Abstract
Hereditary hearing impairment is a common sensory disorder that is genetically and phenotypically heterogeneous. In this study, we used a homozygosity mapping and exome sequencing strategy to study a consanguineous Pakistani family with autosomal recessive severe-to-profound hearing impairment. This led to the identification of a missense variant (p.Ile369Thr) in the LMX1A gene affecting a conserved residue in the C-terminus of the protein, which was predicted damaging by an in silico bioinformatics analysis. The p.Ile369Thr variant disrupts several C-terminal and homeodomain residue interactions, including an interaction with homeodomain residue p.Val241 that was previously found to be involved in autosomal dominant progressive HI. LIM-homeodomain factor Lmxla is expressed in the inner ear through development, shows a progressive restriction to non-sensory epithelia, and is important in the separation of the sensory and non-sensory domains in the inner ear. Homozygous Lmxla mutant mice (Dreher) are deaf with dysmorphic ears with an abnormal morphogenesis and fused and misshapen sensory organs; however, computed tomography performed on a hearing-impaired family member did not reveal any cochleovestibular malformations. Our results suggest that LMX1A is involved in both human autosomal recessive and dominant sensorineural hearing impairment.
Keywords: LMX1A, Autosomal recessive hearing impairment, Deafness, Exome sequencing
Introduction
Hereditary hearing impairment (HI) is a common sensory disorder with a genetically heterogeneous etiology. In the Pakistani population, the prevalence of profound bilateral HI is 1.6 per 1000 (Hussain and Bittles 1998; Elahi et al. 1998). It is estimated that at least 50% of cases with pre-lingual HI have a genetic cause, of which 70% are non-syndromic (NS)HI. Seventy percent of NSHI has an autosomal recessive mode of inheritance (Shearer and Smith 2012). Due to high rates of consanguinity, e.g., −60% of marriages of which-80% are between first cousins (Hussain and Bittles 1998), Pakistan is an ideal population to study to identify autosomal recessive pathogenic variants, including for HI.
Identification of new genes underlying human HI is a crucial step in understanding the complex events of sound coding and the pathogenic mechanisms in HI. Recently, LMX1A was identified as a novel gene implicated in autosomal dominant progressive HI with variable age of onset, (a)symmetry, severity and progression rate, and vestibular dysfunction in half of the affected individuals (Wesdorp et al. 2018). LIM homeobox transcription factor 1 alpha is a transcription factor important for neural progenitor specification and dopamine neurogenesis (Hedlund et al. 2016). The variants causing autosomal dominant HI either affected LMX1A’s homeodomain (p.Val241Leu), which is essential for DNA-binding, or one of the LIM domains (p.Cys97Ser) that are involved in protein-protein interactions (Fig. 1a) (Wesdorp et al. 2018).
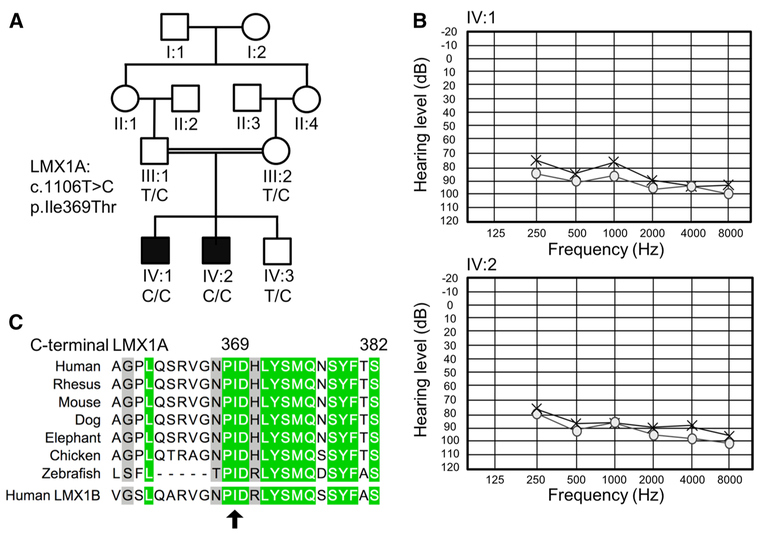
Fig. 1.
Pedigree drawing of family 4755, audiograms for the affected family members, and p.Ile369 residue conservation. a Pedigree of family 4755 with segregation of the c.1106T>C:p.Ile369Thr variant in LMX1A. b Pure-tone audiometry audiograms for affected family members IV:1 (top; age 12) and IV:2 (bottom; age 15); x represents the results for the left ear and o for the right ear. Audiograms show a severe-to-profound hearing impairment with a slight slope with the higher frequencies affected more severely. c Conservation of the C-terminal LMX1A domain and p.Ile369 residue (arrow) amongst species and closest human paralog LMX1B. The p.Ile369 residue is highly conserved across species and human paralog LMX1B. green = high conservation; grey = medium conservation
Several genes causing sensorineural non-syndromic (NS) HI are known to follow both an autosomal dominant and recessive inheritance model depending on the type of variant; for example, MYO7A and GJB2. Variants in these genes that cause autosomal dominant forms of NSHI are often missense variants which exert a dominant negative effect on the activity of the wild-type protein or its close interactors (Liu et al. 1998; del Castillo and del Castillo 2017). The autosomal dominant forms of NSHI are usually less severe, show more variability in phenotype, and are later in onset and progressive in nature compared to their recessive counterparts. The latter often produces a more severe, non-progressive, and pre-lingual HI profile (Liu et al. 1998; del Castillo and del Castillo 2017).
In line with the above, we suggest that different variants in LMX1A underlie both autosomal dominant and recessive forms of HI. Here, we investigated a family with severe-to-profound sensorineural HI from the Khyber Pakhtunkhwa province in Pakistan (Fig. 1).
Materials and methods
Sample collection and clinical evaluation
Institutional review board approval and consent were obtained from the Quaid-i-Azam University and Baylor College of Medicine and Affiliated Hospitals (H-17566). Written informed consent was obtained from all participating family members and peripheral blood samples were collected from five individuals (Fig. 1: III:1, III:2, IV:1; IV:2, IV:3). DNA was extracted using a phenol-chloroform method (Sambrook and Russell 2006). A clinical history was recorded, and other causes of HI, including infections, ototoxic medications, and trauma, were evaluated and excluded. A physical examination was performed to rule out any other additional health problems. Pure-tone audiometry was performed at 250–8000 Hz and testing for gross vestibular defects by a tandem gait test and Romberg test was performed in individuals III:1 (age 43), IV:1 (age 15), IV:2 (age 12), and IV:3 (age 17). The mother (III:2) was unavailable for detailed testing but reported no hearing problems. Additional testing on affected family members (IV:1 and IV:2) to assess cerebellar and cognitive functions included the following: finger-to-finger test, finger-to-nose test, rebound phenomenon test, horizontal gaze nystagmus test, heel-to-shin test, rapid alternating movements of hands, patellar reflex, and the TONI-4 intelligence test (Test of Non-verbal Intelligence-Fourth Edition) (Ritter et al. 2011). We also performed a computed tomography (CT) scan on affected individual IV:1 to identify any cochleovestibular malformations. Prior to exome sequencing and homozygosity mapping, the entire coding region of GJB2, which was a common cause of deafness, was screened by Sanger sequencing with no pathogenic variants identified. Additional common autosomal recessive HI variants in the Pakistani population were excluded as well by Sanger sequencing: i.e., intronic HGF variants (c.482+1986_1988delTGA and c.482+1991_2000delGATGATGAAA), missense variants p.Phe91Ser, and p.Cys99Trp within CIB2 and SLC26A4 missense variants p.Gln446Arg and p.Val239Asp (Schultz et al. 2009; Riazuddin et al. 2012; Shahzad et al. 2013).
Homozygosity mapping
Genotyping of all five DNA samples collected from the family was performed using the Infinium® Human Core-24 v 1.0 BeadChip (Illumina Inc., San Diego, CA, USA). 200 ng of DNA was used to analyze 306,670 single-nucleotide polymorphism (SNP) following the manufacturer’s instructions. Intensities were imported into GenomeStudio version 2011.1 for genotype calling and quality control (Illumina Inc., San Diego, CA, USA). Additional quality control included removing SNPs with: low call rates, i.e., missing > 5% of their genotypes; Mendelian errors [detected with PEDCheck (O’Connell and Weeks 1998)]; double recombination events over short distance that are an indication of genotyping errors [identified with Merlin (Abecasis et al. 2002)]. The genotype data were further analyzed using homozygosity mapping (Homozygosity Mapper) (Seelow and Schuelke 2012).
Exome sequencing
Exome sequencing was performed on affected individual IV:2 using NimbleGen SeqCap EZv.2.0 followed by 70 bp pairedend sequencing on a HiSeq instrument (Illumina Inc., San Diego, CA, USA) with a median read depth of 70×. Reads were aligned to the Human genome (Hg19/GRC37) using the Burrows-Wheeler transform software (BWA-MEM) (Li and Durbin 2010). PCR duplicates were removed with Picard MarkDuplicates, and indel realignment was performed (GATK IndelRealigner). SNPs and small insertion/deletion (Indels) variants were recalibrated with BaseRecalibrator and called jointly with HaplotypeCaller (GATK) (McKenna et al. 2010), and annotated with dbNSFP and ANNOVAR for further filtering and evaluation (Wang et al. 2010; Liu et al. 2011).
Conservation and damaging effects of the variants were evaluated in silico using annotation tools incorporated in dbNSFP and ANNOVAR (Wang et al. 2010; Liu et al. 2011). Variant filtering was performed using the coding region and splice region ± 12 bp based on Ensembl transcripts. For a variant to be retained, it must be a splice-site, missense, nonsense, or indel with a minor allele frequency (MAF) < 0.005 in every population with ≤ 1 homozygous or x-chromosomal hemizygous variant in the Genome Aggregation Database (gnomAD); Greater Middle East (GME) variome database; or the NHLBI Exome Sequencing Project (ESP6500) database, amongst others (Fu et al. 2012; Auton et al. 2015; Lek et al. 2016; Scott et al. 2016). The missense variants must also have a scaled Combined Annotation-Dependent Depletion (CADD) score > 15 (Kircher et al. 2014). We considered several inheritance models possible for this pedigree, i.e., autosomal recessive (homozygous and compound heterozygous) and X-linked, but gave higher priority to homozygous variants within the mapped homozygous regions. Variants of interest, due to their bioinformatics annotation, location, and knowledge of gene function, were verified with Sanger sequencing and tested for segregation using DNA samples from all available family members. To amplify the surrounding region of interest, polymerase chain reaction (PCR) primers were designed using primer3 software (Koressaar and Remm 2007). The PCR amplified products were purified with ExoSAP-IT™ (ThermoFisher Scientific, Sugerland, TX) and sequenced using the BigDye terminator v3.1 cycle sequencing kit on an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA).
3D modeling
The amino acid sequence of LMX1A was retrieved from the NCBI database (UniProtKB-Q8TE12). The protein-protein Basic Local Alignment Search tool (BLASTP) (Altschul et al. 1997) was used to find a template from the Protein Data Bank (PDB). The crystal structure of Fusion of LIM/homeobox protein Lhx3 (PDB ID: 2RGT) with 46.34% with the LMX1A protein was used as a template for modeling (Bhati et al. 2008). Homology modeling techniques were utilized to construct the three-dimensional structure of LMX1A using the MODEL-LER8v1 software (Eswar et al. 2007). The predicted protein model was visualized using UCSF Chimera (Pettersen et al. 2004).
Results
Clinical evaluation
Pure-tone audiometry diagnosed bilateral severe-to-profound HI in both affected individuals (Fig. 1b). The HI in both children was detected before the age of one and was pre-lingual. No gross vestibular dysfunction was observed in both hearing-impaired children, and they reported that they did not suffer from episodes of vertigo. A CT scan was performed in individual IV:1 and revealed normal middle and inner ears with intact cochleae, vestibules, and semicircular canals. Tests to assess cerebellar functioning in the affected individuals were all normal, with the exception of the presence of a pendular knee jerk in both affected individuals. Based on the latter feature alone, however, we cannot conclude the presence of any cerebellar dysfunction. A non-verbal intelligence test (TONI-4) showed a below average intelligence in individual IV:1 (Index score 87) and an average intelligence in individual IV:2 (Index score 92). Physical examinations and recording of clinical history did not reveal any other health problems in the affected children. Individuals III:1 (father), III:2 (mother), and IV:3 (unaffected brother) reported no hearing problems, no signs of vertigo nor any other health problems. Individuals III:1 and IV:3 showed no gross vestibular dysfunction evaluated via a tandem gait test and Romberg test. The unaffected brother (IV:3) showed no hearing impairment, measured via pure-tone audiometry (World Health Organization Classification). The father (III:1) showed normal hearing for the right ear, but did display an elevated hearing threshold for the left ear (mild-to-moderate HI) compared to the International Organization for Standardization (ISO) 7029 standard (95th percentile) for the same age and gender (Supplementary Fig. 1). The cause of his unilateral HI is unclear, although he did report to have been treated 4 years ago with antibiotics for a persistent throat-nose infection. The type of antibiotics was unknown, but commonly used antibiotics in Pakistan include amoxicillin, gentamycin, and streptomycin, some of which can have ototoxic effects (Alharazneh et al. 2011). It is unclear whether the HI is caused by environmental factors such as an infection, noise exposure, or ototoxic drugs or because he is a heterozygous carrier of the missense LMX1A variant.
Homozygosity mapping
Genome-wide genotyping and homozygosity mapping revealed three regions of homozygosity in hearing-impaired family members compared to the three unaffected individuals within the family: chr1:162883337–168460487 (1q23.3–24.2); chr5:78784281–92008283 (5q14.1–14.3); chr5:96856112–148196737(5q15–q32).
Exome sequencing
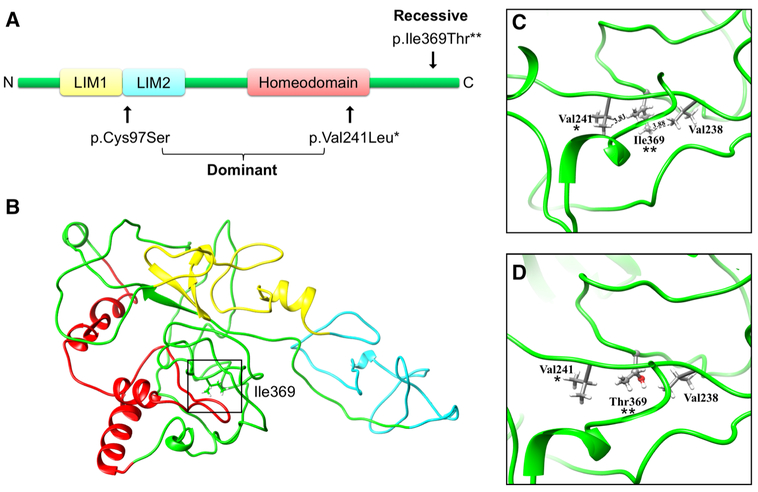
Subsequently, we performed exome sequencing using a DNA sample from hearing-impaired individual IV:2. After variant filtering, only two rare variants in the three mapped regions remained (Supplementary Table 1), a missense variant in APC and LMX1A. A full list of rare-filtered variants that were identified via exome sequencing in individual IV:2 is available in Supplementary Table 1. The APC gene is involved in various forms of cancer (hereditary and somatic) (Aoki and Taketo 2007), and when tested using Sanger sequencing, it did not segregate with the phenotype. The LMX1A gene was of interest as it had previously been implicated in a mouse autosomal recessive deafness phenotype (Koo et al. 2009), and very recently, it has been described in human autosomal dominant hearing impairment (Wesdorp et al. 2018). The missense variant in LMX1A [dbSNP:rs763320093; NM_001174069:c.1106T>C:p.Ile-369Thr], present in the 1q23.3-24.2 homozygous region, using Sanger sequencing, was found to segregate with HI (Fig. 1a). This variant is rare in the current genomic databases with no reported homozygotes [gnomAD allele frequency (AF) = 1.80 × 10−5; GME and South Asian gnomAD AF = 0] (Lek et al. 2016; Scott et al. 2016). It is located in a conserved C-terminal region, changes a hydrophobic residue to a polar neutral residue, and is predicted damaging by various bioinformatic tools (Fig. 1c; CADD = 26.8; GERP = 5.14; Supplementary Table 1) (Liu et al. 2011). 3D protein modeling shows that the C-terminal hydrophobic interactions at residue p.Ile369 with several DNA-binding homeodomain residues in the wild-type protein are lost when a threonine polar side chain is introduced into the loop region (Fig. 2). The variant has been deposited in Clinvar (#SCV000700205) (Landrum et al. 2016).
Fig. 2.
a Schematic representation of the LMX1A protein, which contains two LIM domains and one homeodomain. All reported variants in human HI are indicated on the figure. Autosomal dominant variants causing HI (p.Cys97Ser and p.Val241Leu) were reported in Wesdorp et al. 2018. The variant causing autosomal recessive HI is p.Ile369Thr. Domains were represented as described in the Universal Protein Resource (UniProt) database (Bateman et al. 2017). b Predicted three-dimensional structure of LMX1A. The variant position at residue 369 is highlighted by a box and the three domains are indicated in the same color as the schematic representation in A. c, d Close-up view of wild-type p.Ile369 (c) and mutant p.Thr369 (d) LMX1A. The residue at position 369(**) is represented by a ball-and-stick model, while the nearby residues are represented by a stick model. The hydrophobic interactions with amino acids in the homeodomain seen in the wild-type protein are lost when the threonine, polar side chain is introduced into the loop region. One of these interactions is with the residue (p.Val241*) of the previously reported autosomal dominant HI
Discussion
LMX1A is an evolutionarily conserved transcription factor with an essential function in neural development. It consists of two N-terminal LIM domains, a central DNA-binding homeodomain and a C-terminus (Doucet-Beaupré et al. 2015) (Fig. 2). Dominant LMX1A variants causing non-syndromic dominant HI impact either the homeodo-main or LIM domains (Wesdorp et al. 2018). The variable and progressive HI seen in these cases is believed to be due to insufficient maintenance of cochleovestibular function by one copy of LMX1A (Wesdorp et al. 2018). We show that an in silico predicted damaging autosomal recessive variant in the conserved C-terminal region of LMX1A causes a pre-lingual severe-to-profound HI profile (Figs. 1, 2). Lmx1a is known to bind and transactivate the insulin promoter. Truncation of the C-terminal domain of human paralog LMX1B leads to a 73% significant decrease of this transactivation, which suggests that important domains reside within the C-terminus for homeodomain DNA-binding performance and transcriptional regulation (Dreyer et al. 2000). In addition, we support this hypothesis by showing that the hydrophobic interactions of the p.Ile369 residue in the C-terminal with amino acids in the homeodomain are lost when the threonine, polar side chain is introduced into the loop region in affected individuals. Interestingly, one of these interactions is with homeodomain residue (p.Val241), which was previously reported to result in autosomal dominant progressive HI (Fig. 2) (Wesdorp et al. 2018). The equivalent homeodomain residue in paralog LMX1B (p.Val265Leu) is known to cause nail-patella syndrome (Dunston et al. 2004; Wesdorp et al. 2018). We hypothesize that the interaction of DNA-binding homeodomain residues (including p. Val241) with C-terminus residue p.Ile369 is important for LMX1A’s DNA-binding properties, and that loss of this C-terminal/homeodomain interaction leads to a reduced LMX1A DNA-binding performance and an autosomal recessive phenotype in our affected individuals.
Lmxla expression in the developing mouse is predominantly in the nervous system and otic vesicles which later become restricted to non-sensory epithelia of the ear (Failli et al. 2002; Nichols et al. 2008; Koo et al. 2009). Homozygous Lmx1a mouse mutants (Dreher) have neurological, skeletal, and otic abnormalities (Chizhikov et al. 2006), and exhibit circling behavior, balance abnormalities, and deafness. Heterozygotes littermates display normal hearing (Koo et al. 2009; Steffes et al. 2012). The inner ear in homozygous mice shows impaired morphogenesis/differentiation and mice have fused/misshapen sensory epithelia (Nichols et al. 2008; Steffes et al. 2012). Nichols et al. found that newborn mutant mice only have three sensory epithelia: two enlarged canal cristae and one fused epithelium comprising a combination of the cochlea, saccule, and utricle (Nichols et al. 2008). The same study suggested that Lmx1a is important in defining the non-sensory and sensory epithelium in the inner ear (Nichols et al. 2008). A recent study confirms this role and shows that separation of sensory and non-sensory domains in the inner ear is regulated by mutually antagonistic signals: Notch signaling and Lmx1a (Mann et al. 2017). Lmx1a regulates sensory organ segregation by antagonizing lateral induction and promoting commitment to the non-sensory fate (Mann et al. 2017).
Individuals with autosomal dominant LMX1A variants display progressive HI phenotype with variable age of onset, (a) symmetry, and severity (Wesdorp et al. 2018). The affected individuals in this study, however, display a congenital bilateral severe-to-profound HI profile. This is consistent with knock-out mice (Koo et al. 2009; Steffes et al. 2012), but also similar to other HI genes that express both an autosomal dominant and recessive inheritance (Liu et al. 1998; del Castillo and del Castillo 2017). Compared to the knock-out mouse, we did not observe any vestibulocochlear malformations in affected individual IV:1, assessed via CT-imaging. Heterozygous carriers of the p.Ile369Thr variant in this study do not have hearing impairment with the exception of the father (III:1), who has unilateral elevated hearing thresholds for his age and gender (mild-to-moderate HI; Supplementary Fig. 1), although it is unclear whether this might be due to the p.Ile369Thr variant or environmental reasons.
In addition to inner ear defects, knock-out mice also display cerebellar and alar plate defects, head tossing, and circling behavior, indicative of vestibular defects (Steffes et al. 2012; Glover et al. 2018). We did not observe a gross vestibular, cerebellar, or cognitive phenotype currently present in our patients, although one of the two affected individuals did present with a below average intelligence. This milder phenotype suggests that the p.Ile369Thr may be a hypomorphic and not a full loss-of-function variant. This is similar to what has been suggested for PCDH15, in which hypomorphic alleles cause NSHI with no vestibular dysfunction, while more severe variants of this gene result in Usher syndrome, type 1F (profound congenital deafness, vestibular areflexia, and retinitis pigmentosa) and, in mice, the deaf-circling Ames waltzer (av) (Ahmed et al. 2003; Alagramam et al. 2005). Patients with autosomal dominant LMX1A missense variants also have a normal cognition, but show a vestibular dysfunction in half of the affected individuals (Wesdorp et al. 2018).
Wesdorp et al. suggested that the dominant phenotype associated with missense variants p.Val241Leu and p.Cys97Ser might be due to haploinsufficiency. Haploinsufficiency is also the main hypothesis for dominant nail–patella syndrome due to heterozygous variants in paralog LMX1B (Dunston et al. 2004). However, heterozygous lmx1a mutant mice with null alleles display normal hearing (Steffes et al. 2012), which is not consistent with haploinsuffiency. As mentioned previously, we hypothesize that hypomorphic variants are the basis for an autosomal recessive inheritance and phenotype seen in the family described here. Autosomal dominant inheritance might be due to hypermorphic variants or a more severe level of LMX1A dysfunction compared to the recessive p.Ile369Thr variant. The latter is still inconsistent with null mouse models, where heterozygous mice show normal hearing (Steffes et al. 2012), but it is unknown how other genes and pathways could compensate for LMX1A/lmx1a in humans and mice that might explain this inconsistency.
In conclusion, in line with Dreher knock-out mice and autosomal dominant human HI, we demonstrate a human HI phenotype associated with an autosomal recessive LMX1A missense variant. This variant modifies a conserved residue in the C-terminus and might affect C-terminal/homeodomain interactions and the DNA-binding and transcriptional regulation properties of LMX1A. This finding expands the spectrum of genes involved in HI in the Pakistani population.
Supplementary Material
Acknowledgements
This work was supported by the Higher Education Commission of Pakistan (to WA) and the National Institute on Deafness and Other Communication Disorders grants R01 DC011651 and R01 DC003594 (to SML). Exome sequencing was funded by the National Human Genome Research Institute and the National Heart, Lung and Blood Institute grant HG006493 (to DAN, MJB, and SML). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268201200008I. We would like to acknowledge the entire University of Washington Center for Mendelian Genomics team and their members for exome sequencing and bioinformatics support: Michael J. Bamshad (University of Washington, Seattle Children’s Hospital), Suzanne M. Leal (Baylor College of Medicine), and Deborah A. Nickerson (University of Washington); Peter Anderson (University of Washington), Marcus Annable (University of Washington), Elizabeth E. Blue (University of Washington), Kati J. Buckingham (University of Washington), Imen Chakchouk (Baylor College of Medicine), Jennifer Chin (University of Washington), Jessica X Chong (University of Washington), Rodolfo Cornejo Jr. (University of Washington), Colleen P. Davis (University of Washington), Christopher Frazar (University of Washington), Martha Horike-Pyne (University of Washington), Gail P. Jarvik (University of Washington), Eric Johanson (University of Washington), Ashley N. Kang (University of Washington), Tom Kolar (University of Washington), Stephanie A. Krauter (University of Washington), Colby T. Marvin (University of Washington), Sean McGee (University of Washington), Daniel J. McGoldrick (University of Washington), Karynne Patterson (University of Washington), Sam W. Phillips (University of Washington), Jessica Pijoan (University of Washington), Matthew A. Richardson (University of Washington), Peggy D. Robertson (University of Washington), Isabelle Schrauwen (Baylor College of Medicine), Krystal Slattery (University of Washington), Kathryn M. Shively (University of Washington), Joshua D. Smith (University of Washington), Monica Tackett (University of Washington), Alice E. Tattersall (University of Washington), Marc Wegener (University of Washington), Jeffrey M. Weiss (University of Washington), Marsha M. Wheeler (University of Washington), Qian Yi (University of Washington), and Di Zhang (Baylor College of Medicine).
Footnotes
Online weblinks: ANNOVAR, http://annovar.openbioinformatics. org/, BLASTP, https://blast.ncbi.nlm.nih.gov/Blast.cgi/, Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/, Combined Annotation-Dependent Depletion (CADD), http://cadd.gs.washington.edu/, dbNSFP, https://sites.google.com/site/jpopgen/dbNSFP, dbSNP, https ://www.ncbi.nlm.nih.gov/projects/SNP/, Genome Aggregation Database (gnomAD), http://gnomad.broadinstitute.org/, Genome Analysis Toolkit (GATK), https://software.broadinstitute.org/gatk/, Greater Middle East (GME) Variome Project, http://igm.ucsd.edu/gme, International Organization for Standardization (ISO), https://www.iso.org/, MODELLER8v1 software, https://salilab.org/modeller/, Multiple Alignment tool, http://www.bioinformatics.org/sms/multi_align.html, Picard, http://broadinstitute.github.io/picard/, UCSF Chimera, https ://www.cgl.ucsf.edu/chimera/, and Universal Protein Resource (UniProt), http://www.uniprot.org/.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00439–018-1899–7) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest The authors declare that they have no competing interests.
References
- Ahmed ZM, Riazuddin S, Ahmad J et al. (2003) PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum Mol Genet 12:3215–3223. 10.1093/hmg/ddg358 [DOI] [PubMed] [Google Scholar]
- Alagramam KN, Stahl JS, Jones SM et al. (2005) Characterization of vestibular dysfunction in the mouse model for usher syndrome 1F. J Assoc Res Otolaryngol 6:106–118. 10.1007/s10162-004-5032-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alharazneh A, Luk L, Huth M et al. (2011) Functional hair cell mechanotransducer channels are required for aminoglycoside ototoxicity. PLoS One 6:e22347 https://doi.org/10.1371/journal.pone.00223 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Taketo MM (2007) Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. J Cell Sci 120:3327–3335. 10.1242/jcs.03485 [DOI] [PubMed] [Google Scholar]
- Auton A, Abecasis GR, Altshuler DM et al. (2015) A global reference for human genetic variation. Nature 526:68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Martin MJ, O’Donovan C et al. (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169. https://doi.org/10.1093/nar/gkw 1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhati M, Lee C, Nancarrow AL et al. (2008) Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes. EMBO J 27:2018–2029. 10.1038/emboj.2008.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhikov V, Steshina E, Roberts R et al. (2006) Molecular definition of an allelic series of mutations disrupting the mouse Lmx1a (dreher) gene. Mamm Genome 17:1025–1032. 10.1007/s00335-006-0033-7 [DOI] [PubMed] [Google Scholar]
- del Castillo FJ, del Castillo I (2017) DFNB1 non-syndromic hearing impairment: diversity of mutations and associated phenotypes. Front Mol Neurosci 10:428 10.3389/fnmol.2017.00428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-Beaupré H, Ang S-L, Lévesque M (2015) Cell fate determination, neuronal maintenance and disease state: the emerging role of transcription factors Lmx1a and Lmx1b. FEBS Lett 589:3727–3738. 10.1016/j.febslet.2015.10.020 [DOI] [PubMed] [Google Scholar]
- Dreyer SD, Morello R, German MS et al. (2000) LMX1B transactivation and expression in nail-patella syndrome. Hum Mol Genet 9:1067–1074. 10.1093/hmg/9.7.1067 [DOI] [PubMed] [Google Scholar]
- Dunston JA, Hamlington JD, Zaveri J et al. (2004) The human LMX1B gene: transcription unit, promoter, and pathogenic mutations. Genomics 84:565–576. 10.1016/J.YGENO.2004.06.002 [DOI] [PubMed] [Google Scholar]
- Elahi MM, Elahi F, Elahi A, Elahi SB (1998) Paediatric hearing loss in rural Pakistan. J Otolaryngol 27:348–353 [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA et al. (2007) Comparative protein structure modeling using MODELLER. In: Current protocols in protein science. Wiley, Hoboken, 2.9.1–2.9.31 [DOI] [PubMed] [Google Scholar]
- Failli V, Bachy I, Rétaux S (2002) Expression of the LIM-homeodomain gene Lmxla (dreher) during development of the mouse nervous system. Mech Dev 118:225–228 [DOI] [PubMed] [Google Scholar]
- Fu W, O’Connor TD, Jun G et al. (2012) Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493:216–220. 10.1038/nature11690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JC, Elliott KL, Erives A et al. (2018) Wilhelm His’ lasting insights into hindbrain and cranial ganglia development and evolution. Dev Biol. 10.1016/j.ydbio.2018.02.001 (in Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedlund E, Belnoue L, Theofilopoulos S et al. (2016) Dopamine receptor antagonists enhance proliferation and neurogenesis of midbrain Lmx1a-expressing progenitors. Sci Rep 6:26448 10.1038/srep26448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain R, Bittles AH (1998) The prevalence and demographic characteristics of consanguineous marriages in Pakistan. J Biosoc Sci 30:261–275 [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P et al. (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46:310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo SK, Hill JK, Hwang CH et al. (2009) Lmx1a maintains proper neurogenic, sensory, and non-sensory domains in the mammalian inner ear. Dev Biol 333:14–25. https://doi.org/10.1016/). ydbio.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291. 10.1093/bioinformatics/btm091 [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M et al. (2016) Clin Var: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44:D862–D868. 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. https://doi.org/10.1093/bioinfor matics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Hope C, Walsh J et al. (1998) Mutations in the myosin VIIA gene cause a wide phenotypic spectrum, including atypical Usher syndrome. Am J Hum Genet 63:909–912. 10.1086/302026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E (2011) dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat 32:894–899. 10.1002/humu.21517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann ZF, Gálvez H, Pedreno D et al. (2017) Shaping of inner ear sensory organs through antagonistic interactions between Notch signalling and Lmx1a. Elife 6:e33323 10.7554/eLife.33323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E et al. (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols DH, Pauley S, Jahan I et al. (2008) Lmx1a is required for segregation of sensory epithelia and normal ear histogenesis and morphogenesis. Cell Tissue Res 334:339–358. 10.1007/s00441-008-0709-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC et al. (2004) UCSF Chimera? A visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Riazuddin S, Belyantseva IA, Giese APJ et al. (2012) Alterations of the CIB2 calcium-and integrin-binding protein cause Usher syndrome type 1J and nonsyndromic deafness DFNB48. Nat Genet 44:1265–1271. 10.1038/ng.2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter N, Kilinc E, Navruz B, Bae Y (2011) Test Review: L. Brown, R. J. Sherbenou, & S. K. Johnsen test of nonverbal intelligence-4 (TONI-4). Austin, TX: PRO-ED,2010. J Psychoeduc Assess 29:484–488. 10.1177/0734282911400400 [DOI] [Google Scholar]
- Sambrook J, Russell DW (2006) Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc 2006:pdb.prot4455. 10.1101/pdb.prot4455 [DOI] [PubMed] [Google Scholar]
- Schultz JM, Khan SN, Ahmed ZM et al. (2009) Noncoding mutations of HGF are associated with nonsyndromic hearing loss, DFNB39. Am J Hum Genet 85:25–39. https://doi.org/10.1016/j. ajhg.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EM, Halees A, Itan Y et al. (2016) Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet 48:1071–1076. 10.1038/ng.3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelow D, Schuelke M (2012) HomozygosityMapper2012-bridging the gap between homozygosity mapping and deep sequencing. Nucleic Acids Res 40:W516–W520. 10.1093/nar/gks487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad M, Sivakumaran TA, Qaiser TA et al. (2013) Genetic analysis through OtoSeq of Pakistani families segregating prelingual hearing loss. Otolaryngol Neck Surg 149:478–487. 10.1177/0194599813493075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Smith RJH (2012) Genetics: advances in genetic testing for deafness. Curr Opin Pediatr 24:679–686. 10.1097/MOP.0b013e3283588f5e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffes G, Lorente-Cánovas B, Pearson S et al. (2012) Mutanlallemand (mtl) and belly spot and deafness (bsd) are two new mutations of Lmx1a causing severe cochlear and vestibular defects. PLoS One 7:e51065 10.1371/journal.pone.0051065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164–e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesdorp M, de Koning Gans PAM, Schraders M et al. (2018) Heterozygous missense variants of LMX1A lead to nonsyndromic hearing impairment and vestibular dysfunction. Hum Genet. 10.1007/s00439-018-1880-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.