Abstract
Embryo culture and assisted reproductive technologies have been associated with a disproportionately high number of epigenetic abnormalities in the resulting offspring. However, the mechanisms by which these techniques influence the epigenome remain poorly defined. In this study, we evaluated the capacity of oxygen concentration to influence the transcriptional control of a selection of key enzymes regulating chromatin structure. In mouse embryonic stem cells, oxygen concentrations modulated the transcriptional regulation of the TET family of enzymes, as well as the de novo methyltransferase Dnmt3a. These transcriptional changes were associated with alterations in the control of multiple imprinted genes, including H19, Igf2, Igf2r, and Peg3. Similarly, exposure of in vitro produced bovine embryos to atmospheric oxygen concentrations was associated with disruptions in the transcriptional regulation of TET1, TET3, and DNMT3a, along with the DNA methyltransferase co-factor HELLS. In addition, exposure to high oxygen was associated with alterations in the abundance of transcripts encoding members of the Polycomb repressor complex (EED and EZH2), the histone methyltransferase SETDB1 and multiple histone demethylases (KDM1A, KDM4B, and KDM4C). These disruptions were accompanied by a reduction in embryo viability and suppression of the pluripotency genes NANOG and SOX2. These experiments demonstrate that oxygen has the capacity to modulate the transcriptional control of chromatin modifying genes involved in the establishment and maintenance of both pluripotency and genomic imprinting.
Keywords: Oxidative stress, Assisted reproductive technologies, Genomic imprinting, Histone demethylase, TET, DNMT, DNA methylation, Epigenetics, Developmental programming
1. Introduction
In the past decade, there has been considerable interest in understanding the impact environmental exposures have on long-term health and the incidence of birth defects. Chemical pollutants, dietary deficiencies, embryonic stress and multiple other external factors have all demonstrated long-lasting effects upon development, metabolism, and health (Christian and Stewart, 2010; Barker, 1990; Kinsella and Monk, 2009). However, the underlying mechanisms by which these exposures, some of which are transient, impact development far beyond the period of exposure remain largely unknown.
Some of the first evidence that environmental exposures could influence the long-term health of offspring came from studies employing mammalian embryology. Here, researchers noted an overgrowth phenotype in cattle that was induced by exposure of the embryo to stressors associated with in vitro culture conditions (Young et al., 1998; Farin et al., 2006). These pioneering studies were later followed up by works examining altered DNA methylation, and abnormal patterns of imprinted gene expression arising as a consequence of in vitro embryo culture and somatic cell nuclear transfer (Doherty et al., 2000). Genomic imprinting is an epigenetic mechanism of transcriptional regulation that restricts expression to either the maternally- or paternally-inherited copy of the gene; the opposite parental copy is predominantly silenced via a variety of epigenetic mechanisms (Bartolomei and Tilghman, 1997). A number of studies have suggested that children conceived using Assisted Reproductive Technologies (ARTs) exhibit a higher incidence of imprinting disorders, including Angelman, Prader-Willi and Beckwith-Weidemann syndrome (Powell, 2003; Gosden et al., 2003; DeBaun et al., 2003; Gicquel et al., 2003). Further, increased incidences of congenital malformations, intrauterine growth retardation, premature birth and low birth weights have also been reported in children conceived using ARTs (Arnaud and Feil, 2005; Hansen, 2006; Dean et al., 2005; Wrenzycki et al., 2005; Niemitz and Feinberg, 2004; Kurinczuk et al., 2004). However, the mechanisms by which embryo culture heritably alters phenotype, and the extent to which these changes may be linked to the wider development of environmentally-induced disease remain to be demonstrated.
Studies in multiple species have shown that culturing embryos under atmospheric oxygen concentrations result in a reduced rate of embryo development to the blastocyst stage (Preis et al., 2007; Harvey et al., 2007). Fischer and Bavister demonstrated that intrauterine and oviductal oxygen concentrations range between 2 and 7% oxygen (Fischer and Bavister, 1993). This implies that culturing embryos under low atmospheric oxygen concentrations represents an environment more similar to the in vivo condition. However, embryo handling and best in vitro practices still necessitate transient exposures to atmospheric (20%) oxygen. Recently, a link between components of the oxidative stress pathways and enzymes controlling chromatin structure has been identified (Wang et al., 2011; Delatte et al., 2015). To gain a better understanding of the potential impact of oxygen exposures during in vitro embryo culture on the regulation of chromatin structure, we evaluated the capacity of differing oxygen concentrations to effect the transcriptional control of chromatin modifying genes and the regulation of imprinted gene expression, which previous studies have identified as being sensitive to embryo culture (de Waal et al., 2014). Our data indicate that oxygen has the ability to influence crucial gene regulatory networks involved in the establishment and maintenance of chromatin structure and the control of imprinted genes.
2. Results
2.1. High oxygen concentrations negatively impact embryo development and alter the expression of genes regulating the cellular oxidative stress response and pluripotency
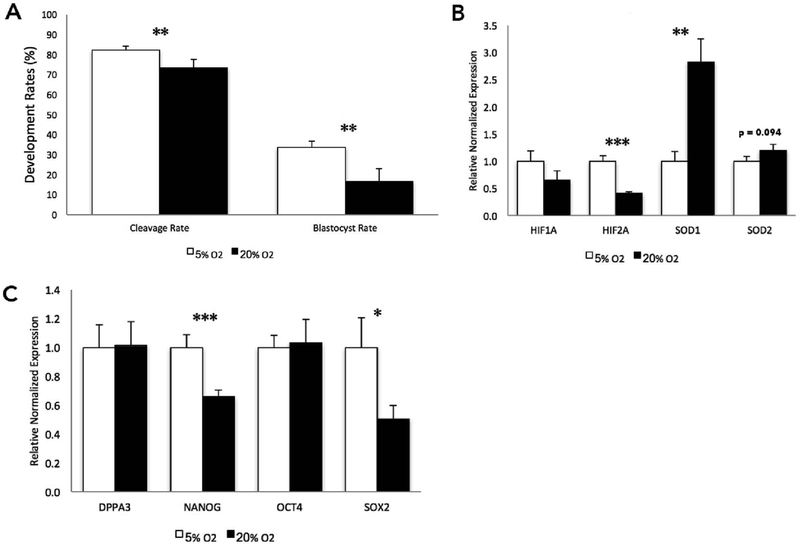
Consistent with previous studies (Harvey et al., 2007; Harvey, 2007; Li et al., 2016), culturing bovine embryos under atmospheric oxygen concentrations (20%), as compared to the industry-standard 5%, resulted in reduced cleavage and blastocyst rates (Fig. 1A). While standard industry practices do not employ atmospheric oxygen concentrations, cultured embryos are frequently exposed to this condition anytime they are removed from the incubators for manipulation. Therefore, we employed a model of constant exposure in an effort to determine the extreme consequence oxygen exposure has on the genetic pathways regulating chromatin structure. To validate our model, we first examined the levels of transcripts encoding genes involved in pathways regulating cellular hypoxia and oxidative stress. The 20% culture group displayed decreased expression of two hypoxia-inducible factors, while an increased abundance of transcripts encoding components of the antioxidant response (Fig. 1B). Specifically, the superoxide dismutase family was significantly up-regulated in the 20% oxygen group, with an almost three-fold increase for SOD1, while SOD2 displayed more modest changes in transcript abundance (Fig. 1B). We next examined transcript levels of genes involved in the maintenance of pluripotency and found a down-regulation of NANOG and SOX2 in the high oxygen (20%) treatment group (Fig. 1C).
Fig. 1.
The impact of oxygen concentrations on bovine preimplantation development and the transcriptional regulation of select genes involved in the oxidative stress response and the control of pluripotency at the blastocyst stage.
A. Cleavage and blastocyst rates of bovine embryos cultured under low (5%) and high (20% - atmospheric) oxygen concentrations (5% oxygen n = 682, 20% oxygen n = 890. from 4 independent replicates). Here, development rates represent the total number of blastocysts on day 7 divided by the total number of presumptive zygotes placed into culture. B. Measurement of transcripts encoding genes involved in oxygen sensing and the oxidative stress response using reverse transcriptase quantitative polymerase chain reaction (RT-qPCR). C. RT-qPCR measurement of transcripts encoding key bovine pluripotency genes. For experiments employing RT-qPCR, measurements were normalized to the geometric mean of GAPDH, SDHA and YWHAZ. n = 6. (P-value: * ≤ 0.05; ** ≤ 0.01; *** ≤ 0.001).
2.2. Atmospheric oxygen levels alter the transcription of the ten-eleven translocation (TET) family and genes regulating DNA methylation
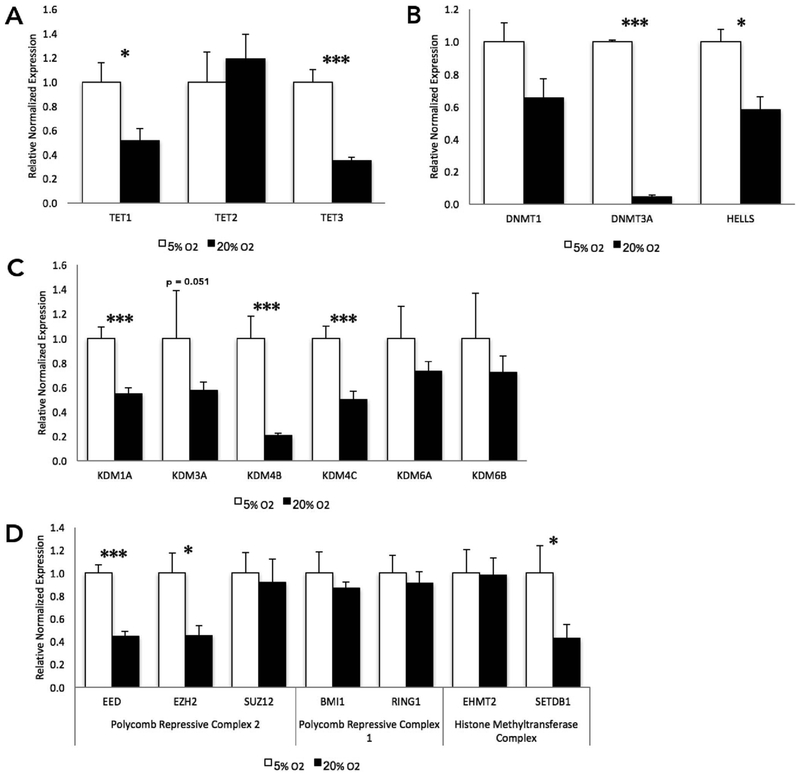
To determine if genes involved in the establishment, maintenance and/or regulation of the epigenome are affected by exposure to atmospheric oxygen during preimplantation development, we examined the levels of transcripts encoding a family of genes that have been found to catalyze the first step in DNA demethylation (Delatte et al., 2014). The ten-eleven translocation (TET) family of genes hydroxylate methylated cystosines in a series of reactions that end with the base-excision repair (BER) machinery removing a 5′-formyl/carboxycystosine, generating a standard, unmethylated cytosine (Kohli and Zhang, 2013). This family of genes, while its precise role remains unclear, has been implicated in the cellular response to oxidative stress (Delatte et al., 2015). During bovine preimplantation development, the TET family of genes are thought to play a role in active genomic demethylation and resetting of the epigenome (Gu et al., 2011). In our experiments, the high-oxygen treatment group exhibited a significant down-regulation of TET1 and TET3 at the blastocyst stage of development, compared to their low oxygen counterparts (Fig. 2A). We next investigated the capacity of atmospheric oxygen to influence the expression of genes regulating DNA methylation. No difference in the levels of transcripts encoding DNMT1 was observed, while the de novo methyltransferase DNMT3A, as well as a gene responsible for the localization of DNMTs to specific loci, HELLS, were both significantly (p < .05) down-regulated in the high atmospheric culture treatment group (Fig. 2B).
Fig. 2.
Oxygen-induced changes in the transcriptional regulation of genes controlling chromatin structure at the blastocyst stage of bovine development.
A. RT-qPCR measurements of transcripts encoding the bovine ten-eleven translocation (TET) gene family. B. Transcript levels of the bovine DNA methyltransferase genes and an associated recruiter, HELLS. C. Measurement of transcripts encoding the bovine histone lysine demethylase family of genes. D. Gene transcript levels of the histone modifying complexes PRC1, PRC2, and histone methyltransferases. RT-qPCR measures were normalized to the geometric mean of GAPDH, SDHA, and YWHAZ. n = 6. (P-value: * ≤ 0.05; ** ≤ 0.01; *** ≤ 0.001).
2.3. Oxygen-induced down-regulation of multiple histone methyltransferase enzymes in cultured bovine blastocysts
Various post-translational histone modifications correlate with distinct transcriptional states and have been associated with the nucleation of repressed or relaxed chromatin structure. These modifications are hypothesized to act in a coordinated context to regulate DNA accessibility (Zhang and Reinberg, 2001). We therefore, examined the levels of transcripts encoding lysine-specific histone demethylase (KDMs) proteins. Specific members of the KDM family have been implicated in the regulation of the cellular response to oxidative stress (Liu et al., 2014). Almost all the candidate genes examined were reduced in the high oxygen treatment group (Fig. 2C). Transcripts encoding KDM1A, 4B and 4C were all significantly reduced while KDM3A was not significantly different (p = .05). The polycomb-repressive complexes (PRCs) are important in establishing and maintaining the transcriptional memory of differentiated cells as well as in the long-term silencing of chromatin (Margueron et al., 2009). Therefore, we examined the abundance of transcripts encoding the core polycomb-repressive complex 1 (PRC1) and polycomb-repressive complex 2 (PRC2) members. EED and EZH2 both displayed a reduction in transcript levels in the 20% oxygen treatment group, while in contrast, BMI1 and RING1, members of the PRC1 complex, showed no signs of altered gene expression (Fig. 2D). We next examined the abundance of transcripts encoding the histone methyltransferases EHMT1 (G9a) and SETDB1. Of these two, only SETDB1 displayed significant decreases in the high-oxygen group (Fig. 2D).
2.4. Oxygen-induced transcriptional changes in a murine embryonic stem cell (mESC) model of development
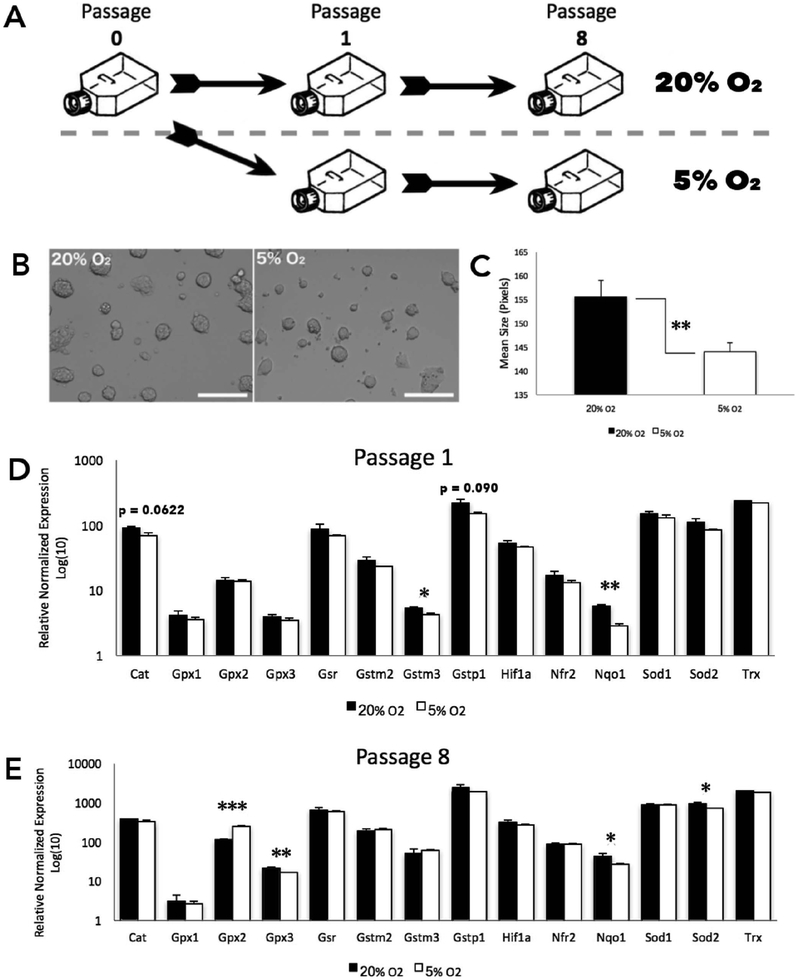
In order to better understand the interactions between oxygen exposure and the transcriptional regulation of factors controlling chromatin structure, we shifted our studies into a murine embryonic stem cell (mESC) model. Although not directly equivalent to cultured embryos, mESCs are an embryonic cell type, which can be cultured with high uniformity in terms of cellular phenotype, chromatin profiles and transcriptional output (Galonska et al., 2015). Therefore, this model allowed us to observe the time course of transcriptional effects that different concentrations of oxygen exert, as well as determine the impact of extended exposures to differing oxygen concentrations. Mouse ESCs are routinely cultured at 20% oxygen due to lack of significant benefits of maintaining the cell lines at reduced oxygen levels (Chen et al., 2009). The mESC line that we employed was of a C57BL/6 (Black6) by castaneus (CAST) cross (Golding et al., 2011). These distinctive genetic backgrounds allow for allele-specific differences in gene expression to be identified through single nucleotide polymorphisms (SNPs). In these experiments, cells were split into two groups: one cultured at 20% oxygen and the other under 5% oxygen, for a duration of 8 passages (Fig. 3A). Within the first 24 h, we noted a smaller colony size in the 5% O2 treatment group as compared to the cells cultured under 20% O2 (Fig. 3B). This change was consistent through to the end of the experiment at passage 8 (Fig. 3C). To measure the impact of oxygen concentrations on patterns of gene expression, we again began by examining the expression of genes associated with the oxidative stress response. Here, RT-qPCR was employed to analyze a panel of 12 well-established oxidative stress response genes (Veazey et al., 2015). After only a single cellular passage, we observed differential expression of Gstm3 and Nqo1, which were significantly reduced in the low oxygen treatment group (Fig. 3D). After 8 passages, Gpx2, Gpx3, and Sod2, which are all involved in cellular detoxification processes, were significantly down-regulated in the low oxygen treatment (Fig. 3E).
Fig. 3.
Oxygen-induced changes in a mouse embryonic stem cells.
A. Schematic representation of the experimental workflow. Cells split from a common flask (Passage 0) were separated into high and low treatment groups. Low (5%) O2 cells were cultured in a “hypoxia chamber”. B. Representative image of murine embryonic stem cells at mid-confluency during experimental treatments. C. Surface area (as calculated using ImageJ software) between the treatment groups. D-E. Measurement of transcripts encoding genes involved in the oxidative stress response at cellular passage 1 and 8. RT-qPCR measures were normalized to the geometric mean of Hprt, Mrpl, Ppia, and Ywhaz. n = 6 (P-value: * ≤ 0.05; ** ≤ 0.01; *** ≤ 0.001).
2.5. Distinct acute and prolonged transcriptional responses of the DNMT and TET families of genes
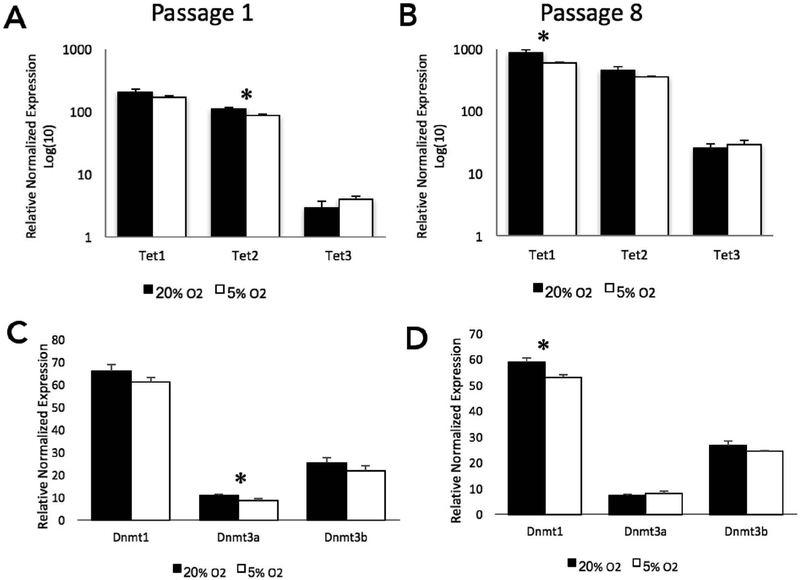
In our studies examining the effects of oxygen concentrations on bovine embryonic development, we noted that select TET genes and DNA methyltransferases were responsive to oxygen concentrations. After a single passage under low oxygen conditions, transcripts encoding Tet2 were significantly down-regulated in the 5% O2 group (Fig. 4A). By the 8th passage, Tet2 was identical to the 20% group, while transcripts encoding Tet1 had become significantly decreased in the 5% O2 group (Fig. 4B). Transcript levels of Dnmt3a were significantly reduced after a single passage (Fig. 4C) while by the 8th passage, transcripts encoding Dnmt1 had become significantly reduced in the low oxygen group (Fig. 4D).
Fig. 4.
Oxygen-induced changes in the transcriptional regulation of genes controlling DNA methylation in mouse embryonic stem cells.
A-B. Transcript levels of the ten-eleven translocation (TET) family of genes at passage 1 and 8. C-D. Transcript levels of the DNA methyltransferase [DNMT] family of genes at passage 1 and 8. RT-qPCR measures were normalized to the geometric mean of Hprt, Mrpl, Ppia, and Ywhaz. n = 6 (P-value: * ≤ 0.05).
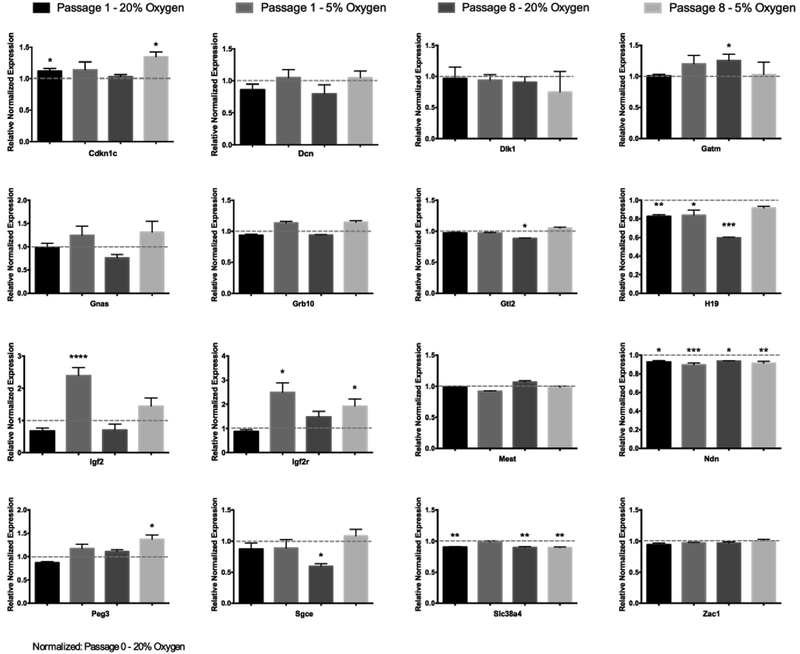
2.6. Oxygen-induced changes in the regulation of Igf2/Igf2r and the allelic expression of multiple imprinted genes
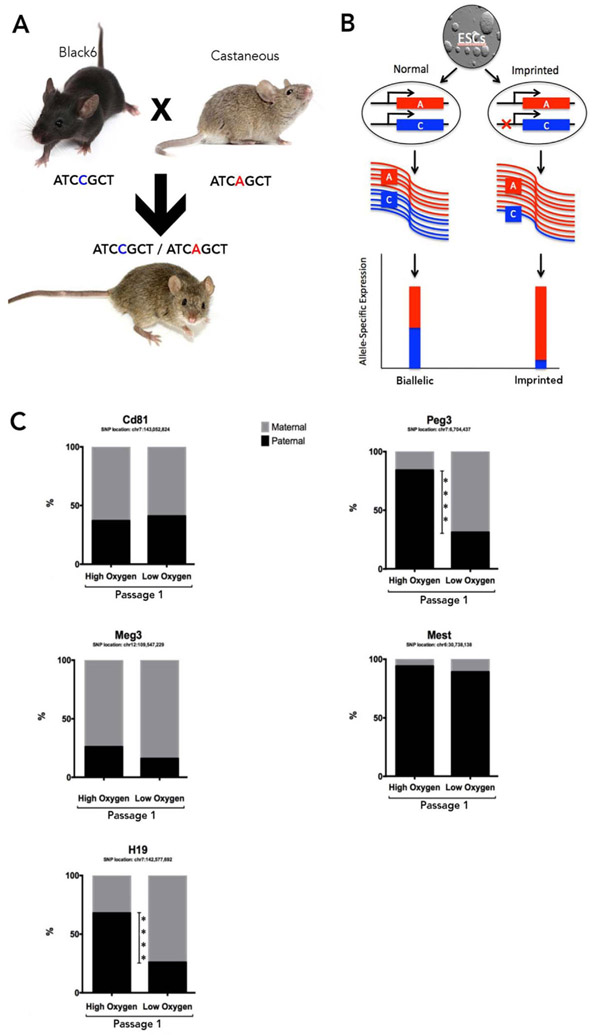
We conducted an analysis of genes regulated through mechanisms involving genomic imprinting. Here, the largest changes were observed for transcripts encoding Igf2 and Igf2r. Igf2 is part of the imprinting locus on mouse chromosome 7 that contains the H19/Igf2 cluster. H19 and Igf2 are reciprocally expressed with H19 being expressed from the maternal allele and Igf2 expressed from the paternal allele (Kaffer et al., 2001). Consistent with this paradigm, Igf2 was up-regulated while transcripts encoding H19 were significantly down-regulated in the 5% oxygen group (Fig. 5). Interestingly, we observed concurrent changes for both Igf2 and Igf2r in the low oxygen group. Lastly, we employed RNA-sequencing to assess single-nucleotide polymorphisms in a panel of known imprinted genes to determine if oxygen concentration could impact allele-specific patterns of gene expression between the treatment groups (Fig. 6A-B). In mESCs, imprinted genes often display leaky expression, where monoallelic expression is defined as ≥ 90% from one parental allele (Golding et al., 2011). After a single passage in either 5% or 20% oxygen, we observed significant shifts between maternal and paternally derived transcripts for H19 and Peg3. Here, H19, which is predominantly expressed from the maternal allele, exhibited a decreased contribution from the paternal allele under low oxygen conditions (Fig. 6C). However, Peg3, a paternally expressed gene, displayed the reverse trend, going from predominantly paternally derived transcripts in the high oxygen treatment to a maternally dominated transcript in the low oxygen group (Fig. 6C). No other imprinted genes exhibited significant shifts in allelic expression.
Fig. 5.
Oxygen-induced changes in the transcriptional regulation of imprinted genes within mouse embryonic stem cells.
Analysis of imprinted gene transcript levels at passage 1 and 8, as determined by RT-qPCR. Measurements were normalized to the geometric mean of the reference genes Gapdh, Hprt, Mrpl and Sdha, and samples graphed relative to Passage 0 (20% oxygen). n = 5 (P-value: * ≤ 0.05; ** ≤ 0.01; *** ≤ 0.001).
Fig. 6.
Analysis of oxygen-induced changes in parent-of-origin expression patterns for select imprinted loci.
A. Visual representation of allelic inheritance from the distinct genetic backgrounds used in this study. This strategy allows tracking of the parental alleles by using single nucleotide polymorphisms. B. Visual representation of balanced, bi-allelically expressed transcripts as well as imbalanced, imprinted genes. C. Allele-specific transcript analysis of a select panel of known imprinted genes, as determined using deep RNA-sequencing. (P-value: **** ≤ 0.0001).
3. Discussion
Prior research has demonstrated that uterine and oviductal oxygen concentrations range between 2 and 7% oxygen, but embryo handling and best in vitro practices still necessitate transient exposures to atmospheric (20%) O2 during assisted reproductive techniques. While culturing embryos under low atmospheric oxygen concentrations represents an environment more similar to the in vivo condition, industry practices fail to fully limit exposures to atmospheric levels of O2 during these procedures. Due to the suggestion that ARTs result in an increased percentage of offspring exhibiting imprinting-related disorders, we wanted to examine whether exposure to atmospheric O2 could act as a stressor during the vulnerable period during preimplantation embryonic development and influence the transcription of genes that play a role in either the establishment, maintenance or regulation of the epigenome. In order to test these effects, we investigated the capacity of distinct levels of atmospheric oxygen to impact embryonic development, the transcriptional control of chromatin modifying genes and the regulation of imprinted gene expression, which previous studies have identified as being sensitive to embryo culture (de Waal et al., 2014).
Culturing bovine embryos under atmospheric oxygen concentrations (20%), as compared to the industry-standard 5% oxygen, resulted in reduced cleavage and blastocyst rates as seen in Fig. 1A. This observation is consistent with previous studies examining the effects of high oxygen levels on preimplantation bovine embryo development (Harvey et al., 2007; Harvey, 2007; Li et al., 2016). While standard industry practices do not employ atmospheric oxygen concentrations, cultured embryos are frequently exposed to this condition anytime they are removed from the incubators for manipulation. Although many oxidative stress response genes are regulated at the level of protein stability, in our experiments, we did observe an up-regulation of the oxidative stress response gene, SOD1, suggesting the mobilization of this cellular factor. In addition, we observed a down-regulation of several genes involved in maintenance of pluripotency. These data indicate that exposure to atmospheric oxygen negatively impacts embryonic development and is broadly associated with a reduction in the level of transcripts encoding the major factors driving embryonic patterning. Under high oxygen, transcripts encoding TET1, TET3, DNMT1, and DNMT3a were significantly down-regulated. In mice, TET3 is essential for embryonic development and the demethylation of the paternal methylome (Gu et al., 2011). Without prior time points of development, our data merely offer a snapshot of these epigenetic processes but do suggest a potential impact on the parental methylome. In order to validate the biological significance of these findings, further experiments that directly examine the impact on both global and gene-specific patterns of DNA methylation will need to be performed.
When we examined the regulation of chromatin modifying genes, we identified alterations in the expression of several candidates with established roles in fetal growth syndromes and the control of cellular differentiation. For example, we found broad down-regulation across the KDM family of lysine demethylases under high oxygen conditions. Altering these genes would likely lead to alterations in histone methylation and hinder the cell's ability to alter these marks. While not directly indicative of errors in the epigenome, the post-translational modifications imparted by these proteins serve as common signals and scaffolding for complexes known to alter the chromatin landscape. To this point, we also observed down-regulation of two important subunits of Polycomb repressive complex 2 (EED and EZH2). This complex is recruited to and silences specific regions of the genome by transferring methyl groups onto lysine 27 of histone H3. This chromatin modification has well-defined roles in the maintenance of the stem cell state and genomic imprinting (Juan et al., 2016; Inoue et al., 2017). Collectively, these observations indicate that oxygen concentrations have a strong influence on the control of chromatin modifying enzymes and significantly impact the regulation of two crucial pluripotency-associated genes. Finally, HELLS, which is known to recruit DNMTs to CDKN1C, displayed significantly decreased transcript levels under increased oxygen concentrations. CDKN1C is an imprinted gene that has been shown to be responsible for sporadic cancers and the overgrowth phenotypes associated with Beckwith-Weidemann syndrome (Algar et al., 1999). Additional studies will be required to determine if oxygen-induced changes in HELLS can be linked to the overgrowth phenotypes observed in large offspring syndrome in cattle.
In order to further our understanding of the effects differing oxygen concentrations have on preimplantation development, we shifted to a model employing a murine embryonic stem cell line. This system allowed us to observe both the acute impacts and prolonged effects of differing oxygen concentrations. While it is known that atmospheric levels of oxygen (20%) during culture negatively affect the ability of the mouse embryo to implant, the mechanisms behind this observation are not well understood (Wale and Gardner, 2013). Culturing these mESCs under differing oxygen levels revealed a restricted growth phenotype in the lower oxygen group. Similar to previous studies, colonies were significantly smaller in size in the low-oxygen group, albeit morphologically normal (Kurosawa et al., 2006). During the transition from atmospheric to low oxygen, we observed a down-regulation of two genes (Gstm3 and Nqo1), which are known to be involved in the cellular response to oxidative stress. Similarly, after 8 passages, Gpx2 and Gpx3, Sod2 and Nqo1 were all significantly different. The general trend of lower transcript levels under the 5% oxygen treatment suggests this condition is associated with a reduced oxidative stress response, with the notable exception of Gpx2. These results, along with previous studies, suggest that transferring mESCs into a low oxygen environment may be more favorable, although the limited experiments done here only suggest this possibility (Kurosawa et al., 2006).
Similar to our work in bovine embryos, studies in mESCs identified alterations in transcripts encoding the Tet family of demethylases. Here, low oxygen conditions were associated with a reduction in transcripts encoding Tet2 but only as an acute response. After 8 passages, no significant differences in Tet2 transcript levels could be detected, while by this point, Tet1 had become significantly down-regulated in the low oxygen group. In mice, Tet1 and Tet2 are hypothesized to have partially overlapping functions, and genetic deletion of both genes is still compatible with life, albeit with a 60% increase in developmental failure (Dawlaty et al., 2013). Therefore, the consequences of oxygen-induced alterations to Tet1 /Tet2 expression are difficult to anticipate and further studies examining global and gene-specific patterns of DNA methylation need to be conducted to verify the biological significance of these transcriptional changes. Unlike our studies in bovine embryos, we did not detect any impact on Tet3 in mESCs under different oxygen atmospheres.
In order to evaluate the downstream effects of these transcriptional changes, we turned our attention to a cohort of genes that are regulated through the processes of genomic imprinting. Our data indicated that several imprinted genes from our panel were dysregulated between our oxygen conditions. One of the more interesting findings was that Igf2 and H19 were dysregulated in a contrasting manner. These two genes are members of an imprinting locus in which Igf2 is paternally expressed while H19 is maternally expressed. In our experiments, Igf2 displayed a 2.5-fold increase in the low oxygen group with an accompanying down-regulation of H19. Further investigation is required to determine the mechanisms by which this occurs and the potential role oxygen concentrations have in modulating the transcriptional regulation of this locus. Another interesting observation to emerge from these studies was the correlative changes in Igf2 and Igf2r. Igf2r binds and sequesters Igf2, leading to a decrease in the amount of Igf2 available to interact with its receptors. Thus, while over-expression of Igf2 has been associated with an overgrowth phenotype (Morison and Reeve, 1998) excessive Igf2r is associated with growth suppression (Rezgui et al., 2008). Thus, due to Igf2r′s affinity for Igf2, it has also been found to regulate growth indirectly by reducing Igf2. In our studies, we observed oxygen-dependent up-regulation of both Igf2 and Igf2r. Here, the increased Igf2r transcript levels could be a compensatory mechanism to combat the elevated levels of Igf2. At this point, we do not know if these alterations in transcription are part of a normal oxidative stress response or the consequence of oxygen-induced alterations in the control of imprinted genes. However, given the established relationship between these proteins and the development of large-offspring syndrome in cattle, further studies into the capacity of oxygen to influence IGF expression are warranted.
Finally, we examined single nucleotide polymorphisms (SNPs) within a cohort of imprinted genes in order to evaluate the acute effects of reduced oxygen concentrations on allele-specific patterns of gene expression. Patterns of imprinted gene expression are established and maintained through epigenetic mechanisms and are often inferred to be good indicators of the “health” of the epigenome (Tang and Ho, 2007). We therefore utilized an established mouse model (Golding et al., 2011), which allowed the identification of parental alleles using single nucleotide polymorphisms, and therefore the capacity to trace the parental origins of mRNA transcripts. Culture under low oxygen conditions induced the H19 transcript pool to revert to a maternally dominated pattern. However, the opposite trend occurred for Peg3, indicating the 5% oxygen treatment was not necessarily more stable, with respect to the regulation of imprinted genes. Collectively, our data suggest that oxygen exposure has the power to influence the transcriptional control of crucial gene networks involved in the establishment and maintenance of chromatin structure and the control of imprinted genes.
4. Experimental procedures
4.1. In vitro fertilization and embryo production
Bovine oocytes were collected at a commercial abattoir (DeSoto Biosciences, Seymour, TN, USA) and shipped in an metal bead incubator (MOFA Global, Verona, WI, USA) at 38.5°C overnight in sealed sterile vials containing 5% CO2 in air-equilibrated maturation medium (Medium 199 with Earle's salts (Invitrogen, Life Technologies Inc., Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA), 1% penicillin–streptomycin (Invitrogen), 0.2-mM sodium pyruvate, 2-mM L-glutamine (Sigma Chemical Co., St. Louis, MO, USA), and 5.0 mg/mL of Folltropin (Vetoquinol, Pullman, WA, USA). The oocytes were matured in this medium for 22–24 h. Matured oocytes were washed twice with warm Tyrode lactate (TL) HEPES (Vetoquinol) supplemented with 0.05 μg/μL of gentamicin (Invitrogen) while being handled on a 38.5C stage warmer (Minitube). In vitro fertilization was conducted using a 2-h pre-equilibrated modified TL medium supplemented with 250-mM sodium pyruvate, 1% penicillin–streptomycin, 6 mg/mL of fatty acid–free BSA (Sigma), 20-mM penicillamine, 10-mM hypotaurine, and 10 μg/mL of heparin (Sigma) at 38.5°C, 5% CO2 in a humidified air incubator. Frozen semen was thawed at 35°C for 1 min, then separated by centrifugation at 200g for 20 min in a density gradient medium (Isolate; Irvine Scientific, Santa Ana, CA, USA) 50% upper and 90% lower. The supernatant was removed; the sperm pellet was resuspended in 2-mL modified Tyrode's medium and centrifuged at 200 g for 10 min to wash. The sperm pellet was then removed and placed into a warm 0.65-mL microtube (VWR Scientific, Pittsburg, PA, USA) before mixing in Nunc four-well multidishes (VWR)) containing up to 50 matured oocytes per well at a concentration of 1.0 × 106 sperm/mL. Sixteen to 18 h after insemination, oocytes were cleaned of cumulus cells by a 2-min vortex in 0.45-mL TL HEPES in a 0.65-mL microtube (VWR), washed in TL HEPES, and then randomly assigned to the different treatment groups: 5% oxygen controls and 20% atmospheric oxygen treatment and cultured in Synthetic Oviductal Fluid until the blastocyst stage. Cleavage rates were recorded on Day 2, at which point viable embryos were separated from nonviable embryos. Embryos were monitored daily for morphologic progression, and blastocyst rates were recorded on Day 8 after IVF.
4.2. Cell culture
Primary mESC cells were derived from B6XCAST F1 embryos as previously described (Golding et al., 2010). ESC cultures were maintained using 2i medium (Galonska et al., 2015) composed of: DMEM (Sigma D5671) supplemented with 10 μg/mL LIF (Sigma L5158), BIO (Sigma B1686), 1 μM PD0325901 (Sigma PZ0162), 50 mg/ml penicillin/streptomycin (Sigma P4333-100), 100 μM β-mercaptoethanol (Sigma M3148), 2mM L-Glutamine (Sigma G7513-100), MEM nonessential amino acids (Invitrogen 11140-050), and 15% Hyclone ESC grade fetal bovine serum (Fisher SH30070.03E) under high (20%) and low (5%) oxygen conditions for duration of 8 passages. Low oxygen culture conditions were maintained by using a modular incubation chamber perfused with premixed 5% O2, 5% CO2 and 90% N2 gas. Images of cell cultures were taken using an inverted Nikon microscope (Eclipse TE300) running NIS Elements and mESC size was measured using ImageJ software (Schneider et al., 2012). DNA and RNA were isolated during each passage using TRIzol (Life Technologies).
4.3. RNA-seq library preparation and data analysis
Total RNA was purified with an RNeasy Mini kit (Qiagen). RNA samples were analyzed for quality using a BioAnalyzer 2100 (Agilent). Once quality was verified, RNA samples were shipped to Global Biologics LLC (Columbia, Missouri) for library preparation and sequencing (1 μg RNA in 20 μL nuclease free water). Libraries were pooled and sequenced on an Illumina HISEQ 2500 (paired-end, 2 × 100 bp). The RNAseq reads were assessed for quality with FastQC. Adapters were removed with Cutadapt (Martin, 2011) and processed for quality with FastX-toolkit. Mapping was performed using Tophat2 and assembly by Cufflinks using the UCSC mm10 annotation (Trapnell et al., 2009, 2012). RNA expression levels were determined with Cuffmerge and Cuffdiff (Trapnell et al., 2013). Statistical analysis and visualization was performed with R, using RStudio running cummeRbund.
4.4. RNA isolation and RT-qPCR
Bovine blastocyst stage embryos were pooled in groups of ten and RNA extracted using the miRNeasy Micro kit (Qiagen), according to the manufacturer's instructions. cDNA was generated and pre-amplified with the RT2 PreAMP cDNA Synthesis kit (Qiagen). cDNA was quantified on a custom RT2 Profiler PCR Array: CAPB11472 (Qiagen). This array was run on the CFX384 Real-Time System (Bio-Rad). For the studies examining bovine embryos, the utilized reference genes were GAPDH (NM_001034034), SDHA (NM_174178) and YWHAZ (NM_174814). The stability of these reference genes was validated using the BestKeeper software package (Pfaffl et al., 2004). Murine embryonic stem cell samples were lysed and total RNA isolated with TRizol (Life technologies). cDNA was generated using random primer sets and the Superscript II first-strand synthesis system (Invitrogen). Real-time qPCR reactions were performed on a CFX384 Real-Time System (Bio-Rad) using SYBR green (Applied Biosystems). For work in embryonic stem cells, transcripts encoding Hprt (NM_013556), Mrpl (NM_053158) Ppia (NM_008907), and Ywhaz (NM_011740) served as reference genes. The stability of these transcripts across treatment groups was verified using the RNA-sequencing datasets. For each experiment, the reference genes used are indicated in the figure legend. Relative gene expression levels were calculated using comparative Ct values, in which Ct is the cycle threshold number normalized to the geometric mean of the reference genes. Primer sequences can be found in Supplemental Table 1 Primer Sequences.
4.5. Statistical analysis
For all experiments, statistical significance was set at alpha = 0.05. For analysis of gene expression, the replicate cycle threshold (Ct) values for each transcript were compiled and normalized to the geometric mean of the indicated reference genes. Normalized expression levels were calculated using the ddCt method described previously (Schmittgen and Livak, 2008). Relative fold change values from a minimum of three biological replicates were transferred into the statistical analysis program GraphPad (GraphPad Software, Inc., La Jolla, CA) where datasets were first verified for normality using the Brown-Forsythe test. For single comparisons, an unpaired student's t-test was applied. In all instances, we have marked statistically significant differences with an asterisk (*). In our mouse model, distinct single nucleotide polymorphisms between the maternal (C57BL/6J) and paternal (Mus muscuhis castaneus) strains (Mann et al., 2003) allowed us to track allelic patterns of gene transcription for multiple imprinted genes. For RNA sequence-based comparisons of allelic patterns of imprinted gene expression, the proportion of identified single nucleotide polymorphisms were analyzed using either Chi-Square or, if read counts were less than 5, a Fisher's Exact test.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health, Grant 1R24RR032683-01, 1R21HD055631-01A2 (CRL) and 1R01HD058969 (MEW). W.M.S. was supported through the Texas A&M University College of Veterinary Medicine and Biomedical Sciences Graduate Student Research Trainee Award. The authors gratefully acknowledge the technical contributions of Gayle Williamson and Chelsie Steinhauser to the experiments employing bovine embryo culture.
Abbreviations:
- ART
Assisted Reproductive Techniques
- DNMT
DNA Methyltransferase
- mESC
Murine Embryonic Stem Cell
- PRC
Polycomb-Repressive Complex
- SNPs
Single Nucleotide Polymorphisms
- TET
Ten-Eleven Translocation
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.gep.2018.01.001.
References
- Algar EM, Deeble GJ, Smith PJ, 1999, Jul. CDKN1C expression in beckwith-wiedemann syndrome patients with allele imbalance. J. Med. Genet 36 (7), 524–531. [PMC free article] [PubMed] [Google Scholar]
- Arnaud P, Feil R, 2005, Jun. Epigenetic deregulation of genomic imprinting in human disorders and following assisted reproduction. Birth Defects Res. Part C Embryo Today - Rev 75 (2), 81–97. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Tilghman SM, 1997. Genomic imprinting in mammals. Annu. Rev. Genet 31 (1), 493–525. [DOI] [PubMed] [Google Scholar]
- Barker DJ, 1990. November 17. The fetal and infant origins of adult disease. BMJ 301 (6761), 1111 PMCID: PMC1664286; PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-F, Kuo H-C, Chen W, Wu F-C, Yang Y-S, Ho H-N, 2009, Jan. A reduced oxygen tension (5%) is not beneficial for maintaining human embryonic stem cells in the undifferentiated state with short splitting intervals. Hum. Reprod 24 (1), 71–80. [DOI] [PubMed] [Google Scholar]
- Christian P, Stewart CP, 2010, Mar. Maternal micronutrient deficiency, fetal development, and the risk of chronic disease. J. Nutr 140 (3), 437–445. [DOI] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, et al. , 2013, February 11. Combined deficiency of Tet1 and tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell 24 (3), 310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waal E, Mak W, Calhoun S, Stein P, Ord T, Krapp C, et al. , 2014, Feb. In vitro culture increases the frequency of stochastic epigenetic errors at imprinted genes in placental tissues from mouse concepti produced through assisted reproductive technologies. Biol. Reprod 90 (2), 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean W, Lucifero D, Santos F, 2005, Jun. DNA methylation in mammalian development and disease. Birth Defects Res. Part C Embryo Today - Rev 75 (2), 98–111. [DOI] [PubMed] [Google Scholar]
- DeBaun MR, Niemitz EL, Feinberg AP, 2003, Jan. Association of in vitro fertilization with beckwith-wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am. J. Hum. Genet 72 (1), 156–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte B, Deplus R, Fuks F, 2014. June 2. Playing tetris with DNA modifications. EMBO J 33 (11), 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte B, Jeschke J, Defrance M, Bachman M, Creppe C, Calonne E, et al. , 2015, August 4. Genome-wide hydroxymethylcytosine pattern changes in response to oxidative stress. Sci. Rep 5, 12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM, 2000, Jun. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol. Reprod 62 (6), 1526–1535. [DOI] [PubMed] [Google Scholar]
- Farin PW, Piedrahita JA, Farin CE, 2006, January 7. Errors in development of fetuses and placentas from in vitro-produced bovine embryos. Theriogenology 65 (1), 178–191. [DOI] [PubMed] [Google Scholar]
- Fischer B, Bavister BD, 1993, November. Oxygen tension in the oviduct and uterus of rhesus monkeys, hamsters and rabbits. J. Reprod. Fertil 99 (2), 673–679. [DOI] [PubMed] [Google Scholar]
- Galonska C, Ziller MJ, Karnik R, Meissner A, 2015, October 1. Ground state conditions induce rapid reorganization of core pluripotency factor binding before global epigenetic reprogramming. Cell Stem Cell 17 (4), 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gicquel C, Gaston V, Mandelbaum J, Siffroi J-P, Flahault A, Le Bouc Y, 2003, May. In vitro fertilization may increase the risk of beckwith-wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am. J. Hum. Genet 72 (5), 1338–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding MC, Zhang L, Mann MRW, 2010, May 7. Multiple epigenetic modifiers induce aggressive viral extinction in extraembryonic endoderm stem cells. Cell Stem Cell 6 (5), 457–467. [DOI] [PubMed] [Google Scholar]
- Golding MC, Magri LS, Zhang L, Lalone SA, Higgins MJ, Mann MRW, 2011, Sep. Depletion of kcnq1ot1 non-coding RNA does not affect imprinting maintenance in stem cells. Development 138 (17), 3667–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosden R, Trasler J, Lucifero D, Faddy M, 2003, June 7. Rare congenital disorders, imprinted genes, and assisted reproductive technology. Lancet 361 (9373), 1975–1977. [DOI] [PubMed] [Google Scholar]
- Gu T-P, Guo F, Yang H, Wu H-P, Xu G-F, Liu W, et al. , 2011, September 4. The role of tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477 (7366), 606–610. [DOI] [PubMed] [Google Scholar]
- Hansen JM, 2006, Dec. Oxidative stress as a mechanism of teratogenesis. Birth Defects Res. Part C Embryo Today - Rev 78 (4), 293–307. [DOI] [PubMed] [Google Scholar]
- Harvey AJ, Kind KL, Thompson JG, 2007, Jul. Regulation of gene expression in bovine blastocysts in response to oxygen and the iron chelator desferrioxamine. Biol. Reprod 77 (1), 93–101. [DOI] [PubMed] [Google Scholar]
- Harvey AJ, 2007, Mar. The role of oxygen in ruminant preimplantation embryo development and metabolism. Anim. Reprod. Sci 98 (1–2), 113–128. [DOI] [PubMed] [Google Scholar]
- Inoue A, Jiang L, Lu F, Suzuki T, Zhang Y, 2017, July 27. Maternal h3k27me3 controls DNA methylation-independent imprinting. Nature 547 (7664), 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Wang S, Ko KD, Zare H, Tsai P-F, Feng X, et al. , 2016, October 25. Roles of h3k27me2 and h3k27me3 examined during fate specification of embryonic stem cells. Cell Rep 17 (5), 1369–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffer CR, Grinberg A, Pfeifer K, 2001, Dec. Regulatory mechanisms at the mouse igf2/H19 locus. Mol. Cell Biol 21 (23), 8189–8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella MT, Monk C, 2009, Sep. Impact of maternal stress, depression and anxiety on fetal neurobehavioral development. Clin. Obstet. Gynecol 52 (3), 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y, 2013, October 24. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502 (7472), 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurinczuk JJ, Hansen M, Bower C, 2004. The risk of birth defects in children born after assisted reproductive technologies. Curr. Opin. Obstet. Gynecol 16 (3), 201–209. [DOI] [PubMed] [Google Scholar]
- Kurosawa H, Kimura M, Noda T, Amano Y, 2006, Jan. Effect of oxygen on in vitro differentiation of mouse embryonic stem cells. J. Biosci. Bioeng 101 (1), 26–30. [DOI] [PubMed] [Google Scholar]
- Li W, Goossens K, Van Poucke M, Forier K, Braeckmans K, Van Soom A, Peelman LJ, 2016. Jun. High oxygen tension increases global methylation in bovine 4-cell embryos and blastocysts but does not affect general retrotransposon expression. Reprod. Fertil. Dev 28 (7), 948–959. [DOI] [PubMed] [Google Scholar]
- Liu X, Greer C, Secombe J, 2014, Oct. KDM5 interacts with foxo to modulate cellular levels of oxidative stress. PLoS Genet 10 (10), e1004676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann MRW, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS, 2003, Sep. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod 69 (3), 902–914. [DOI] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, et al. , 2009, October 8. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461 (7265), 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J 17 (1), 10. [Google Scholar]
- Morison IM, Reeve AE, 1998, Mar. Insulin-like growth factor 2 and overgrowth: molecular biology and clinical implications. Mol. Med. Today 4 (3), 110–115. [DOI] [PubMed] [Google Scholar]
- Niemitz EL, Feinberg AP, 2004, Apr. Epigenetics and assisted reproductive technology: a call for investigation. Am. J. Hum. Genet 74 (4), 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP, 2004. Mar. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – excel-based tool using pair-wise correlations. Biotechnol. Lett 26 (6), 509–515. [DOI] [PubMed] [Google Scholar]
- Powell K, 2003, April 17. Fertility treatments: seeds of doubt. Nature 422 (6933), 656–658. [DOI] [PubMed] [Google Scholar]
- Preis KA, Seidel GE, Gardner DK, 2007, Jul. Reduced oxygen concentration improves the developmental competence of mouse oocytes following in vitro maturation. Mol. Reprod. Dev 74 (7), 893–903. [DOI] [PubMed] [Google Scholar]
- Rezgui D, Williams C, Savage SA, Prince SN, Zaccheo OJ, Jones EY, et al. , 2008, December 29. Structure and function of the human gly1619arg polymorphism of M6P/IGF2R domain 11 implicated in IGF2 dependent growth. J. Mol. Endocrinol 42 (4), 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ, 2008, Jun. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc 3 (6), 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW, 2012. NIH image to imagej: 25 years of image analysis. Nat Methods 9 (7), 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W-Y, Ho S-M, 2007, Jun. Epigenetic reprogramming and imprinting in origins of disease. Rev. Endocr. Metab. Disord 8 (2), 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL, 2009. TopHat: discovering splice junctions with rna-seq. Bioinformatics 25 (9), 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. , 2012, Mar. Differential gene and transcript expression analysis of rna-seq experiments with tophat and cufflinks. Nat. Protoc 7 (3), 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L, 2013, Jan. Differential analysis of gene regulation at transcript resolution with rna-seq. Nat. Biotechnol 31 (1), 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey KJ, Parnell SE, Miranda RC, Golding MC, 2015. Dose-dependent alcohol-induced alterations in chromatin structure persist beyond the window of exposure and correlate with fetal alcohol syndrome birth defects. Epigenet. Chromatin 8, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wale PL, Gardner DK, 2013, Sep. Oxygen affects the ability of mouse blastocysts to regulate ammonium. Biol. Reprod 89 (3), 75. [DOI] [PubMed] [Google Scholar]
- Wang L, Chia NC, Lu X, Ruden DM, 2011, July 1. Hypothesis: environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics 6 (7), 853–856. [DOI] [PubMed] [Google Scholar]
- Wrenzycki C, Herrmann D, Lucas-Hahn A, Gebert C, Korsawe K, Lemme E, et al. , 2005, Mar. Epigenetic reprogramming throughout preimplantation development and consequences for assisted reproductive technologies. Birth Defects Res. Part C Embryo Today - Rev 75 (1), 1–9. [DOI] [PubMed] [Google Scholar]
- Young LE, Sinclair KD, Wilmut I, 1998, Sep. Large offspring syndrome in cattle and sheep. Rev. Reprod 3 (3), 155–163. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D, 2001, September 15. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15 (18), 2343–2360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.