Abstract
There is abundant variation in gene expression between individuals, populations, and species. The evolution of gene regulation and expression within and between species is thought to frequently contribute to adaptation. Yet considerable evidence suggests that the primary evolutionary force acting on variation in gene expression is stabilizing selection. We review here the results of recent studies characterizing the evolution of gene expression occurring in cis (via linked polymorphisms) or in trans (through diffusible products of other genes) and their contribution to adaptation and response to the environment. We review the evidence for buffering of variation in gene expression both at the level of transcription and translation, and the possible mechanisms for this buffering. Lastly, we summarize unresolved questions about the evolution of gene regulation.
Keywords: cis, trans; regulatory evolution; stabilizing selection; buffering
Evolution of gene expression between and within species
Regulation of gene expression is complex, and involves interactions between DNA, RNA, proteins, and the environment. Variation in gene expression is ubiquitous within populations and between species, however discerning the functional implications of that variation remains a challenge. Interpreting the functional implications of variation requires understanding how variation in gene expression propagates through gene regulatory networks, and ultimately results in complex phenotypes and disease. A common approach to analyzing the evolution of gene expression is to break it into its cis and trans components. That is, gene expression differences in cis that are due to linked polymorphisms (allele-specific and local to the affected gene) and differences in gene expression in trans, or due to diffusible products that needn’t be linked with the affected gene (in diploids, in the absence of cis-regulatory differences, trans-regulatory changes are expected to affect both alleles equally) (Fig. 1). Studies on the evolution of gene expression have found abundant variation within and between natural populations for both cis and trans-regulation of gene expression [1-12]. Both of these modes of gene expression regulation likely contribute to adaptation and divergence, however evidence to date suggests they may have different genetic and evolutionary properties.
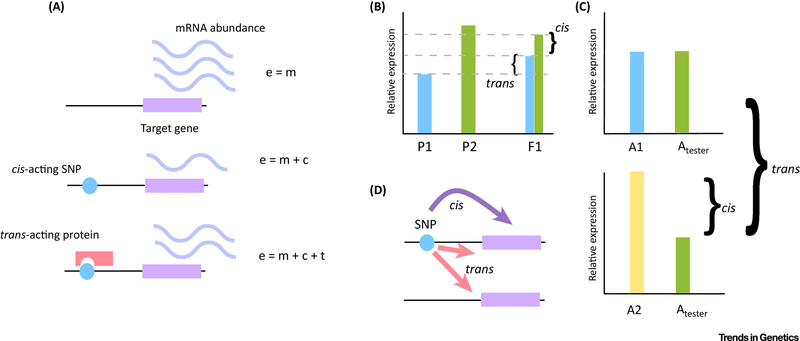
Figure 1:
cis and trans differences in gene regulation and the ways that cis and trans differences can be experimentally detected a) An allele is shown with average expression due to the presence of wild-type cis-regulatory modules and trans-regulatory factors. When these cis-regulatory modules are mutated, this will cause deviation from normal expression by a value c – cis factor – with the expression level m + c. This is shown in the second portion of the panel, where a cis-acting SNP is shown as a blue circle that alters the regulatory output of the associated gene, indicated as periwinkle squiggles. This same mutant allele might be represented in a genetic background where it is additionally affected in trans, with the resulting expression level m + c + t. In the example on the figure, the c is negative, but the t is positive. This is represented by a cis-mutation in blue, with a trans-acting protein interacting with the SNP in coral. Because c is negative and t is positive overall expression is closer to the average expression represented in the first panel. b) The earliest approach to cis-trans decomposition was to characterize expression of parental alleles in an F1 hybrid, often using techniques such as pyrosequencing. In the F1 hybrid differences in the expression of only one allele relative to the other allele is a cis effect. trans effects can be detected by comparing the expression ratio of each allele in hybrids to the ratio of expression between parents - if the ratio of expression is different in the F1 hybrid this is a trans effect. Here the bracket labeled cis indicates the differences between the two alleles in the F1. The trans bracket measures the amount of expression which is altered relative to the expression of the parental alleles. c) In the common reference design, a panel of individuals from a population sample are crossed to a single ‘tester’ strain (shown in green as Atester). Differences between the expression of the ‘tester’ allele and the population alleles (A1 and A2, blue and yellow) within an individual are cis effects, while differences in the expression of the tester allele between individuals are trans effects. In the top panel, there is no cis effect as the Atester and A1 allele are expressed at the same level. In the bottom panel, there is a cis effect between A2 and Atester, and there is a trans effect because the Atester allele is expressed at a different level in different individuals (between the A1 and A2 background). Note that this does not measure all trans effects, only those originating from the A1 and A2 backgrounds but not the Atester background. This section of the figure is based off of a figure from Fear et al. (2016). d) The eQTL approach to cis-trans decomposition. In eQTL studies the characterization of cis and trans is somewhat different, often meaning that the SNP effecting expression is either linked to the locus it effects or unlinked, though in some cases the contribution of specific alleles to eQTLs has been decomposed. In general, a SNP that has an effect of a gene expression phenotype will be mapped, and if it is within the gene that it effects it is called a cis-eQTL. If it is unlinked, these are termed trans-eQTLs. When the effect of individual alleles on expression is characterized the definition is the same as for F1 crosses.
When intra- and interspecific variation in gene expression is decomposed to cis and trans components, trans factors have often been found to make larger intraspecific contributions [12-16]. The larger contribution of trans factors within species is sometimes attributed to trans factors having a larger mutational target, thus being more likely to arise in comparison with cis regulatory mutations. As genes affect each other through many genetic and metabolic networks, trans factors could potentially arise anywhere in the genome (though the actual number of positions that could affect a particular gene in trans is smaller than the entire genome) [12,16,17]. Between species cis-regulatory differences are thought to have a greater contribution to divergence, suggesting that under selection they preferentially accumulate overtime [14,18-20]. A greater contribution of cis factors between species could be due to a number of reasons, such as trans factors having more deleterious pleiotropic-side effects, or because trans effects are more frequently recessive [21,22].
In several systems, and with different experimental approaches, work on cis-trans decomposition has found that cis and trans regulatory differences often influence the same gene, and when they do more often than not they act in opposite directions [4,5,7,11,12,23,24]. While both cis and trans factors might destabilize the transcriptome, cis-trans compensation – if common – will serve to re-stabilize the overall expression level of genes despite the presence of segregating – and putatively adaptive – regulatory variation [4,7]. If cis-trans compensation stabilizes overall gene expression this is consistent with other work which suggests that stabilizing selection is the predominant mode of evolution for gene expression (see Glossary) [14,25]. Overall, recent work on cis and trans differences in gene expression within populations, between species, and in different environments is creating a general picture of the importance of each type of regulatory difference in adaptation and speciation; providing a framework for advancing the understanding of evolution of gene regulation and expression.
Experimental approaches
There are several approaches to characterize the evolution of gene expression at single loci due to factors in cis and in trans. In diploids, cis differences will be found local to the affected gene and be allele specific, while trans changes can be linked or unlinked, but affect both alleles. There are three main approaches to characterizing cis and trans variation, two of which are closely related: comparing allele-specific expression between homozygote parents and their F1 hybrids and a common reference design (Fig. 1). In F1 hybrids cis and trans effects are partitioned by comparing the expression level of individual alleles in the hybrid (cis) and overall expression of both alleles compared between the parental strains (cis + trans) [16]. The common reference design is similar, but involves crossing a panel of genotypes to a common strain (i.e. creating a panel of F1 individuals where ½ of their genome is identical across all crosses). Within this cross design cis components of expression variation are quantified by comparing between the common allele and the population allele within a genotype, while comparing expression of the common allele across the population’s genomic backgrounds estimates variation due to trans factors [26,27]. These approaches are different in that the F1 hybrid approach relies upon a comparison with parental expression levels while the common reference design uses population samples. Both approaches are agnostic with regard to the source of trans differences. While these approaches have been used extensively for individual genes, we will focus primarily on genome-wide studies. Genome-wide, these designs have been successfully used to characterize cis and trans differences in yeast, plants, Drosophila, birds, and mice [1,2,10,28-31].
The third approach to understanding cis and trans variation is eQTL analysis, which correlates a molecular phenotype such as gene expression with genetic variation [11,32-35]. However, the definition of cis and trans in eQTL versus allele-specific expression studies is not entirely the same as eQTLs primarily characterize physical proximity (Fig. 1) [11,36-38]. The strength of the eQTL approach is in the ability to approximate the number of regulatory differences involved in changes in mRNA expression (though the authors note that a single eQTL does not imply a single genetic change), and to estimate the effect sizes of these regulatory differences. However, the eQTL approach has less power to detect cis-eQTLs that have cis and trans effects in the opposite direction. Further, because mapping trans factors requires correcting for a larger number of statistical tests, it has less power in comparison with mapping cis factors [39]. One exception is finding ‘trans hotspots’, i.e. genomic regions that appear to affect a disproportionate number of genes [39,40].
One approach to the study of cis-trans differences in gene expression that the authors believe warrants further investigation is to understand their distribution within gene regulatory networks. Very little is known about how cis and trans differences are distributed throughout gene regulatory networks, though several approaches are available to analyze expression variation in the context of gene regulatory networks, for example Structural Equation Models (see [26] for review). However, understanding these patterns requires detailed knowledge of the gene regulatory networks that does not exist for more than a handful of cases, and there has not yet been any systematic investigation of the distribution of cis and trans changes within networks across multiple systems. One noteworthy attempt found that the number of trans regulators negatively correlates with the evolution of cis regulation, though the phenotype was limited to the number of upstream and downstream targets [39]. We do not generally know how regulatory perturbations are distributed throughout gene regulatory networks, if these effects are local or do they propagate through gene networks, or if they are frequently dampened or amplified. Indeed, there are many unanswered questions about the evolution of gene expression, and it remains an area with abundant opportunities for future research.
Adaptation in cis and trans
Adaptive evolution of individual or limited groups of genes affecting expression in cis and trans has been investigated extensively, more so than can be summarized here, and a number of excellent reviews are available [22,40-42]. There is considerable support for expression of individual genes often evolving in cis, though the exact adaptive significance is not always established [32,43-47]. In general, it is thought that cis-regulatory changes will be less pleiotropic and more commonly found at ‘structural’ genes that do not directly impact the expression of other genes (meaning they are involved in catalyzing enzymatic reactions or other structural roles rather than regulating other genes) [41,43,46,48]. However, such genes still belong to gene regulatory networks, and thus directly or indirectly are expected to affect the expression of other genes. In plants, for example, the structural genes required for producing the compounds that create flower color also produce other compounds necessary for proper organismal function, while the transcription factors that activate them are tissue specific, thus even though they are structural genes evolution of flower color is biased towards trans-regulatory changes [49]. Therefore, there is still a lot to understand about the importance of cis-regulatory changes within gene networks, and how adaptive differences are spread throughout gene networks.
In genome-wide surveys of gene expression, adaptation is often inferred as an excess of either eQTLs that change gene expression in the same direction or cis-regulatory differences biased towards the alleles of one parent [50-53]. In the case of cis-regulatory expression this is limited to particular pathways or functionally enriched categories, and can also include an excess of cis/trans pairs that are in the same direction, if trans differences are investigated [51]. If the eQTLs or cis-regulatory differences are neutral then changes in the direction of expression or differences between parental alleles would be expected to be roughly equal. For example, a recent application of this method to yeast eQTLs found an excess of up-regulated biofilm suppressor genes associated with adaptation to human hosts [54]. In addition, by expanding these types of analyses to outgroup species lineage specific selection can be inferred [52,55]. It is important to note that while some genome-wide studies have found support for the contribution of cis-factors to divergence and adaptation, in many cases trans-factors are not investigated [56-59]. There is some evidence that they contribute to adaptation, for instance, in a study on the evolution of cichlid opsins trans differences were found to contribute more to adaptation between species than cis differences [60]. Note that these inferences of adaptation generally apply to particular subsets of genes and eQTLs, and do not necessarily imply that the general mode of evolution of gene expression is adaptive. It is also possible that many of the observed differences in cis and trans are due to context specific effects, as the environment will invariably effect gene expression in some way (Box 1).
Box 1 Environmental effects on cis-trans variation.
Gene expression robustness in the face of environmental variation can be an important component of system homeostasis, while gene regulatory differences in response to the environment can be an important component of adaptation, and potential for adaptation. It is currently unclear how important cis and trans effects are for the response to environmental differences, and whether they are maintained within populations due to gene by environment interactions. In yeast these questions have been approached in a variety of ways, for example in interspecific yeast crosses local cis effects tended to be less condition dependent than trans effects [99]. In another case, allele specific expression was examined in normal and heat-stressed environments for two yeast species, one of which was adapted to higher temperatures, and abundant cis-regulatory divergence was uncovered between species [3]. However, these cis effects were not environment dependent and were not adaptive for thermal tolerance [3]. A recent study in yeast that did not examine trans factors found that induced mutations in cis had different effects depending upon the environment, but their effect on fitness and relevance to adaptation are not known [100]. These conclusions are corroborated in other systems, for example in C. elegans while abundant cis effects were detected between strains, they were not environment dependent and the response to heat stress was mediated largely by trans-effects [38]. Several studies have found that cis effects are concordant between environments in intra-specific crosses of Drosophila and inter-specific differences in Arabidopsis [27,101]. In contrast to these studies, in grass while cis-effects changed in magnitude but not direction with treatment (drought stress) very few trans effects were detected, though this may be largely related to issues of power [102]. Overall, the current evidence suggests, but is not conclusive, that trans regulation might be more important for environment dependent differences in gene expression, and it is unclear what the relationship is between these trans differences and long term adaptation to the environment [15,27].
Stabilizing selection on gene expression
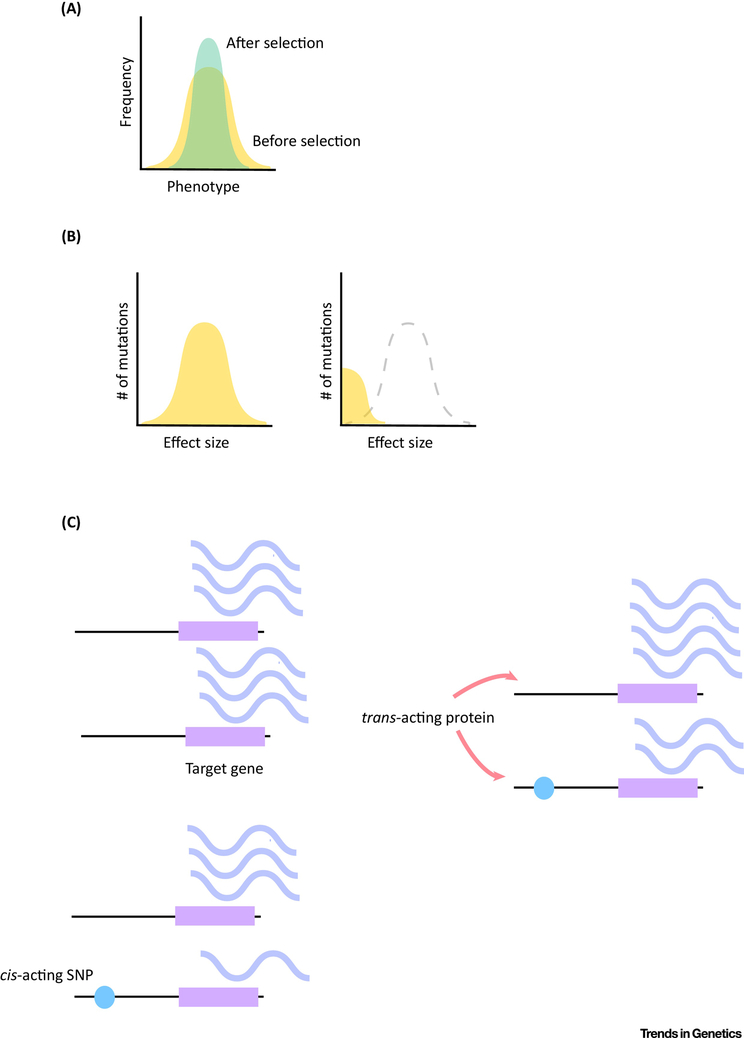
Due to the frequency with which compensatory cis-trans pairs are observed in genome-wide studies, it has been theorized that in general selection on gene expression may be stabilizing. Stabilizing selection requires the maintenance of a mean, non-extreme phenotype in a population or species (Fig. 2a). There has been abundant evidence for stabilizing selection on gene expression, in a number of model and non-model systems, however the relative contribution of cis and trans differences, or compensatory cis-trans effects, have not been explicitly examined. We will summarize recent evidence for stabilizing selection on gene expression, and note that the contribution of cis-trans compensation to stabilizing selection is an area of potential interest. In Drosophila, comparisons between three species with different divergence times found that while the number of genes with evidence for cis regulatory divergence increased over divergence time, the number of differences in total expression did not [14]. This would imply that cis regulatory differences are being compensated in some fashion. In addition, in this study less than 2.2% of genes showed differences in expression consistent with genetic drift [14]. Both of these observations are consistent with widespread stabilizing selection on gene expression levels. Without decomposing the contribution of cis or trans this type of approach was extended to larger phylogenetic distances and similar results were found, though with the additional caveat that at shorter time scales both types of evolution of gene expression will appear neutral, and that our ability to detect both stabilizing and directional selection is dependent upon the scale of divergence at which it is measured [19]. Between flycatcher species variance in gene expression was correlated with between species divergence, which also suggests stabilizing and/or neutral evolution in gene expression [61]. Between humans and primates the amount of inter-species variation in gene expression can be explained by variation in gene expression within species – which is consistent with stabilizing selection on gene expression [62-64].
Figure 2).
Selection and cis-trans compensation a) Stabilizing selection on phenotypes within a population. Prior to selection the distribution of phenotypes is broader (shown in yellow), and after selection extreme phenotypes have been selected out (green). b) A hypothetical illustration of purifying selection, with the effect size of mutations on the x axis and the number of mutations with that effect size on the y. The distribution of mutations with different effect sizes is approximated in yellow. The panel on the left illustrates essentially neutral evolution, as mutations with larger effect sizes (and, by inference, more deleterious effects) are not being selected out of the population. On the right is an example of purifying selection, in which mutations of larger effect size are selected out of the population. This is one potential way to for stabilizing selection to act on gene expression. c) cis-trans compensation. The upper left panel represents a wild type individual with equal expression between the two alleles at a single gene, with the regulatory region shown as a black bar and the coding region a purple square. Relative transcript abundance is illustrated by the periwinkle squiggles. The lower left panel shows how a cis-acting SNP could potentially appear on one of the alleles, reducing gene expression allele-specifically. This SNP is illustrated with a blue circle. In the panel on the right, a trans-acting mechanism upregulates both alleles to preserve the total mRNA output from the gene (compensation). In this panel, the arrows indicate that a trans acting protein is interacting with the regulatory regions of both copies of the gene.
Another approach to evaluating the prevalence of stabilizing selection is to investigate the distribution of mutational effects in standing variation relative to new mutations [65-67]. For example, under the house-of-cards theory of stabilizing selection mutations are expected to be infrequent, and the effects of mutations are expected to far exceed standing variation [25,68]. The house-of-cards theory of stabilizing selection is referred to as such because new mutations are thought to disrupt multiple processes and are subject to strong selection, compared to related models in which new mutations have weak effects. In contrast, neutral evolution would predict that selection is negligible and equilibrium genetic variance is a balance between mutation and drift. The predictions for house-of-cards stabilizing selection were found to best fit the distribution of mutational effects in D. melanogaster, S. cerevisiae, and C. elegans [25]. Many lines of evidence thus suggest that stabilizing selection on gene expression may be an important factor influencing inter and intra-species gene expression evolution. Stabilizing selection on gene expression can occur through different molecular mechanisms, including purifying selection – wherein deleterious mutations are selected against (Fig. 2b), and/or compensatory evolution – where small mutations affecting gene expression in cis (trans) are compensated for through trans (cis) factors that stabilize overall gene expression level (Fig. 2c). Below we will focus on the latter of these two potential hypotheses.
Buffering gene regulatory differences with cis-trans compensation
Between species
Between species cis-trans effects that compensate one another for individual genes are more commonly observed than those with the same direction of effect. However, despite common observation, the potential significance of this patterns in terms of selection and/or adaptation has not been investigated. Fixation of cis-trans factors between species could occur as a result of stabilizing selection on overall gene expression or to ameliorate the negative pleiotropic side effects of a selected mutation. Stabilizing selection on overall gene expression could result in the fixation of compensatory cis (trans) factors in response to mildly deleterious trans (cis) factors to maintain gene expression levels. It is also possible that compensatory mutations are selected for to mitigate the side effects of an otherwise beneficial mutation, as has been observed in the evolution of antibiotic resistance [69-74]. It is clear from a number of different studies and systems that compensatory cis-trans pairs are more common than those that are not compensatory. For example, when two divergent yeast species were crossed, 67% of genes with both cis-trans effects were compensatory, consistent with buffering of regulatory divergence [75]. In a different study that focused on genome-wide cis and trans effects for a single gene in yeast, compensatory effects were two to three times more likely than to have cis and trans effects with the same direction [18]. In interspecific crosses between D. simulans / D. sechellia and D. melanogaster / D. simulans, compensatory cis/trans pairs accounted for 73% and 87% of cases respectively [14]. In house mice hybrids the majority of cis-trans pairs were compensatory [9], thus from yeast to vertebrates cis-trans pairs are more likely to be compensatory. It is possible that this type of cis-trans compensation results from co-adaptation within species, and contributes to reproductive barriers. For a thorough review of the potential for co-adapted gene complexes to contribute to speciation see [76].
Within species
While between species cis-trans differences that are compensatory will (or can) be fixed, within species cis-trans differences that compensate one another will be polymorphic and not necessarily co-inherited. However, it has been recognized that within species there are abundant coupled cis-trans factors contributing to buffering of gene expression variation, though again the importance of this pattern for selection or adaptation is not known [2,11,27]. For example, in conifers, abundant cis-trans compensation was observed within a population using an eQTL study design that characterized local trans effects and cis-regulatory differences [11]. Although the study was not designed to detect unlinked trans effects, the presence of linked compensatory trans effects lead the authors to suggest that this compensation was due to self-regulation or closely linked trans modifiers. Linkage disequilibrium is high in organisms such as conifers, and it is unclear how likely it is that linkage between trans modifiers and cis variants would be maintained in more readily recombining species such as Drosophila. However, cis-trans compensation has been characterized in studies of within species variation in Drosophila. One study used a common reference design to characterize cis and trans variation within D. melanogaster, and found that 85% of cis-trans combinations are compensatory [27]. The combination of many studies finding cis-trans compensation within species suggests that cis-trans compensation can accumulate intra-specifically, though it does little to suggest a potential mechanism for this compensation.
Buffering gene regulatory differences at the level of translation
While differences in the cis and trans regulation of gene expression are abundant, it is also possible that these differences have little functional implication beyond the level of translation. Buffering at the level of translation could be another source of what is essentially cis-trans compensation if changes in transcription in cis are countered by changes in translation in trans. Translational buffering could be regulated in cis, trans, or both, though most commonly the source is not mapped. This would be an interesting area of future research. At the level of translation, it was found that in hybrids between two species of yeast, differences in translation efficiency and transcription occurred seven times more frequently in opposite directions – essentially cis-trans compensation at the level of translation [75]. Due to this, 20-80% of non-conserved transcription was buffered at the level of translation [75]. The observation of buffering at the level of translation is similar to earlier reports in two yeast species that found that aberrant expression of mRNA in hybrids was not reflected in ribosome occupancy [77]. However, another study found no evidence of buffering at the level of translational efficiency including when they reanalyzed the earlier referenced data [78]. In another study on yeast the results were more mixed, as there was some concordance between eQTLs and protein QTLs but the relationship was variable. Two large eQTL hotspots that overlapped with protein QTL found effects in opposite directions which would serve to buffer the overall level of gene expression/protein, but this effect was not general [79,80]. In a study on specific genes in snakes no evidence was found of post-transcriptional buffering of mRNA expression differences, rather a high concordance was found between mRNA abundance and the proteome [81]. Overall, the evidence for posttranscriptional compensation for mutations affecting transcription remains inconclusive. It maybe be that buffering at the level of translation is important for particular subsets of genes or developmental pathways, and that a more detailed understanding of translational dynamics will illuminate these differences.
Mechanisms underlying the appearance of compensation
Inherent mutational and inference biases
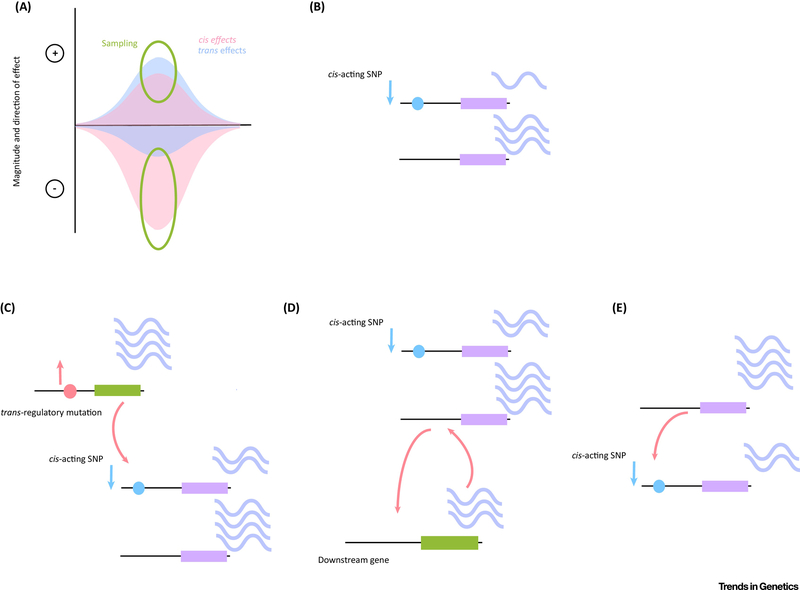
It is possible that the appearance of cis-trans compensation is due to biases in mutation or ascertainment. A recent study found that there was no bias in the frequency of cis or trans effects in either direction (increase or decrease), however overall cis regulatory differences have a larger effect size [12]. In addition, for cis regulatory mutations, variants that decreased expression had a larger magnitude of effect than those that increased expression. The opposite was true for trans regulatory mutations, with larger effect sizes when they increased expression [12]. These results are from a screening for cis and trans effects on a single gene (genome-wide) so it is not known whether these results are generalizable. If these patterns are generalizable, it will have two effects, first that we will be more able to detect cis effects than trans effects, because larger effect sizes are easier to detect. This is also true because it is more difficult to detect effects that are distal, or not linked to the affected gene, in eQTL studies due to the larger burden of multiple testing. This is because the potential location for a proximal eQTLs is limited, while the potential location of a distal eQTL is the entire genome. We will also more frequently detect trans effects that increase expression, and cis effects that decrease expression (Fig. 3a). Which is to say that the observed patterns of cis-trans compensation may be due to the analysis method used and its sensitivity to detect changes and/or mutational bias rather than selection, which is especially true for eQTL studies (Fig. 3a). In one instance in which this could be investigated however, the opposite was true, and 65% of cis/trans pairs had a positive cis effect and a negative trans effect [27]. Additional problems might result from confounding sampling error or from estimation biases with either cis or trans effects. However, every method of examining cis-trans differences finds an excess of compensatory effects, in every system, lending some confidence to the pattern.
Figure 3).
Potential causes and mechanisms for cis-trans compensation. a) It is possible that the apparent widespread prevalence of cis and trans effects that compensate one another is due to ascertainment bias. Large cis effects are more commonly negative, and large trans effects are more commonly positive. Thus, given that we are biased towards detecting differences of large effect, it is possible that it is a methodological artifact. In this figure, the frequency of cis and trans differences are shown, where the frequency distribution of cis effects is shown in pink and trans effects in blue. The green circles indicate what is being sampled by any given study from the distribution. b) A wild type allele (bottom) where the number of transcripts is illustrated as an arbitrary number of periwinkle squiggles. An allele with a cis-regulatory mutation (top) is shown, where the mutation is illustrated as a blue dot. The total number of transcripts produced is reduced by 2/3. This illustration will serve as a baseline for c) through e). c) It is possible that the observation of cis-trans compensation is due to the accumulation of effects in cis (trans) that are then compensated for by a mutation in trans (cis). Here a trans regulatory difference is shown at the gene in green, where a SNP illustrated by the pink dot results in upregulation and an increase in the number of transcripts. This acts on its downstream target, the gene shown in purple, which has a SNP in cis illustrated by the blue circle that downregulates the gene. The two mutations together stabilize the overall level of gene expression. Note that the authors are agnostic as to whether cis or trans mutations appear first, this is one example. d) It is also possible that cis-trans compensation is caused by gene regulatory network feedback (which could be positive or negative, shown here is positive). In this case activation of a downstream gene may feedback on the target gene and normalize gene expression levels in spite of a cis-regulatory mutation. Gene network feedback can happen locally, through self-regulation, or essentially as a trans effect that does not require a mutation. Here the downstream gene is shown in green, and there is no mutation at the locus. Feedback on the upstream gene with a cis-acting SNP shown in blue serves to stabilize gene expression. e) Transvection is the most hypothetical explanation for cis-trans compensation, and it is also the least well understood. It has been observed across species and genes that the regulatory information from one copy of an allele has the potential to regulate the other copy. Enhancers show cis-preference, but if for any reason the enhancer on the other allele was not regulating its target as expected they can act in trans. Here the genes in purple are two copies of the same locus, one of which has a mutation in cis that is shown as a blue circle. Communication between each copy of the allele stabilizes gene expression output (periwinkle squiggles).
Compensatory mutations
It has been hypothesized that a trans mutation spreads jointly with a cis mutation to compensate for their slightly deleterious effects (Fig. 3b, c) [7]. While the joint spread of cis-trans mutations is possible in some species with extensive linkage disequilibrium, or if mutations within the target genes act in trans, as a general mechanism these factors will not be co-transmitted within species [11]. For example in Drosophila even when co-localized on the same arm long-range linkage disequilibrium in flies is not a general feature [82]. Such a strong excess of ~80% compensatory cis-trans interactions within species cannot be explained by co-evolution without co-inheritance.
If a trans mutation downregulates an allele only in the presence of a cis mutation, with the cis effects compensated, it is possible that these cis-trans mutations may exist jointly and underlie widespread compensation. Indeed, multiple studies have detected abundant cis-trans epistasis [83-86], however the statistical methodology has not been developed to test whether those interaction terms are of a compensatory nature [87,88]. Conceptually, this scenario is akin to Wright’s arguments on the evolution of dominance, where a slightly deleterious mutation (in cis) is allowed to segregate due to a secondary mutation (in trans) that renders the first mutation recessive. Population genetic analyses have established that the strength of selection for such secondary modifiers is most frequently vanishingly small [89]. Given that the actual genes or mechanism of trans effects is generally not known, these patterns warrant further investigation.
Gene network feedback
It is possible that many cis-trans compensatory interactions are due to feedback within gene regulatory networks (Fig. 3a, d). There is some support for this in single gene and genome-wide studies, though there is room for this to be more extensively investigated. For example, two single gene studies in yeast found support for gene-network feedback: at ROX1 where negative feedback confers robustness to the expression despite naturally occurring allelic variants [90], and at AMN1 where a local trans-eQTL was found to operate through a regulatory feedback loop involving several additional genes [91]. Genome-wide, one study on buffering by feedback found that approximately 15% of allelic differences are compensated through this mechanism [78], and work in conifers also found that 10% of local eQTLs acted in trans, suggesting they may work through self-regulation [11]. The potential for gene network feedback to stabilize overall gene expression levels is likely to be important for at least some genes, and investigating its importance more broadly is in interesting avenue for future research.
Transvection
In both mammals and dipterans, a type of inter-chromosomal communication has been observed, termed transvection, where the regulatory information on one chromosome can be used to regulate the expression of the allele on the other chromosome (Fig. 3a, e) [92-96]. Initially considered an oddity, transvection is now understood to be widespread [92,93,95]. It involves both up and down regulation of alleles through coordination of expression between alleles [97], and it affects a large majority of tested regulatory regions [95]. The effect of transvection on natural transcriptome variation has never been tested, but given the evidence for its ability to work in trans and compensate for deficiency mutations it is a candidate mechanism for compensation. Some instances of transvection require pairing between homologous chromosomes, a peculiar feature of insects, but other instances appear to be independent of pairing [92,97]. While the principles of chromosome architecture are an area of intense research at the moment, it is unclear how important transvection is for gene regulation in different systems. However, there is evidence that insulator proteins involved in establishing inter-chromosomal contacts in mammals and insects may facilitate transvection [98-100]. This is an interesting area for future research, as there is currently very little understanding of the role of transvection in natural populations, and no understanding of its potential role in cis/trans effects.
Concluding Remarks and Future Perspectives
While many individual cases of the evolution of gene expression have been investigated, a broad view of the evolution of gene regulation remains to be formulated. Genome-wide approaches have failed to create a consensus about how gene expression evolves in general, in part because there are likely different answers for different subsets of genes or systems (see Outstanding Questions). The path by which any particular gene is going to evolve is going to be affected by its role within its gene network and its developmental context. However, both gene networks and developmental context are known for only a very few systems, making it difficult to place any patterns that are found in the evolution of gene expression into a larger picture. For example, genes with structural roles within networks may have inherently different evolutionary dynamics than those involved in transcription, or those with physiological effects. As more gene regulatory networks are characterized in non-model systems and we gain a better understanding of their developmental role we foresee many historically observed patterns fitting into new paradigms. Emerging technologies make this an exciting time to study the evolution of gene expression, including new techniques to plumb the effect of chromatin organization, gene neighborhood, and cell or tissue specific differences in gene regulation. In the coming years, these emerging technologies and the increasing tractability of non-model systems promises many new and exciting insights about the evolution of gene expression.
Outstanding Questions.
How are cis and trans effects distributed within gene regulatory networks, and what implications does their position have for their effect and potential to contribute to adaptation?
How do cis and trans effects, including those that respond to the environment, contribute to the long-term divergence and adaptation of species?
How important are different sources of buffering for variation in gene expression?
What mechanism is responsible for the wide-spread cis-trans compensation observed across techniques and organisms – is it an artifact of our methods, due to mutational compensation, gene network feedback, or transvection?
Trends Box.
cis regulatory differences appear to be more commonly responsible for adaptive evolution, though there are exceptions that illustrate the importance of gene network context in the path by which evolution proceeds.
Current evidence supports the supposition that genome-wide gene expression evolves under stabilizing selection. There is limited evidence that some of this stabilizing selection is due to compensatory cis-trans evolution, but more research is needed.
Overall when cis-trans contributions to gene expression differences are investigated there is an excess of compensatory cis-trans pairs.
The observation of an excess of cis-trans pairs that are compensatory could be due to mutational and ascertainment bias, selection for compensatory mutations, buffering from gene network feedback, or potentially communication between alleles (transvection).
Acknowledgements
The authors would like to thank J. Butler for assistance in the production of this manuscript, as well as Trisha Wittkopp and three anonymous reviewers for comments on the manuscript. Work on this manuscript was supported with funding from the National Institutes of Health, grants GM103804 and MH091561
Glossary
- Adaptive evolution
Evolutionary changes that increase survivorship or reproduction.
- eQTLs
Expression quantitative trait loci, regions of the genome that contribute to variation in expression levels of RNA.
- Linkage disequilibrium
The non-random association of alleles within a population.
- Pleiotropic
A gene is referred to as pleiotropic if it effects more than one phenotype.
- Stabilizing selection
The favoring of individuals in the population with mean, rather than extreme, phenotypes. Stabilizing selection generally reduces existing phenotypic variation, and is measured at the level of phenotypes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shi X et al. (2012) cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat. Comm 3, 950–9 [DOI] [PubMed] [Google Scholar]
- 2.Osada N et al. (2017) cis- and trans-regulatory effects on gene expression in a natural population of Drosophila melanogaster. Genetics 206, 2139–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li XC and Fay JC (2017) cis-regulatory divergence in gene expression between two thermally divergent yeast species. Genome Biol. Evol 9, 1120–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landry CR et al. (2005) Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171, 1813–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goncalves A et al. (2012) Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome Res. 22, 2376–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graze RM et al. (2009) Regulatory divergence in Drosophila melanogaster and D. simulans, a genomewide analysis of allele-specific expression. Genetics 183, 547–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahasi KR et al. (2011), Two types of cis-trans compensation in the evolution of transcriptional regulation. Proc. Natl. Acad. Sci 108, 15276–15281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meiklejohn CD et al. (2014) The roles of cis- and trans-regulation in the evolution of regulatory incompatibilities and sexually dimorphic gene expression. Genome Res. 24, 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mack KL et al. (2016) Gene regulation and speciation in house mice. Genome Res. 26, 451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tirosh I et al. (2009) A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324, 659–662 [DOI] [PubMed] [Google Scholar]
- 11.Verta J-P et al. (2016) Dissection of expression-quantitative trait locus and allele specificity using a haploid/diploid plant system - insights into compensatory evolution of transcriptional regulation within populations. New Phytol. 211, 159–171 [DOI] [PubMed] [Google Scholar]
- 12.Metzger BPH et al. (2016) Contrasting frequencies and effects of cis- and trans-regulatory mutations affecting gene expression. Mol. Biol. Evol 33, 1131–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhoné B et al. (2017) No excess of cis-regulatory variation associated with intra-specific selection in wild pearl millet (Cenchrus americanus). Genome Biol. Evol 9, 388–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coolon JD et al. (2014) Tempo and mode of regulatory evolution in Drosophila. Genome Res. 24, 797–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J et al. (2015) Temperature stress mediates decanalization and dominance of gene expression in Drosophila melanogaster. PLoS Genet. 11, e1004883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wittkopp PJ et al. (2004) Evolutionary changes in cis and trans gene regulation. Nature 430, 85–88 [DOI] [PubMed] [Google Scholar]
- 17.Gruber JD et al. (2012) Contrasting properties of gene-specific regulatory, coding, and copy number mutations in Saccharomyces cerevisiae: Frequency, effects, and dominance. PLoS Genet. 8, e1002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzger BPH et al. (2017) Evolutionary dynamics of regulatory changes underlying gene expression divergence among Saccharomyces species. Genome Biol. Evol 9, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nourmohammad A et al. (2017) Adaptive evolution of gene expression in Drosophila. Cell Reports 20, 1385–1395 [DOI] [PubMed] [Google Scholar]
- 20.Gordon KL and Ruvinsky I (2012) Tempo and mode in evolution of transcriptional regulation. PLoS Genet. 8, e1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemos B et al. (2008) Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc. Nat. Acad. Sci. USA 105, 14471–14476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prud’homme B et al. (2007) Emerging principles of regulatory evolution. Proc. Nat. Acad. Sci. USA 104, 8605–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romero IG et al. (2012) Comparative studies of gene expression and the evolution of gene regulation. Nat. Rev. Genet 13, 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brawand D et al. (2104) The evolution of gene expression levels in mammalian organs. Nature 478, 343–348 [DOI] [PubMed] [Google Scholar]
- 25.Hodgins-Davis A et al. (2015) Gene expression evolves under a House-of-Cards Model of stabilizing selection. Mol. Biol. Evol 32, 2130–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nuzhdin SV et al. (2012) Genotype–phenotype mapping in apost-GWAS world. Trends Genet. 28, 421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fear JM et al. (2016) Buffering of genetic regulatory networks in Drosophila melanogaster. Genetics 203, 1177–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Springer NM and Stupar RM (2007) Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. The Plant Cell 19, 2391–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wittkopp PJ et al. (2008) Regulatory changes underlying expression differences within and between Drosophila species. Nature Genet. 40, 346–350 [DOI] [PubMed] [Google Scholar]
- 30.Wang X et al. (2016) Allele-specific transcriptome and methylome analysis reveals stable inheritance and cis-regulation of DNA methylation in Nasonia. PLoS Biol. 14, e1002500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang M et al. (2017) Bayesian inference of allele-specific gene expression indicates abundant cis-regulatory variation in natural flycatcher populations. Genome Biol. Evol 9, 1266–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishikawa A et al. (2017) Different contributions of local- and distant-regulatory changes to transcriptome divergence between stickleback ecotypes. Evolution 71, 565–581 [DOI] [PubMed] [Google Scholar]
- 33.Zhu Z et al. (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nature Genet. 48, 481–487 [DOI] [PubMed] [Google Scholar]
- 34.Albert FW and Kruglyak L (2015) The role of regulatory variation in complex traits and disease. Nat. Rev. Genet 16, 197–212 [DOI] [PubMed] [Google Scholar]
- 35.Nica AC and Dermitzakis ET (2013) Expression quantitative trait loci: present and future. Philos. Trans. R. Soc. B 368, 20120362–20120362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knowles DA et al. (2015) Allele-specific expression reveals interactions between genetic variation and environment. Nature Methods 14, 699–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crowley JJ et al. (2015) Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nature Genet. 47, 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snoek B et al. (2017) Contribution of trans-regulatory eQTL to cryptic genetic variation in C. elegans. bioRxiv DOI: 10.1101/120147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang B and Wittkopp PJ (2017) Structure of the transcriptional regulatory network correlates with regulatory divergence in Drosophila. Mol. Biol. Evol 34, 1352–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin A and Orgogozo V (2013) The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution 67, 1235–1250 [DOI] [PubMed] [Google Scholar]
- 41.Stern DL and Orgogozo V (2008) The loci of evolution: how predictable is genetic evolution? Evolution 62, 2155–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rebeiz M and Williams TM (2016) Using Drosophila pigmentation traits to study the mechanisms of cis-regulatory evolution. Curr. Opin. Insect Sci 19, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Signor SA et al. (2016) Genetic convergence in the evolution of male-specific color patterns in Drosophila. Curr. Biol 26, 2423–2433 [DOI] [PubMed] [Google Scholar]
- 44.Yassin A et al. (2016) Ancient balancing selection at tan underlies female colour dimorphism in Drosophila erecta. Nat. Comm 7, 10400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rebeiz M et al. (2011), Evolutionary origin of a novel gene expression pattern through co-option of the latent activities of existing regulatory sequences. Proc. Nat. Acad. Sci. USA 108, 10036–10043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yassin A et al. pdm3 is responsible for recurrent evolution of female-limited color dimorphism in Drosophila. Curr. Biol 26, 2412–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Brown NM et al. (2015) A recurrent regulatory change underlying altered expression and Wnt response of the stickleback armor plates gene EDA. eLife 4, e05290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romero IG et al. (2012) Comparative studies of gene expression and the evolution of gene regulation. Nat. Rev. Genet 13, 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Streisfeld MA and Rausher MD (2009) Altered trans-regulatory control of gene expression in multiple Anthocyanin genes contributes to adaptive flower color evolution in Mimulus aurantiacus. Mol. Biol. Evol 26, 433–444 [DOI] [PubMed] [Google Scholar]
- 50.Naranjo S et al. (2015) Dissecting the genetic basis of a complex cis-regulatory adaptation. PLoS Genet. 11, e1005751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fraser HB et al. (2010) Evidence for widespread adaptive evolution of gene expression in budding yeast. Proc. Nat. Acad. Sci. USA 107, 2977–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fraser HB et al. (2011) Systematic detection of polygenic cis-regulatory evolution. PLoS Genet. 3, e1002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Artieri CG and Fraser HB (2014) Evolution at two levels of gene expression in yeast. Genome Res. 24, 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kita R et al. (2017) High-resolution mapping of cis-regulatory variation in budding yeast. Proc. Nat. Acad. Sci. USA 114, E10736–E10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riedel N et al. (2015) Multiple-line inference of selection on quantitative traits. Genetics 201, 305–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arunkumar R et al. (2016) Recent mating-system evolution in Eichhorniais accompanied by cis-regulatory divergence. New Phytol. 211, 697–707 [DOI] [PubMed] [Google Scholar]
- 57.Bell GDM et al. (2013) RNA-seq analysis of allele-specific expression, hybrid effects, and regulatory divergence in hybrids compared with their parents from natural populations. Genome Biol. Evol 5, 1309–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lemmon ZH et al. (2014) The role of cis regulatory evolution in maize domestication. PLoS Genet. 10, e1004745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Josephs EB et al. (2015) Association mapping reveals the role of purifying selection in the maintenance of genomic variation in gene expression. Proc. Nat. Acad. Sci. USA 112, 15390–15395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Quin KE et al. (2012) Evolution of cichlid vision via trans-regulatory divergence. BMC Evol. Biol 12, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uebbing S et al. (2016) Divergence in gene expression within and between two closely related flycatcher species. Mol. Ecol 25, 2015–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Somel M et al. (2014) Transcriptomic insights into human brain evolution: acceleration, neutrality, heterochrony. Curr. Opin. Genet. Dev 29, 110–119 [DOI] [PubMed] [Google Scholar]
- 63.Khaitovich P et al. (2006) Evolution of primate gene expression. Nat. Rev. Genet 7, 693–702 [DOI] [PubMed] [Google Scholar]
- 64.Blekhman R et al. (2010) Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 20, 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Denver DR et al. (2005) The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nature Genet. 37, 544–548 [DOI] [PubMed] [Google Scholar]
- 66.Metzger BPH et al. (2015) Selection on noise constrains variation in a eukaryotic promoter. Nature 521, 344–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith JD et al. (2013) A novel test for selection on cis-regulatory elements reveals positive and negative selection acting on mammalian transcriptional enhancers. Mol. Biol. Evol 30, 2509–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turelli M (1985) Effects of pleiotropy on predictions concerning mutation-selection balance for polygenic traits. Genetics 111, 165–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.San Millan A et al. (2014) Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Comm 5, 5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maisnier-Patin S and Andersson DI (2004) Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. R. Microbiol 155, 360–369 [DOI] [PubMed] [Google Scholar]
- 71.Hall AR and MacLean RC (2011) Epistasis buffers the fitness effects of rifampicin- resistance mutations in Pseudomonas aeruginosa. Evolution 65, 2370–2379 [DOI] [PubMed] [Google Scholar]
- 72.Angst DC and Hall AR (2013) The cost of antibiotic resistance depends on evolutionary history in Escherichia coli. BMC Evol. Biol 13, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brandis G et al. (2012) Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Molecular Microbiology 85, 142–151 [DOI] [PubMed] [Google Scholar]
- 74.Brandis G and Hughes D (2013) Genetic characterization of compensatory evolution in strains carrying rpoB Ser531Leu, the rifampicin resistance mutation most frequently found in clinical isolates. J. Antimicrob. Chemother 68, 2493–2497 [DOI] [PubMed] [Google Scholar]
- 75.Wang Z et al. (2015) Evolution of gene regulation during transcription and translation. Genome Biol. Evol 7, 1155–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mack KL and Nachman MW (2017) Gene regulation and speciation. Trends Genet. 33, 68–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McManus CJ et al. (2014) Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res. 24, 422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bader DM et al. (2015) Negative feedback buffers effects of regulatory variants. Mol. Syst. Biol 11, 785–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Albert FW et al. (2017) Genetics of trans-regulatory variation in gene expression. biorXiv 1–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Albert FW et al. (2014) Genetics of single-cell protein abundance variation in large yeast populations. Nature 506, 494–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rokyta DR et al. (2015) Post-transcriptional mechanisms contribute little to phenotypic variation in snake venoms. G3 5, 2375–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Langley CH et al. (2012) Genomic variation in natural populations of Drosophila melanogaster. Genetics 192, 533–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mackay TFC (2013) Epistasis and quantitative traits: using model organisms to study gene–gene interactions. Nat. Rev. Genet 15, 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mackay TF and Moore JH (2014) Why epistasis is important for tackling complex human disease genetics. Genome Med 6, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vonesch SC et al. (2016) Genome-wide analysis reveals novel regulators of growth in Drosophila melanogaster. PLoS Genet. 12, e1005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.He X et al. (2016) Epistatic partners of neurogenic genes modulate Drosophila olfactory behavior. Genes Brain. Behav 15, 280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wayne ML et al. (2007) Simpler mode of inheritance of transcriptional variation in male Drosophila melanogaster. Proc. Nat. Acad. Sci. USA 104, 18577–18582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Genissel A et al. (2007) cis and trans regulatory effects contribute to natural variation in transcriptome of Drosophila melanogaster. Mol. Biol. Evol 25, 101–110 [DOI] [PubMed] [Google Scholar]
- 89.Bourguet D (1999) The evolution of dominance. Heredity 83, 1–4 [DOI] [PubMed] [Google Scholar]
- 90.Denby CM et al. (2012) Negative feedback confers mutational robustness in yeast transcription factor regulation. Proc. Nat. Acad. Sci. USA 109, 3874–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ronald J et al. (2005) Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet. 1, e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Duncan IW (2002) Transvection effects in Drosophila. Annu. Rev. Genet 36, 521–556 [DOI] [PubMed] [Google Scholar]
- 93.Mellert DJ and Truman JW (2012) Transvection is common throughout the Drosophila genome. Genetics 191, 1129–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ou SA et al. (2009) Effects of chromosomal rearrangements on transvection at the yellow gene of Drosophila melanogaster. Genetics 183, 483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blick AJ et al. (2016) The capacity to act in trans varies among Drosophila enhancers. Genetics 203, 203–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goldsborough AS and Kornberg TB (1996) Reduction of transcription by homologue asynapsis in Drosophila imaginal discs. Nature 381, 807–810 [DOI] [PubMed] [Google Scholar]
- 97.Johnston RJ and Desplan C (2014) Interchromosomal communication coordinates intrinsically stochastic expression between alleles. Science 343, 661–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fujioka M et al. (2016) Determinants of chromosome architecture: insulator pairing in cis and in trans. PLoS Genet. 12, e1005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dekker J and Mirny L (2016) The 3D genome as moderator of chromosomal communication. Cell 164, 1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ali T et al. (2016) Insulators and domains of gene expression. Curr. Opin. Genet. Dev 37, 17–26 [DOI] [PubMed] [Google Scholar]
- 101.Smith EN and Kruglyak L (2008) Gene–environment interaction in yeast gene expression. PLoS Biol. 6, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Duveau F et al. (2017) Effects of mutation and selection on plasticity of a promoter activity in Saccharomyces cerevisiae. Proc. Nat. Acad. Sci. USA 114, E11218–E11227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.He F et al. (2016) The footprint of polygenic adaptation on stress-responsive cis-regulatory divergence in the Arabidopsis genus. Mol. Biol. Evol 33, 2088–2101 [DOI] [PubMed] [Google Scholar]
- 104.Lovell JT et al. (2016) Drought responsive gene expression regulatory divergence between upland and lowland ecotypes of a perennial C4 grass. Genome Res. 26, 510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]