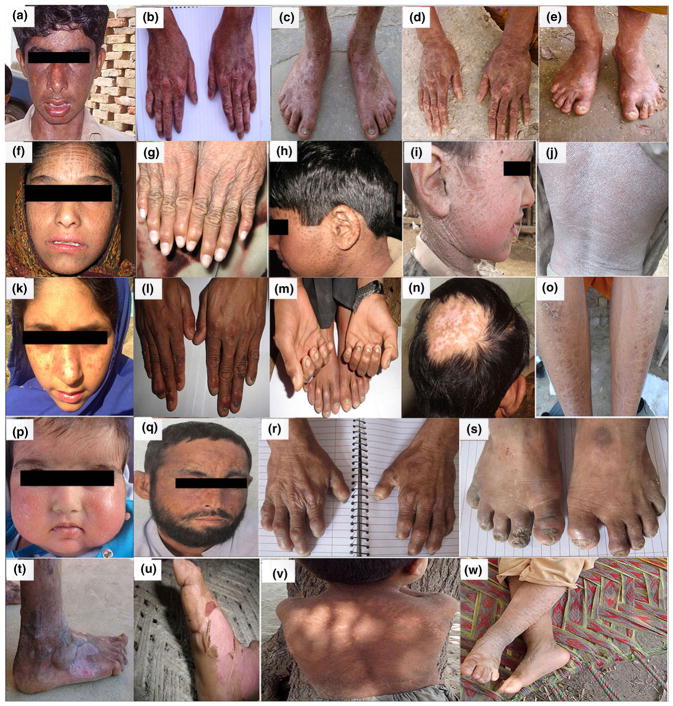
Figure 2.
Affected members in the 10 families ED112, ED113, ED129, ED156, ED172, ED178, ED188, ED212, ED236, and ED238. (a–c) Clinical phenotypes observed in affected members in family ED112 consistent with Kindler syndrome: (a) Hypo- and hyperpigmented patches over the face of affected individual IV-6; (b, c) Cutaneous atrophy and poikiloderma over the dorsum of hand and foot in affected individual IV-4. (d, e) Cutaneous atrophy, hypo- and hyperpigmentation, and telangiectasia on the hand and foot of affected individual IV-1 from family ED113 with Kindler syndrome. (f-h) Family ED129 with Chanarin-Dorfman syndrome: (f) Brownish scales and black spots on the facial area of affected individual V-12; (g) Hands of affected individual V-3 with brownish scales; (h) Black spots on face, neck, and ear of affected individual V-2. (i, j) Individuals with ichthyosis vulgaris in family ED156: (i) Large, adhesive, brown-colored scales on neck and entire lateral aspect of face; (j) Fine white scales and generalized dryness of skin on posterior side of trunk in affected individual IV-1 in family ED156. (k) Hyperpigmentation over the face of individual IV-1 of family ED172, who has xeroderma pigmentosum. (l–n) Family ED178 with recessive dystrophic epidermolysis bullosa: (l, m) Nails of affected individuals IV-3 and IV-4 show micronychia; (n) Skin blister on the scalp with hair loss in affected individual IV-4. (o–p) Individuals with lamellar ichthyosis from family ED188: (o) Mild scales on shin of affected individual V-6; (p) Fine white scales on the face of affected individual V-1. (q–s) Family members of ED212 affected with variegate porphyria: (q) Hyperpigmentation, scar, and crust on the facial region and ear of affected individual V-1; (r) Short fingers with small nails and scar over dorsum of hands in affected individual V-1; (s) Short toes with dystrophic nails and blister in affected individual V-2. (t, u) Epidermolysis bullosa simplex in family ED236: (t) Blisters, scars, and fresh wounds over foot of affected individual IV-2; (u) Scars on the foot of affected individual IV-3. (v, w) Clinical features of Sjögren-Larsson syndrome in affected individuals of family ED238: (v) Hyperkeratosis and scales on the back and neck of affected member IV-1: (w) Large polygonal scales on lower limbs of affected member IV-3
Table 1.
Clinical features of 10 families presented here
| Phenotypes | ED112 | ED113 | ED129 | ED156 | ED172 | ED178 | ED188 | ED212 | ED236 | ED238 |
|---|---|---|---|---|---|---|---|---|---|---|
| Skin | ||||||||||
| Pigmentation | Hyper/ Hypo | Hyper/ Hypo | Normal | Normal | Hyper | Hyper/Hypo | Normal | Hyper | Hyper | Normal |
| Blisters | Mild | Mild | − | − | − | Mild | − | Mild | Severe | − |
| Scares | + | + | − | − | − | + | − | + | + | − |
| Crust | − | − | − | − | − | − | − | + | − | − |
| Scales | − | − | Brownish/ Black | White/ Brown | − | − | White/ Brown | − | − | Brown |
| Photosensitivity | + | + | − | − | + | − | − | + | − | − |
| Erythroderma | + | + | − | − | − | − | − | − | + | − |
| Ectropion | − | − | − | − | − | − | + | − | − | − |
| Nail | Normal | Normal | Normal | Normal | Normal | Small and deformed | Normal | Small and deformed | Onycholysis | Normal |
| Hair | Normal | Normal | Normal | Normal | Normal | Hair loss from scalp | Normal | Normal | Normal | Normal |
| Teeth | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal |
| Skeletal Structure | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Brachydactyly | Normal | Normal |
| Hearing | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal |
| Vision | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal |
| Growth | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal |
| Gestation | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal |
| Mental Health | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Neurodevelopmental delays |
Family ED129
In this large consanguineous family, there are six affected members including three males and three females (Fig. 1c). Clinical features including congenital ichthyosiform erythroderma, dry and brownish scaly skin over most part of the body, and black spots as a result of deposition of neutral lipids were consistent with Chanarin-Dorfman syndrome (Fig. 2f–h; Table 1). Three affected individuals (V-2, V-3, and V-11) had more severe phenotypes. Other ectodermal structures (hair, nails, and teeth) were normal.
Family ED156
This family had three affected members (Fig. 1d). Clinical evaluation of the affected members (IV-1, IV-2, and IV-3) revealed generalized dryness of skin and scales of variable size and intensity ranging from discrete white to large hyperkeratotic scales on the body, and hyperlinearity and mild erythroderma of the palms (Fig. 2i, j; Table 1) consistent with ichthyosis vulgaris. Other dermatologic manifestations, including palmoplantar keratoderma, ectropion, and eclabium, were not present.
Family ED172
The two affected siblings in this consanguineous family (Fig. 1e) were born to unaffected parents. Hyperpigmentation restricted to the facial region (Fig. 2k; Table 1), consistent with xeroderma pigmentosum, was observed in both affected children. The older affected sister (IV-1) had slightly more severe hyperpigmentation than her younger brother (IV-2). No hyperpigmentation was observed elsewhere.
Family ED178
This family included three affected children resulting from a first-cousin marriage (Fig. 1f). All affected individuals exhibited recessive dystrophic epidermolysis bullosa (RDEB) phenotypes (Fig. 2l–n; Table 1). Disease in family ED178 was milder than previously reported RDEB phenotypes.7 Both affected brothers (IV-3 and IV-4) exhibited sparse to absent scalp hair, skin blisters restricted to the scalp, and micronychia of fingernails and toenails. Their affected sister (IV-1) had generalized skin blisters over the extremities, but scalp and nails were normal. No affected individuals had intellectual disability, dental anomalies, syndactyly, or any systemic manifestations.
Family ED188
This six-generation pedigree included four affected members (V-4, V-6, V-10, and VI-1) (Fig. 1g). They were born as collodion babies, compatible with the phenotype of lamellar ichthyosis, with gradual shedding of encasement followed by fine white scales, palmoplantar keratoderma, and erythroderma (Fig. 2o, p; Table 1).
Family ED212
This family had two children affected with variegate porphyria (V-1 and V-2) (Fig. 1h) who were born to healthy consanguineous parents. Both affected children had similar phenotypes that included hyperpigmentation, scarring, crusted erosions, and photosensitivity. Skin creases were more prominent in facial regions and extremities. Their hands and feet were small and hyperpigmented with brachydacyly variable nail dystrophy, and short misshapen terminal phalanges (Fig. 2q–s; Table 1).
Family ED236
This family included three affected individuals (IV-2, IV-3, and IV-6) (Fig. 1i) with typical features of epidermolysis bullosa simplex. All three affected individuals exhibited large bullae, pustules, blisters, and erosion, more prominent on the dorsa of hands and feet and milder over the limbs, trunk, and palmoplantar skin (Fig. 2t, u; Table 1). The blisters appeared after mild mechanical stress and healed with mild scarring. The nails had a form of onycholysis and detached easily from the nail bed. No palmoplantar hyperkeratosis was observed. None of the normal siblings or parents exhibited any skin or nail symptoms, including skin fragility.
Family ED238
Affected individuals in this family were diagnosed with Sjogren-Larsson syndrome. They had diffuse generalized ichthyosis on the neck, trunk, and extremities with variable intensity (Fig. 1j). Scales were not present on the face. In all three affected individuals, hyperkeratosis was more prominent as compared to scales on their trunks. Affected individuals had early global neurodevelopmental delay, particularly of speech, language, and motor milestones. Receptive language appeared better than expressive language. All three affected family members had lower limb spasticity with variable severity. Subject IV-1 did not walk until age 2.5 years and thereafter walked with difficulty. Subject IV-2 did not sit until 12 months and cannot stand independently. The youngest affected family member was unable to sit and stand at the time of examination. Forelimbs of affected individuals were fully functional. Hair, nails, and teeth were normal (Fig. 2v, w; Table 1).
Genotyping and screening of genes
DNA samples of all available members of five families (ED112, ED113, ED129, ED172, and ED212) were genotyped using the Infinium® HumanCoreExome Chip, and in two other families (ED156 and ED236), STR markers were genotyped. Homozygosity mapping and linkage analysis using whole-genome SNP genotypes revealed single (ED112, ED113, and ED129) and multiple (ED172 and ED212) large homozygous regions that were shared among affected members within the same family.
Families ED112 and ED113
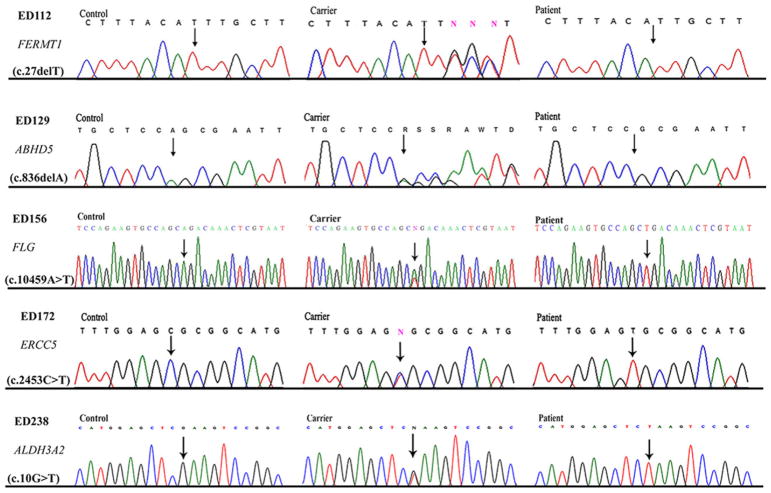
A single homozygous region of 17.46 Mb (chr20:0.63–17.52 Mb) with a maximum multipoint LOD score of 2.65 was identified on chromosome 20p13-p12.1. Within the same locus, a 3.25 Mb region (chr20:4.82–8.07 Mb) was mapped for family ED113, with maximum multipoint LOD score 3.31. Exome sequence data from affected member IV-3 of family ED112 revealed a novel homozygous deletion, chr20:6100175delA (c.27delT; p.Phe9-Leufs*23), in the FERMT1 gene (Fig. 3). Sanger sequencing of FERMT1 in family ED113 identified a homozygous splice site variant, chr20:6064685A>G (c.1718+2A>G), which was previously reported in another family with Kindler syndrome (MIM 173650) by Fassihi et al. (2005).8 Cosegregation of the respective sequence variants with Kindler syndrome in families ED112 and ED113 was verified by Sanger sequencing of DNA from all available family members.
Figure 3.
Sequence chromatograms of novel sequence variants in FERMT1, ABHD5, FLG, ERCC5, and ALDH3A2 identified in families ED112, ED129, ED156, ED172, and ED238, respectively. DNA sequences from control individuals, unaffected heterozygous carriers, and affected homozygous individuals are shown. Sequence variants are indicated by arrows
Family ED129
A single large homozygous region of 9.71 Mb (chr3:40.50–50.21 Mb) was identified on chromosome 3p22.1-p21.31, with significant maximum multipoint LOD score 5.01. Analysis of the exome sequence data identified a novel homozygous deletion, chr3:43759225delA (c.836delA; p.Gln279Argfs*14), which results in a frameshift in the ABHD5 gene (Fig. 3). Sanger sequencing of the variant in all available DNA samples from family ED129 verified its cosegregation with Chanarin-Dorfman syndrome (MIM 275630) in this family.
Family ED156
Because of the ichthyosis vulgaris phenotype, genotyping was carried out using STR markers closely linked to the FLG gene on chromosome 21q21.3. Haplotype analyses showed cosegregation of STR haplotypes with ichthyosis in the family. The entire coding sequence and intron-exon boundaries of FLG was sequenced in seven individuals, including three affected (IV-1, IV-2, and IV-3) family members. Sanger sequencing revealed a novel FLG mutation, chr1:152276903T>A (c.10459A>T; p.Arg3487*) that cosegregated with ichthyosis vulgaris (MIM 146700) in this family (Fig. 3).
Family ED172
As a result of the small size of this family, we identified three large homozygous regions, including: a 35.12 Mb region (chr1:32.13–67.25 Mb) on chromosome 1p35.2-p31.3; a 16.82 Mb region (chr8:0.16–16.99 Mb) on 8p23.3-p22; and a 14.71 Mb region (chr13:100.38–115.09 Mb) on 13q32.3-q34. A maximum LOD score of 1.93 was obtained at each of these three homozygous regions. Analysis of the exome sequence data of affected member IV-1 identified a homozygous mis-sense variant, chr13:103519115C>T (c.2453C>T; p.Ala818Val), in the ERCC5 gene within the homozygous linkage interval on chromosome 13q32.3-q34. This missense variant showed cosegregation with xeroderma pigmentosum in the family (Fig. 3). Polyphen-2, SIFT, and MutationTaster predicted the change to likely be damaging, deleterious, and disease-causing. This is consistent with a diagnosis of xeroderma pigmentosum complementation group G (XPG) (MIM 278780) in this family.
Family ED178
Genome-wide SNP analysis revealed a 25.80 Mb homozygous region (chr3:26.25–52.06 Mb) on chromosome 3p24.2-p21.2, with maximum multipoint LOD score 2.53. Analysis of the exome data revealed a previously reported missense variant chr3:48616704G>A (c.5314C>T; p.Arg1772Trp), in the COL7A1 gene, located on chromosome 3p21.31,9 within the homozygous region. This is consistent with a diagnosis of dystrophic EB (MIM 226600) in this family.
Family ED188
Exome sequencing of DNA from affected family member V-4 revealed a recurrent nonsense variant, chr17:8011840G>A (c.1630C>T; p.Gln544*), in exon 13 of the gene ALOXE3.10 Sanger sequencing revealed cosegregation of the ALOXE3 variant with lamellar ichthyosis (MIM 606545) in this family.
Family ED212
Genome-wide linkage mapping revealed three homozygous regions, including an 8.08 Mb region (chr1:159.51–167.58 Mb) on chromosome 1q23.3-q24.2; a 16.01 Mb region (chr2:137.75–153.76 Mb) on 2q22.1-q23.3; and a 5.26 Mb region (chr5:25.48–30.74 Mb) on 5p14.1-p13.3. A maximum multipoint LOD score of 2.91 was obtained for each homozygous region. Analysis of the exome data identified a previously reported mis-sense variant, chr1:161138252G>A (c.502C>T; p.Arg168Cys), in the PPOX gene, located on chromosome 1q23.3-q24.2.11 This is consistent with the diagnosis of variegate porphyria (MIM 176200) in this family.
Family ED236
Because of the apparent autosomal recessive epidermolysis bullosa simplex (EBS) phenotype, genotyping was performed with microsatellite markers closely linked to genes reported for similar phenotypes. Haplotype analysis demonstrated cosegregation of EBS with haplotypes spanning the KRT14 gene on chromosome 17q21.2. The entire coding sequence and intron-exon boundaries of KRT14 was sequenced in seven individuals including three affected (IV-2, IV-3, and IV-6) family members. Sanger sequencing revealed a previously known single-base deletion, chr17:39742995delA (c.92delT; p.Ile31Thrfs*87), that results in a premature termination codon in exon 1 of KRT14. This is consistent with the diagnosis of EBS generalized intermediate type (MIM 131900) in this family.
Family ED238
Exome sequencing was performed using DNA from five family members, including three affected (IV-1, IV-2, and IV-3) and two unaffected (III-2 and III-3) individuals. Autozygosity mapping of the exome sequence data using AgileVCFMapper revealed a 16.17 Mb homozygous region on chromosome 17p13.2-p11.2 (chr17:4.43–21.14 Mb). Across the 22 autosomes, we identified five exome variants that were: (i) homozygous in the three affected individuals and heterozygous in the two obligate heterozygotes, and (ii) had minor allele frequency (MAF) <0.01 in the Exome Aggregation Consortium (ExAC) database; all were absent in 400 ethnically matched control chromosomes. Of the five variants, only variant chr17:19552294G>T was located within the 17p13.2-p11.2 homozygous region, a nonsense alteration, c.10G>T (p.Glu4*), in ALDH3A2 gene. This variant is predicted to result in premature termination of the ALDH3A2 protein. Cosegregation of the variant with the disease phenotypes was verified by Sanger sequencing. This is consistent with the diagnosis of Sjogren-Larsson syndrome (MIM 270200) in this family.
Discussion
Genodermatoses can be collectively termed as disorders of the skin that occur on a genetic basis, in some instances with concurrent systemic manifestations.2 In this study, 10 families with different types of genetic skin disorders were included. Microsatellites and SNP-based linkage analysis accompanied by exome and Sanger sequencing were used for the diagnosis of autosomal recessive phenotypes in these families.
This analysis demonstrates the utility of this approach, particularly for diagnosis of recessive phenotypes in consanguineous families.
In families ED112 and ED113, we respectively identified a novel pathologic frameshift (c.27delT; p.Phe9Leufs*23) and a previously reported variant (c.1718+2A>G) in FERMT1, encoding the kindlin-1 protein.12,13 Homozygous mutations in FERMT1 lead to the skin fragility disorder known as Kindler syndrome.13
In family ED129, we identified a novel pathogenic frameshift deletion (c.836delA) in ABHD5, encoding a/b-hydrolase domain containing protein 5. Mutations in ABHD5 result in Chanarin-Dorfman syndrome.
In family ED156, we identified a known pathologic nonsense variant (c.10459A>T) in FLG, encoding profilaggrin.14 Mutations in FLG result in ichthyosis vulgaris.
In family ED172, we identified a novel pathogenic missense sequence variant (c.2453C>T; p.Ala818Val) in ERCC5. Mutations in ERCC5 cause xeroderma pigmentosum complementation group G (XPG) and Cockayne syndrome.15
In family ED178, we identified a known missense variant (c.5314C>T; p.Arg1772Trp) in the COL7A1 gene, encoding collagen VII alpha.16,17 Loss-of-function COL7A1 mutations result in dystrophic epidermolysis bullosa.18
In family ED188, we identified a known variant in ALOXE3 (c.1630C>T [p.Gln544*]),10 encoding epidermal lipoxygenase-3. 19 Mutations in ALOXE3 cause lamellar ichthyosis.10
In family ED212, the first with variegate porphyria (VP) phenotype reported from Pakistan, we identified a known missense variant (c.502C>T; p.Arg168Cys) in PPOX gene, encoding pro-toporphyrinogen oxidase. Mutations in PPOX cause variegate porphyria (MIM 176200).
In family ED236, we identified a known homozygous frame-shift in KRT14 (c.92delT), encoding keratin 14. This same mutation was previously found in another consanguineous Pakistani family with epidermolysis bullosa simplex (EBS).20
In family ED238, we identified a nonsense mutation (c.10G>T; p.Glu4*) in ALDH3A2, encoding fatty aldehyde dehydrogenase (FALDH). Mutations in ALDH3A2 result in Sjogren-Larsson syndrome (MIM 270200), consistent with the phenotype in this family but for which diagnosis was not considered a priori.
Taken together, our findings demonstrate the utility of exome sequencing coupled with SNP genotyping to identify disease-causing variants in diverse types of genodermatoses, particularly in consanguineous families.
Acknowledgments
We highly appreciate participation of the family members in the present study. This work was supported by the Higher Education Commission (HEC) of Pakistan; Skin Diseases Research Center grant AR057212 from the National Institutes of Health (Molecular Genetic Analysis Core; to R.A.S.); and funding to the University of Washington Center for Mendelian Genomics from the National Human Genome Research Institute and National Heart, Lung, and Blood Institute grant U54HG006493 (to D.A.N., S.M.L. and M.J.B).
University of Washington Center for Mendelian Genomics (UW CMG): Michael J. Bamshad1,2, Suzanne M. Leal3, Deborah A. Nickerson1, Peter Anderson1, Marcus Annable1, Elizabeth Marchani Blue1, Kati J. Buckingham1, Jennifer Chin1, Jessica X Chong1, Rodolfo Cornejo Jr.1, Colleen P. Davis1, Christopher Frazar1, Zongxiao He3, Gail P. Jarvik1, Guillaume Jimenez1, Eric Johanson1, Tom Kolar1, Stephanie A. Krauter1, Daniel Luksic1, Colby T. Marvin1, Sean McGee1, Daniel J. McGoldrick1, Karynne Patterson1, Marcos Perez1, Sam W. Phillips, Jessica Pijoan1, Peggy D. Robertson1, Regie Santos-Cortez3, Aditi Shankar1, Krystal Slattery1, Kathryn M. Shively1, Deborah L. Siegel1, Joshua D. Smith1, Monica Tackett1, Gao Wang3, Marc Wegener1, Jeffrey M. Weiss1, Riana I. Wernick1, Marsha M. Wheeler1, Qian Yi1; 1University of Washington; 2Seattle Children’s Hospital; 3Baylor College of Medicine.
Footnotes
Conflict of interest: We declare no conflict of interest.
References
- 1.Lim X, Nusse R. Wnt signaling in skin development, homeostasis, and disease. Cold Spring Harb Perspect Biol. 2013;5:a008029. doi: 10.1101/cshperspect.a008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jen M, Nallasamy S. Ocular manifestations of genetic skin disorders. Clin Dermatol. 2016;34:242–275. doi: 10.1016/j.clindermatol.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Scriver CR. Henry Friesen Award Lecture. Work, the clinician-scientist and human biochemical genetics. Clin Invest Med. 2001;24:179–195. [PubMed] [Google Scholar]
- 4.Seelow D, Schuelke M, Hildebrandt F, et al. HomozygosityMapper–an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–W599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abecasis GR, Cherny SS, Cookson WO, et al. Merlin rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 6.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serafi R, Jelani M, Almramhi MM, et al. Identification of two homozygous sequence variants in the COL7A1 gene underlying dystrophic epidermolysis bullosa by whole-exome analysis in a consanguineous family. Ann Hum Genet. 2015;79:350–356. doi: 10.1111/ahg.12123. [DOI] [PubMed] [Google Scholar]
- 8.Fassihi H, Wessagowit V, Jones C, et al. Neonatal diagnosis of Kindler syndrome. J Dermatol Sci. 2005;39:183–185. doi: 10.1016/j.jdermsci.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Whittock NV, Ashton GH, Mohammedi R, et al. Comparative mutation detection screening of the type VII collagen gene (COL7A1) using the protein truncation test, fluorescent chemical cleavage of mismatch, and conformation sensitive gel electrophoresis. J Invest Dermatol. 1999;113:673–686. doi: 10.1046/j.1523-1747.1999.00732.x. [DOI] [PubMed] [Google Scholar]
- 10.Eckl KM, Krieg P, Küster W, et al. Mutation spectrum and functional analysis of epidermis-type lipoxygenases in patients with autosomal recessive congenital ichthyosis. Hum Mutat. 2005;26:351–361. doi: 10.1002/humu.20236. [DOI] [PubMed] [Google Scholar]
- 11.Warnich L, Kotze MJ, Groenewald IM, et al. Identification of three mutations and associated haplotypes in the protoporphyrinogen oxidase gene in South African families with variegate porphyria. Hum Mol Genet. 1996;5:981–984. doi: 10.1093/hmg/5.7.981. [DOI] [PubMed] [Google Scholar]
- 12.Siegel DH, Ashton GH, Penagos HG, et al. Loss of kindlin-1, a human homolog of the Caenorhabditis elegans actin-extracellularmatrix linker protein UNC-112, causes Kindler syndrome. Am J Hum Genet. 2003;73:174–187. doi: 10.1086/376609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herz C, Aumailley M, Schulte C, et al. Kindlin-1 is a phosphoprotein involved in regulation of polarity, proliferation, and motility of epidermal keratinocytes. J Biol Chem. 2006;281:36082–36090. doi: 10.1074/jbc.M606259200. [DOI] [PubMed] [Google Scholar]
- 14.Harding CR, Aho S, Bosko CA. Filaggrin–revisited. Int J Cosmetic Sci. 2013;35:412–423. doi: 10.1111/ics.12049. [DOI] [PubMed] [Google Scholar]
- 15.Lalle P, Nouspikel T, Constantinou A, et al. The founding members of xeroderma pigmentosum group G produce XPG protein with severely impaired endonuclease activity. J Invest Dermatol. 2002;118:344–351. doi: 10.1046/j.0022-202x.2001.01673.x. [DOI] [PubMed] [Google Scholar]
- 16.Christiano AM, Greenspan DS, Hoffman GG, et al. A missense mutation in type VII collagen in two affected siblings with recessive dystrophic epidermolysis bullosa. Nat Genet. 1993;4:62–66. doi: 10.1038/ng0593-62. [DOI] [PubMed] [Google Scholar]
- 17.Christiano AM, McGrath JA, Tan KC, et al. Glycine substitutions in the triple-helical region of type VII collagen result in a spectrum of dystrophic epidermolysis bullosa phenotypes and patterns of inheritance. Am J Hum Genet. 1996;58:671–681. [PMC free article] [PubMed] [Google Scholar]
- 18.Kon A, Nomura K, Pulkkinen L, et al. Novel glycine substitution mutations in COL7A1 reveal that the Pasini and Cockayne-Touraine variants of dominant dystrophic epidermolysis bullosa are allelic. J Invest Dermatol. 1997;109:684–687. doi: 10.1111/1523-1747.ep12338093. [DOI] [PubMed] [Google Scholar]
- 19.Schneider C, Pratt DA, Porter NA, et al. Control of oxygenation in lipoxygenase and cyclooxygenase catalysis. Chem Biol. 2007;14:473–488. doi: 10.1016/j.chembiol.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batta K, Rugg EL, Wilson NJ, et al. A keratin 14 ‘knockout’ mutation in recessive epidermolysis bullosa simplex resulting in less severe disease. Br J Dermatol. 2000;143:621–627. doi: 10.1111/j.1365-2133.2000.03722.x. [DOI] [PubMed] [Google Scholar]