Abstract
To date, the expression of recombinant proteins in transgenic plants is becoming a powerful alternative to classical expression methods. Special efforts are directed to the development of contained cultivation systems based on cell culture or rhyzosecretion, which reliably prevents the heterologous DNA releasing into the environment. A promising object for the development of such systems is the tiny aquatic plant of Wolffia arrhiza, which can be used as a dipped culture in bioreactors. Herein we have expressed the human granulocyte colony-stimulating factor (hG-CSF) in nuclear-transformed Wolffia. The nucleotide sequence of hG-CSF was optimized for expression in Wolffia and cloned into the vector pCamGCSF downstream of double CaMV 35S promoter. Wolffia plants were successfully transformed and 34 independent transgenic lines with hG-CSF gene were obtained, PCR and Southern blot analysis confirmed the transgenic origin of these lines. Western blot analysis revealed accumulation of the target protein in 33 transgenic lines. Quantitative ELISA of protein extracts from these lines showed hG-CSF accumulation up to 35.5 mg/kg of Wolffia fresh weight (0.194% of total soluble protein). This relatively high yield holds promise for the development of Wolffia-based expression system in strictly controlled format to produce various recombinant proteins.
Keywords: biopharming, hG-CSF, recombinant proteins, transgenic duckweed, transgene expression system, Wolffia arrhiza
Introduction
Nowadays a large number of expression systems for recombinant proteins production have been developed. Recombinant proteins are produced in the cells of bacteria, yeast, mammals, and insects (Walsh, 2014; Rader and Langer, 2015; Sysuev and Pokrovskaya, 2015). However, along with the advantages these systems have a number of substantial drawbacks, in particular post-translation modifications and correct folding of many eukaryotic proteins do not occur in bacterial and yeast cells (Jensen, 2006; Merlin et al., 2014; Tschofen et al., 2016). Animal cells are devoid of such shortcomings but their use as bioproducers is limited by reason of recombinant proteins high production cost (Demain and Vaishnav, 2009). Thus, the development of expression systems combining the advantages of microbial systems (high expression level, relatively low production cost) and systems based on animal cell culture (the complete identity of recombinant protein and its properties to native counterpart) is still urgent.
Apparently the advantages of microbial and animal cell culture expression systems are combined in plant-based platforms to the greatest extent. Basically the protein glycosilation and folding in the higher plants are similar to those in mammalian cells (Strasser, 2016). Unlike animals, plant cells do not contain viruses and microorganisms pathogenic for humans, that substantially simplifies the purification of recombinant proteins for medical purposes (Daniell et al., 2001). Plant cultivation does not require expensive equipment, wherein the agricultural scale of the production guarantees the availability of recombinant proteins in the quantities quite enough for wide therapeutical use (Kaufman, 2011; Wilken and Nikolov, 2012).
The most common current expression systems are based on well-studied agricultural plants, such as tobacco, rice, corn etc. The main problem of these systems is the probability of uncontrolled transfer of heterologous DNA into the environment by seeds, pollen or plant residues during the cultivation of transgenic plant in the field or greenhouse. The fear of this greatly complicates the commercialization of plant-based expression systems. Considerable effort, therefore, has been focused on the development of contained cultivation systems, such as axenic culture of suspension cells or hairy roots (Georgiev et al., 2012; Santos et al., 2016). The contained plant systems can be cultivated in bioreactors, entirely preventing the accidental release of genetically modified plant material to the environment.
Another approach to the development of contained cultivation systems is the using of small aquatic plants—duckweed [Lemna minor (L.) and Wolffia arrhiza (L.) Horkel ex Wimm.] as recombinant proteins producers. Duckweed are small monocotyledonous plants with a high content of total protein (up to 45% of DW), demonstrating high growth rate (doubling of biomass in 1–6 days) and almost exclusively vegetative reproduction (Armstrong and Thorne, 1984). These plants are capable to photosynthesis in vitro and can be effectively and inexpensively cultivated in bioreactors of various types (Gasdaska et al., 2003; Khvatkov et al., 2013).
In Lemnaceae family, Wolffia arrhiza is the most evolutionary advanced secondary primitively organized species (Wolff, 1992). Plant bodies are tiny; generally globoid to ovoid-ellipsoid or cylindrical; 0.4–1.3 mm long and 0.2–1.0 mm wide; floating on or partially below water surface (Wayne and Thorne, 1984). The most important Wolffia's feature is, that unlike other Lemnoideae, it does not have a root system, this property allows the cultivating of Wolffia in bioreactors using submergence, which in turn could considerably increase the profitability of recombinant proteins production.
To date, there are already examples of successful expression of commercially important proteins in duckweed plants: the industrial enzyme endoglucanase E1 from Acidothermus cellulolyticus (Sun et al., 2007), anti-CD20, anti-CD30, and anti-interferon a2b monoclonal antibodies (Cox et al., 2006; Biolex Therapeutics website), and the protective antigens of influenza virus - peptide M2e (Firsov et al., 2015, 2018), and hemagglutinin (Bertran et al., 2015). Despite that Wolffia is very promising plant for biotechnology, yet there is no reports of its use for recombinant proteins expression. The main problem of using Wolffia for recombinant proteins expression is the absence of a method for its genetic transformation. The development of such a method allows to start the studies on the development of Wolffia—based expression system.
The aim of this study is to explore the feasibility of recombinant proteins expression in nuclear-transformed Wolffia arrhiza plants by the example of human granulocyte colony-stimulating factor (hG-CSF). hG-CSF consists of 207 amino acid residues, including a signal peptide (aa 1–29). Mature hG-CSF has a length of 178 aa, it is stabilized by 2 disulfide bonds and glycosylated at position 166. At the present time recombinant hGCSF is widely used for management of various etiology neutropenias in the therapy of patients with HIV, aplastic anemia, pneumonia, leukemia, and others diseases (Harousseau et al., 2000; Babalola et al., 2004; Dale, 2010).
Here, we report the successful Agrobacterial transformation of Wolffia and expression of human granulocyte colony-stimulating factor in transgenic plants. Recombinant GCSF accumulated at a high level, sufficient for further studies to create Wolffia-based expression system to produce the offered and other recombinant proteins.
Materials and methods
Gene synthesis and plasmid construction
The amino acid sequence of hG-CSF corresponding to the drug Filgrastim (DrugBank: DB00099) was selected for expression in Wolffia. The N-terminal signal peptide hG-CSF has been replaced by a corresponding peptide of rice α-amilase (GenBank: AAA33897.1). To optimize the codon composition a table of codon usage in Lemna gibba was used (http://www.kazusa.or.jp/codon). The optimization of hG-CSF nucleotide sequence was performed by Gene Composer software (Lorimer et al., 2009). The set of overlapping oligonucleotides was designed using Gene2Oligo (Rouillard et al., 2004).
Nucleotide sequence of hG-CSF was synthesized by PCR (Rouillard et al., 2004) and cloned into sites HindIII and EcoRI of plasmid pUC18 followed by sequencing. Then, hG-CSF (in Appendix) sequence was cut out from pUC18 and, using the same sites, cloned into plant expression vector pCamPPVcp, replacing the plum pox virus coat protein gene (Dolgov et al., 2010). The resulting plasmid pCamGCSF was transferred into A. tumefaciens strain EHA105 (Hood et al., 1993) and used for Wolffia transformation.
Tissue culture and Agrobacterium-mediated transformation of W. arrhiza
For research, we obtained a population of Wolffia arrhiza (L.) Horkel ex Wimm from the collection of Main Botanic Garden of the Russian Academy of Sciences (RDSC Clone Wolffia 5564; Supplementary Table 1) (Figure 1).
Figure 1.
Images of characteristic morphological features of Wolffia arrhiza. Dorsal view showing several flat-topped, dark green plants and side view of W. arrhiza budding plant showing flattened, dorsal surface of daughter plant.
An aseptic population of W. arrhiza plants was cultivated on SH medium (Schenk and Hildebrandt, 1972) supplemented with 2% (w/v) sucrose and 0.7% (w/v) agar (Panreac, Spain). 10 plants were placed on each Petri dish filled with culture medium. The cluster structures formed after 4 months of pre-cultivation on MS2G media containing 5.0 mg l−1 2,4-Dichlorophenoxyacetic acid (2,4-D) and 0.5 mg l−1 N6-Benzyladenine (BA) (Khvatkov et al., 2015b), were used for transformation. The explants developed at light intensity of 65 μmol m−2 s−1, day photoperiod 16 h and temperature 21 ± 1°C.
W. arrhiza was transformed as described by Khvatkov et al. (2015a). Explants were co-cultivated with A. tumefaciens containing the plasmid pCamGCSF, with subsequent transfer to SH medium containing 2.0 mg l−1 2,4-D, 2.0 mg l−1 BA, 5.0 mg l−1 hygromycin B (Hyg) and 100 mg l−1 timentin. After2 weeks the explants were transferred to SH medium lack of growth regulators for regeneration. Each newly emerged Hyg-resistant plant was placed into a separate culture tube with W3M media (Dolgov et al., 2013) supplemented with 5.0 mg l−1 Hyg for further development.
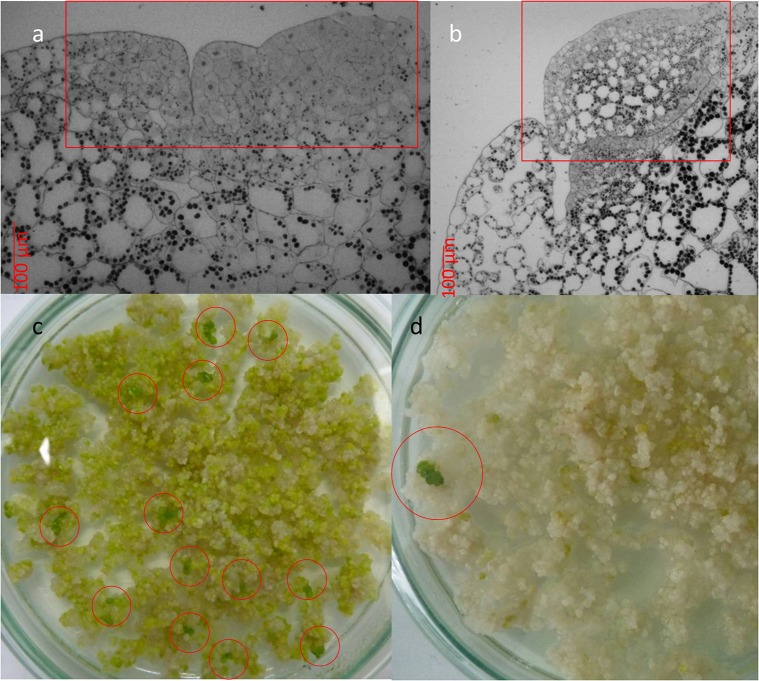
The inspection of the selected plant material for their regenerative capacity [viz. the development of meristematic tissue (Figure 3a) and formation of plant regenerants (Figure 3b)] was performed by light microscopy. Preparation of samples and microscopic examination of plant material was carried out according to the procedure described in Miroshnichenko et al. (2017).
Figure 3.
Development of transgenic Wolffia plants in the presence of 5.0 mg l−1 Hyg in the culture media. (a) Meristematic area (squared in red) on the cluster surface which developed after 6 weeks exposure to selective media. (b) Globular structures (squared in red) on the cluster surface which developed after 8–10 weeks exposure to selective media. (c, d) Multiple (c) or single (d) formation of whole plants (circled in red) in the presence of 5.0 mg l−1 Hyg in the culture media.
Selection of pure transgenic lines was carried out as described by Khvatkov et al. (2015a). Transformation efficiency was determined by dividing the number of explants which formed transgenic populations upon the total number of explants in each experiment (percentagewise).
PCR and southern blot analysis of transformants
The Wolffia genomic DNA was isolated from transformed and non-transformed plants according to the method of Dellaporta et al. (1983). The respective forward and reverse primer sequences for hptII were hptf: 5′-acattgttggagccgaaatc-3′ and hptr: 5′-gacattggggagtttagcga-3′; those for hG-CSF were G1for: 5′-gtccaagcttatggcgaagaggatcgcc-3′ and G2rev: 5′-atgaattctcacggttgggcgagatg-3′; those for virC were virC1: 5′-gcactatctacctaccgctacgtcatc-3′ and virC2: 5′-gttgtcgatcgggactgtaaatgtg-3′.
For Southern blot analysis, genomic DNA from transformed and non-transfomed Wolffia plants in amount of 50 μg was digested overnight at 37°C with 100 U HindIII which cuts the T-DNA of pCamGCSF at a single position. After agarose gel electrophoresis (0.8%), the products of digestion were transferred and immobilized onto Hybond+ membrane (Amersham, USA) according to the manufacturer's instructions. The DNA probe was constructed by PCR using plasmid pCamGCSF as the template, and primers G1for and G1rev. Probe DNA (600 bp) was labeled with alkaline phosphatase using the AlkPhos Direct Labeling Kit (Amersham Bioscience, USA). Prehybridization, hybridization (overnight at 60°C) with alkaline phosphatase-labeled probe, and subsequent washings of the membrane were carried out according to the AlkPhos Direct Labeling Kit protocol. Detection was performed using CDP-Star detection reagent according to the manufacturer's instructions (Amersham Bioscience).
Western blot analysis
Transgenic and non-transformed Wolffia plants (in an amount of 1 g) were grinded in liquid nitrogen and resuspended in 3x volume extraction buffer (50 mM Tris-HCl (pH7.6), 150 mM NaCl, 5 mM EDTA, 5 mM β-merkaptoethanol, 0.62 μM aprotinin, 8.4 μM leupeptin). Extraction was performed during 40 min at 4°C followed by centrifugation for 20 min at 16,000 g; supernatant was collected for subsequent analysis. Protein concentration in the samples was determined by Bradford method (Bradford, 1976).
Protein electrophoresis (70 μg/lane) was carried out in 10–25% gradient SDS-PAGE. The separated proteins were transfered onto a nitrocellulose membrane (BioRad, USA). Rabbit polyclonal antibody against hG-CSF (diluted 1:1,000; Abcam, UK) and anti-rabbit IgG conjugated with alkaline phosphatase (1:3,000; Pierce, USA) were used. Recombinant hG-CSF (Abcam, UK) served as the positive control. The blots were visualized using chromogenic substrate BCIP/NBT (Fermentas, Lithuania).
ELISA quantification of hG-CSF accumulation
The total protein was extracted as described above. The protein samples were serially diluted in PBS and loaded into 96-well ELISA plate (0.25, 0.5, 1.0, and 2.0 μg of TSP per well), using recombinant hG-CSF (Abcam, UK) as the reference standard.
The plates were incubated for 2 h at RT, then washed three times with PBST (PBS with 0.1% Tween 20) for 5 min each and blocked in PBST containing 2% bovine serum albumin (1 h at RT). Hybridization with the primary antibody was performed overnight at 4°C. After washing, the secondary antibody was added and plates were incubated for 1 h at RT followed by the washing. Rabbit polyclonal antibody against hG-CSF (Abcam, UK) was diluted at 1:1,000, anti-rabbit IgG conjugated with alkaline phosphatase (Pierce, USA)—at 1:2,000. The plates were developed for 30 min at RT using TMB Peroxidase EIA substrate (BioRad). The plates were read at 405 nm and the amount of plant—expressed hG-CSF was estimated based on reference standards.
Statistical analysis
The significance of differences in hG-CSF accumulation between Wolffia transgenic lines were analyzed by analysis of variance (ANOVA) followed by multiple comparisons of individual averages and evaluation by Duncan's test (Statistica 6.1 software; StatSoft Inc).
Results
Vector construction and Agrobacterium-mediated transformation of Wolffia arrhiza
We used the data on codon usage in Lemna gibba, which belongs to the same Lemnaceae family, as W. arrhiza, to optimize the codon composition of hG-CSF nucleic acid sequence. We assumed that the codon usage do not differ significantly in these two species. Nucleic sequence of hG-CSF with the codon composition optimized for the expression in Lemnaceae was synthesized and cloned under the control of the CaMV double 35S promoter. The resulting plasmid pCamGCSF (Figure 2) was used for hG-CSF expression in Wolffia plants, following its Agrobacterium-mediated transformation.
Figure 2.

Schematic presentation of pCamGCSF plasmids used for Wolffia transformation. d35S, double 35S RNA Cauliflower Mosaic Virus promoter; GCSF, human granulocyte colony-stimulating factor coding sequence; alpha-amilase SP, nucleotide sequence of rice alpha-amylase signal peptide; hptll, hygromycin phosphotransferase (HPT) coding sequence; pACaMV, 35S RNA Cauliflower Mosaic Virus terminator with polyadenylation signal; RB, right border; LB, left border; npt, neomycin phosphotransferase (NPT) coding sequence allowing for A. tumefaciens selection.
In experiments on Agrobacterium-mediated transformation, transgenic tissue is distinguishable after 1.5 months under selection on SH medium in the presence of 5.0 mg l−1 Hyg. Green meristematic area appeared on cluster surface after 6 weeks of cultivation on SH medium lack of growth regulators (Figure 3a, squared in red). Emerald green globular structures were originally developed from meristematic area (Figure 3a, squared in red) followed by the arising of the whole plants (Figures 3c,d, circled in red). Regeneration of various cultures on the hormone-free medium is a rare but fairly well-known phenomenon. Thus, in addition to the Wolffia, the ability to regenerate on hormone-free media is capable, for example, of carrots (Górecka et al., 2009), lotus (Rybczynski and Badzian, 1987), flax (Aycan et al., 2014), parsley (Masuda et al., 2006), beets (Doley and Saunders, 1989), and etc. Obviously, hormone enriched media during the cultivation of explants of these cultures in vitro are required solely to maintain them in the state of the callus structures, and not for regeneration.
The morphological differences between Hyg-resistant Wolffia plants and non-transformed ones were not observed. In liquid culture the development and growth rate of transgenic plants did not differ from the corresponding characteristics of the non-transformed control plants. In experiments on Agrobacterium-mediated transformation of Wolffia, 8100 explants were used on the whole; the transformation frequency was 0.42%. In total, 34 independent Wolffia lines which developed without signs of hygromycin toxic effect were obtained; these lines were used for further analysis.
PCR and southern blot analysis of transformants
The fragments of the expected size were amplified from the DNA of all analyzed putatively transgenic Wolffia lines. As it was shown by PCR using virC1 and virC2 primers, all lines were free of agrobacterial contamination (data not shown). Thus, according to PCR analysis, all of 34 Hyg-resistant Wolffia lines contained selective hptII gene and target hG-CSF sequence.
The selected transgenic lines with different hG-CSF expression were analyzed by Southern blot. The obtained results confirmed integration of hG-CSF gene into Wolffia genome (Figure 4). Based on the hybridization profile, there were one (lines 2, 8, 24, and 27), two (lines 17, 19, 28), and four or more insertions (lines 4, 6, 32, and 33) of the transgene into the obtained lines, which is typical for Agrobacterium-mediated transformation. The DNA from non-transformed plants failed to hybridize to the probe.
Figure 4.

Southern hybridization analysis of some EHA105/pCamGCSF-transformed Wolffia plants. DNA (10 μg) digested with HindIII and hybridized with a 600-bp hG-CSF probe. pCamGCSF, pDNA (pCamGCSF/HindIII); WG, DNA of transgenic Wolffia lines digested with HindIII; Wild-type, genomic DNA of wild-type Wolffia plants digested with HindIII.
Analysis of hG-CSF expression in transgenic Wolffia plants
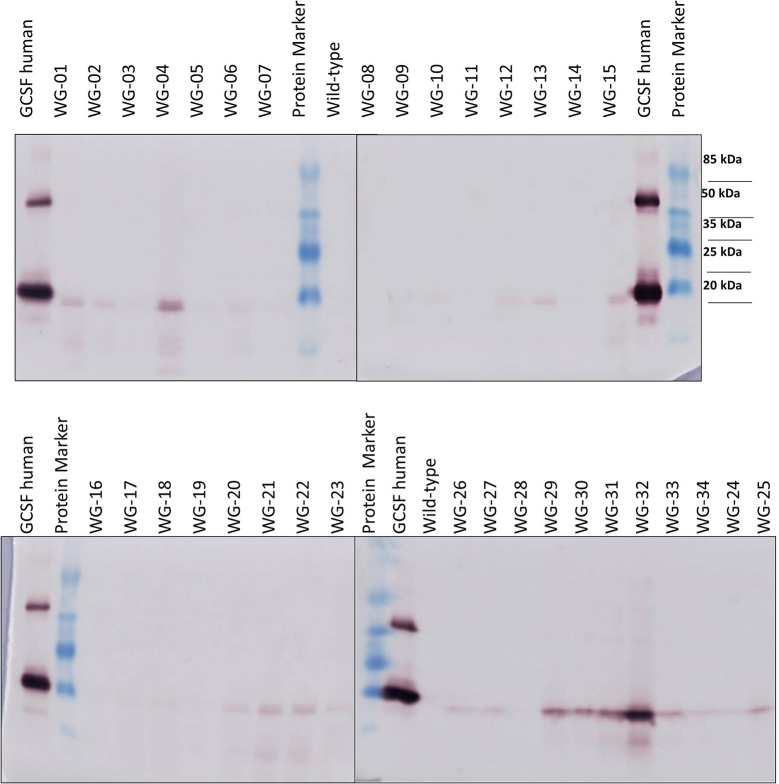
Western blot analysis using anti-hG-CSF antibody has revealed the presence of a 20-kDa band corresponding to the recombinant hG-CSF in 33 transgenic lines out of 34 examined (Figure 5). hG-CSF from the transgenic plants migrated at the same rate as the control protein. This result confirms the cleavage of the rice α-amylase N- terminal signal peptide, i.e., the correct processing of pre-hG-CSF in Wolffia plants. Immunoreactive bands of similar weight were not observed in the protein samples from non-transformed control plants. It should be noted, that there was no noticeable degradation of the recombinant hG-CSF in transgenic plants. In some lines (№ 3, 5, 7, 8, 11, 14, 16, and 19), the target protein was detected at low level; the immunoreactive bands were weak (Figure 5). The maximum hG-CSF accumulation was observed in lines WG-04 and WG-32, in others lines the accumulation of the target protein was lower.
Figure 5.
Western blot analysis of hG-CSF expression in some transgenic Wolffia lines. Protein Marker, molecular size marker TS 26612 Protein MW Marker; GCSF human, recombinant human G-CSF (200 ng; AbCam, UK).
According to ELISA, 33 out of 34 transgenic lines expressed hG-CSF in plant tissues (Table 1). Quantification of the target protein in transgenic Wolffia plants using anti-hG-CSF antibody showed accumulation from 0.36 to 35.5 μg of hG-CSF per 1 g fresh weight (FW) of Wolffia, corresponding to 0.002–0.194% of total soluble protein (TSP). The highest levels of hG-CSF accumulation were observed in lines WG-04 and WG-32 (33.9 and 35.5 μg g−1 FW; 0.191 and 0.194% of TSP, respectively), and the lowest in lines WG-09, WG-13 and WG-14 (0.36, 0.53 and 0.36 μg g−1 FW; 0.002, 0.003, and 0.002% of TSP, respectively) (Figure 6).
Table 1.
Quantitative indicators of hG-CSF expressed in W.arrhiza transgenic plants.
| Name of transgenic line | Saturation with hG-CSF inside plant tissues | |
|---|---|---|
| Target protein per TSP (%) | mg kg−1 of FW | |
| WG-01 | 0.005 ± 0.002 | 0.95a |
| WG-02 | 0.037 ± 0.017 | 6.57ab |
| WG-03 | 0.014 ± 0.006 | 2.49ab |
| WG-04 | 0.191 ± 0.052 | 33.85de |
| WG-05 | 0.011 ± 0.006 | 1.95a |
| WG-06 | 0.087 ± 0.070 | 15.45bc |
| WG-07 | 0.008 ± 0.004 | 1.42a |
| WG-08 | 0.009 ± 0.001 | 1.60a |
| WG-09 | 0.002 ± 0.001 | 0.36a |
| WG-10 | 0.004 ± 0.003 | 0.71a |
| WG-11 | 0.007 ± 0.002 | 1.24a |
| WG-12 | 0.004 ± 0.003 | 0.71a |
| WG-13 | 0.003 ± 0.001 | 0.53a |
| WG-14 | 0.002 ± 0.001 | 0.36a |
| WG-15 | 0.007 ± 0.004 | 1.24a |
| WG-16 | 0.008 ± 0.002 | 1.42a |
| WG-17 | 0.008 ± 0.004 | 1.42a |
| WG-18 | 0.005 ± 0.002 | 0.89a |
| WG-19 | 0.006 ± 0.001 | 1.06a |
| WG-20 | 0.005 ± 0.003 | 0.89a |
| WG-21 | 0.005 ± 0.002 | 0.89a |
| WG-22 | 0.006 ± 0.001 | 1.07a |
| WG-23 | 0.006 ± 0.004 | 1.0a |
| WG-24 | 0.033 ± 0.021 | 5.86ab |
| WG-25 | 0.008 ± 0.001 | 1.42a |
| WG-26 | 0.005 ± 0.003 | 0.89a |
| WG-27 | 0.010 ± 0.001 | 1.78a |
| WG-28 | 0.000 ± 0.000 | 0.00a |
| WG-29 | 0.010 ± 0.004 | 1.78a |
| WG-30 | 0.028 ± 0.008 | 4.97ab |
| WG-31 | 0.006 ± 0.004 | 1.07a |
| WG-32 | 0.194 ± 0.060 | 35.45e |
| WG-33 | 0.119 ± 0.023 | 15.13c |
| WG-34 | 0.004 ± 0.003 | 0.71a |
Different letters in a column indicate significant differences in variant data according by Duncan's test.
Figure 6.
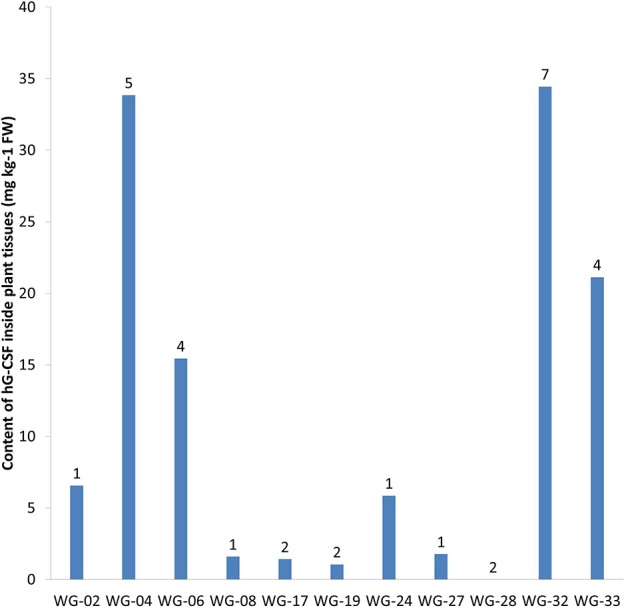
The result of ELISA of 11 transgenic Wolffia lines containing hG-CSF gene. Numbers designate the number of hG-CSF gene insertions into transgenic line genome.
As a result of our study, our research team was the first to receive transgenic Wolffia plants expressing the recombinant human G-CSF. In two lines—WG-04 and WG-32—the target protein was accumulated at a high level—approximately 35 mg of recombinant protein per 1 kg of Wolffia FW. Thus, by the example of hG-CSF we have demonstrated the potential of Wolffia as a producer of recombinant proteins.
Discussion
We have obtained 34 different lines of Wolffia with confirmed transgenic status; the target protein hG-CSF was detected in 33 lines (Table 1). Morphologically transgenic Wolffia plants did not differ from their non-transformed counterparts. Expression of the foreign protein did not affect the rate of reproduction in liquid culture or plant TSP content. According to ELISA accumulation of recombinant hG-CSF ranged from 0.36 to 35.5 μg g−1 FW of Wolffia (0.002–0.2% of TSP).
In our study, we identified almost 100-fold variation in target protein accumulation among the transgenic lines. Variations in recombinant protein expression between independently derived transgenic lines are common (Richter et al., 2000; Sharma and Sharma, 2009). They are often related to differences in the number of transgene copies or in the position within the genome into which the foreign DNA was integrated (Dolgova et al., 2015). Both of these factors might be applied to the transgenic W. arrhiza lines that were obtained (Figure 6). The Southern blot analysis showed notable differences between the studied lines both as in the number of transgene inserts and in the sites of their integration into the plant genome.
Currently the recombinant hG-CSF used in medicine is produced either in E. coli expression system (Filgrastim medication; Vacchelli et al., 2013) or in CHO cell culture (Lenograstim; Serova et al., 2012). The recombinant GCSF manufactured in the bacterial system is non-glycosylated; GCSF produced in CHO cells is glycosylated and does not differ from G-CSF naturally made in humans. Both forms of recombinant hG-CSF are almost similar in pharmacological properties, although there are reports that the glycosylated form has a higher efficiency of mobilizing stem cells than Filgrastim and is more effective at lower doses (Ria et al., 2010; Sari et al., 2013). At that, the cost of GCSF production in CHO culture is significantly higher than in E. coli system. Therefore, efforts have been applied to develop alternative expression systems for recombinant GCSF production, including plant- based.
Recombinant GCSF was expressed in plastids of lettuce (Sharifi et al., 2013) and in nuclear-transformed tobacco plants (Sharifi et al., 2012). The level of GCSF accumulation in these studies is not reported. The hG-CSF was also successfully expressed in suspension - cultured tobacco and rice cells. In tobacco BY-2 cells, recombinant hG-CSF was expressed at 17.8 mg/l, the hG-CSF accumulation has been determined in the tissues of initial transgenic callus lines (Nair et al., 2014). Hong et al. (2006) produced hG-CSF in suspension culture of transgenic rice cells (Oryza sativa cv. Dong-jin). The expression of recombinant hG-CSF in callus lines amounted up to 185 μg g−1 of callus FW. The recombinant GCSF was able to secrete into the culture medium, its accumulation reached 2.5 mg l−1 of medium. Recombinant hG-CSF obtained in both studies retained specific biological activity, as shown in cell assays.
In our study, the accumulation of hG-CSF in the most productive lines WG-04 and WG-32 reaches 0.2% of TSP. Such a level of recombinant protein expression (a few of tenths of a percent) is common for nuclear-transformed plants (Lico et al., 2012; Shahid and Daniell, 2016). However, due to the high content of total protein in Wolffia, the yield of hG-CSF amounted up to 35 mg kg−1 of fresh weight, and this is a fairly good level. It should be mentioned that the cultivation conditions of transgenic Wolffia plants have not yet been fully optimized, so under the optimal conditions the hG-CSF yield could be significantly increased. In addition, it is of great interest to study the Wolffia recombinant proteins secretion into the culture medium on the example of GCSF; hence this will be the aim of our further research.
In our experiments we have expressed the human G-CSF in nuclear-transformed Wolffia plants without impact on plant morphology or growth rate. The accumulation of target hG-CSF protein in the best transgenic line reached 35 mg kg−1 FW. In conclusion the present study distinctly demonstrates the feasibility of recombinant proteins expression in nuclear-transformed Wolffia plants and opens the way for development of Wolffia-based expression systems.
Author contributions
PK, AS, and MC: Obtaining transgenic lines of wolffia; AF, LS, and OK: Western blot analysis and ELISA of transgenic plants; AP Southern blot analysis; IT: Gene synthesis and plasmid construction; IC: Light microscopy; SD: Scientific adviser of this project.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The work was supported by a grant from the Russian Science Foundation (contract no. 14-50-00079).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2018.00304/full#supplementary-material
Appendix
acgcctctgggccccgcctcctccctcccccagtccttcctcctgaagtgtctggagcaggtcaggaagatccaaggcgatggagccgccctccaggagaagctgtgcgcgacctacaagctgtgccaccccgaggaactcgtgctcctgggccattctctcggcattccgtgggcccccctgtcgtcttgtccttcgcaagccctgcagctcgccggatgcctctcccagctgcactcgggactcttcctctaccagggactcctccaagcgctcgagggcatctctcccgaactcggacctacgctcgacaccctccaactggacgtggcggatttcgccacgaccatctggcaacaaatggaagaactcggaatggcgcccgccctgcagcccactcaaggggcgatgcccgcgttcgcctccgcctttcagcggcgggccgggggagtgctggtggcgtcgcacctccagtcttttctcgaagttagctatcgcgtcctcaggcatctcgcccaaccgtga
Nucleotide sequence of hG-CSF for expression in W. arrhiza (the sequence of the signal peptide is highlighted in yellow, stop codon is italicized).
References
- Armstrong W. P., Thorne R. F. (1984). The genus Wolffia (Lemnaceae) in California. Madrono 31, 171–179. [Google Scholar]
- Aycan M., Kayam M., Darcin E. S., Kom D., Yildiz M. (2014). Shoot regeneration from hypocotyl explants of flax (Linum usitatissimum L.) on Hormone-Free Growth Medium, in Conference: 3rd International Molecular Biology and Biotechnology Congress (Sarajevo: ), 47. [Google Scholar]
- Babalola C., Nightingale C. P., Nicolau D. (2004). Adjunctive efficacy of granulocyte colony-stimulating factor on treatment of Pseudomonas aeruginosa pneumonia in neutropenic and non-neutropenic hosts. J. Antimicrob. Chemother. 53, 1098–1100. 10.1093/jac/dkh237 [DOI] [PubMed] [Google Scholar]
- Bertran K., Thomas C., Guo X., Bublot M., Pritchard N., Regan J. T., et al. (2015). Expression of H5 hemagglutinin vaccine antigen in common duckweed (Lemna minor) protects against H5N1 high pathogenicity avian influenza virus challenge in immunized chickens. Vaccine 33, 3456–3462. 10.1016/j.vaccine.2015.05.076 [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Cox K. M., Sterling J. D., Regan J. T. (2006). Glycan optimization of a human monoclonal antibody in the aquatic plant Lemna minor. Nat. Biotechnol. 24, 1591–1597. 10.1038/nbt1260 [DOI] [PubMed] [Google Scholar]
- Dale D. C. (2010). Neutropenia and neutrophilia, in Williams Hematology, eds Lichtman M. A., Kipps T. J., Kaushansky K., Beutler B., Seligsohn U., Prchal J. T. (New York, NY: Mcgraw-Hill; ), 939–950. [Google Scholar]
- Daniell H., Streatfieldb S. J., Wycoff K. (2001). Medical molecular farming: production of antibodies, biopharmaceuticals and edible vaccines in plants. Trends Plant Sci. J. 6, 219–226. 10.1016/S1360-1385(01)01922-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S. L., Wood J., Hicks J. B. (1983). A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1, 19–21. 10.1007/BF02712670 [DOI] [Google Scholar]
- Demain A. L., Vaishnav P. (2009). Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 27, 297–306. 10.1016/j.biotechadv.2009.01.008 [DOI] [PubMed] [Google Scholar]
- Doley W. P., Saunders J. W. (1989). Hormone-free medium will support callus production and subsequent shoot regeneration from whole plant leaf explants in some sugarbeet (Beta vulgaris L.) populations. Plant Cell Rep. 8, 222–225. 10.1007/BF00778538 [DOI] [PubMed] [Google Scholar]
- Dolgov S., Mikhaylov R., Serova T., Shulga O., Firsov A. (2010). Pathogen-derived methods for improving resistance of transgenic plums (Prunus domestica L.) for Plum pox virus infection. Julius Kühn Arch. 427, 133–140. [Google Scholar]
- Dolgov S. V., Chernobrovkina M. A., Khvatkov P. A. (2013). Composition of Medium for Culturing Duckweed Plants (Wolffia arrhiza) in Vitro. Russian Patent RU2472338C1. Moscow: Rospatent. [Google Scholar]
- Dolgova A. S., Dolgov S. V., Nazipova N. N., Maksimenko O. G., Georgiev P. G. (2015). Arabidopsis termination elements increase transgene expression in tobacco plants. Plant Cell Tissue Organ Cult. 120, 1107–1116. 10.1007/s11240-014-0667-1 [DOI] [Google Scholar]
- Firsov A., Tarasenko I., Mitiouchkina T., Ismailova N., Shaloiko L., Vainstein A., et al. (2015). High-yeld expression of M2e peptide of avian influenza virus H5N1 in transgenic duckweed plants. Mol. Biotechnol. 57, 653–661. 10.1007/s12033-015-9855-4 [DOI] [PubMed] [Google Scholar]
- Firsov A., Tarasenko I., Mitiouchkina T., Shaloiko L., Kozlov O., Vinokurov L., et al. (2018). Expression and immunogenicity of M2e peptide of avian influenza virus H5N1 fused to ricin toxin b chain produced in duckweed plants. Front Chem. 6:22. 10.3389/fchem.2018.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasdaska J. R., Spenser D., Dickey L. (2003). Advantages of therapeutic protein production in the aquatic plant Lemna. Biol. Process. J. 2, 49–56. 10.12665/J22.Gasdaska [DOI] [Google Scholar]
- Georgiev M. I., Agostini E., Ludwig-Müller J., Xu J. (2012). Genetically transformed roots: from plant disease to biotechnological resource. Trends Biotechnol. 30, 528–537. 10.1016/j.tibtech.2012.07.001 [DOI] [PubMed] [Google Scholar]
- Górecka K., Krzyzanowska D., Kiszczak W., Kowalska U. (2009). Plant regeneration from carrot (Daucus carota L.) anther culture derived embryos. Acta Physiol. Plant. 31, 1139–1145. 10.1007/s11738-009-0332-1 [DOI] [Google Scholar]
- Harousseau J. L., Witz B., Lioure B., Berger M., Desablens B., Delain M., et al. (2000). Granulocyte colony-stimulating factor after intensive consolidation chemotherapy in acute myeloid leukemia: results of a randomized trial of the group Ouest-Est leucémies aigues myeloblastiques. J. Clin. Oncol. 18, 780–787. 10.1200/JCO.2000.18.4.780 [DOI] [PubMed] [Google Scholar]
- Hong S. Y., Kwon T. H., Jang Y. S., Kim S. H., Yang M. S. (2006). Production of bioactive human granulocyte-colony stimulating factor in transgenic rice cell suspension. Protein Express Purif. 47, 68–73. 10.1016/j.pep.2005.09.028 [DOI] [PubMed] [Google Scholar]
- Hood E. E., Gelvin S. B., Melchers L. S., Hoekema A. (1993). New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2, 208–218. 10.1007/BF01977351 [DOI] [Google Scholar]
- Jensen O. N. (2006). Interpreting the protein language using proteomics. Nat. Rev. Mol. Cell Biol. 7, 391–403. 10.1038/nrm1939 [DOI] [PubMed] [Google Scholar]
- Kaufman J. (2011). The economic potential of plant-made pharmaceuticals in the manufacture of biologic pharmaceuticals. J. Commer. Biotechnol. 17, 173–182. 10.1057/jcb.2010.37 [DOI] [Google Scholar]
- Khvatkov P. A., Chernobrovkina M. A., Sinyov V. V., Dolgov S. V. (2013). Study on conditions for Wolffia arrhiza (L.) Horkel ex Wimm submerged culturing in a modified bioreactor. Biotechnology 6, 51–56. [Google Scholar]
- Khvatkov P., Chernobrovkina M., Okuneva A., Pushin A., Dolgov S. (2015a) Transformation of Wolffia arrhiza (L.) Horkel ex Wimm. Plant Cell Tissue Organ Cult. 123, 299–307. 10.1007/s11240-015-0834-z [DOI] [Google Scholar]
- Khvatkov P., Chernobrovkina M., Okuneva A., Shvedova A., Chaban I., Dolgov S. (2015b) Callus induction regeneration in Wolffia arrhiza (L.) Horkel ex Wimm. Plant Cell Tissue Organ Cult. 120, 263–273. 10.1007/s11240-014-0603-4 [DOI] [Google Scholar]
- Lico C., Santi L., Twyman R. M., Pezzotti M., Avesani L. (2012). The use of plants for the production of therapeutic human peptides. Plant Cell Rep. 31, 439–451. 10.1007/s00299-011-1215-7 [DOI] [PubMed] [Google Scholar]
- Lorimer D., Raymond A., Walchli J., Mixon M., Barrow A., Wallace E., et al. (2009). Gene Composer: database software for protein construct design, codon engineering, and gene synthesis. BMC Biotechnol. 9:36. 10.1186/1472-6750-9-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K., Kodayasunori Y., Okazawa K. (2006). Callus formation and embryogenesis of endosperm tissues of parsley seed cultured on hormone-free medium. Physiol. Plant. 41, 135–138. 10.1111/j.1399-3054.1977.tb05544.x [DOI] [Google Scholar]
- Merlin M., Gecchele E., Capaldi S., Pezzotti M., Avesani L. (2014). Comparative evaluation of recombinant protein production in different biofactories: the green perspective. Biomed Res. Int. 2014:136419 10.1155/2014/136419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroshnichenko D., Chaban I., Chernobrovkina M., Dolgov S. (2017). Protocol for efficient regulation of in vitro morphogenesis in einkorn (Triticum monococcum L.), a recalcitrant diploid wheat species. PLoS ONE. 12:e0173533 10.1371/journal.pone.0173533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair N. R., Chidambareswaren M., Manjula S. (2014). Enhanced heterologous expression of biologically active human granulocyte colony stimulating factor in transgenic tobacco BY-2 cells by localization to endoplasmic reticulum. Mol. Biotechnol. 56, 849–862. 10.1007/s12033-014-9765-x [DOI] [PubMed] [Google Scholar]
- Rader R. A., Langer E. S. (2015). Biopharmaceutical manufacturing: historical and future trends in titers, yields, and efficiency in commercial-scale bioprocessing. Bioprocess J. 13, 47–54. 10.12665/J134.Langer [DOI] [Google Scholar]
- Ria R., Gasparre T., Mangialardi G., Bruno A., Iodice G., Vacca A., et al. (2010). Comparison between filgrastim and lenograstim plus chemotherapy for mobilization of PBPCs. Bone Marrow Transplant. 45, 277–281. 10.1038/bmt.2009.150 [DOI] [PubMed] [Google Scholar]
- Richter L. J., Thanavala Y., Arntzen C. J., Mason H. S. (2000). Production of hepatitis B surface antigen in transgenic plants for oral immunization. Nat. Biotechnol. 18, 1167–1171. 10.1038/81153 [DOI] [PubMed] [Google Scholar]
- Rouillard J. M., Lee W., Truan G., Gao X., Zhou X., Gulari E. (2004). Gene2Oligo: oligonucleotide design for in vitro gene synthesis. Nucleic Acids Res. 32, 176–180. 10.1093/nar/gkh401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybczynski J., Badzian T. (1987). High regeneration potential of root segments of Lotus corniculatus L. seedlings on hormone free media. Plant Sci. 51, 239–244. 10.1016/0168-9452(87)90198-1 [DOI] [Google Scholar]
- Santos R. B., Abranches R., Fischer R., Sack M., Holland T. (2016). Putting the spotlight back on plant suspension cultures. Front. Plant Sci. 7:297. 10.3389/fpls.2016.00297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sari N., Dalva K., Ilhan I. E. (2013). Comparison of filgrastim and lenograstim in pediatric solid tumors. Pediatr. Hematol. Oncol. 7, 655–661. 10.3109/08880018.2013.828144 [DOI] [PubMed] [Google Scholar]
- Schenk R. U., Hildebrandt A. C. (1972). Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant cell cultures. Can. J. Bot. 50, 199–204. 10.1139/b72-026 [DOI] [Google Scholar]
- Serova I. A., Dvoryanchikov G. A., Andreeva L. E., Burkov I. A., Dias L. P. B., Battulin N. R., et al. (2012). A 3,387 bp 50-flanking sequence of the goat alpha-S1-casein gene provides correct tissue-specific expression of human granulocyte colony-stimulating factor (hG-CSF) in the mammary gland of transgenic mice. Transgenic Res. 21, 485–498. 10.1007/s11248-011-9547-1 [DOI] [PubMed] [Google Scholar]
- Shahid N., Daniell H. (2016). Plant based oral vaccines against zoonotic and non zoonotic diseases. Plant Biotechnol. J. 14, 2079–2099. 10.1111/pbi.12604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi T. M., Habashi A. A., Rajabi M. H. (2013). Human granulocyte colony-stimulating factor (hG-CSF) expression in plastids of Lactuca sativa. Iran. Biomed. J. 17, 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi T. M., Solouki M., Tohidfar M., Sadeghizadeh M. (2012). Expression of human granulocyte-colony stimulating factor (hG-CSF) gene in tobacco (Nicotiana tabacum). Austr. J. Crop Sci. 6, 135–140. [Google Scholar]
- Sharma A. K., Sharma M. K. (2009). Plants as bioreactors: recent developments and emerging opportunities. Biotechnol. Adv. 27, 811–832. 10.1016/j.biotechadv.2009.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R. (2016). Plant protein glycosylation. Glycobiology 26, 926–939. 10.1093/glycob/cww023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Cheng J. J., Himmel M. E., Skory C. D., Adney W. S., Thomas S. R., et al. (2007). Expression and characterization of Acidothermus cellulolyticus E1 endoglucanase in transgenic duckweed Lemna minor 8627. Bioresour. Technol. 98, 2866–2872. 10.1016/j.biortech.2006.09.055 [DOI] [PubMed] [Google Scholar]
- Sysuev B. B., Pokrovskaya J. S. (2015). Recombinant microorganisms and cell culture in the technology of protein preparations. Dev. Regist. Drugs 4, 96–109. [Google Scholar]
- Tschofen M., Knopp D., Hood E., Stöger E. (2016). Plant molecular farming: much more than medicines. Annu. Rev. Anal. Chem. 9, 271–294. 10.1146/annurev-anchem-071015-041706 [DOI] [PubMed] [Google Scholar]
- Vacchelli E., Eggermont A., Fridman W. H., Galon J., Zitvogel L., Kroemer G., et al. (2013). Trial Watch: immunostimulatory cytokines. Oncoimmunology 2:e24850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh G. (2014). Biopharmaceutical benchmarks 2014. Nature Biotechnol. 32, 992–1002. 10.1038/nbt.3040 [DOI] [PubMed] [Google Scholar]
- Wayne A. P., Thorne R. F. (1984). The genus Wolffia (Lemnaceae) in California. Madro-o 31, 171–179. [Google Scholar]
- Wilken L. R., Nikolov Z. L. (2012). Recovery and purification of plant-made recombinant proteins. Biotechnol Adv. 30, 419–433. 10.1016/j.biotechadv.2011.07.020 [DOI] [PubMed] [Google Scholar]
- Wolff P. (1992). Les lentilles d'eau de l'Alsace. – Bull. Assoc. Amis Jard.bot. Col de Saveme, in 60e anniversaire 1932/1992 (Saverne: ), 25–33. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.