Abstract
The biological relevance of non-protein coding RNAs in the regulation of critical plant processes has been firmly established in recent years. This has been mostly achieved with the discovery and functional characterization of small non-coding RNAs, such as small interfering RNAs and microRNAs (miRNAs). However, recent next-generation sequencing techniques have widened our view of the non-coding RNA world, which now includes long non-coding RNAs (lncRNAs). Small and lncRNAs seem to diverge in their biogenesis and mode of action, but growing evidence highlights their relevance in developmental processes and in responses to particular environmental conditions. Light can affect MIRNA gene transcription, miRNA biogenesis, and RNA-induced silencing complex (RISC) activity, thus controlling not only miRNA accumulation but also their biological function. In addition, miRNAs can mediate several light-regulated processes. In the lncRNA world, few reports are available, but they already indicate a role in the regulation of photomorphogenesis, cotyledon greening, and photoperiod-regulated flowering. In this review, we will discuss how light controls MIRNA gene expression and the accumulation of their mature forms, with a particular emphasis on those miRNAs that respond to different light qualities and are conserved among species. We will also address the role of small non-coding RNAs, particularly miRNAs, and lncRNAs in the regulation of light-dependent pathways. We will mainly focus on the recent progress done in understanding the interconnection between these non-coding RNAs and photomorphogenesis, circadian clock function, and photoperiod-dependent flowering.
Keywords: microRNAs, long non-coding RNAs, light, circadian clock, photoperiod
Introduction
Light is fundamental for plant life. It not only provides the energy necessary for photosynthesis but also functions as an indicator of time and place. Light quality and duration inform a plant about its neighbors in a process described as shade avoidance, which ultimately shapes its architecture and development. Light cues also function as indicators of time of the day and season [e.g., short days (SDs) in autumn and winter versus long days (LDs) in spring and summer]. Last but not least, light, together with temperature, is an input to the circadian clock, providing circadian entrainment and contributing to maintain rhythmicity and robustness of the clock. Proper clock resetting is critical for the adequate timing of biological processes, which will then reflect in plant fitness.
In parallel with seasonal and positional information, light also provides important developmental cues. For instance, during photomorphogenesis, when the young seedling emerges through the soil reaching for light, hypocotyl elongation is inhibited, while roots grow, cotyledons open and expand, and the first true leaves develop (Leivar and Monte, 2014). These morphological changes are accompanied by the development of chloroplasts and chlorophyll accumulation. Later in development, light will also regulate the time of flowering. Depending on each species requirements, specific photoperiods (the relative duration of the daily light and dark periods) will provide molecular cues, such as the accumulation of specific regulators, required for the transition of shoot apical meristems into inflorescence meristems (Andrés and Coupland, 2012).
Therefore, considering all light-dependent and/or light-regulated processes, it is not surprising that light signals promote massive transcriptional changes affecting both protein coding genes as well as non-protein coding transcripts (López-Juez et al., 2008; Wang H. et al., 2014). Although light regulation of (protein-coding) gene expression has been studied in extreme detail, the role of light in the regulation of non-protein coding RNAs is less known.
Non-coding RNAs have been divided into different functional groups based on the size of their active forms. Non-coding RNAs shorter than 200 base pairs (bp) fall within the category of small non-coding RNAs, whereas longer ones constitute long non-coding RNAs (lncRNAs). Historically, small non-coding RNAs were the first identified and characterized in great detail. Within this group, microRNAs (miRNAs) are transcribed from MIRNA genes as longer precursors (primary miRNAs, pri-miRNAs) by RNA polymerase II. Plant pri-miRNAs fold into stem-loop structures and are processed by a complex containing DICER-LIKE1 (DCL1), HYPONASTIC LEAVES1 (HYL1), and SERRATE (SE), which will first release the stem-loop (pre-miRNA) structure, and then cleave it giving rise to a mature miRNA:miRNA∗ duplex, normally 20–22 nucleotides (nt) long, with the rarer cases of 23–25 nt, with 2-nt 3′ overhangs (Voinnet, 2009; Borges and Martienssen, 2015; Yu et al., 2017). This duplex is methylated at its 3′ ends by the methyltransferase HUA ENHANCER1 (HEN1) and this methylation is necessary to stabilize miRNAs (Yu et al., 2005). The mature miRNA strand is then selectively bound by ARGONAUTE1 (AGO1) and loaded into the RNA-induced silencing complex (RISC), which recognizes transcripts with full or partial complementarity to the miRNA and cleaves and/or represses translation of these miRNA target transcripts, therefore silencing them (Voinnet, 2009; Yu et al., 2017) (Figure 1). miRNAs act in particular developmental processes and/or in response to certain environmental cues, sometimes in specific tissues where they accumulate at lower levels than their targets, protein-coding transcripts.
FIGURE 1.
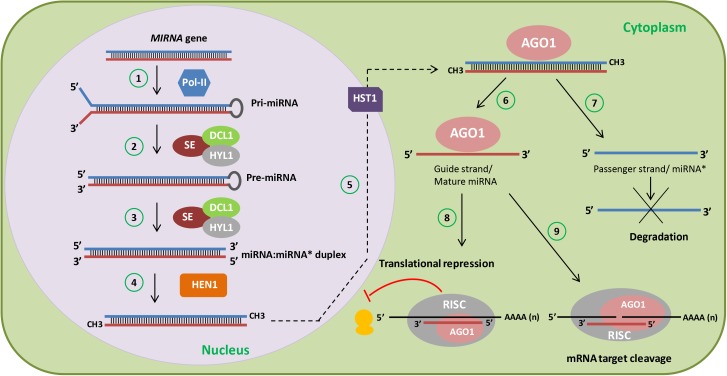
miRNA biogenesis in plants. Cartoon depicting a simplified scheme of miRNA biogenesis in plants. MIRNA genes are first transcribed by RNA polymerase II (Pol-II) into primary-miRNAs (pri-miRNAs) (1). Then, the dicing complex involving DICER-LIKE1 (DCL1), HYPONASTIC LEAVES1 (HYL1), and SERRATE (SE) generate the precursor-miRNA (pre-miRNA) (2) after capping, polyadenylation, and splicing. Subsequent cleavage of the pre-miRNA gives rise to the miRNA:miRNA∗ duplex (3). Afterward, HUA ENHANCER 1 (HEN1) stabilizes the miRNA:miRNA∗ duplex through 3′ methylation (4) and HASTY1 (HST1) (5) exports it to the cytoplasm. During ARGONAUTE1 (AGO1) loading, one strand is selected (guide strand or mature miRNA) (6) and the other strand (passenger strand or miRNA∗) is degraded (7). The miRNA strand is then loaded into the RNA-induced silencing complex (RISC) containing AGO1, which recognizes transcripts with complementarity to the miRNA and represses the translation (8) or cleaves (9) these miRNA targets.
lncRNAs, on the other hand, can exert their biological function without further processing and accumulate to similar levels as miRNAs (Liu et al., 2012). lncRNAs are classified according to their genomic location into intergenic (expressed in between coding regions), intronic (expressed from introns), or natural antisense transcripts (NATs, expressed from the complementary strand to a protein-coding gene or another lncRNA; Liu et al., 2012; Wang H. et al., 2014). Comprehensive screening approaches have revealed thousands of lncRNAs that display spatial-, temporal-, and developmental-specific patterns of expression. Interestingly, among the few lncRNAs whose biological function is known, two are involved in light-regulated processes. HIDDEN TREASURE 1 (HID1) is involved in photomorphogenesis and seedling greening (Wang Y. et al., 2014; Wang et al., 2018), whereas CDF5 LONG NON-CODING RNA (FLORE) mediates the circadian regulation of flowering time (Henriques et al., 2017).
Here, we review the role of miRNAs and lncRNAs associated with, or regulated by light-dependent pathways. We will discuss light regulation of miRNA expression, biogenesis, processing, and function, as well as their role in light-related processes, such as photomorphogenesis and photoperiod-dependent flowering. We will also discuss the available reports characterizing lncRNA function in light-dependent pathways.
MicroRNAs
Light regulation of plant miRNAs can occur through at least three different mechanisms: regulation of MIRNA gene transcription and its levels, regulation of miRNA processing via effects on components of the miRNA biogenesis machinery, and regulation of miRNA function through control of RISC factors. In turn, miRNAs can target genes encoding components of the light signaling pathways, thereby affecting light responses such as photomorphogenesis and photoperiod-dependent flowering. Below we discuss the recent progresses made in these fields of research. Due to the interconnection between light and the circadian clock, we will also discuss the available reports on circadian-regulated ncRNAs.
Light Regulation of miRNA Levels
Light, photoperiod, and the circadian clock modulate the expression levels of MIRNA genes. This regulation is achieved by the presence of light-responsive elements in their promoters, and can be triggered by specific light qualities (red, blue, far-red, UV-A, and UV-B). To identify the miRNA families under this control, several screening approaches have been developed, resulting in extensive listings of light-responsive miRNAs.
For instance, the comparison of miRNAs between wild-type rice (Oryza sativa) plants and a phytochrome B (phyB) mutant identified 135 differentially expressed miRNAs, of which 97 were upregulated and 38 were downregulated. These miRNAs include conserved and novel miRNAs, with sizes varying between 21-nt and 24-nt, such as miR156, miR166, miR171, and miR408 (Sun et al., 2015). Differently from a previous report showing that miR172 is regulated by PHYB in potato (Martin et al., 2009), Sun et al. (2015) did not find any miR172 family member that responded to PHYB in rice, suggesting that PHYB-mediated regulation of miRNAs may differ between plant species. Using degradome sequencing, it was found that 70 rice genes were targeted by 32 differentially expressed miRNAs between the wild-type and the phyB mutant (Sun et al., 2015). A large proportion of them (42%) encode transcription factors, suggesting that regulation of gene expression by miRNA target genes may play an important role in PHYB-mediated light signaling. Members of the transcription factor families identified by Sun et al. (2015) are involved in light signaling in Arabidopsis. It is worth noting that one of the identified miRNA target genes is a homolog of the Arabidopsis circadian-regulated TANDEM ZINC KNUCKLE PROTEIN (TZP) gene, which is involved in blue light-associated growth (Loudet et al., 2008). However, further research is required to fully understand the role of these rice miRNAs and target genes in light-regulated processes (Sun et al., 2015).
Moreover, ELONGATED HYPOCOTYL 5 (HY5), a major regulator of photomorphogenesis that acts downstream of several photoreceptors, was shown to bind to the promoters and regulate the expression of several MIRNA genes, such as MIR156D, MIR402, MIR408, and MIR858A in Arabidopsis (Zhang et al., 2011). This, together with the effect of PHYB on miRNA levels (Sun et al., 2015), strongly suggests that light affects miRNA abundance. In support of this, small RNA sequencing indicates that the miRNA population of the seedling apical hook changes after treating dark-grown soybean (Glycine max) seedlings with far-red light. The levels of several miRNAs (miR166, miR167, miR168, miR394, miR396, miR530, miR1507, miR1508, miR1509, and miR2218) respond to far-red light in different parts of the soybean seedling, in most cases showing upregulation by far-red light (Li et al., 2014). Therefore, light seems to upregulate miRNAs in the apical hook, leading to downregulation of target genes that presumably repress apical hook and cotyledon opening.
Light has strong effects on plant metabolism, especially in the regulation of fundamental plant processes such as photosynthesis (Kooke and Keurentjes, 2012). To understand the involvement of miRNAs in light regulation of potato metabolism, Qiao et al. (2017) used high-throughput sequencing to study the expression levels of miRNAs and mRNAs in dark-treated and red-light-treated leaves and detached tuber skin in potato. Among the 69 known and novel miRNAs that were differentially regulated, most were upregulated in leaves, whereas in tuber skin, about half of them were upregulated and half downregulated (Qiao et al., 2017). Notably, the miR399 family and the novel miRNA miRn55 showed opposite light responses in leaves (upregulation) and tuber skin (downregulation). General gene expression analyses revealed that most differentially expressed mRNAs (67%) were upregulated in tuber skin, whereas in leaves, about half were upregulated and half downregulated. Only 14% of differentially expressed miRNAs and 12% of differentially expressed genes were common to leaves and tuber skin, indicating that the molecular response to red light differs between these tissues. Nevertheless, there was an effect of light on genes involved in primary and secondary metabolism in both tissues, especially in carbohydrate metabolism, which is downregulated, and alkaloid metabolism, which is upregulated. Repression of carbohydrate metabolism was supported by anti-correlation between miRNAs and mRNAs, which suggests downregulation of putative miRNA targets involved in this process (Qiao et al., 2017). Although confirmation of these findings using additional methods is required, this work points to a role of miRNAs in the regulation of metabolic pathways affected by red light in potato.
Additional support to the effect of red light on miRNA levels has been obtained in Arabidopsis. In this species, the most differentially expressed miRNAs in red light versus darkness are miR160, miR163, miR319, miR394, miR779, miR851, miR854, and miR2111 (Sun et al., 2018). At least part of this effect is mediated by PHYTOCHROME-INTERACTING FACTOR4 (PIF4), a transcription factor that is a negative regulator of PHYB-mediated red light signaling. In a pif4 mutant, 22 mature miRNAs, representing 16 miRNA families, show altered levels relative to wild-type plants under red light. PIF4 promotes the expression of genes encoding miR156/157, miR160, miR165/166, miR167, miR170/171, and miR394 and represses the expression of genes encoding miR172 and miR319 by binding to the promoters of several of these genes. In addition to acting as a transcription factor for MIRNA genes, PIF4 regulates miRNA processing, as discussed below (Sun et al., 2018).
Besides light exposure, photoperiod can also control miRNA accumulation. Li et al. (2015) used high-throughput sequencing to identify miRNAs regulated by photoperiod in soybean. Comparing miRNA levels in seedlings grown under LDs and SDs revealed 37 miRNA families to be day-length-responsive. Five members of the miR156 family were induced under LD conditions and this correlated with downregulation of four miR156-target SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) genes. Given that miR156 overexpression delays flowering in soybean (Cao et al., 2015), lower miR156 levels under SD than LD may explain the fact that soybean flowering is induced by SD. Conversely, the expression of six members of the miR172 family was induced under SD conditions and this was coordinated with the repression of ten predicted targets belonging to the APETALA2 (AP2)-like family (Li et al., 2015). In addition to miR156 and miR172, other conserved miRNAs, such as miR159/319, miR166, miR167, miR395, and miR408 were also shown to be photoperiod regulated in soybean.
The simplified model of the circadian clock places light as an input that confers clock entrainment. Similarly to light, the clock also controls the expression of a vast set of genes, especially at the dark/light transition (Michael et al., 2008), thus allowing plants to anticipate certain daily processes. Interestingly, the circadian clock can also regulate the expression of protein-coding as well as non-coding transcripts (Hazen et al., 2009). To uncover circadian-regulated miRNAs, Siré et al. (2009) compared mature and pri-miRNA transcript levels under light/dark cycles or under free-running (continuous light) conditions for two consecutive days. It was found that miR167, miR168, miR171, and miR398 levels oscillated under light/dark cycles, peaking around the light/dark transition. However, for certain miRNAs, the waving pattern was not accompanied by a clear antiphasic expression of their targets but rather by a phase delay. This was the case of miR171/SCARECROW-LIKE 6 (SCL6, also named SCL6-IV, HAM3, and LOM3) and miR398/Cu/Zn SUPEROXIDE DISMUTASE 2 (CSD2) pairs, where target gene expression is reduced as the miRNA levels increase. However, in the case of the miR168/AGO1 and miR167/AUXIN RESPONSE FACTOR 6 (ARF6) pairs, an in-phase pattern of accumulation was found, suggesting that miRNAs could regulate their targets by incorporating feedback loops. However, when these modules were tested under free-running conditions, a clear oscillation was not found, an indication that these miRNAs are most likely light regulated.
In another report, Li et al. (2016) addressed the mechanism by which the circadian clock regulates carbon and nitrogen metabolism in rice plants grown in field conditions. Using a comprehensive approach, the authors investigated how light/dark cycles control certain biological processes. They investigated metabolic and enzymatic activities, as well as gene expression and alternative splicing events that could show rhythmic behavior. In their search for oscillatory miRNAs associated with carbon and/or nitrogen metabolism, Li et al. (2016) uncovered miR1440b, miR1877, miR2876-5p, and miR5799 as possible rice miRNA candidates. These miRNAs oscillate under light/dark cycles and negatively correlate with their targets at the majority of time points analyzed. However, it still remains to be seen whether these are bona fide circadian miRNAs oscillating under free-running conditions.
UV-Responsive miRNAs
Since different light qualities can trigger diverse developmental responses, some of which can be detrimental, several screening approaches have been designed to understand how light, especially high light, UV-A, and UV-B radiation can modulate accumulation of miRNAs and their targets. Zhou et al. (2016), using seedlings from Brassica rapa exposed to blue light and UV-A, identified 15 conserved and 226 novel differentially expressed miRNAs that could target genes encoding regulators of plant growth, development, and photomorphogenesis (Zhou et al., 2016). Blue light and UV-A exposure slightly downregulated miR156/157 expression, which correlated with upregulation of their targets, SPL9 (Bra004674) and SPL15 (Bra003305), which encode transcription factors involved in the regulation of many processes, including the juvenile-to-adult transition, flowering, and secondary metabolism, especially anthocyanin biosynthesis (Wu et al., 2009; Gou et al., 2011; Wang and Wang, 2015). In Arabidopsis, miR156 and SPLs are involved in complex feedback loops (Wu et al., 2009; Yant et al., 2010). As plants age, miR156 levels decrease and SPLs (e.g., SPL9) accumulate, inhibiting anthocyanin biosynthesis (Gou et al., 2011). Zhou et al. (2016) hypothesize that blue and UV-A light activate genes involved in light signaling and some of them promote anthocyanin biosynthesis. When anthocyanin levels increase, then the regulatory feedback loops involving miR156/157 would act to balance its metabolism.
Similar to UV-A, UV-B radiation affects plant growth, physiology, and metabolism (Dotto and Casati, 2017). In one of the first reports on the effect of light on miRNAs, Zhou et al. (2007) used a computational approach to predict MIRNA genes induced by UV-B light in Arabidopsis. miRNAs belonging to 11 miRNA families were identified as putatively upregulated by UV-B light (Zhou et al., 2007). Although these results have not been experimentally confirmed in Arabidopsis, several reports have shown the effect of UV-B on miRNA levels in aspen (Populus tremula), maize (Zea mays), and wheat (Triticum aestivum). Jia et al. (2009) found that the level of eight miRNA families increased and that of seven families decreased in response to UV-B light stress in aspen. In maize, from the 16 UV-B-regulated miRNA families detected, 6 were upregulated and 10 downregulated (Casati, 2013). UV-B-treated wheat plants showed upregulation of three and downregulation of another three known miRNAs, in addition to a novel wheat miRNA, which was slightly upregulated shortly after UV-B exposure and then was downregulated (Wang et al., 2013). Thirteen UV-B-responsive miRNA families, i.e., miR156/157, miR159, miR160, miR164, miR165/166, miR167, miR169, miR170/171, miR172, miR393, miR395 miR398, and miR399, are common to at least two plant species, indicating substantial conservation in the miRNAs that respond to UV-B light. Although the direction of the effect (upregulation or downregulation) on several miRNAs differs between species, others are consistently promoted (miR165/166, miR167, and miR398) or repressed (miR395) by UV-B light in three species. This conservation probably reflects important roles of these miRNAs in plant responses to UV-B radiation.
The upstream regulatory regions of the UV-B-regulated MIRNA genes contain numerous light-related motifs, such as GT-1 sites, G boxes, I-boxes, CCAAT-boxes, and GATA-boxes (Zhou et al., 2007; Jia et al., 2009; Wang et al., 2013), consistent with the effect of light on these genes. Stress-responsive cis-elements were also found in several of those promoters (Jia et al., 2009; Wang et al., 2013). Given that some of these miRNAs are involved in responses to abiotic and biotic stresses (Casati, 2013; Li et al., 2017), we hypothesize that these miRNAs might coordinate adaptive responses to diverse threats.
Among the targets of the UV-B-regulated miRNAs are transcription factors, several factors involved in auxin signaling and factors involved in responses to other stresses (Zhou et al., 2007; Jia et al., 2009; Casati, 2013). In most cases, as would be expected, there is an inverse correlation between the effect of UV-B on miRNAs and on their corresponding target transcripts (Zhou et al., 2007; Jia et al., 2009; Casati, 2013; Wang et al., 2013). For instance, in maize, an increase in miR165/166 correlated with inhibition of their target gene, encoding a HD-ZIP III transcription factor, ROLLED LEAF1 (RLD1; Casati, 2013). In agreement with the role of these miRNAs in delimitating HD-ZIP III transcripts to the adaxial side of leaf primordia (Juarez et al., 2004), UV-B-dependent repression of RLD1 was associated with leaf arching, suggesting a role for miR165/166 in this UV-B response (Casati et al., 2006). In aspen and wheat, miR395 downregulation is associated with upregulation of its targets encoding ATP sulfurylases (APS), enzymes involved in inorganic sulfate assimilation (Jia et al., 2009; Wang et al., 2013). miR395 and APS are inversely regulated by sulfate starvation, and miR395 overexpression reduces APS transcript levels and affects the response of Arabidopsis to abiotic stress conditions (Jones-Rhoades and Bartel, 2004; Kim et al., 2010), linking again a UV-B-regulated miRNA with other stress responses.
This is also the case of miR164, whose downregulation in UV-B-treated maize leaves is associated with increased levels of two stress-responsive NAC-DOMAIN PROTEIN target transcripts, as well as EXOSTOSIN PROTEIN-LIKE and an ASPARTYL PROTEASE (Hegedus et al., 2003; Casati, 2013). Interestingly, other miRNAs (miR171, miR172, miR156, and miR529) that target genes involved in critical developmental transitions (juvenile-to-adult transition and/or flowering) were also downregulated by UV-B light (Lauter et al., 2005; Xie et al., 2006; Chuck et al., 2008; Cuperus et al., 2011). This suggests that UV-B exposure promotes miRNA-mediated responses that delay certain developmental transitions, allowing plants either to repair UV-B-induced damage or to adapt to these stressful conditions.
Overall, the analysis of UV-B-regulated miRNAs in several plant species indicates that there is a core set of conserved UV-B-responsive miRNAs. Nevertheless, the findings also suggest that different species may recruit distinct miRNAs in order to cope with UV-B stress. Understanding the role of these miRNAs in UV-B-regulated processes awaits further investigation.
miRNAs Accumulated in High Light Conditions
Besides UV light, high light can also be detrimental for the development of different plant species. To address how light, especially high light, controls miRNA expression in ma bamboo (Dendrocalamus latiflorus) grown under LD conditions, Zhao et al. (2013) generated small RNA libraries and determined how miRNA families were regulated. Several conserved miRNAs were identified, miR168 being the most abundant family, followed by miR156/157, miR535, miR165/166, and miR167. Interestingly, miR172 was not found, probably due to the unique flowering cycle of ma bamboo. Using stem-loop RT-qPCR, Zhao et al. (2013) determined the level of novel miRNAs in plants grown under regular white light, high light, or dark. However, the role of these miRNAs in high-light-mediated processes still remains to be determined.
A global analysis of the light-regulated miRNAs reported so far reveals that numerous miRNA families are affected by diverse light conditions (Table 1). Some of them, such as miR156/157, miR159/319, miR164, miR165/miR166, miR167, miR170/171, miR172, miR395, miR398, miR399, and miR408, have been identified as responsive to several light conditions in at least five plant species, strongly suggesting that they may play roles in light responses (Table 1). However, light could also modulate miRNA processing and/or activity toward its targets. Below we present the most recent reports addressing this type of regulation.
Table 1.
miRNA families differentially regulated by light.
In this table, we present a summary of the different miRNAs that respond to two or more light treatments. 1miRNA families reported to be regulated by light in at least two papers have been included. miRNA families are ordered according to the number of papers reporting their light regulation. 2W, white light; FR, far-red light; R, red light; B, blue light; L/D, light/dark; LD/SD, long days/short days. 3Ath, Arabidopsis thaliana; Gma, Glycine max (soybean); Mdo, Malus x domestica (apple); Osa, Oryza sativa (rice); Ppy, Pyrus pyrifolia (Chinese sand pear); Ptr, Populus tremula (aspen); Stu, Solanum tuberosum (potato); Tae, Triticum aestivum (wheat); Zma, Zea mays (maize).
Regulation of miRNA Biogenesis, Processing, and Function by Light
Besides regulating MIRNA gene expression, light can also modulate the levels and the activity of mature miRNAs. This can be achieved by a direct connection between light and regulators of the miRNA biogenesis pathway. HYL1 is a RNA-binding protein involved in miRNA processing (Yu et al., 2017). In Arabidopsis, HYL1 protein levels are regulated by CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), an E3 ubiquitin ligase that mediates the proteasomal degradation of light signaling factors, such as photoreceptors (Cho et al., 2014). It was found that the reduction of miRNA levels in cop1 mutants relative to wild-type plants greatly overlapped with that observed in the hyl1 mutant. Further analyses showed that COP1 regulates HYL1 protein levels but not through direct ubiquitination. Combining protein synthesis blockers with either autophagy or 26S proteasome inhibitors revealed that HYL1 was degraded by an unknown protease, which removed its N-terminal fragment, where two RNA-binding domains, essential for miRNA processing, are located (Cho et al., 2014). Light regulates COP1 nucleocytoplasmic shuttling (Jang et al., 2010) and, therefore, during the day, light stabilizes HYL1 due to cytoplasmic accumulation of COP1, which would target the protease acting on HYL1 degradation (Figure 2). This regulation could control miRNA processing and thus help understand the light/dark accumulation pattern of several mature miRNAs, such as miR167, miR168, miR171, and miR398, previously described by Siré et al. (2009).
FIGURE 2.
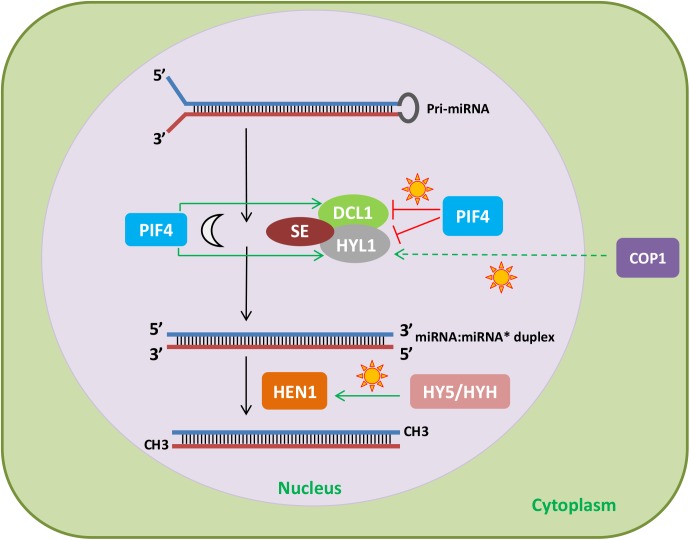
Effect of light signaling components on the miRNA biogenesis and processing machinery. The light signaling factors COP1 and PIF4 regulate the stability of HYL1. In the light, COP1, probably through degradation of an unknown protease, increases HYL1 protein levels. In addition, PIF4 stabilizes HYL1 and DCL1 in the dark and destabilizes them under red light. Finally, HY5 and HYH promote HEN1 expression in response to light. Therefore, light can affect miRNA levels by regulating light signaling components that modulate the abundance of miRNA biogenesis and processing factors.
HYPONASTIC LEAVES1 protein levels are also controlled by PIF4, another protein involved in photomorphogenesis (Sun et al., 2018). Both HYL1 and DCL1 interact with PIF4, which destabilizes them under red light and stabilizes them in the dark. The mechanism by which PIF4 controls the levels of HYL1 and DCL1 is unknown, but it does not involve transcriptional regulation or the ubiquitin-proteasome pathway. Given that PIF4 interacts with the photoreceptor PHYB and the miRNA processing factors DCL1 and HYL1, the question arises as to whether DCL1 and HYL1 affect light responses (Figure 2). Indeed, dcl1 and hyl1 mutants show shorter hypocotyls than wild-type plants under red light, indicating that DCL1 and HYL1 play negative roles in photomorphogenesis (Sun et al., 2018).
Another miRNA processing factor regulated by light signaling pathways is HEN1. Light induction of HEN1 expression results from the coordinated action of HY5 and HY5 homolog (HYH; Figure 2), which positively regulate HEN1 downstream of several photoreceptors (Tsai et al., 2014). Consistent with this regulation, HEN1 and HYL1 are negative regulators of photomorphogenesis, in the case of HEN1, this is due to its role as a repressor of key transcription factors of this process (Tsai et al., 2014). In addition, alternative splicing of transcripts encoding several miRNA biogenesis and processing factors, such as DCL1, HYL1, and HEN1, is affected by different light qualities, adding a new layer to the regulation of miRNAs by light (reviewed in Hernando et al., 2017).
The de-etiolation process can also affect miRNA activity toward its targets. Lin et al. (2017) matched small RNA profiling with degradome sequencing in Arabidopsis and found that, although light exposure did not change dramatically the level of most miRNAs, it could modulate their target cleavage activity. In fact, with the exception of miR163, most miRNAs showed little fluctuations in their amount upon light transition. This did not correspond with the analysis of degradome signatures, implying that de-etiolation especially promoted the degradation of miR156/157 and miR396 targets, and thus suggesting higher cleavage activity for these families (Lin et al., 2017). However, although Lin et al. (2017) report that miR168 regulates AGO1 levels under light and could modulate RISC activity and the efficiency of miRNA target cleavage, the mechanisms underlying this regulation by light still need to be demonstrated.
Interestingly, another report has shown AGO1 involvement in light regulation of adventitious rooting and hypocotyl elongation. Arabidopsis ago1 mutants are hypersensitive to light, probably due to deregulation of the PHYA-dependent light signal transduction pathway (Sorin et al., 2005). Moreover, de-etiolation of an Arabidopsis hypomorphic ago1-27 mutant in far-red light is impaired, indicating that normal photomorphogenesis requires AGO1 (Li et al., 2014). Given that AGO1 is an essential component of the RISC, these findings suggest that miRNA function is involved in far-red light responses.
Therefore, several components of the miRNA machinery are involved in certain light responses. On the other hand, light modulates MIRNA gene expression, accumulation of mature miRNAs by regulating components of the biogenesis pathway, as well as miRNA activity. How miRNAs participate in light as well as photoperiod signaling events will be discussed next.
miRNAs Involved in Light and Photoperiod Signaling
Our analysis revealed several miRNA families that accumulate upon certain light treatments in different species. Most of these miRNAs mediate critical biological processes necessary for plants to adjust to these changes. They can regulate metabolism, plant development, and certain responses to stress.
miR396 and UV-B Regulation of Cell Proliferation
For instance, UV-B radiation affects cell proliferation in several species (Dotto and Casati, 2017). In Arabidopsis, UV-B light upregulates miR396 levels, leading to repression of the miR396 target genes GROWTH REGULATING FACTOR1 (GRF1), GRF2, and GRF3, with consequent inhibition of cell proliferation in leaves (Casadevall et al., 2013). In fact, either reducing miR396 activity or expressing miR396-resistant forms of GRF3 or GRF2 results in reduced sensitivity of leaf growth to UV-B irradiation, confirming that GRFs mediate this phenotype. Nevertheless, downregulation of GRF3 by UV-B light also seems to occur through miR396-independent mechanisms. In addition, miR396-mediated repression of leaf cell proliferation by UV-B radiation is dependent on the mitogen-activated protein kinase MPK3, known to be involved in the response to UV-B stress (Casadevall et al., 2013). Interestingly, in UV-B-treated maize leaves, miR396 inhibition had the opposite effect on GRFs, which were upregulated (Casati, 2013). This could be due to the different developmental stages analyzed in Arabidopsis and maize, although further studies are necessary to clarify the role of miR396 in the regulation of UV-B-dependent responses in dicots and monocots.
miRNAs Regulating Metabolism
As mentioned before, light controls many metabolic pathways in plants. Within this context, light-regulated miRNAs were shown to modulate metabolic events, such as methylation of specific signaling molecules (e.g., hormones), nutrient allocation, and pigment synthesis/accumulation.
miR163 as well as pri-miR163 were recently shown to be highly induced by light in Arabidopsis (Chung et al., 2016; Lin et al., 2017). Moreover, miR163 may target PXMT1 (At1g66700), a neighbor gene encoding a putative 1,7-paraxanthine methyltransferase that has been implicated in the methylation of natural chemicals such as hormones (Chung et al., 2016). Whereas pri-miR163 is induced in red, blue, and white light, and PXMT1 expression is inhibited upon light exposure, light does not affect pri-miR163 maturation and processing. Both pri-miR163 and its target PXMT1 accumulate in roots, where miR163 accumulation increases root length, especially in the elongation/differentiation zone, most likely by inhibiting PXMT1. Interestingly, the miR163/PXMT1 pair also seems to regulate germination, probably by controlling the early stages of plant development since radicle emergence to seedling de-etiolation. However, to fully understand the role of miR163 in this process, a more extensive functional characterization of PXMT1 would be necessary.
Plant metabolism requires proper nutrient allocation, especially in the case of nutrients that constitute co-factors and are present in different proteins and biological processes. This is the case of copper, since it is required for electron transport in photosynthesis, defense against reactive-oxygen species as well as ethylene perception (Zhang et al., 2014). Maintenance of copper homeostasis is critical to photosynthesis efficiency and thus plant growth. This is achieved by the SPL7 transcription factor, which functions as a copper sensor able to adjust target gene expression upon changing copper levels. Surprisingly, light and copper seem to regulate gene expression of many shared biological processes, suggesting a direct connection between the transcriptional regulators SPL7 and HY5 (Zhang et al., 2014). Using chromatin immunoprecipitation and RNA sequencing, Zhang et al. (2014) characterized the SPL7 regulon and found it to overlap with HY5. This analysis revealed MIR408 as a SPL7/HY5 common target, with SPL7 being the predominant transcription factor determining MIR408 levels, thus confirming previous work showing that HY5 regulates MIR408 expression (Zhang et al., 2011). Nevertheless, HY5/SPL7 coordinated regulation allowed miR408 accumulation upon copper deficiency and high light. Confirming this regulation, MIR408 silencing phenotypes were similar to those of the hy5 spl7 double mutant, whereas miR408 constitutive expression partially rescued them, suggesting that miR408 is a critical component of the SPL7/HY5 network (Zhang et al., 2014). Further analysis of the expression levels of the miR408 targets, LACCASE12 (LAC12) and LAC13, showed that the SPL7/HY5/miR408 module allows differential regulation of very closely related genes. Therefore, this signaling network constitutes a molecular mechanism that integrates light and copper signals to maintain copper homeostasis and consequently efficient photosynthetic rates that promote plant growth.
miRNAs and pigment synthesis
Flavonoids are a group of phenolic compounds that include flavonols, flavones, isoflavones, and anthocyanins (Sharma et al., 2016). Due to their molecular diversity, flavonoids can confer protection against biotic and abiotic stress, as well as regulate plant growth and development. Anthocyanins, for instance, include a wide range of pigments (blue, red, and purple), which can not only protect against UV radiation but can also attract pollinators (Wang et al., 2016). Due to their relevance to plant life, the biosynthesis and regulation of all these compounds have been extensively studied. The transcription factors MYB-like 2 (MYBL2) and SPL9 are negative regulators of anthocyanin biosynthesis in Arabidopsis (Dubos et al., 2008; Matsui et al., 2008; Gou et al., 2011). On the other hand, overexpression of miR156 increases anthocyanin levels in Arabidopsis, since some miR156 target genes, including SPL9, repress anthocyanin biosynthesis (Gou et al., 2011; Cui et al., 2014). In Chinese sand pear (Pyrus pyrifolia) fruits, bagging and subsequent re-exposure to light causes anthocyanin accumulation in fruit peel, giving red color to the fruits (Qian et al., 2013). Qian et al. (2017) hypothesized that miR156 and SPLs would also be involved in anthocyanin accumulation in pears in response to bagging treatments. They detected miR156 in peels of bagging-treated pear fruits and found that the promoters of two pear PpMIR156 genes contained light-responsive elements. After bag removal, miR156 levels increased, whereas PpSPL transcripts decreased. This was accompanied by upregulation of homologs of genes encoding transcription factors that control anthocyanin biosynthesis, and followed by upregulation of anthocyanin biosynthesis genes (Qian et al., 2017). Although functional analyses of miR156 and PpSPL genes in pear have not yet been reported, these results point to a possible role of miR156 and PpSPL proteins in light-induced anthocyanin biosynthesis in bagging-treated pear fruits.
In a related report, Qu et al. (2016) investigated the miRNA profiling of “Granny Smith” apple peels upon re-exposure to sunlight after fruit bagging, using high-throughput sequencing of small RNA libraries. They found that over 67% of differentially expressed miRNAs were downregulated in the bagged group as compared to the unbagged control. These findings suggest the existence of a light-regulated pool of miRNAs in apple peel. miR156, miR160, miR171, miR172, miR395, and miR398 were differentially expressed upon sunlight re-exposure. Interestingly, these miRNAs families had previously been identified as UV-B-regulated miRNAs in several plant species (see Section “UV-Responsive miRNAs”). In Arabidopsis, in addition to miR156, miR858 positively regulates anthocyanin biosynthesis by targeting specific repressors of this process (Gou et al., 2011; Qian et al., 2013; Cui et al., 2014; Wang et al., 2016). In tomato, miR858 also promotes anthocyanin biosynthesis (Jia et al., 2015). Interestingly, miR156, miR828, and miR858 expression levels varied upon apple fruit debagging, suggesting their involvement in anthocyanin biosynthesis in apple peels. In fact, miR156 correlated with anthocyanin accumulation only in “Granny Smith,” whereas miR5072 did it only in the “Starkrimson” cultivar. These results highlight the high specificity of certain miRNA families to control particular responses, such as pigment accumulation to protect against light stress.
As previously mentioned, miR858 is a positive regulator of anthocyanin biosynthesis (Wang et al., 2016). In Arabidopsis, MIR858A expression is induced by light, especially high light, in a HY5-dependent manner. Upon transfer from dark to high light, MIR858A level oscillates, suggesting also circadian regulation. Interestingly, miR858a overexpression partially complements the long hypocotyl phenotype of the hy5 mutant and rescues the anthocyanin levels of this mutant. miR858a and miR858b can promote anthocyanin biosynthesis in two ways: (1) by indirectly inducing the expression of genes encoding components of the anthocyanin biosynthetic pathway and (2) by inhibiting protein accumulation of their target gene encoding the repressor MYBL2 protein, most likely at the translational level. HY5 directly binds and represses the MYBL2 promoter probably by a mechanism that involves the loss of H3K9Ac and H3K4me3-positive histone marks (Wang et al., 2016). Therefore, HY5 has a dual effect in promoting anthocyanin biosynthesis: it binds and represses MYBL2, and it promotes MIR858A expression in response to light (e.g., high light) signals.
It has been shown that hen1 mutants, which have reduced levels of mature miRNAs, accumulate high anthocyanin levels (Tsai et al., 2014), a result somehow contradictory with miR156 and miR858 promoting anthocyanin biosynthesis. However, Sharma et al. (2016) propose a different function for miR858 in the regulation of flavonoid biosynthesis. They found that miR858 targets the positive regulators of flavonoid biosynthesis MYB11, MYB12, and MYB111. Moreover, transcriptional profiling of miR858-overexpressing lines (miR858-OX) revealed MYB20, MYB42, and MYB83 as new targets of miR858. Since these transcription factors regulate several growth responses, the authors characterized growth phenotypes of miR858-OX and miR858a target mimic lines (MIM858), in which miR858a activity is silenced. While miR858-OX plants showed enhanced growth, early flowering, and an increase in seed size, MIM858 plants displayed reduced growth, late flowering, and smaller seeds (Sharma et al., 2016). The metabolic profiling of these lines confirmed their opposite effect on flavonoid accumulation, since miR858 depletion resulted in the accumulation of major flavonoids, whereas they were significantly reduced in miR858-OX. This was accompanied by an opposite effect on lignin biosynthesis, since miR858-OX showed increased lignification and upregulation of lignin biosynthetic genes. These results indicate that miR858 could differently regulate anthocyanin, other flavonoids, and lignin biosynthesis. This could be achieved by targeting different MYB transcription factors in response to distinct light conditions (white light versus low or high light). This differential regulation could then help maintain a metabolic flux balance between the different biosynthetic pathways.
Chlorophylls are critical plant pigments involved in light absorption and energy transfer during photosynthesis. However, this fundamental process for plant life also generates reactive oxygen species, which are detrimental for plant growth and development. Considering that chlorophyll biosynthesis precursors are also sources for reactive oxygen species, it is of paramount importance to tightly regulate this pathway. One of such mechanisms includes miR171 and miR171-targeted SCL genes, also known as HAIRY MERISTEM (HAM) or LOST MERISTEMS (LOM). In Arabidopsis, miR171 positively regulates chlorophyll biosynthesis in the light through downregulation of miR171-targeted SCLs, leading to upregulation of protochlorophyllide oxidoreductase (POR), a key enzyme in chlorophyll biosynthesis (Ma et al., 2014). MIR171C-overexpressing plants and scl6 scl22 scl27 mutants show higher chlorophyll and POR levels than wild-type plants, whereas plants expressing a miR171-resistant form of SCL27 (rSCL27) show reduced chlorophyll and POR levels (Wang et al., 2010; Ma et al., 2014). On the other hand, downregulation of POR expression reduces the chlorophyll content of wild-type, MIR171C-OX, and scl triple mutants (Ma et al., 2014), indicating that the role of miR171c and SCLs on chlorophyll regulation is mediated by PORC. SCL27 represses PORC expression by directly binding and inhibiting its promoter.
Moreover, the miR171-SCL module also mediates gibberellin-dependent effects on chlorophyll biosynthesis due to its regulation of DELLA proteins and PORC expression in the light, but not in the dark. In fact, SCL27 interacts with the DELLA protein RGA and this interaction reduces the ability of SCL27 to bind to the PORC promoter. Finally, SCLs induce the expression of MIR171 genes, revealing the existence of a regulatory feedback loop (Ma et al., 2014). Ma et al. (2014) propose that this feedback loop may also contribute to maintain the diurnal oscillation of miR171.
Taken together, these findings emphasize the role of miRNAs in diverse light-regulated processes. In addition, miRNAs can also directly target major regulators of the light signaling pathways and/or the photoperiod pathway, thus modulating photomorphogenesis or the time to flower, respectively. The most recent findings on miRNAs involved in these processes are discussed below.
Photomorphogenesis
Within the group of light-responsive miRNAs, the miR156/157 family occupies a prominent position (Table 1). In Arabidopsis, miR157d and miR319 promote the degradation of their target transcripts, encoding the positive and negative key photomorphogenesis regulators HY5 and TCP (TEOSINTE BRANCHED1, CYCLOIDEA, and PCF) transcription factors, respectively. Interestingly, both induction and stabilization of miR157d and miR319 were shown to depend on HEN1, with HEN1 accumulation increasing miR157d and miR319 levels in de-etiolating seedlings (Tsai et al., 2014). In turn, HY5 induces HEN1 expression in a light-dependent manner. Therefore, HEN1 and HY5 constitute a negative feedback regulatory loop that is mediated by miR157, since this miRNA will ultimately target HY5 transcripts for cleavage. In a hen1 mutant, both HY5 transcript and protein levels are increased, probably due to the decrease in miR157d, resulting in a light-hypersensitive phenotype. Oppositely, miR157 constitutive expression reduced HY5 transcript and protein levels, resulting in seedlings displaying a light hyposensitivity phenotype. Nevertheless, the light-hypersensitive phenotype displayed by hen1 mutants may also be dependent on miR319, which promoted the cleavage of TCP mRNAs and repressed hypocotyl elongation (Tsai et al., 2014). The role of miR319 in photomorphogenesis has been confirmed using a loss of function mutant, which shows longer hypocotyls than the wild type under red light (Sun et al., 2018). Perhaps, the HEN1/HY5 feedback loop may help explain the inconsistencies found in the effect of HEN1, miR156, and miR858 on anthocyanin accumulation.
Three additional miRNAs, miR160, miR167, and miR848, affect Arabidopsis hypocotyl elongation under red light (Sun et al., 2018). It is worth mentioning that miR160 and miR167 respond to different light conditions in several species (Table 1). Further understanding of the role of these miRNAs in light responses will undoubtedly provide novel insights into how miRNAs contribute to adaptation to different light environments.
Besides the early stages of photomorphogenesis, when the seedling reaches for light and hypocotyl elongation is critical, miRNAs regulate other light responses during plant adult life, such as the perception of their surroundings and neighbors or the time to flower, for instance. When in isolation, plants receive light with a high red to far-red (R:FR) ratio. However, when they grow under a canopy or in close proximity to neighboring plants, the R:FR ratio is lower and this triggers diverse morphological and physiological responses that allow the plants to adapt to these light conditions (Ballaré and Pierik, 2017). When Arabidopsis plants grown under light/dark cycles are treated with a pulse of far-red light at the end of the light period (end-of-day far-red, EOD-FR), they show earlier flowering, increased petiole elongation, and a reduction in the number of rosette branches in comparison with plants grown under normal white-light/dark cycles (WL; Xie et al., 2017). EOD-FR treatment causes a decrease in mature miR156 levels by downregulating the expression of several MIR156 genes. Consistent with this, there is upregulation of several miR156-target SPL genes in EOD-FR-treated plants (Xie et al., 2017). This suggests that the miR156-SPL module might be involved in EOD-FR responsiveness. Indeed, when plants with reduced miR156 activity (MIM156 plants) are grown under WL, they show a constitutive EOD-FR response (Xie et al., 2017). These findings suggest that a reduction in miR156 levels is required for this response, and place miR156 as a repressor of this process.
PHYTOCHROME-INTERACTING FACTORS (PIFs) mediate the effect of phytochromes on numerous light responses (Leivar and Monte, 2014). PIF protein abundance increases under low R:FR light conditions (Lorrain et al., 2008; Leivar et al., 2012; Xie et al., 2017), and PIFs act as positive regulators of EOD-FR responses, showing therefore an opposite behavior to miR156. Further substantiating a previous report suggesting that MIR156E is a direct target gene of PIF5 (Hornitschek et al., 2012), Xie et al. (2017) showed that several PIFs repress transcription of five MIR156 genes, including MIR156E, by directly binding to PIF-binding sites present in their promoters. This causes a reduction in mature miR156 levels and a concomitant increase in miR156-target SPL transcript abundance. Moreover, genetic analyses show that miR156 acts downstream of PIF5 (Xie et al., 2017). Altogether, these results demonstrate that miR156 responds to EOD-FR treatments and mediates, probably through downregulation of SPL genes, the effect of PIFs on at least part of the low R:FR light responses. It will be interesting to determine whether these findings can be reproduced under low R:FR conditions more closely resembling canopy or proximity shade. Therefore, not only do miR156/157 respond to light but also affect several light-regulated processes.
miRNAs and Photoperiod-Regulated Processes
Photoperiod affects many plant metabolic, physiological, and developmental processes. It also affects the levels of some miRNAs, as we have already discussed (Li et al., 2015). An example is miR156, which is regulated by photoperiod in potato and soybean, although not in Arabidopsis (Jung et al., 2012; Bhogale et al., 2014; Li et al., 2015). miR156, via its target genes, downregulates miR172 in several plant species (Chuck et al., 2007; Wu et al., 2009; Bhogale et al., 2014). In Arabidopsis, mature miR172 levels are higher under LD than SD and this difference does not seem to be driven by transcription, given that the abundance of at least two miR172 primary transcripts is lower under LD than SD (Jung et al., 2007). Rather, it probably results from regulation of miR172 processing. A gigantea (gi) mutant, which shows a reduced response to photoperiod and reduced expression of two genes involved in miRNA processing, DCL1 and SE, shows lower mature miR172, but not pri-miR172, levels than the wild type. However, additional factors must contribute to the photoperiodic regulation of miR172, given that this miRNA still responds to photoperiod in the gi mutant (Jung et al., 2007). Potato and soybean also show different miR172 levels under LD and SD (Martin et al., 2009; Li et al., 2015), suggesting that the photoperiodic regulation of this miRNA may be evolutionarily conserved.
Interestingly, at least four photoreceptors, phytochrome A, phytochrome B, and cryptochrome 1 and 2, regulate miR172 levels (Jung et al., 2007; Martin et al., 2009; Zhao et al., 2015). In agreement with this, red light downregulates miR172 and blue light upregulates it in Arabidopsis. Therefore, both light quality and duration affect miR172 abundance. In addition, TIMING OF CAB EXPRESSION1 (TOC1), a component of the circadian clock, reduces mature miR172 levels, but despite this, mature miR172 levels do not show daily oscillations in Arabidopsis (Jung et al., 2007). Further research is still needed to determine whether the circadian clock regulates miR172.
In Arabidopsis, flowering is regulated by photoperiod, such that plants flower earlier under LD than SD. By contrast, soybean flowering and potato tuber formation are induced by SD. Overexpression of miR172 promotes flowering in Arabidopsis and tuberization in potato and reduces the photoperiodic tuberization response (Aukerman and Sakai, 2003; Chen, 2004; Jung et al., 2007; Martin et al., 2009), strongly suggesting that miR172 is involved in the regulation of photoperiodic processes. miR172 targets a subfamily of AP2-like genes, including AP2 itself, which partially redundantly repress flowering (Zhu and Helliwell, 2011). In Arabidopsis, and probably in soybean, alteration of miR172-target gene expression or function leads to a reduced response of flowering to photoperiod, confirming the role of the miR172/AP2-like module in the photoperiodic regulation of flowering (Mathieu et al., 2009; Yant et al., 2010; Zhao et al., 2015). In Arabidopsis plants grown under LD, GI, and probably several photoreceptors, upregulates miR172, which negatively regulates its target genes, which in turn directly repress the expression of several genes promoting flowering, including FLOWERING LOCUS T (FT), and promote the expression of genes repressing flowering (Figure 3) (Jung et al., 2007; Mathieu et al., 2009; Yant et al., 2010). Downregulation of miR172 target genes, thus, accelerates flowering under LD conditions. This photoperiodic flowering pathway is independent of the photoperiodic flowering regulator CONSTANS (CO) in Arabidopsis (Jung et al., 2007; Mathieu et al., 2009; Yant et al., 2010), although it has been proposed to involve a CO-like gene in soybean (Zhao et al., 2015).
FIGURE 3.
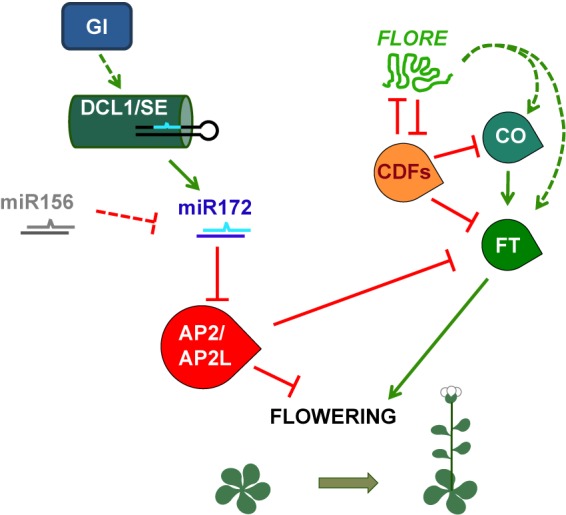
Simplified model of the photoperiodic regulation of Arabidopsis flowering time mediated by the miRNA miR172 and the lncRNA FLORE. GI regulates the DCL1/SE complex and thus miR172 accumulation, which will in turn modulate the protein levels of the flowering repressors AP2 and AP2-like. On the other hand, light-responsive miR156 is able to repress miR172. Although miR156 is regulated by photoperiod in soybean and potato, in Arabidopsis this regulation in not clear. AP2 and AP2-like exert their function by inhibiting FT transcription, which is positively regulated by CO. The CO-FT module is also negatively targeted by the CDFs. In addition, FLORE is able to inhibit these transcription factors and thus promote an increase in CO and FT transcript levels, most likely in an indirect way. This model provides a glimpse of the different layers of regulation required to ensure that Arabidopsis plants flower under the most favorable photoperiodic conditions, which occur when day length increases and short days give way to longer days.
Given that miR156 responds to light in several species, to photoperiod in potato and soybean, is involved in flowering control, and negatively regulates miR172 (Chuck et al., 2007; Wang et al., 2009; Wu et al., 2009; Bhogale et al., 2014; Li et al., 2015) (Table 1), it seems likely that miR156 may play a role in photoperiodic flowering. However, whether this is the case still needs to be proved. In addition to light quality and photoperiod, miR156 and miR172 levels respond to other environmental and endogenous signals and play roles in processes other than flowering and tuberization (Zhu and Helliwell, 2011; Han et al., 2013; Nova-Franco et al., 2015; Ripoll et al., 2015; Wang and Wang, 2015; Huo et al., 2016; Li et al., 2017; Díaz-Manzano et al., 2018; Luan et al., 2018), indicating that miR156 and miR172 influence many aspects of plant biology.
The miR170/171 family has been shown to be responsive to different light conditions in several species (Table 1). In addition, the rice OsMIR171C promoter contains several elements putatively involved in light responses (Fan et al., 2015). Consistent with this, OsMIR171C transcript levels show a diurnal oscillation with a peak early in the morning, similar to the oscillation of mature miR171 in Arabidopsis (Fan et al., 2015). Conversely, transcript levels of four rice miR171 target genes, OsHAM1 to OsHAM4, accumulate from the evening until early morning under light/dark cycles. Although it is not clear whether these rice oscillation patterns are regulated by light or by the circadian clock, miR171c levels respond to photoperiod, with higher levels under LD than SD (Fan et al., 2015). miR171c upregulation in the delayed heading (dh) mutant, which carries a T-DNA insertion in the OsMIR171C promoter, leads to late flowering (heading), strongly suggesting that miR171c represses flowering (Fan et al., 2015). Moreover, the upregulation of miR171c by LD, which are non-inductive conditions for rice flowering, fits with a role in delaying flowering. However, further research is still required to determine if miR171 affects the photoperiodic regulation of flowering in rice. Nevertheless, the expression pattern of miR171c and OsHAMs in the shoot apex of wild-type plants is consistent with a role in regulating the floral transition. This is also supported by the downregulation of three key positive regulators of flowering (Ehd1, Hd3a, and RFT1) in the dh mutant in comparison with the wild type (Fan et al., 2015). Interestingly, the dh mutant also shows upregulation of miR156 (Fan et al., 2015), a flowering repressor miRNA in several species (Wang, 2014), and of OsPHYC, which encodes a photoreceptor that also delays flowering in rice (Takano et al., 2005; Fan et al., 2015). Therefore, miR171 is regulated by light and, in turn, it may be involved in regulating light responses mediated by OsPHYC.
Another miRNA that responds to day length and regulates photoperiodic flowering is miR5200, a miRNA specific of the Pooideae family. In a Brachypodium distachyon accession that flowers much earlier under LD than SD, mature miR5200, as well as pri-miR5200a and pri-miR5200b, is present at much higher levels under SD than LD and its pri-miRNAs follow a daily rhythm under SD (Wu et al., 2013). The expression pattern of MIR5200A and MIR5200b correlates with active histone marks in these genes under SD and repressive histone marks under LD. Another five Pooideae species accumulate more miR5200 under SD than LD, revealing conservation of this photoperiodic regulation among these grass species (Wu et al., 2013). In B. distachyon, miR5200 targets two FT-like genes, FTL1 and FTL2, whose transcripts are more abundant under long than short photoperiods. B. distachyon plants overexpressing this miRNA flower much later than wild-type plants under LD, whereas plants with reduced miR5200 activity (MIM5200) flower earlier than the wild type under SD, and like the wild type under LD, showing therefore a reduced response to photoperiod. Consistently, FTL1 and FTL2 mRNA levels are increased in MIM5200 plants under SD, but unchanged under LD in comparison with wild-type plants (Wu et al., 2013). Therefore, miR5200 plays an important role in the photoperiodic regulation of flowering in B. distachyon by delaying flowering under SD.
Long Non-Coding RNAs
Next-generation sequencing techniques have unraveled a whole new transcriptional landscape that includes lncRNAs. Several screening approaches have been designed to identify novel plant lncRNAs that would accumulate in response to certain environmental cues, in specific organs/tissues or at particular developmental stages. However, although lncRNA listings are available, the functional characterization of most of these transcripts is yet to be done. Most of the knowledge on plant lncRNA function is associated with FLOWERING LOCUS C (FLC), a negative regulator of flowering. In fact, several lncRNAs arising from this locus were shown to repress FLC transcript levels upon vernalization and thus promote flowering. COLD OF WINTER-INDUCED NON-CODING RNA FROM THE PROMOTER (COLDWRAP) is transcribed from the FLC promoter region, whereas COLD-ASSISTED INTRONIC NON-CODING RNA (COLDAIR) is expressed from the first intron. ANTISENSE LONG (ASL) and COLD-INDUCED LONG ANTISENSE INTRAGENIC RNA (COOLAIR) both originate from the FLC 3′UTR region and show partial overlap. The combined action of the three lncRNAs was shown to silence FLC expression, and therefore to promote flowering, through the accumulation of repressive histone marks (reviewed in Chekanova, 2015; Whittaker and Dean, 2017). Although most of the research on these lncRNAs pertains to vernalization-regulated flowering, and therefore is outside the scope of this review, there are reports connecting FLC with photoperiod and circadian regulation. FLC lengthens the circadian period, especially at high temperature (Swarup et al., 1999; Edwards et al., 2006), but shortening of the circadian period by vernalization is not mediated by FLC (Salathia et al., 2006). Therefore, whether the regulation of FLC by vernalization-associated lncRNAs is linked to its role in circadian periodicity is yet to be determined.
Several FLC clade members were shown to regulate flowering both under LDs and SDs. Protein levels of one of them, MAF3 oscillated throughout the day, suggesting regulation by the photoperiod or the circadian clock (Gu et al., 2013). In fact, FLC clade members could form multi-protein complexes and regulate flowering depending on photoperiod and temperature. However, further research is needed to determine whether vernalization-associated lncRNAs would also participate in these processes. Therefore, in this section, we will discuss the available reports focused on light and circadian-responsive lncRNAs.
lncRNAs as Regulators of Light-Dependent Processes
One of the screening approaches that have allowed the identification of novel lncRNA transcriptional units used a reproducibility-based tiling-array analysis strategy (RepTAS) previously described by Liu et al. (2012). Thus, Wang H. et al. (2014) identified a total of 37,238 light-responsive lncNAT pairs (NAT pairs where one of the components is an lncRNA) in Arabidopsis. These NAT pairs were afterward verified by different methods, including the screening of a novel ATH (Arabidopsis thaliana) NAT custom array, strand-specific RNA-seq (ssRNA-seq), and quantitative RT-PCR (RT-qPCR). To identify bona fide light-regulated lncRNAs, the expression levels of these antisense and sense transcripts were determined in etiolated seedlings and seedlings undergoing de-etiolation that were exposed to continuous white light for 1 and 6 h. Not only did this approach allow the identification of light-responsive lncNATs but also revealed their preferential accumulation in cotyledons compared to hypocotyls, and roots. Interestingly, the number of light-responsive lncNAT pairs was much higher after 6 h of treatment than at 1 h, indicating spatial-, developmental-, and temporal-specific patterns of accumulation. When the expression levels of both the sense and antisense components of each NAT pair was determined, these lncNAT pairs were classified into two groups: light-responsive concordant NAT pairs and light-responsive discordant NAT pairs. In order to further characterize their expression pattern, a parallel analysis was performed matching specific histone modifications with their transcriptional changes. This approach showed that transcription changes of these light-regulated NATs were preferentially correlated with histone H3 acetylation (H3K9ac and/or H3K27ac), H3K9ac being the predominant positive histone mark, suggesting its involvement in regulating light-responsive NAT pairs (Wang H. et al., 2014). Although the functional characterization of most of these candidates is yet to be done, these results provide the first evidence that light is a major shaper of the plant non-coding transcriptional landscape, including both long and small non-coding RNAs.
HIDDEN TREASURE 1 was the first lncRNA described as a positive regulator of photomorphogenesis under continuous red light. This is achieved by HID1 ability to downregulate the expression levels of PIF3, a negative regulator of this process (Wang Y. et al., 2014). In agreement with this, PIF3 mRNA levels were increased in hid1 mutants that displayed elongated hypocotyls under red light, a typical phenotype of PIF accumulation. Functional characterization of HID1 revealed that this lncRNA assembles in nuclear protein–RNA complexes where it is able to associate with PIF3 5′UTR and repress its expression. Therefore, HID1 would act through PIF3 to modulate hypocotyl elongation. Sequence homology searches revealed that HID1 is present in other plant species. Confirming its conservation, Arabidopsis hid1 mutants expressing OsHID1, a rice homolog, could be rescued of their elongated hypocotyl phenotype (Wang Y. et al., 2014). These results suggest that lncRNAs may be conserved between species and regulate similar biological processes, similar to the families of conserved miRNAs.
Interestingly, besides its role as a positive regulator of photomorphogenesis in red light, HID1 can also act as a negative regulator of cotyledon greening during de-etiolation of Arabidopsis in white light (Wang et al., 2018). Within the several steps of this process, the reduction of protochlorophyllide (Pchlide) to chlorophyllide via POR is fundamental for cotyledon greening to occur. Confirming its repressor role in this process, dark-grown hid1 mutants transferred to white light presented a higher rate of cotyledon greening compared to Arabidopsis wild-type plants. Moreover, HID1 knockdown led to a reduction of Pchlide content in the dark, as well as higher POR mRNA levels, indicating that HID1 plays a role in cotyledon greening by promoting Pchlide accumulation and repressing POR transcription (Wang et al., 2018). To uncover the HID1/PIF3 relationship in cotyledon greening, double hid1 pif3 mutants were generated. PIF3 was previously shown to regulate seedling greening by preventing Pchlide over-accumulation (Leivar and Monte, 2014). However, hid1 pif3 double mutants greening rate was comparable to that of hid1, suggesting that HID1 would act downstream of PIF3 (Wang et al., 2018). Since PIF3 does not regulate HID1 expression, the exact mechanism by which HID1 and PIF3 regulate greening still remains to be determined (Wang et al., 2018). Together, these results suggest that depending on the light-mediated developmental process, HID1 could associate with different factors allowing it to perform different functions.
Circadian-Regulated lncRNAs in Photoperiod-Dependent Flowering
Long non-protein coding RNAs can also display circadian regulation. This is characterized by their ability to maintain an oscillatory pattern of expression even in the absence of environmental cues (e.g., under constant conditions). Several screening approaches allowed the identification of circadian ncRNAs in fungi, mammals, and Arabidopsis, some of them being lncRNAs that were also NATs (Kramer et al., 2003; Hazen et al., 2009; Coon et al., 2012; Xue et al., 2014). However, even if non-coding NATs were reported for components of the Arabidopsis oscillator (Hazen et al., 2009), their function had not been investigated. Recently, we characterized a circadian-regulated Arabidopsis lncRNA, FLORE, which is a NAT to the CYCLING DOF FACTOR 5 (CDF5) transcriptional regulator (Henriques et al., 2017). FLORE and CDF5 displayed an antiphasic pattern of expression that resulted in part from a mutual inhibitory function, as well as an opposite biological function (Figure 3). CDFs have been characterized as repressors of photoperiodic flowering due to their ability to repress CO and FT transcription (Song et al., 2015). FLORE inhibits CDF5 accumulation and contributes to maintain its proper oscillatory pattern. Therefore, due to downregulation of CDF5, FLORE overexpression in the vascular tissue promotes flowering. On the other hand, CDF5 will also regulate FLORE transcript levels, thus contributing to its oscillation. These findings indicate that the circadian clock is able to control the expression of both coding and non-coding transcripts, in this case, CDF5 and FLORE (Henriques et al., 2017). However, the mutual regulation within this NAT pair seems to be required to ensure the timely accumulation of each transcript. Further research is still necessary to uncover the molecular mechanisms that account for this regulation.
Final Conclusions
Both small and lncRNAs have been associated with specific biological processes that can occur at particular developmental stages, often in very precise locations within certain organs and tissues. Light is one of the major environmental cues for plants and essential for their survival. Therefore, it is not surprising that light modulates the expression, processing, and activity of these non-coding RNAs. It became apparent that, in the case of specific miRNAs, a correlation pattern emerges where the accumulation of specific families associates with particular light conditions in different species. Moreover, this seems to be a two-way regulation since miRNAs can also target basic components of light and photoperiod signaling, thus being also required for the proper functioning of these pathways. Although some of the miRNAs that are consistently revealed as light-regulated in diverse plant species have not been shown to play functional roles in light responses so far, they are excellent candidates for performing such roles. lncRNAs have recently become the subject of scrutiny, but the few reports available prevent the establishment of any specific trend. Sequencing and bioinformatics efforts led to the construction of a plant-specific lncRNA database (Jin et al., 2013). However, other approaches combining transcriptomics, epigenetic, and functional assays would provide further insights into the biological role of plant lncRNAs. Nevertheless, it is noteworthy mentioning that, from the reduced group of studied lncRNAs, two of them associate with light-dependent processes. These findings do show, however, that light is a massive re-programmer of non-coding transcription able to set in motion-specific fine-tuning mechanisms with the ultimate goal of adjusting plant development to their surroundings.
Author Contributions
All the authors read the cited publications, prepared summaries, and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. RH group was supported by grants from the Spanish “Ministerio de Economía y Competitividad” (MINECO) (BIO2015-72161-EXP, BIO2015-70812-ERC, and RYC-2011-09220) and by the European Commission (PCIG2012-GA-2012-334052). PS-L acknowledges financial support from the MINECO (Grant BFU2015-64409-P). We also acknowledge the financial support of the “Severo Ochoa Program for Centres of Excellence in R&D” 2016-2019, SEV-2015-0533) and the Generalitat de Catalunya (CERCA Program).
References
- Andrés F., Coupland G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13 627–639. 10.1038/nrg3291 [DOI] [PubMed] [Google Scholar]
- Aukerman M. J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15 2730–2741. 10.1105/tpc.016238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré C. L., Pierik R. (2017). The shade-avoidance syndrome: multiple signals and ecological consequences. Plant Cell Environ. 40 2530–2543. 10.1111/pce.12914 [DOI] [PubMed] [Google Scholar]
- Bhogale S., Mahajan A. S., Natarajan B., Rajabhoj M., Thulasiram H. V., Banerjee A. K. (2014). MicroRNA156: a potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 164 1011–1027. 10.1104/pp.113.230714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F., Martienssen R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16 727–741. 10.1038/nrm4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Li Y., Wang J., Nan H., Wang Y., Lu S., et al. (2015). GmmiR156b overexpression delays flowering time in soybean. Plant Mol. Biol. 89 353–363. 10.1007/s11103-015-0371-5 [DOI] [PubMed] [Google Scholar]
- Casadevall R., Rodriguez R. E., Debernardi J. M., Palatnik J. F., Casati P. (2013). Repression of growth regulating factors by the MicroRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. Plant Cell 25 3570–3583. 10.1105/tpc.113.117473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P. (2013). Analysis of UV-B regulated miRNAs and their targets in maize leaves. Plant Signal. Behav. 8:e26758. 10.4161/psb.26758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P., Stapleton A. E., Blum J. E., Walbot V. (2006). Genome-wide analysis of high-altitude maize and gene knockdown stocks implicates chromatin remodeling proteins in response to UV-B. Plant J. 46 613–627. 10.1111/j.1365-313X.2006.02721.x [DOI] [PubMed] [Google Scholar]
- Chekanova J. A. (2015). Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 27 207–216. 10.1016/j.pbi.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303 2022–2025. 10.1126/science.1088060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. K., Chaabane S. B., Shah P., Poulsen C. P., Yang S. W. (2014). COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat. Commun. 5:5867. 10.1038/ncomms6867 [DOI] [PubMed] [Google Scholar]
- Chuck G., Cigan A. M., Saeteurn K., Hake S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39 544–549. 10.1038/ng2001 [DOI] [PubMed] [Google Scholar]
- Chuck G., Meeley R., Hake S. (2008). Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135 3013–3019. 10.1242/dev.024273 [DOI] [PubMed] [Google Scholar]
- Chung P. J., Park B. S., Wang H., Liu J., Jang I.-C., Chua N.-H. (2016). Light-inducible miR163 targets PXMT1 transcripts to promote seed germination and primary root elongation in Arabidopsis. Plant Physiol. 170 1772–1782. 10.1104/pp.15.01188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon S. L., Munson P. J., Cherukuri P. F., Sugden D., Rath M. F., Müller M., et al. (2012). Circadian changes in long noncoding RNAs in the pineal gland. Proc. Natl. Acad. Sci. U.S.A. 109 13319–13324. 10.1073/pnas.1207748109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L.-G., Shan J.-X., Shi M., Gao J.-P., Lin H.-X. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80 1108–1117. 10.1111/tpj.12712 [DOI] [PubMed] [Google Scholar]
- Cuperus J. T., Fahlgren N., Carrington J. C. (2011). Evolution and functional diversification of MIRNA genes. Plant Cell 23 431–442. 10.1105/tpc.110.082784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Manzano F. E., Cabrera J., Ripoll J.-J., del Olmo I., Andrés M. F., Silva A. C., et al. (2018). A role for the gene regulatory module microRNA172/TARGET OF EARLY ACTIVATION TAGGED 1/FLOWERING LOCUS T (miRNA172/TOE1/FT) in the feeding sites induced by Meloidogyne javanica in Arabidopsis thaliana. New Phytol. 217 813–827. 10.1111/nph.14839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto M., Casati P. (2017). Developmental reprogramming by UV-B radiation in plants. Plant Sci. 264 96–101. 10.1016/j.plantsci.2017.09.006 [DOI] [PubMed] [Google Scholar]
- Dubos C., Le Gourrierec J., Baudry A., Huep G., Lanet E., Debeaujon I., et al. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55 940–953. 10.1111/j.1365-313X.2008.03564.x [DOI] [PubMed] [Google Scholar]
- Edwards K. D., Anderson P. E., Hall A., Salathia N. S., Locke J. C. W., Lynn J. R., et al. (2006). FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18 639–650. 10.1105/tpc.105.038315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T., Li X., Yang W., Xia K., Ouyang J., Zhang M. (2015). Rice osa-miR171c mediates phase change from vegetative to reproductive development and shoot apical meristem maintenance by repressing four OsHAM transcription factors. PLoS One 10:e0125833. 10.1371/journal.pone.0125833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J.-Y., Felippes F. F., Liu C.-J., Weigel D., Wang J.-W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23 1512–1522. 10.1105/tpc.111.084525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Le C., Wang Y., Li Z., Jiang D., Wang Y., et al. (2013). Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 4:1947. 10.1038/ncomms2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Zhang X., Wang Y., Ming F. (2013). The Suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS One 8:e73541. 10.1371/journal.pone.0073541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen S., Naef F., Quisel T., Gendron J., Chen H., Ecker J., et al. (2009). Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol. 10:R17. 10.1186/gb-2009-10-2-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus D., Yu M., Baldwin D., Gruber M., Sharpe A., Parkin I., et al. (2003). Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol. Biol. 53 383–397. 10.1023/B:PLAN.0000006944.61384.11 [DOI] [PubMed] [Google Scholar]
- Henriques R., Wang H., Liu J., Boix M., Huang L.-F., Chua N.-H. (2017). The antiphasic regulatory module comprising CDF5 and its antisense RNA FLORE links the circadian clock to photoperiodic flowering. New Phytol. 216 854–867. 10.1111/nph.14703 [DOI] [PubMed] [Google Scholar]
- Hernando C. E., Garcia C., Mateos J. L. (2017). Casting away the shadows: elucidating the role of light-mediated posttranscriptional control in plants. Photochem. Photobiol. 93 656–665. 10.1111/php.12762 [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M. V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., et al. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71 699–711. 10.1111/j.1365-313X.2012.05033.x [DOI] [PubMed] [Google Scholar]
- Huo H., Wei S., Bradford K. J. (2016). DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. U.S.A. 113 E2199–E2206. 10.1073/pnas.1600558113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I., Henriques R., Seo H., Nagatani A., Chua N. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22 2370–2383. 10.1105/tpc.109.072520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X., Ren L., Chen Q.-J., Li R., Tang G. (2009). UV-B-responsive microRNAs in Populus tremula. J. Plant Physiol. 166 2046–2057. 10.1016/j.jplph.2009.06.011 [DOI] [PubMed] [Google Scholar]
- Jia X., Shen J., Liu H., Li F., Ding N., Gao C., et al. (2015). Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta 242 283–293. 10.1007/s00425-015-2305-5 [DOI] [PubMed] [Google Scholar]
- Jin J., Liu J., Wang H., Wong L., Chua N.-H. (2013). PLncDB: plant long non-coding RNA database. Bioinformatics 29 1068–1071. 10.1093/bioinformatics/btt107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M. W., Bartel D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14 787–799. 10.1016/j.molcel.2004.05.027 [DOI] [PubMed] [Google Scholar]
- Juarez M. T., Kui J. S., Thomas J., Heller B. A., Timmermans M. C. P. (2004). microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428 84–88. 10.1038/nature02363 [DOI] [PubMed] [Google Scholar]
- Jung J.-H., Ju Y., Seo P. J., Lee J.-H., Park C.-M. (2012). The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 69 577–588. 10.1111/j.1365-313X.2011.04813.x [DOI] [PubMed] [Google Scholar]
- Jung J.-H., Seo Y.-H., Seo P. J., Reyes J. L., Yun J., Chua N.-H., et al. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19 2736–2748. 10.1105/tpc.107.054528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y., Lee H. J., Jung H. J., Maruyama K., Suzuki N., Kang H. (2010). Overexpression of microRNA395c or 395e affects differently the seed germination of Arabidopsis thaliana under stress conditions. Planta 232 1447–1454. 10.1007/s00425-010-1267-x [DOI] [PubMed] [Google Scholar]
- Kooke R., Keurentjes J. J. B. (2012). Multi-dimensional regulation of metabolic networks shaping plant development and performance. J. Exp. Bot. 63 3353–3365. 10.1093/jxb/err373 [DOI] [PubMed] [Google Scholar]
- Kramer C., Loros J. J., Dunlap J. C., Crosthwaite S. K. (2003). Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 421 948–952. 10.1038/nature01427 [DOI] [PubMed] [Google Scholar]
- Lauter N., Kampani A., Carlson S., Goebel M., Moose S. P. (2005). microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. U.S.A. 102 9412–9417. 10.1073/pnas.0503927102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E. (2014). PIFs: systems integrators in plant development. Plant Cell 26 56–78. 10.1105/tpc.113.120857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J. M., Cohn M. M., Monte E., Al-Sady B., Erickson E., et al. (2012). Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24 1398–1419. 10.1105/tpc.112.095711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liang Z., Ding G., Shi L., Xu F., Cai H. (2016). A natural light/dark cycle regulation of Carbon-Nitrogen metabolism and gene expression in rice shoots. Front. Plant Sci. 7:1318. 10.3389/fpls.2016.01318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Castillo-González C., Yu B., Zhang X. (2017). The functions of plant small RNAs in development and in stress responses. Plant J. 90 654–670. 10.1111/tpj.13444 [DOI] [PubMed] [Google Scholar]
- Li W., Wang P., Li Y., Zhang K., Ding F., Nie T., et al. (2015). Identification of MicroRNAs in response to different day lengths in soybean using high-throughput sequencing and qRT-PCR. PLoS One 10:e0132621. 10.1371/journal.pone.0132621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Varala K., Hudson M. E. (2014). A survey of the small RNA population during far-red light-induced apical hook opening. Front. Plant Sci. 5:156. 10.3389/fpls.2014.00156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.-C., Tsai H.-L., Lim S.-L., Jeng S.-T., Wu S.-H. (2017). Unraveling multifaceted contributions of small regulatory RNAs to photomorphogenic development in Arabidopsis. BMC Genomics 18:559. 10.1186/s12864-017-3937-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Jung C., Xu J., Wang H., Deng S., Bernad L., et al. (2012). Genome-wide analysis uncovers regulation of Long Intergenic Noncoding RNAs in Arabidopsis. Plant Cell 24 4333–4345. 10.1105/tpc.112.102855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Juez E., Dillon E., Magyar Z., Khan S., Hazeldine S., de Jager S. M., et al. (2008). Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell 20 947–968. 10.1105/tpc.107.057075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P. D., Whitelam G. C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53 312–323. 10.1111/j.1365-313X.2007.03341.x [DOI] [PubMed] [Google Scholar]
- Loudet O., Michael T. P., Burger B. T., Le Metté C., Mockler T. C., Weigel D., et al. (2008). A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105 17193–17198. 10.1073/pnas.0807264105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Y., Cui J., Li J., Jiang N., Liu P., Meng J. (2018). Effective enhancement of resistance to Phytophthora infestans by overexpression of miR172a and b in Solanum lycopersicum. Planta 247 127–138. 10.1007/s00425-017-2773-x [DOI] [PubMed] [Google Scholar]
- Ma Z., Hu X., Cai W., Huang W., Zhou X., Luo Q., et al. (2014). Arabidopsis miR171-targeted scarecrow-like proteins bind to GT cis-elements and mediate gibberellin-regulated chlorophyll biosynthesis under light conditions. PLoS Genet. 10:e1004519. 10.1371/journal.pgen.1004519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Adam H., Díaz-Mendoza M., Żurczak M., González-Schain N. D., Suárez-López P. (2009). Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 136 2873–2881. 10.1242/dev.031658 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Yant L. J., Mürdter F., Küttner F., Schmid M. (2009). Repression of flowering by the miR172 Target SMZ. PLoS Biol. 7:e1000148. 10.1371/journal.pbio.1000148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K., Umemura Y., Ohme-Takagi M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55 954–967. 10.1111/j.1365-313X.2008.03565.x [DOI] [PubMed] [Google Scholar]
- Michael T., Mockler T., Breton G., McEntee C., Byer A., Trout J., et al. (2008). Network discovery pipeline elucidates conserved time-of-day–specific cis-regulatory modules. PLoS Genet. 4:e14. 10.1371/journal.pgen.0040014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nova-Franco B., Íñiguez L. P., Valdés-López O., Alvarado-Affantranger X., Leija A., Fuentes S. I., et al. (2015). The Micro-RNA172c-APETALA2-1 node as a key regulator of the common Bean-Rhizobium etli nitrogen fixation symbiosis. Plant Physiol. 168 273–291. 10.1104/pp.114.255547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian M., Ni J., Niu Q., Bai S., Bao L., Li J., et al. (2017). Response of miR156-SPL module during the red peel coloration of bagging-treated Chinese sand pear (Pyrus pyrifolia Nakai). Front. Physiol. 8:550. 10.3389/fphys.2017.00550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian M., Zhang D., Yue X., Wang S., Li X., Teng Y. (2013). Analysis of different pigmentation patterns in ‘Mantianhong’ (Pyrus pyrifolia Nakai) and ‘Cascade’ (Pyrus communis L.) under bagging treatment and postharvest UV-B/visible irradiation conditions. Sci. Hortic. 151 75–82. 10.1016/j.scienta.2012.12.020 [DOI] [Google Scholar]
- Qiao Y., Zhang J., Zhang J., Wang Z., Ran A., Guo H., et al. (2017). Integrated RNA-seq and sRNA-seq analysis reveals miRNA effects on secondary metabolism in Solanum tuberosum L. Mol. Genet. Genomics 292 37–52. 10.1007/s00438-016-1253-5 [DOI] [PubMed] [Google Scholar]
- Qu D., Yan F., Meng R., Jiang X., Yang H., Gao Z., et al. (2016). Identification of microRNAs and their targets Associated with fruit-bagging and subsequent sunlight re-exposure in the “Granny Smith” apple exocarp using high-throughput sequencing. Front. Plant Sci. 7:27. 10.3389/fpls.2016.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll J. J., Bailey L. J., Mai Q.-A., Wu S. L., Hon C. T., Chapman E. J., et al. (2015). microRNA regulation of fruit growth. Nat. Plants 1:15036. 10.1038/nplants.2015.36 [DOI] [PubMed] [Google Scholar]
- Salathia N., Davis S. J., Lynn J. R., Michaels S. D., Amasino R. M., Millar A. J. (2006). FLOWERING LOCUS C-dependent and -independent regulation of the circadian clock by the autonomous and vernalization pathways. BMC Plant Biol. 6:10. 10.1186/1471-2229-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Tiwari M., Pandey A., Bhatia C., Sharma A., Trivedi P. K. (2016). MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiol. 171 944–959. 10.1104/pp.15.01831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siré C., Moreno A. B., Garcia-Chapa M., López-Moya J. J., Segundo B. S. (2009). Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS Lett. 583 1039–1044. 10.1016/j.febslet.2009.02.024 [DOI] [PubMed] [Google Scholar]
- Song Y. H., Shim J. S., Kinmonth-Schultz H. A., Imaizumi T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66 441–464. 10.1146/annurev-arplant-043014-115555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C., Bussell J. D., Camus I., Ljung K., Kowalczyk M., Geiss G., et al. (2005). Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 17 1343–1359. 10.1105/tpc.105.031625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Xu X. H., Wu X., Wang Y., Lu X., Sun H., et al. (2015). Genome-wide identification of microRNAs and their targets in wild type and phyB mutant provides a key link between microRNAs and the phyB-mediated light signaling pathway in rice. Front. Plant Sci. 6:372. 10.3389/fpls.2015.00372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Li M., Zhou Y., Guo T., Liu Y., Zhang H., et al. (2018). Coordinated regulation of Arabidopsis microRNA biogenesis and red light signaling through Dicer-like 1 and phytochrome-interacting factor 4. PLoS Genet. 14:e1007247. 10.1371/journal.pgen.1007247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K., Alonso-Blanco C., Lynn J. R., Michaels S. D., Amasino R. M., Koornneef M., et al. (1999). Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J. 20 67–77. 10.1046/j.1365-313X.1999.00577.x [DOI] [PubMed] [Google Scholar]
- Takano M., Inagaki N., Xie X., Yuzurihara N., Hihara F., Ishizuka T., et al. (2005). Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17 3311–3325. 10.1105/tpc.105.035899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H.-L., Li Y.-H., Hsieh W.-P., Lin M.-C., Ahn J. H., Wu S.-H. (2014). HUA ENHANCER1 is involved in posttranscriptional regulation of positive and negative regulators in Arabidopsis photomorphogenesis. Plant Cell 26 2858–2872. 10.1105/tpc.114.126722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136 669–687. 10.1016/j.cell.2009.01.046 [DOI] [PubMed] [Google Scholar]
- Wang B., Sun Y., Song N., Wang X., Feng H., Huang L., et al. (2013). Identification of UV-B-induced microRNAs in wheat. Genet. Mol. Res. 12 4213–4221. 10.4238/2013.October.7.7 [DOI] [PubMed] [Google Scholar]
- Wang H., Chung P. J., Liu J., Jang I.-C., Kean M. J., Xu J., et al. (2014). Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 24 444–453. 10.1101/gr.165555.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang H. (2015). The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol. Plant 8 677–688. 10.1016/j.molp.2015.01.008 [DOI] [PubMed] [Google Scholar]
- Wang J.-W. (2014). Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 65 4723–4730. 10.1093/jxb/eru246 [DOI] [PubMed] [Google Scholar]
- Wang J.-W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138 738–749. 10.1016/j.cell.2009.06.014 [DOI] [PubMed] [Google Scholar]
- Wang L., Mai Y.-X., Zhang Y.-C., Luo Q., Yang H.-Q. (2010). MicroRNA171c-Targeted SCL6-II, SCL6-III, and SCL6-IV Genes regulate shoot branching in Arabidopsis. Mol. Plant 3 794–806. 10.1093/mp/ssq042 [DOI] [PubMed] [Google Scholar]
- Wang Y., Fan X., Lin F., He G., Terzaghi W., Zhu D., et al. (2014). Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. U.S.A. 111 10359–10364. 10.1073/pnas.1409457111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J., Deng X.-W., Zhu D. (2018). Arabidopsis noncoding RNA modulates seedling greening during deetiolation. Sci. China Life Sci. 61 199–203. 10.1007/s11427-017-9187-9 [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Song Z., Zhang H. (2016). Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis. Mol. Plant 9 1395–1405. 10.1016/j.molp.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Whittaker C., Dean C. (2017). The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33 555–575. 10.1146/annurev-cellbio-100616-060546 [DOI] [PubMed] [Google Scholar]
- Wu G., Park M. Y., Conway S. R., Wang J.-W., Weigel D., Poethig R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138 750–759. 10.1016/j.cell.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Liu D., Wu J., Zhang R., Qin Z., Liu D., et al. (2013). Regulation of FLOWERING LOCUS T by a microRNA in Brachypodium distachyon. Plant Cell 25 4363–4377. 10.1105/tpc.113.118620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Wu C., Xiong L. (2006). Genomic organization, differential expression, and interaction of SQUAMOSA Promoter-Binding-Like transcription factors and microRNA156 in rice. Plant Physiol. 142 280–293. 10.1104/pp.106.084475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Liu Y., Wang H., Ma X., Wang B., Wu G., et al. (2017). Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat. Commun. 8:348. 10.1038/s41467-017-00404-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z., Ye Q., Anson S. R., Yang J., Xiao G., Kowbel D., et al. (2014). Transcriptional interference by antisense RNA is required for circadian clock function. Nature 514 650–653. 10.1038/nature13671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T. T., Ott F., Lanz C., Wollmann H., et al. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22 2156–2170. 10.1105/tpc.110.075606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R. W., et al. (2005). Methylation as a crucial step in plant microRNA biogenesis. Science 307 932–935. 10.1126/science.1107130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Jia T., Chen X. (2017). The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 216 1002–1017. 10.1111/nph.14834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., He H., Wang X., Wang X., Yang X., Li L., et al. (2011). Genome-wide mapping of the HY5-mediated genenetworks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65 346–358. 10.1111/j.1365-313X.2010.04426.x [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhao X., Li J., Cai H., Deng X. W., Li L. (2014). MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant Cell 26 4933–4953. 10.1105/tpc.114.127340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Chen D., Peng Z., Wang L., Gao Z. (2013). Identification and characterization of MicroRNAs in the leaf of Ma Bamboo (Dendrocalamus latiflorus) by deep sequencing. PLoS One 8:e78755. 10.1371/journal.pone.0078755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Cao D., Huang Z., Wang J., Lu S., Xu Y., et al. (2015). Dual functions of GmTOE4a in the regulation of photoperiod-mediated flowering and plant morphology in soybean. Plant Mol. Biol. 88 343–355. 10.1007/s11103-015-0322-1 [DOI] [PubMed] [Google Scholar]
- Zhou B., Fan P., Li Y., Yan H., Xu Q. (2016). Exploring miRNAs involved in blue/UV-A light response in Brassica rapa reveals special regulatory mode during seedling development. BMC Plant Biol. 16:111. 10.1186/s12870-016-0799-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Wang G., Zhang W. (2007). UV-B responsive microRNA genes in Arabidopsis thaliana. Mol. Syst. Biol. 3:103. 10.1038/msb4100143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q.-H., Helliwell C. A. (2011). Regulation of flowering time and floral patterning by miR172. J. Exp. Bot. 62 487–495. 10.1093/jxb/erq295 [DOI] [PubMed] [Google Scholar]