Abstract
There is tremendous enthusiasm for the microbiome in academia and industry. This Perspective argues that in order to realize its potential, the field needs to focus on establishing causation and molecular mechanism, with an emphasis on phenotypes that are large in magnitude, easy to measure, and unambiguously driven by the microbiota.
The microbiome research community is producing exciting results at a rapid clip. Many of these early findings are correlative and associative, but phenomenology often precedes mechanism in a new area; by uncovering important phenotypes impacted by the microbiome, these studies illustrate why we should delve deeper. Indeed, there is broad agreement that the microbiome is ready to mature into a mechanism-based discipline, and a small but growing number of studies have pioneered the quest for molecular understanding in this space.
In industry, a raft of early entrants are seeking to develop therapies based on tantalizing observations in the areas of infectious disease, cancer immunotherapy, and immune modulation (Atarashi et al., 2013; Gopalakrishnan et al., 2018; Matson et al., 2018; Routy et al., 2018; van Nood et al., 2013). However, high-profile failures (Ratner, 2016) have led to hand-wringing about the feasibility of drug discovery and development in a murky space with few hard landmarks.
Although there is an abundance of enthusiasm for the microbiome, the approaches most likely to yield impactful molecular mechanisms are still topics of intense debate. The central argument of this Perspective is that academic and industrial efforts should focus on causality and mechanism, with an eye toward phenotypes that are large in magnitude, easy to measure, and unambiguously driven by the microbiota. I examine two areas ripe for mechanistic inquiry, small molecule production and immune modulation by the microbiome, and then consider two topics critical to studying and exploiting these phenotypes: the role of individual bacterial species and molecules in complex mixtures and synthetic ecology as a tool for studying and developing therapeutics from the microbiome.
Pursue causation and mechanism.
Many microbiota-host interaction studies share a common format: They start with a phenotype (often linked to disease) and seek to understand the microbial taxa or genes responsible for it. This is akin to forward genetics, with three important distinctions (Figure 1): The phenotype and genotype are in different organisms (host and microbe, respectively), the primary technique is differential sequence analysis rather than genetics, and the outcome is usually a bacterial taxon associated with a phenotype rather than a gene responsible for the phenotype.
Figure 1:
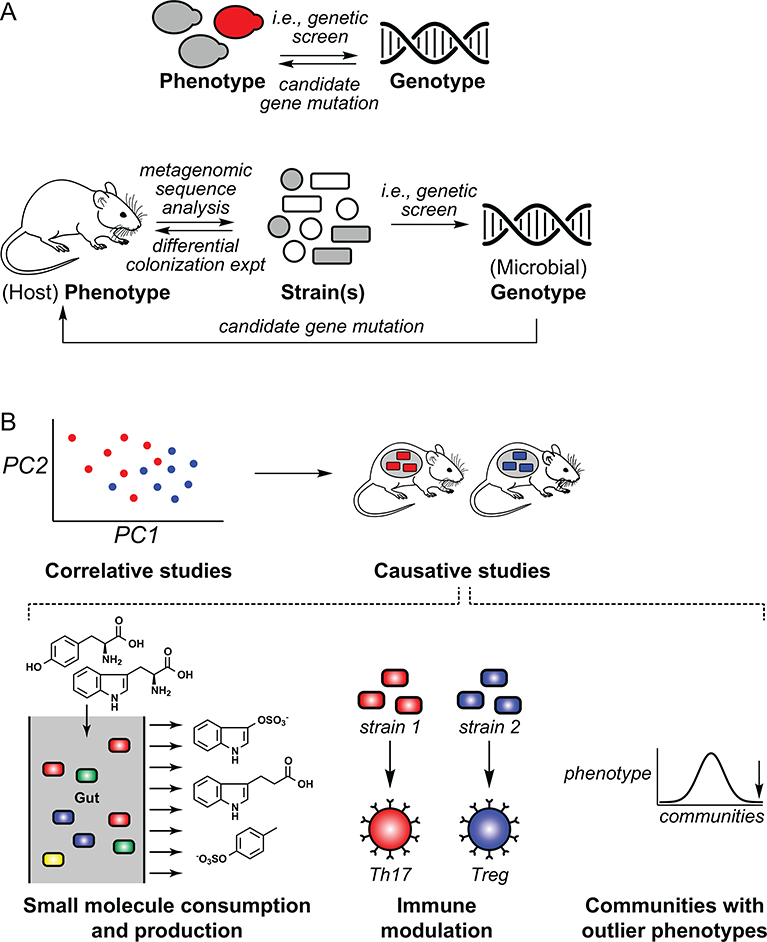
Challenges and approaches to decipher mechanism in microbiome research. (A) In classical genetics, the phenotype and genotype are in the same organism. Genetic studies in the microbiome have two important distinctions: the phenotype and genotype are in different organisms (host and microbe, respectively), and the process of determining which microbial species in a complex mixture are responsible for a phenotype is a formidable challenge. (B) Two areas in which studies of causality and mechanism have gained momentum are small molecule consumption/production and immune modulation. Communities with outlier phenotypes are an important vista for understanding and translating gut microbiome function.
The forward genetic approach has had notable successes, which can be grouped into three categories (referenced examples throughout are meant to be illustrative, not exhaustive): 1) Comparative sequence analyses of disease cohorts that result in taxonomic associations (Gevers et al., 2014; Qin et al., 2012); 2) Studies that show a phenotype can be transferred by microbiome transplant in germ-free mice (Gopalakrishnan et al., 2018; Matson et al., 2018; Ridaura et al., 2013; Routy et al., 2018; Thaiss et al., 2016); and 3) Efforts that narrow down to one or a few species that modulate a host phenotype (Atarashi et al., 2013; Buffie et al., 2015; Buffington et al., 2016; Surana and Kasper, 2017). Two challenges stand out: First, given the large number of strains in a comparative sequence analysis, the process of narrowing down species that are causally linked to a phenotype can suffer from the statistical problems of cherry picking. Given the practical challenges of testing every possible strain, strains that are merely correlated are often assumed to be causative. Second, even when a causative strain is identified, it remains difficult to establish the mechanistic link to phenotype.
In contrast, an approach analogous to reverse genetics has been used less often in microbiome studies. Two related experimental formats are most common: 1) colonizing mice with communities that differ only in the presence of a single strain and comparing the outcome (Buffie et al., 2015; Faith et al., 2015; Romano et al., 2017), or 2) colonizing mice with wild-type versus mutant versions of a strain (Cullen et al., 2015; Dodd et al., 2017; Round et al., 2011). Although it can suffer from the problem of cherry picking, reverse genetics has the advantage that it is a simpler entry point to establish causality and enable studies of mechanism. More complex model microbiomes, improvements in genetic tools for prominent gut bacterial strains, methods to cultivate previously uncultured bacterial strains (Browne et al., 2016; Lagier et al., 2016), new technologies that facilitate colonization in the background of a complex community (Shepherd et al., 2018), and better animal models of microbiome-related disease will strengthen both forward and reverse approaches in addressing causality and mechanism more directly.
Future technologies – e.g., the microbiome equivalents of whole-genome knockout and CRISPR screens – will blur the distinction between forward and reverse genetics. The important thing is not the difference between these approaches but, for a given situation, whether they can reveal causation and molecular mechanism. In the remainder of this section, we consider two areas in which inroads into causality and mechanism seem likely: 1) production of small molecules and 2) modulation of the host immune response. Readers are referred to recent literature in a third area ripe for mechanistic investigation: colonization resistance against pathogens (Pamer, 2016; Panigrahi et al., 2017). We then raise the speculative possibility of outlier phenotypes with large effect sizes that, like rare human genetic variants, could hold great therapeutic promise.
Small molecule production and consumption
Given the prominent role of small molecules in mediating signaling interactions, the fact that the gut microbiota produce a pool of molecules tens of millimolar in concentration is of particular interest. One familiar example is the short-chain fatty acids (SFCAs) that accumulate at 150–600 mmol/day (Wilson, 2005) and signal through G-protein coupled receptors 41 and 43 (GPR41 and GPR43) to modulate the host immune and metabolic systems (Ulven, 2012). Another is trimethylamine N-oxide (TMAO), which derives from gut microbial metabolism of choline and carnitine and is present at 2–6 μM in plasma (Tang et al., 2013). It is proposed to be not just a marker of cardiovascular disease but itself proatherogenic (Brown and Hazen, 2014) and prothrombotic (Zhu et al., 2016) in mouse models of disease. Although SFCAs and TMAO are heavily studied, the microbiota produce many other small molecules at concentrations comparable to those achieved by drugs used in the clinic. Aromatic amino acid metabolites are common, including the tryptophan metabolite indole, which is converted in the liver to indoxyl sulfate (IS) (10–130 mg/day) (Patel et al., 2012); the tyrosine metabolite p-cresol, which the host transforms into p-cresol sulfate (PCS) (20–230 mg/day) (Patel et al., 2012); and the phenylalanine metabolite phenylacetic acid, which the host conjugates to glutamine to form phenylacetylglutamine (PAG) (4.5–70 μm/mm creatinine) (Bouatra et al., 2013). TMAO, IS, and PCS accumulate in patients with renal failure (Sirich et al., 2013; Tang et al., 2015), suggesting that a microbiota-targeted therapy to eliminate their production could be a new therapeutic opportunity in renal disease.
An important source of small molecules is common dietary input, which be converted by gut bacterial species into a variety of metabolic end products with different biological activities. For example, tryptophan can be converted to indoxyl sulfate, indolepropionic acid, indoleacetic acid, and tryptamine. The levels of these metabolites vary widely among individuals, presumably reflecting differences in metabolic gene content among their gut communities. Thus, given a common diet, divergent microbiota can generate a metabolic output that can vary widely among individuals. This paradigm raises questions that are relevant to basic research and industry alike: Which molecules are beneficial, and which others deleterious? What is the optimal gut metabolic output, and how will it vary based on disease susceptibility? Can robust, resilient gut communities be designed to produce (and not produce) specific sets of molecules?
Modulation of the host immune response
Several studies have demonstrated that the microbiota directly modulates the host immune response. Two key observations suggest that immune modulation by the microbiota might be far more specific than had previously been recognized. First, Littman and coworkers showed that the TH17 cells induced in response to segmented filamentous bacterium (SFB), a well-studied mouse symbiont, express a T-cell receptor specific for an SFB antigen (Yang et al., 2014). Although it was previously known that SFB induces TH17 cells, it is striking that a gut colonist – without breaching the intestinal epithelium – can ‘program’ a population of immune cells that are specific to it. This paradigm has been extended by recent work on Helicobacter hepaticus (Hh), which induces Hh-specific regulatory T cells (Xu et al., 2018). Belkaid and coworkers have further demonstrated the generality of this result by showing that the CD8+ T cell response to strains of the skin commensal Staphylococcus epidermidis is not only specific but lasts for months, indicating that the ‘programming’ effect is potent and persistent (Naik et al., 2015).
This unanticipated degree of specificity in the mucosal immune response to the commensal microbiota has far-reaching implications, and raises several key questions: 1) How many gut commensals induce their own corresponding immune cell population? Can a designed or engineered gut consortium serve as a tool for programming the host T cell response? 2) Does this phenomenon explain the profound influence of the microbiome on the efficacy of anti-PD1 therapy in melanoma and other cancers? (Gopalakrishnan et al., 2018; Matson et al., 2018; Routy et al., 2018) 3) What are the presumptive microbiota-derived or induced molecules that determine T cell fate – and the cell types and signaling pathways through which they act? 4) If commensal-induced T cells express a TCR specific for a bacterial antigen, how do they – by cross-reactivity, proximity, or otherwise – modulate immune response against host tissues? And can this antigen-specific response be re-directed by expressing host antigens in bacterial strains?
One goal, perhaps realizable in the near-term, would be the use commensals or commensal-derived molecules as the basis of a new class of immunotherapies. One great advantage of this approach would be its selectivity, which could lead to a cleaner side-effect profile than systemic immunotherapies. For example, fine control over the local T cell response could enable an enhanced CD8+ T cell response to potentiate checkpoint blockade for melanoma or colorectal adenocarcinoma, without the risks of systemic immune stimulation. Likewise, local Treg stimulation could suppress immune activity in the small and large intestine (for inflammatory bowel disease) or skin (for psoriasis) without the downsides of general immune suppression. One could even imagine engineering a Treg-inducing bacterial strain to express a host autoantigen to reverse autoimmune disease.
Outlier microbiome phenotypes
One of the most direct benefits of DNA sequencing to human medicine has been to identify genetic outliers who harbor a rare mutation that confers protection against disease. For example, nonsense mutations in the proprotein convertase PCSK9 lower LDL-C and are protective against cardiovascular disease (Cohen et al., 2005), loss of function of Nav1.7 confers insensitivity to pain (Cox et al., 2006), and CCR5 deficiency protects against HIV infection (Liu et al., 1996; Samson et al., 1996). All three proteins have become validated drug targets, and new therapeutics targeting each are now approved or in clinical trials.
Might the same concept apply to the microbiome? Unlike genome-wide and microbiome-wide association studies (Gilbert et al., 2016), which often reveal common variants with a small effect size, the goal would be to look for rare communities (harboring unusual bacterial species, or common species in unusual ratios) with large, protective effects against a disease of interest. For example, do there exist rare individuals whose gut bacteria are exceptionally efficient at harvesting calories, and therefore protect against metabolic disease? Might there be people with risk alleles for Crohn’s disease who are protected against disease by a gut community that suppresses inflammation or prevents a bad actor from blooming? How about hospital workers who, despite constant exposure to Staphylococcus aureus, are not colonized due to a rare, protective skin community? Phenotypes like these would be a great setting in which to explore mechanism, and the transplantability of the microbiome could make it possible to endow millions of people with a rare disease-preventive gut or skin community.
Individual organisms and molecules can have a big impact.
The sheer complexity of a host-associated community – hundreds of microbial species, thousands of molecules – raises the question of how much difference a single organism or molecule can make. Are most microbiota-related phenomena the result of dozens of microbes or molecules acting in concert, irreducible to the effects of individual actors?
Early evidence suggests otherwise. The specific T cell responses to SFB and S. epidermidis are examples of how a single bacterial species can exert a clear effect on the host, but several others are worth noting. Faecalibacterium prausnitzii (Miquel et al., 2013) and Parabacteroides distasonis (Kverka et al., 2011) have been linked to the suppression of intestinal inflammation, specific strains of Lactobacillus alter behavior and cognition in mice (Bravo et al., 2011; Buffington et al., 2016), and Bacteroides fragilis has been shown, inter alia, to counter the neurodevelopmental and gut barrier defects of mice in the maternal immune activation model of autism spectrum disorder (Hsiao et al., 2013). Though not an individual organism, a mixture of 17 anaerobic Firmicutes potently induces regulatory T cells (Atarashi et al., 2013).
Although these interactions are not yet understood at the level of molecular mechanism, individual molecules from the microbiota are known to have a signal that rises well above background. In addition to the well-documented examples of SCFAs and TMAO (see above), polysaccharide A (Mazmanian et al., 2008) and α-galactosylceramide from B. fragilis (An et al., 2014) are potent immune modulators that act through TLR2 and CD1d, respectively; heat-stable enterotoxin, from which linaclotide is derived, induces GI motility via guanylate cyclase 2C (Schulz et al., 1990); and secondary bile acids, including deoxycholic acid and ursodeoxycholic acid, have a wide range of effects on host metabolic and immune activity as well as colon carcinogenesis (Wahlström et al., 2016). Moreover, individual host factors such as RegIIIγ and nitrate exert large effects on the composition and function of the gut community (Vaishnava et al., 2011; Winter et al., 2013). Given that the hunt for microbiota-derived metabolites of consequence is still in its infancy (Sharon et al., 2014) these examples are likely to be the first of many.
To place these findings in context, cell and developmental biologists have long understood that individual molecules can exert phenotypes that rise above the background of a complex pool. Mammals are complex mixtures of thousands of chemicals, yet signaling molecules such as serotonin (Canli and Lesch, 2007), oxytocin (Kosfeld et al., 2005), sphingosine-1-phosphate (Spiegel and Milstien, 2003), and estradiol (Balthazart and Ball, 2006) have potent, unambiguous activities, both at the level of cells and physiology. There is every reason to think that the paradigm of dominant activity for individual molecules will hold for an important subset of molecules from the microbiota. If so, it will be far easier than currently anticipated to understand the mechanisms of microbe-host interactions and exploit them therapeutically.
Develop the basic science underlying synthetic ecology.
Developments in academic and industrial microbiome research are creating a need for a mechanistic understanding of microbial communities. In academia, although germ-free mouse mono-colonization experiments have yielded tremendous mechanistic insights into microbe-host interactions, experiments with very large, defined communities that approach native-like complexity would capture a great deal of biology missing in the simpler format (e.g., the effects of tens of millimolar pool of molecules, perhaps tens to hundreds of distinct immune modulatory events, etc.). Experiments in which one strain is removed (or, in the extreme, one gene from one strain) and an effect on the host is observed are more likely to stand the test of time.
On the industry side, the striking efficacy of fecal microbiota transplant (FMT) in recurrent Clostridium difficile infection (van Nood et al., 2013), combined with a surprisingly low rate of acute adverse events (for exceptions, see (Alang and Kelly, 2015; Quera et al., 2014)) and promising early data on engraftment (Smillie et al., 2018), has spurred efforts to test FMT in other indications. However, there are two key challenges in generalizing fecal transplant, both of which suggest important limitations of feces as a source of material for community transplants: (i) Its supply is inherently limited and difficult to scale. The need for safety, reproducibility and ease of manufacture will favor a transition from donor feces to bacterial communities of defined composition. (ii) A ‘lead’ fecal sample cannot be optimized in the sameway that medicinal chemistry or protein engineering can improve a small molecule or biologic lead. A defined community would enable the kind of tinkering that is critically important in drug discovery.
The academic and industrial challenges can both be addressed by developing new methods for constructing and interrogating synthetic communities that are defined but consist of hundreds of strains, approaching the level of complexity of a native fecal sample. Although there are many practical challenges to overcome, early work in the plant microbiome community has begun demonstrating the value of this approach (Bai et al., 2015). Such a system could open the door to studying an array of fundamental basic science questions: How many meaningful strain-strain interactions exist in a community of hundreds of strains? What constitutes a niche, and how do strains map to niches? What are the molecular correlates of stability, and how does a community reconfigure in response to a perturbation? Most importantly, it would enable landmark studies of causation and mechanism in the microbiome, bringing a near atomistic level of control to a system that could benefit tremendously from more reductionist inquiry.
Acknowledgments
I am indebted to members of the Fischbach Group, Sandy Johnson (UCSF), Elizabeth Sattely (Stanford), Dylan Dodd (Stanford), Justin Sonnenburg (Stanford), KC Huang (Stanford), Dan Littman (NYU), Yasmine Belkaid (NIH), and James Fraser (UCSF) for helpful discussions. This work was supported by an HHMI-Simons Faculty Scholar Award, a Fellowship for Science and Engineering from the David and Lucile Packard Foundation, an Investigators in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Foundation, an award from BASF, an award from the Leducq Foundation, and NIH grants DK110174, DK113598, DK101674, and AI101018. The author is on the board of directors of Achaogen, the scientific advisory boards of NGM Biopharmaceuticals and Indigo Agriculture, and a co-founder of Revolution Medicines.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alang N, and Kelly CR (2015). Weight gain after fecal microbiota transplantation. Open Forum Infectious Diseases 2, ofv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, and Kasper DL (2014). Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236. [DOI] [PubMed] [Google Scholar]
- Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, Dombrowski N, Münch PC, Spaepen S, Remus-Emsermann M, et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. [DOI] [PubMed] [Google Scholar]
- Balthazart J, and Ball GF (2006). Is brain estradiol a hormone or a neurotransmitter? Trends in Neurosciences 29, 241–249. [DOI] [PubMed] [Google Scholar]
- Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, et al. (2013). The Human Urine Metabolome. PLoS ONE 8, e73076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, and Cryan JF (2011). Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proceedings of the National Academy of Sciences of the United States of America 108, 16050–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, and Hazen SL (2014). Metaorganismal nutrient metabolism as a basis of cardiovascular disease. Current Opinion in Lipidology 25, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, and Lawley TD (2016). Culturing of “unculturable” human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, and Costa-Mattioli M (2016). Microbial Reconstitution Reverses Maternal Diet-Induced Social and Synaptic Deficits in Offspring. Cell 165, 1762–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canli T, and Lesch KP (2007). Long story short: the serotonin transporter in emotion regulation and social cognition. Nature Neuroscience 10, 1103–1109. [DOI] [PubMed] [Google Scholar]
- Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, and Hobbs HH (2005). Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nature Genetics 37, 161–165. [DOI] [PubMed] [Google Scholar]
- Cox JJ, Reimann F, Nicholas AK, Thornton G, Roberts E, Springell K, Karbani G, Jafri H, Mannan J, Raashid Y, et al. (2006). An SCN9A channelopathy causes congenital inability to experience pain. Nature 444, 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, and Goodman AL (2015). Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science (New York, N.Y 347, 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, et al. (2017). A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Colombel J-F, and Gordon JI (2015). Identifying strains that contribute to complex diseases through the study of microbial inheritance. Proceedings of the National Academy of Sciences of the United States of America 112, 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vázquez-B aeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. (2014). The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host & Microbe 15, 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, Jansson JK, Dorrestein PC, and Knight R (2016). Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 535, 94–103. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. (2018). Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science (New York, N.Y 359, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. (2013). Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosfeld M, Heinrichs M, Zak PJ, Fischbacher U, and Fehr E (2005). Oxytocin increases trust in humans. Nature 435, 673–676. [DOI] [PubMed] [Google Scholar]
- Kverka M, Zakostelska Z, Klimesova K, Sokol D, Hudcovic T, Hrncir T, Rossmann P, Mrazek J, Kopecny J, Verdu EF, et al. (2011). Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin Exp Immunol 163, 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier J-C, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, Caputo A, Cadoret F, Traore SI, Hadji Seck El, et al. (2016). Culture of previously uncultured members of the human gut microbiota by culturomics. Nature Microbiology 2016. 1:12 1, 16203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, and Landau NR (1996). Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86, 367–377. [DOI] [PubMed] [Google Scholar]
- Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, Luke JJ, and Gajewski TF (2018). The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science (New York, N.Y 359, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, and Kasper DL (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625. [DOI] [PubMed] [Google Scholar]
- Miquel S, Martin R, Rossi O, Bermudez-Humaran LG, Chatel JM, Sokol H, Thomas M, Wells JM, and Langella P (2013). Faecalibacterium prausnitzii and human intestinal health. Current Opinion in Microbiology 16, 255–261. [DOI] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, Conlan S, Himmelfarb S, Byrd AL, Deming C, et al. (2015). Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamer EG (2016). Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science (New York, N.Y 352, 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi P, Parida S, Nanda NC, Satpathy R, Pradhan L, Chandel DS, Baccaglini L, Mohapatra A, Mohapatra SS, Misra PR, et al. (2017). A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 548, 407–412. [DOI] [PubMed] [Google Scholar]
- Patel KP, Luo FJ, Plummer NS, Hostetter TH, and Meyer TW (2012). The production of p-cresol sulfate and indoxyl sulfate in vegetarians versus omnivores. Clinical Journal of the American Society of Nephrology : CJASN 7, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. [DOI] [PubMed] [Google Scholar]
- Quera R, Espinoza R, Estay C, and Rivera D (2014). Bacteremia as an adverse event of fecal microbiota transplantation in a patient with Crohn’s disease and recurrent Clostridium difficile infection. Journal of Crohn’s and Colitis 8, 252–253. [DOI] [PubMed] [Google Scholar]
- Ratner M (2016). Seres’s pioneering microbiome drug fails mid-stage trial. Nature Biotechnology 34, 1004–1005. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science (New York, N.Y 341, 1241214–1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano KA, Martínez-del Campo A, Kasahara K, Chittim CL, Vivas EI, Amador-Noguez D, Balskus EP, and Rey FE (2017). Metabolic, Epigenetic, and Transgenerational Effects of Gut Bacterial Choline Consumption. Cell Host & Microbe 22, 279–290.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, and Mazmanian SK (2011). The Toll-Like Receptor 2 Pathway Establishes Colonization by a Commensal of the Human Microbiota. Science (New York, N.Y 332, 974–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2018). Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science (New York, N.Y 359, 91–97. [DOI] [PubMed] [Google Scholar]
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, et al. (1996). Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382, 722–725. [DOI] [PubMed] [Google Scholar]
- Schulz S, Green CK, Yuen PS, and Garbers DL (1990). Guanylyl cyclase is a heat-stable enterotoxin receptor. Cell 63, 941–948. [DOI] [PubMed] [Google Scholar]
- Sharon G, Garg N, Debelius J, Knight R, Dorrestein PC, and Mazmanian SK (2014). Specialized metabolites from the microbiome in health and disease. Cell Metabolism 20, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, and Sonnenburg JL (2018). An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 557, 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirich TL, Funk BA, Plummer NS, Hostetter TH, and Meyer TW (2013). Prominent Accumulation in Hemodialysis Patients of Solutes Normally Cleared by Tubular Secretion. J. Am. Soc. Nephrol 25, ASN.2013060597–ASN.2013060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie CS, Sauk J, Gevers D, Friedman J, Sung J, Youngster I, Hohmann EL, Staley C, Khoruts A, Sadowsky MJ, et al. (2018). Strain Tracking Reveals the Determinants of Bacterial Engraftment in the Human Gut Following Fecal Microbiota Transplantation. Cell Host & Microbe 23, 229–240.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel S, and Milstien S (2003). Sphingosine-1-phosphate: an enigmatic signalling lipid. Nature Reviews 4, 397–407. [DOI] [PubMed] [Google Scholar]
- Surana NK, and Kasper DL (2017). Moving beyond microbiome-wide associations to causal microbe identification. Nature 552, 244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WHW, Wang Z, Kennedy DJ, Wu Y, Buffa JA, Agatisa-Boyle B, Li XS, Levison BS, and Hazen SL (2015). Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res 116, 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, and Hazen SL (2013). Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. The New England Journal of Medicine 368, 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss CA, Itav S, Rothschild D, Meijer M, Levy M, Moresi C, Dohnalová L, Braverman S, Rozin S, Malitsky S, et al. (2016). Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature 540, 544–551. [DOI] [PubMed] [Google Scholar]
- Ulven T (2012). Short-chain free fatty acid receptors FFA2/GPR43 and FFA3/GPR41 as new potential therapeutic targets. Frontiers in Endocrinology 3, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, and Hooper LV (2011). The Antibacterial Lectin RegIIIγ Promotes the Spatial Segregation of Microbiota and Host in the Intestine. Science (New York, N.Y 334, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al. (2013). Duodenal infusion of donor feces for recurrent Clostridium difficile. The New England Journal of Medicine 368, 407–415. [DOI] [PubMed] [Google Scholar]
- Wahlström A, Sayin SI, Marschall H-U, and B äckhed F (2016). Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metabolism 24, 41–50. [DOI] [PubMed] [Google Scholar]
- Wilson M (2005). Microbial inhabitants of humans : their ecology and role in health and disease (New York: Cambridge University Press; ). [Google Scholar]
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, et al. (2013). Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science (New York, N.Y 339, 708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, Belkaid Y, Bonneau R, and Littman DR (2018). c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. (2014). Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, et al. (2016). Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell 165, 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]


