Abstract
In Brief
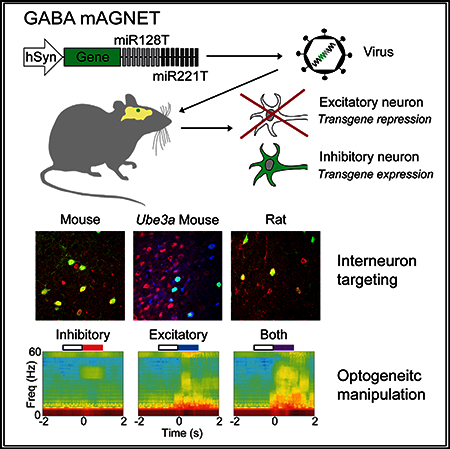
Viral targeting of neuron subtypes is desirable for neuroscience research. Keaveney et al. developed a microRNA-based viral tool for labeling cortical interneurons. They demonstrate its utility via neuron subtype labeling in a murine disease model and in rats and through dual-color optogenetic control of two neuron types.
SUMMARY
More specific and broadly applicable viral gene-targeting tools for labeling neuron subtypes are needed to advance neuroscience research, especially in rodent transgenic disease models and genetically intractable species. Here, we develop a viral vector that restricts transgene expression to GABAergic interneurons in the rodent neocortex by exploiting endogenous microRNA regulation. Our interneurontargeting, microRNA-guided neuron tag, ‘‘GABA mAGNET,’’ achieves >95% interneuron selective labeling in the mouse cortex, including in a murine model of autism and also some preferential labeling of interneurons in the rat brain. We demonstrate an application of our GABA mAGNET by performing simultaneous, in vivo optogenetic control of two distinct neuron subtypes. This interneuron labeling tool highlights the potential of microRNA-based viral gene targeting to specific neuron subtypes.
Graphical Abstract
INTRODUCTION
A wide variety of genetically encoded sensors and actuators are revolutionizing neuroscience research. Over the years, whole-animal transgenic techniques have been successful in mice for targeted gene expression in specific neuron subtypes (Tamamaki et al., 2003; Taniguchi et al., 2011). For other experimental mammalian models and for human gene therapy, transgenic strategies remain limited or impractical, and viral gene delivery remains an effective, time-efficient strategy to transduce post-mitotic neurons (Blessing and Déglon, 2016). Additionally, when used in transgenic lines, viral gene delivery further expands the capacity to express different molecules in neuron subtypes. A major challenge in developing viral gene delivery tools is the limited DNA packaging capacity of the most commonly used viral vectors, lentivirus (LV) and adeno-associated virus (AAV) (Gray et al., 2010; Blessing and Déglon, 2016). Thus, the use of cell-type-specific promoter and enhancer elements for viral gene delivery has been mainly limited to a few neuron types, as the large size and the complexity of these natural elements often leads to complications with viral packaging or transgene expression (Dittgen et al., 2004; Nathanson, et al., 2009a).
We recently explored the use of microRNA (miRNA) regulation, a gene regulation mechanism orthogonal to promoter elements, to restrict gene expression to specific subsets of neurons and illustrated the synthetic biology design principles involved (Sayeg et al., 2015). This strategy is uniquely suited for virally delivered genetic classifiers because miRNA recognition sites are small (~22 nt) and can be easily packaged in viral vectors. Additionally, miRNA targeting is highly engineerable: several miRNAs each with multiple recognition site repeats can be combined to regulate the expression of a single gene. Finally, miRNAs are particularly enriched in the mammalian brain compared to other organs (Nelson et al., 2008), making them highly suitable for neuroscience applications.
Inhibitory neurons (~20% of cortical neurons) play an essential role in regulating neural network activity, and their dysfunction has been implicated in various brain disorders (Marín, 2012). For example, imbalanced excitation and inhibition have been linked to autism spectrum disorders (ASD) (Nelson and Valakh, 2015). While transgenic mouse lines have revolutionized our ability to target specific interneuron types (Taniguchi et al., 2011), it remains difficult to cross-breed transgenic driver lines with transgenic disease models. Virally targeting interneurons has been demonstrated using shortened or non-mammalian promoters (Nathanson et al., 2009a; Delzor et al., 2012) or short cell-type-specific enhancer elements (Lee et al., 2014; Vogt et al., 2014; Dimidschstein et al., 2016). miRNA regulation is orthogonal to promoter and enhancer regulatory elements and represents an additional strategy to achieve interneuron targeting from viral vectors.
In this study, we developed a miRNA-guided neuron tag (‘‘mAGNET’’) to restrict transgene expression to cortical inhibitory (GABA+) neurons in the mouse neocortex (GABA mAGNET). GABA mAGNET achieves 98% cortical interneuron targeting selectivity in mice. To illustrate the utility of this tool, we demonstrated neuron subtype labeling in a Ube3a (2xTg) transgenic mouse model of autism, established some cross-species functionality in the rat cortex and hippocampus, and performed dual-color optogenetic manipulation of distinct neuron subtypes in non-transgenic mice. This work highlights the promise of miRNA-based gene targeting for basic brain research and translational applications.
RESULTS
hSyn-Driven Lentiviral GABA mAGNET Targets Interneurons with 91% Selectivity in Mouse Cortex
Figure 1A illustrates the mAGNET gene-targeting strategy, where we can utilize a constitutive promoter to drive gene expression non-specifically and encode cellular specificity by including ‘‘signature’’ miRNA target or recognition sites (miRT) within the 3’ untranslated region of the gene. Because miRNA regulation is an inhibitory mode of gene expression control, signature miRNAs are chosen based on their higher expression levels in off-target cells compared to the on-target cells (Sayeg et al., 2015). The designed mAGNET vector, when packaged as a virus and injected at a specific location in the brain, leads to the production of transcripts in all infected cells. However, signature miRNAs in off-target cells inhibit gene expression by hybridizing to their complementary target sites within the mAGNET transcript. Conversely, the low level of signature miRNAs in the on-target cell type permits the mAGNET transcripts to be translated and expressed.
Figure 1. mAGNET Gene-Targeting Strategy and LV-GABA-mAGNET Targeting in the Mouse Cortex:
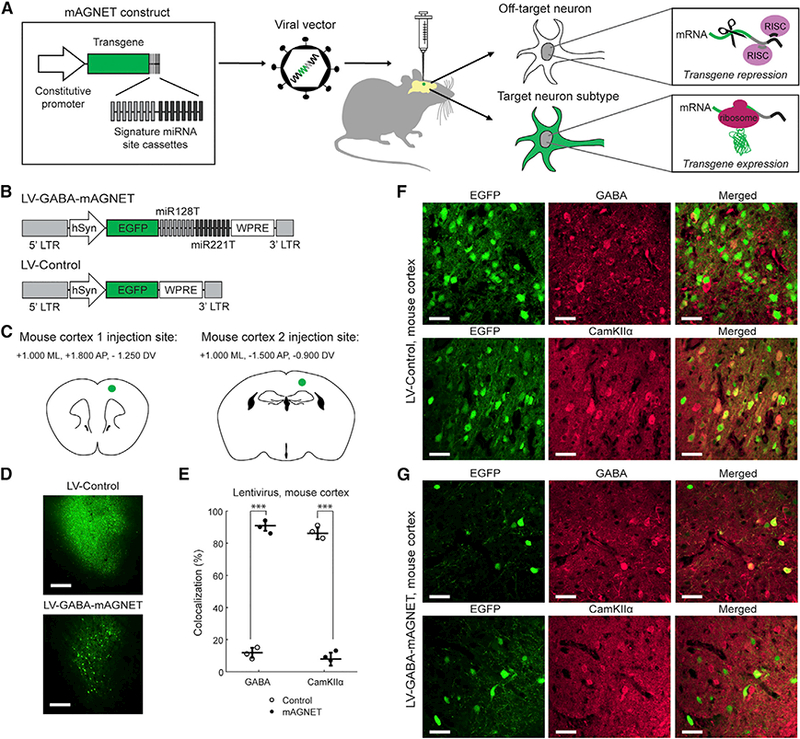
(A) Schematic of viral, microRNA-guided neuron tag (mAGNET) gene-targeting strategy.
(B) LV-GABA-mAGNET and LV-Control lentiviral vector designs; LTR = long terminal repeat, hSyn = human synapsin promoter, EGFP = enhanced green fluorescent protein, and WPRE = woodchuck hepatitis virus post-regulatory element. miR128T/miR221T refer to cassettes of eight respective miRNA target sites - note that these sites are not shown to scale—they are enlarged for visualization purposes.
(C) Schematics of stereotaxic virus injection sites in mouse cortex.
(D) Confocal microscopy images showing comparison of lentivirus transduction in WT mouse cortex (cortex 1 injection site). Scale bars, 250 μm.
(E) Colocalization of EGFP+ cells with inhibitory (GABA) and excitatory (CamKIIα) cell markers in the WT mouse cortex (n = 3 mice each). Scatterplot shows individual animals (circles) and mean (horizontal bar) ± SD. ***p < 0.005, Pearson’s chi-square test.
(F and G) Representative confocal images of EGFP+ cortical cells transduced with LV-Control (F) or LV-GABA-mAGNET (G), immunofluorescence of inhibitory (GABA) and excitatory (CamKIIα) cell markers, and colocalization. Scale bars, 35 μm.
See also Figure S1 and Tables S1 and S2.
To label GABA+ interneurons in the mouse cortex, we selected two signature miRNAs with both high expression levels in off-target excitatory neurons and the highest ratios of expression from excitatory to inhibitory neurons: mir-128 and mir-221 (He et al., 2012). Mir-128 is one of the most highly expressed miRNAs in the mammalian brain (Lagos-Quintana et al., 2002) and is known to be an essential regulator of genes involved in neuronal excitability (Tan et al., 2013). Mir-221 is known to play a role in tumor angiogenesis (Santhekadur et al., 2012), although its function in brain tissue is unknown. Our previous work demonstrated that eight sites for mir-128 and mir-221 each resulted in some restriction of gene expression to interneurons (Sayeg et al., 2015).
To generate a highly specific interneuron mAGNET, we explored orthogonal elements of the gene delivery system, starting with the constitutive promoter that drives mAGNET expression. Our previous designs utilized a strong EF1α promoter (Sayeg et al., 2015). However, we reasoned that this strong promoter that drives transcription at a high rate, may hinder effective regulation by endogenous neural microRNAs in off-target cells. We therefore employed the pan-neuronal human synapsin 1 promoter (hSyn, 480 bp) (Figure 1B), which mediates neuron-specific, long-term transgene expression from viral vectors (Kügler et al., 2003). We packaged the hSyn-driven LV-GABA-mAGNET and LV-Control vectors as lentiviruses pseudotyped with vesicular stomatitis virus glycoprotein (VSVG) envelope protein and injected each at two locations in the adult, wild-type (WT) mouse cortex (n = 3 mice per virus) (Figure 1C). Fluorescence image analysis at two weeks post-injection revealed that virally transduced EGFP+ cells in the mAGNET-infected mice were more sparsely distributed compared to the control (Figure 1D). Furthermore, individual EGFP+ neurons were clearly visible without immunostaining and exhibited no observable difference in fluorescence intensity between the LV-GABA-mAGNET and LV-Control or compared to previous EF1α-driven lentiviruses (Sayeg et al., 2015). These results suggest that LV-GABA-mAGNET virus infected cells within a comparable volume to the LV-Control virus and the sparse distribution of EGFP+ cells is consistent with the distribution of inhibitory neurons in the mammalian neocortex.
Examining brain slices, we assessed interneuron targeting selectivity-defined as the percentage of EGFP+ neurons that colocalize with a GABA immunofluorescence marker for cortical inhibitory neurons. We also independently quantified EGFP colocalization with a CamKIIα immunofluorescence marker for cortical excitatory neurons (off-target cells) as a complimentary measure. The LV-Control vector (with no miRNA sites) exhibited ~12% GABA targeting selectivity (and ~86% colocalization with CamKIIα immunofluorescence) (Figures 1E and 1F; Table S1), consistent with previous observations of a slight tropism toward excitatory neurons with VSVG-pseudotyped lentivirus (Nathanson et al., 2009b; Sayeg et al., 2015). In contrast, the LV-GABA-mAGNET predominantly labeled interneurons, exhibiting 91 ± 3% target selectivity (Figures 1E and 1G; Table S1). Targeting was consistent at both cortical injection locations (Figure S1A).
AAV Packaging Improves GABA mAGNET Targeting Selectivity to 98%
We next explored whether adeno-associated virus (AAV) packaging would further improve interneuron targeting selectivity, given the excitatory-neuron tropism of VSVG-pseudotyped lentivirus (Nathanson et al., 2009b; Sayeg et al., 2015). AAV is commonly used in biomedical research and in human gene therapy (Xu et al., 2001; Nathanson, et al., 2009b; Aschauer, et al., 2013; Maguire et al., 2014; Watakabe et al., 2015). Recently, a systematic comparison of AAV serotypes 1, 2, 5, 6, 8, and 9 in the mouse brain demonstrated that AAV serotype 9 is most efficient for transducing cortical neurons, with slight tropism toward cortical inhibitory neurons (Aschauer et al., 2013). We thus packaged the hSyn-driven GABA mAGNET as AAV2.9 (using AAV-2 genomic backbone pseudotyped with AAV-9 capsid protein) to further improve interneuron targeting (Figure 2A). Because AAVs have a small packaging capacity (~5 kb) compared to lentivirus (~10 kb), the compact size of the miRNA recognition sites, as well as, the short hSyn promoter were especially important for efficient packaging in AAVs.
Figure 2. Selective Interneuron Labeling with rAAV2.9 Packaged GABA mAGNET in the Mouse Cortex:
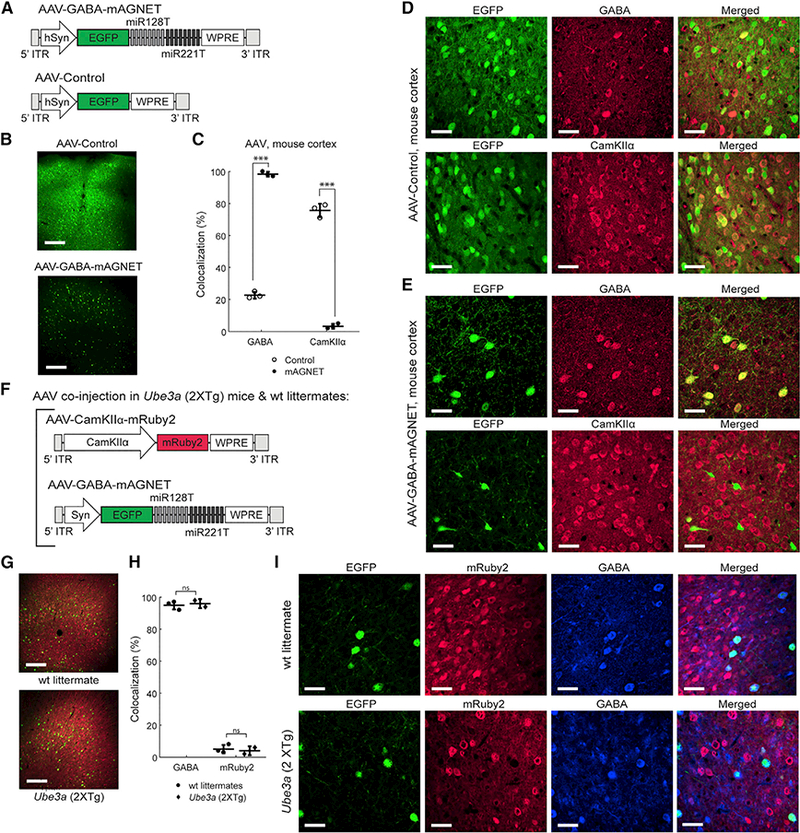
(A) AAV-GABA-mAGNET and AAV-Control vector designs.
(B) Confocal microscopy images showing comparison of AAV transduction in C57bL/6 WT mouse cortex (cortex 1 site). Scale bars, 250 μm.
(C) Colocalization of EGFP+ cells with inhibitory and excitatory marker immune stains in the C57bL/6 WT mouse cortex (n = 3 mice each). Scatterplot shows individual animals (circles) and mean (horizontal bar) ± SD. ***p < 0.005, Pearson’s chi-square test (see Experimental Procedures).
(D-E) Representative confocal images of EGFP+ cortical cells transduced with AAV-Control (D) or AAV-GABA-mAGNET(E), immunofluorescence of inhibitory (GABA) and excitatory (CamKIIα) cell markers, and colocalization. Scale bars, 35 mm.
(F)Co-injected AAV-CamKIIa-mRuby2 and AAV-GABA-mAGNET viral vector designs.
(G) Confocal microscopy images showing cells transduced by AAV-CamKIIα-mRuby2 (red) and AAV-GABA-mAGNET (green) in the cortex (cortex 1 site) of a Ube3a (2xTg) autism model mouse versus FVP WT littermate. Scale bars, 250 μm.
(H) Colocalization of EGFP+ cells (transduced by AAV-GABA-mAGNET) with GABA immunofluorescence or mRuby2 (transduced by AAV-CamKIIα-mRuby2) in Ube3a (2xTg) mice (n = 2) and WT littermates (n = 3). Scatterplot shows individual animals (circles) and mean (horizontal bar) ± SDns = no significance, Pearson’s chi-square test.
(I) Representative confocal images ofEGFP+ cortical cellstransduced with AAV-GABA-mAGNET, mRuby2 fluorescencefrom cortical cellstransduced with AAV CamKIIα mRuby2, immunofluorescence of an inhibitory (GABA) cell marker, and colocalization from a Ube3a (2xTg) mouse (bottom) and WT littermate (top). Scale bars, 35 μm.
See also Figures S1 and S2 and Tables S1 and S2.
We injected AAVs at the cortex 1 injection site (Figure 1C) in adult, WT mice (n = 3 mice per virus), since we previously observed no differences in interneuron targeting across cortical locations (Figure S1). We processed the brain tissue at three weeks post-injection (Figure 2B) and found that the AAVs (Figure 2B) infected a broader region than lentivirus (Figure 1D), consistent with the ability to package AAVs at higher titers (AAV titer: ~2E12 vg/mL, ~0.5 μL injected versus LV titer: ~1E9 infectious particles/mL, ~1 μL injected). Of the AAV-Control transduced EGFP+ cells, 23 ± 2% are GABA+ cells (76 ± 4% are CamKIIa+) (Figures 2C and 2D; Table S1), consistent with the general observation that AAV2.9 exhibits a slight transduction bias toward cortical inhibitory neurons. With this transduction bias, AAV-GABA-mAGNET exhibits 98 ± 2% targeting selectivity for cortical interneurons (Figures 2C and 2E; Table S1), achieving the cell-type targeting standards set by transgenic animals (Tamamaki et al., 2003; Taniguchi et al., 2011).
To determine whether AAV-GABA-mAGNET preferentially labeled a particular interneuron subtype, we performed immunohistochemistry with non-overlapping interneuron subtype markers, parvalbumin (PV), and calretinin (CR). We found that EGFP+ cells expressed these markers in the proportions corresponding to their expected distributions in mouse cortex (Xu et al., 2006; Gonchar et al., 2008; Rudy et al., 2011; Tremblay et al., 2016) (Figure S2). Thus, AAV-GABA-mAGNET is highly specific for cortical interneurons without a labeling bias between interneuron subtypes.
AAV GABA mAGNET Targets Cortical Interneurons with 95% Selectivity in a Transgenic Autism Mouse Model
Viral vector-mediated gene targeting is especially useful in transgenic disease models because it offers the opportunity of cell-type-specific investigation without the need of crossbreeding with cell-type-specific transgenic drive lines. Accordingly, we demonstrated the utility of our AAV-GABA-mAGNET in a mouse autism model with overexpression of the autism risk gene Ube3a (Smith et al., 2011). To examine the targeting selectivity of GABA mAGNET in Ube3a (2XTg) mice, we co-injected AAV-GABA-mAGNET with AAV-CamKIIα-mRuby2 (Figure 2F), a virus that uses a shortened CamKIIα promoter (Dittgen et al., 2004) to drive expression of the red fluorescent mRuby2 reporter in cortical excitatory neurons. These AAVs were co-injected at the cortex 1 location (Figure 1C) in adult Ube3a (2xTg) mice and WT FVP littermate controls. At three weeks post-injection, we found that the total number of transduced EGFP+ neurons was similar between the Ube3a (2XTg) mice and WT littermates (Figure 2G). Of the transduced EGFP+ cells, 96 ± 2% are GABA+ in the Ube3a (2xTg) mice and similarly, 95 ± 3% in the WT littermates (Figures 2H and 2I; Table S1). Together, these results suggest that signature miRNA expression is similar between the WT and autistic transgenic mice and between the FVP and C57b/L6 genetic background. Furthermore, we achieved viral-mediated labeling of two neuron subtypes (inhibitory and excitatory neurons) (Figures 2G–2I), illustrating how our GABA mAGNET will allow further characterization of disease mechanisms and therapeutic strategies in disease models.
Interneuron Targeting with GABA mAGNET in the Rat Brain
Rats are another preferred animal model but with limited genetargeting tools available. Because there is a high degree of miRNA sequence conservation across mammals and especially between rodents (Bak et al., 2008; Minami et al., 2014), we hypothesized that mouse-derived GABA mAGNETs might achieve some interneuron selectivity in the rat brain. We injected GABA mAGNET and control viruses in adult, WT rats (n = 3 per virus) at two cortical sites equivalent to the coordinates in the mice (see Supplemental Experimental Procedures). We again observed that labeled neurons were much sparser in the mAGNET animals compared to the controls (Figures 3A and 3E). LV-Control and AAV-Control virus primarily labeled excitatory neurons with limited interneuron target selectivity (16% for LV-Control [Figures 3B and 3C; Table S1] and 21% for AAV-Control [Figures 3G and 3F; Table S1]). The LV-GABA-mAGNET achieved 74 ± 3% interneuron targeting selectivity (Figures 3B, 3D, and S1B; Table S1), and AAV-GABA-mAGNET exhibited a higher interneuron targeting selectivity of 82 ± 2% (Figures 3F and 3H; Table S1). When tested in the rat hippocampus (Figure S3A), AAV-GABA-mAGNET achieved 80 ± 5% interneuron targeting selectivity (Figures S3C and S3E; Table S1). Together, these results demonstrate that miRNA-based genetargeting exhibits some cross-species and cross-brain region functionality, and further miRNA profile data in rats could facilitate the design of mAGNETs with improved targeting selectivity.
Figure 3. Interneuron Targeting with GABA mAGNETs in the Rat Cortex:
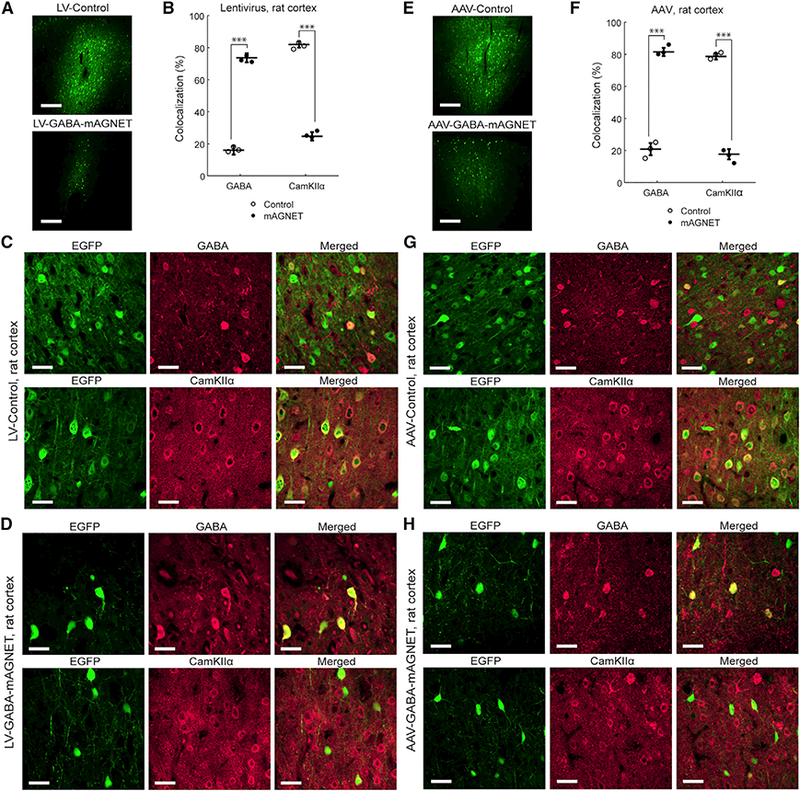
(A) Confocal microscopy images showing comparison of lentivirus transduction in rat cortex (cortex 1 site). Scale bars, 250 μm.
(B) Colocalization of EGFP+ cells with inhibitory and excitatory marker immune stains in rat cortex (n = 3 rats each). Scatterplot shows individual animals (circles) and mean (horizontal bar) ± SD. ***p < 0.005, Pearson’s chi-square test.
(C) Representative confocal images of EGFP+ cortical cells transduced with LV-Control, immunofluorescence of inhibitory (GABA) and excitatory (CamKIIα) cell markers, and colocalization. Scale bars, 35 μm.
(D) Same as (C), for cells transduced with LV-GABA-mAGNET.
(E-H) Same as (A-D), for AAV-Control and AAV-GABA-Magnet
See also Figures S1 and S3 and Tables S1 and S2.
GABA mAGNET Enables Dual-Color Optogenetic Manipulation of Two Neuron Types
To demonstrate an application of our optimized GABA mAGNET, we performed dual-color optogenetic manipulation of two neuron types in the WT mouse cortex using Jaws and ChR2 — rhodopsins that have minimal spectral overlap (Chuong et al., 2014; Klapoetke et al., 2014). We co-injected two AAVs: AAV-Jaws-GABA-mAGNET, which mediates Jaws-mRuby2 expression to inhibitory neurons and allows them to be silenced with red light, and AAV-CamKIIα-ChR2, which mediates ChR2-EGFP expression in CamKIIα+ excitatory neurons and allows these cells to be activated with blue light (Figure 4A). Immuno-staining results verified that interneuron targeting selectivity with the AAV-Jaws-GABA-mAGNET remained high (95 ± 3%).
Figure 4. Simultaneous Optogenetic Manipulation of Two Neuron Subtypes In Vivo with AAV-GABA-Magnet.
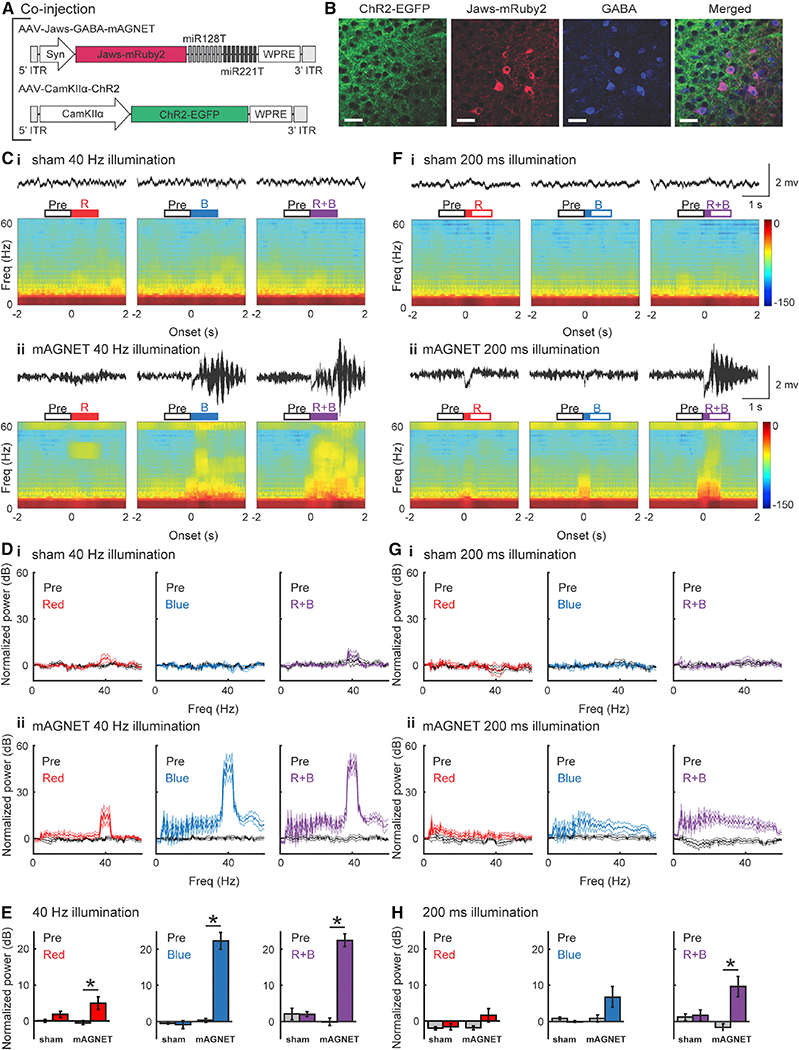
(A) AAVvectordesignsto selectively express Jaws-mRuby2 in cortical inhibitory neuronsforsilencing (AAV-Jaws-GABA-mAGNET), and ChR2-EGFP in cortical excitatory neurons for activation (AAV-CamKIIα-ChR2).
(B) Representative confocal images of CHR2-EGFP expression, Jaws-mRuby2 expression, GABA immunostaining, and colocalization. Scale bars, 35 μm.
(C) Examples of local field potential (LFP) recordings and spectrograms from a sham (non-injected, WT) mouse (Ci) and from a mAGNET (virus-injected, WT) mouse (Cii) during 40 Hz pulse train laser illumination. Both LFP trace and spectrogram were aligned to illumination onset. Open boxes above indicate the time window for calculating oscillation power and solid parts indicate illumination duration: pre-illumination (Pre), red laser illumination (R), blue laser illumination (B), and simultaneous red and blue laser illumination (R+B).
(D) Normalized oscillation power during 40 Hz pulse train laser illumination across animals (sham: n = 5, mAGNET: n = 6).
(E) Gamma oscillations (30–50 Hz) induced by 40 Hz pulse train laser illumination with red laser (left), blue laser (middle), or both (right) in sham and mAGNET mice (*p < 0.05, Wilcoxon rank-sum test). Plot shows mean ± SEM.
(F-H) Same as (C-E), for 200 ms pulse laser illumination.
See also Table S2.
Four weeks after injection, we recorded local field potentials (LFPs) while silencing inhibitory neurons (expressing Jaws) with red light or activating excitatory neurons (expressing ChR2) with blue light, or both simultaneously. We applied two illumination patterns: a 1 s long pulse train at 40 Hz or a single 200-ms-long pulse. In mAGNET mice, 40 Hz illumination with either the red or the blue light alone was sufficient to induce oscillations centered at 40Hz and concurrent illumination with both blue and red light induced stronger oscillations (Figure 4C). At a population level, across all animals, we found that silencing the inhibitory neurons with 40 Hz red light led to a significant increase in LFP oscillation power, centered at 40 Hz (n = 6 mice) (Figures 4Dii and 4E, left [p < 0.05, Wilcoxon rank-sum test]). Activating the excitatory neurons with 40 Hz blue laser illumination, either alone or simultaneously with silencing the inhibitory neurons, produced stronger oscillations (Figures 4Dii and 4E, middle and right [p < 0.05]). These results demonstrate that both Jaws and ChR2 were expressed at functional levels in separate cell types and can be effectively optogenetically controlled; therefore, miRNA regulatory cassettes did not hinder rhodopsin expression in the on-target cells. The same 40 Hz laser illuminations produced no change in sham mice (no virus injection) as expected (n = 5) (Figures 4Ci and 4Di).
With 200 ms pulse illumination, red or blue light alone only elicited small changes in the LFPs, but simultaneous illumination with red and blue light consistently enhanced power at higher frequencies (Figure 4F). At a population level, across all animals, we found that silencing the inhibitory neurons or activating the excitatory neurons with a 200 ms pulse of red light produced little effect (n = 6) (Figure 4Gii, left and middle). Simultaneous inhibitory silencing and excitatory activation induced increases in LFP power over a wide frequency range (Figure 4Gii, right). At gamma frequencies, neither inhibitory silencing nor excitatory activation alone was sufficient to produce significant changes, but simultaneous silencing of inhibitory neurons and activation of excitatory neurons led to significantly stronger gamma oscillations (p < 0.05, Wilcoxon rank-sum test) (Figure 4H). This summation phenomenon confirms successful optogenetic control of both neuron types. 200 ms laser illuminations in sham mice (without virus injection) produced no effects (Figures 4Fi and 4Gi). Together, these results suggest that activation of excitatory neurons and inhibition of inhibitory neurons have an additive effect on network dynamics, when each perturbation produces small changes (Figure 4H [with 200ms long light illumination]). However, when excitatory neurons were driven strongly by ChR2, silencing of inhibitory neurons failed to further enhance oscillation power at gamma frequencies (Figure 4E [1 s light illumination at 40 Hz]).
DISCUSSION
In this study, we have developed GABA mAGNET: a miRNA-based, viral gene delivery tool that restricts transgene expression to cortical inhibitory neurons in mice. Integrating synthetic biology principles on designing signature miRNA binding site cassettes and promoter and viral packaging designs, we report a GABA mAGNET with >95% targeting selectivity toward cortical inhibitory neurons, comparable to that achieved with whole-animal transgenic techniques (Tamamaki et al., 2003; Taniguchi et al., 2011). This interneuron targeting tool remains effective at labeling cortical interneurons in Ube3a 2X Tg transgenic murine model of autism, where transgenic cell-type targeting is difficult. We also demonstrate that GABA mAGNET exhibits ≥80% interneuron specificity in the rat cortex and hippocampus. Finally, to illustrate the possible application of the GABA mAGNET targeting tool, we used it to perform a dual-color optogenetic manipulation of distinct neuron subtypes in non-transgenic mice, highlighting the promise of miRNA-based gene targeting in basic brain research and translational applications.
The mAGNET strategy offers several unique advantages for gene delivery in the brain. First, miRNAs are particularly enriched in the mammalian brain (Nelson et al., 2008), are important in regulating neural activity (Tan et al., 2013), and could potentially serve as biomarkers for brain diseases (Reed et al., 2018). Second, the small footprint of miRNA sites facilitates packaging into virus, an important mode of gene delivery to post-mitotic neurons. Finally, the combinatorial nature of miRNA regulation allows for the potential to engineer selectivity. For example, GABA mAGNETs could be designed to further specify targeting to subtypes of inhibitory neurons, such as parvalbumin, calretinin, or somatostatin neurons, by incorporating specific miRNA cassettes. mAGNETs are orthogonal to other gene regulatory strategies and thus can be easily combined with other cell targeting techniques. For example, short Dlx enhancer elements taken from the distal-less homeobox genes have been shown to be effective in targeting interneurons across several brain regions and species (Lee et al., 2014; Vogt et al., 2014; Dimidschstein et al., 2016). Incorporating the 400-bp-long mDlx enhancer in front of the short hSyn promoter in our AAV-GABA-mAGNET vector may further improve interneuron targeting selectivity in the rat brain.
Viral gene-targeting tools are especially valuable in murine disease models due to challenges in cross-breeding disease model animals with other transgenic cell-type-targeting lines. Our AAV co-delivery experiment demonstrates that AAV-GABA-mAGNET could be applied with other viral cell-type-labeling tools in a mouse model of autism to study the molecular mechanisms underlying excitation/inhibition (E/I) imbalance, a known feature of the disorder (Lee et al., 2017). In fact, this gene-targeting tool could be applied in a variety of murine transgenic disease models to study disease mechanisms (Marín 2012).
The mouse-derived GABA mAGNETs developed here also exhibited interneuron preference in the rat cortex (~74%−82% [Figure 3]) and hippocampus (~80% [Figure S3]). This suggests that miRNA-based targeting has potential in other animal models, including species where transgenic targeting is not possible or more difficult. Although genetic engineering techniques such as CRISPR are yielding promising results in rats (Li et al., 2013) and nonhuman primates (Sato and Sasaki 2018), transgenic targeting has historically been difficult in these species. While the GABA mAGNET was not as specific in the rat brain as in the mouse, it may provide a useful interim research tool. In the future, systematic analysis of cell-type- and brain-region-specific miRNA profiles in other species would greatly advance the design of mAGNETs for use in species where transgenic options are less readily available.
With advancements in genetically encoded neurotechnologies, a need has arisen for methods of genetically targeting independent neuron subtypes in the same animal. Here, we showed that mAGNETs can be used in conjunction with other promoter-based labeling techniques to optogenetically manipulate separate neuron subtypes in wild-type animals. This approach could also be implemented within transgenic animals (Reijmers et al., 2007) to further expand the number of cell types targeted. The results of our experiment stimulating excitatory neurons at 40 Hz are consistent with previous reports that periodic stimulation with blue laser light can activate and entrain ChR2-expressing neurons (Boyden et al., 2005) and LFP power at those frequencies (Iaccarino et al., 2016). Interestingly, we also found that periodic silencing of the inhibitory neurons could also entrain cortical rhythms, likely through disinhibition of the excitatory neurons and/or post-illumination rebound of inhibitory neurons (Chuong et al., 2014) or both. The observation that silencing inhibitory neurons produced less impact than activating excitatory neurons could potentially be due to the fact that inhibitory neurons only constitute a small fraction of the neural population (< 20%).
In the future, miRNA-based genetic classifiers could play a critical role in both basic neuroscience research and human gene therapy. Further cell-type-specific sequencing of miRNAs (Alberti et al., 2018) will allow the design of mAGNETs to target a greater repertoire of neuron types across more species. In addition to targeting known cell types, mAGNETs could also be useful for defining new neuron subtypes. The recent development of single-cell transcriptomics has shed light on the genetic diversity among neurons (Zeisel et al., 2015; Tasic, et al., 2016), suggesting that neuron identity may be too complex to be defined by the expression of one marker gene. Furthermore, mAGNETs may have significance for human gene therapy. Virus is currently the most promising method of therapeutic gene delivery in the central nervous system (Maguire et al., 2014), and the ability to virally target therapeutic transgenes to specific neuronal populations in the human brain would aid the development of better treatments for neurodegenerative and neuropsychiatric disorders. MicroRNAs are already being explored for knocking down gene therapies in off-target tissues (Geisler and Fechner 2016); thus. it’s feasible that the mAGNET approach illustrated here for mice could be further developed to target neuron subtypes in the human brain. The GABA mAGNET presented here provides a means of virally labeling interneurons in the rodent brain and opens the door for the development of many more miRNA-based tools to advance neuroscience research and possibly human gene therapy.
EXPERIMENTAL PROCEDURES
Detailed descriptions are provided in the Supplemental Information. All procedures were in accordance with the NIH Guide for Laboratory Animals and approved by the Boston University Animal Care and Use and Biosafety Committees. Adult female wild-type C57BL/6J mice, FVP/NJ-Tg(Ube3a)1Mpan/J autism model mice, and adult male Sprague-Dawley wild-type rats were used. High-titer replication-incompetent lentivirus and AAVs were prepared and injected into the cortex and the hippocampus via custom hardware. Optogenetic control experiments were performed when mice were awake and head-fixed. LFP recordingswere madewith a glass electrode, and laser illumination was delivered through optical fibers coupled with the glass electrode. Immunohistology was performed with antibodies against GABA, CamKIIα, followed by secondary antibodies and ToPRO-3 nuclear dye forvisualization. Brain slices were imaged on an Olympus FV1000 scanning confocal microscope, and co-localization was quantified manually. Two statistical tests were used throughout this work. Interneuron targeting selectivity was assessed with a Pearson’s chi-square test of independence with one degree of freedom (as cortical neurons examined are either excitatory or inhibitory). Optogenetic control-induced LFP power changes were assessed with a Wilcoxon rank-sum test.
Supplementary Material
Highlights.
Developed GABA mAGNET, a miRNA-based viral gene delivery tool for interneurons
GABA mAGNET achieves >95% cortical interneuron targeting in mice
GABA mAGNET works in a mouse autism model and exhibits some functionality in rats
GABA mAGNET enables viral-mediated optogenetic manipulation of two neuron subtypes
ACKNOWLEDGMENTS
X.H. acknowledges funding from NIH Director’s New Innovator Award 1DP2NS082126, NINDS 1R01NS087950–01, Pew Foundation, Alfred P. Sloan Foundation, and Boston University Biomedical Engineering Department. M.K.K. acknowledges NIH F31NS095465–02. H.Y.M. acknowledges NIH R01MH079407. We thank Shamsh Shaikh and Kimberly Ching for helping to optimize immunohistology and confocal imaging. We thank Yuda Huo for providing instruction on producing AAV viruses and Zachary Gardner for breeding Ube3a (2xTg) autism mice.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
DATA AND SOFTWARE AVAILABILITY
The accession numbers for the viral vector sequences reported in this paper are GenBank: (LV-GABA-mAGNET) MH458076, (LV-Control) MH458077, (AAV-GABA-mAGNET) MH458078, (AAV-Control) MH58079, (AAV-CamKIIalpha-mRuby2) MH58080, (AAV-Jaws-GABA-mAGNET) MH458081, (AAV-CamKIIalpha-ChR2) MH458082.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, three figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.06.049.
REFERENCES
- Alberti C, Manzenreither RA, Sowemimo I, Burkard TR, Wang J, Mahofsky K, Ameres SL, and Cochella L (2018). Cell-type specific sequencing of microRNAs from complex animal tissues. Nat. Methods 15, 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschauer DF, Kreuz S, and Rumpel S (2013). Analysis of transduction efficiency, tropism, and axonal transport of AAV serotypes 1,2, 5,6, 8 and 9 in the mouse brain. PLoS ONE 8, e76310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak M, Silahtaroglu A, M0ller M, Christensen M, Rath MF, Skryabin B, Tommerup N, and Kauppinen S. (2008). MicroRNA expression in the adult mouse central nervous system. RNA 14, 432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blessing D, and Deglon N (2016). Adeno-associated virusand lentivirusvectors: a refined toolkit for the central nervous system. Curr. Opin. Virol 21, 61–66. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, and Deisseroth K (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci 8, 1263–1268. [DOI] [PubMed] [Google Scholar]
- Chuong AS, Miri ML, Busskamp V, Matthews GA, Acker LC, Sørensen AT, Young A, Klapoetke NC, Henninger MA, Kodandaramaiah SB, et al. (2014). Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat. Neurosci 17, 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delzor A, Dufour N, Petit F, Guillermier M, Houitte D, Auregan G, Brouillet E, Hantraye P, and Déglon N (2012). Restricted transgene expression in the brain with cell-type specific neuronal promoters. Hum. Gene Ther. Methods 23, 242–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimidschstein J, Chen Q, Tremblay R, Rogers SL, Saldi GA, Guo L, Xu Q, Liu R, Lu C, Chu J, et al. (2016). A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci 19, 1743–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittgen T, Nimmerjahn A, Komai S, Licznerski P, Waters J, Margrie TW, Helmchen F, Denk W, Brecht M, and Osten P (2004). Lentivirus-based genetic manipulations of cortical neurons and their optical and electro-physiological monitoring in vivo. Proc. Natl. Acad. Sci. USA 101,18206–18211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler A, and Fechner H (2016). MicroRNA-regulated viral vectors forgene therapy. World J. Exp. Med 6, 37–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonchar Y, Wang Q, and Burkhalter A (2008). Multiple distinct subtypes of GABAergic neurons in mouse visual cortex identified bytriple immunostaining. Front. Neuroanat 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SJ,Woodard KT, and Samulski RJ (2010). Viral vectors and delivery strategies for CNS gene therapy. Ther. Deliv 1, 517–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, and Huang ZJ (2012). Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron 73, 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- laccarino HF, Singer AC, Martorell AJ, Rudenko A, Gao F, Gillingham TZ, Mathys H, Seo J, Kritskiy O, Abdurrob F, et al. (2016). Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature 540, 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, et al. (2014). Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler S, Kilic E, and Bahr M (2003). Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther. 10, 337–347. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, and Tuschl T (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol 12, 735–739. [DOI] [PubMed] [Google Scholar]
- Lee AT, Gee SM, Vogt D, Patel T, Rubenstein JL, and Sohal VS (2014). Pyramidal neurons in prefrontal cortex receive subtype-specific forms of excitation and inhibition. Neuron 81, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Lee J, and Kim E (2017). Excitation/Inhibition Imbalance in Animal Models of Autism Spectrum Disorders. Biol. Psychiatry 81, 838–847. [DOI] [PubMed] [Google Scholar]
- Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, et al. (2013). Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol 31, 681–683. [DOI] [PubMed] [Google Scholar]
- Maguire CA, Ramirez SH, Merkel SF, Sena-Esteves M, and Breakefield XO (2014). Gene therapy for the nervous system: challenges and new strategies. Neurotherapeutics 11, 817–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O (2012). Interneuron dysfunction in psychiatric disorders. Nat. Rev. Neurosci 13, 107–120. [DOI] [PubMed] [Google Scholar]
- Minami K, Uehara T, Morikawa Y, Omura K, Kanki M, Horinouchi A, Ono A, Yamada H, Ohno Y, and Urushidani T (2014). miRNA expression atlas in male rat. Sci. Data 1, 140005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson JL, Jappelli R, Scheeff ED, Manning G, Obata K, Brenner S, and Callaway EM (2009a). Short Promoters in Viral Vectors Drive Selective Expression in Mammalian Inhibitory Neurons, but do not Restrict Activity to Specific Inhibitory Cell-Types. Front. Neural Circuits 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson JL, Yanagawa Y, Obata K, and Callaway EM (2009b). Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience 161, 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SB, and Valakh V (2015). Excitatory/Inhibitory Balance and Circuit Homeostasis in Autism Spectrum Disorders. Neuron 87, 684–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Wang WX, and Rajeev BW (2008). MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 18, 130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed ER, Latourelle JC, Bockholt JH, Bregu J, Smock J, Paulsen JS, and Myers RH; PREDICT-HD CSF ancillary study investigators (2018). MicroRNAs in CSF as prodromal biomarkers for Huntington disease in the PREDICT-HD study. Neurology 90, e264–e272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmers LG, Perkins BL, Matsuo N, and Mayford M (2007). Localization of a stable neural correlate of associative memory. Science 317, 1230–1233. [DOI] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, and Hjerling-Leffler J (2011). Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev. Neurobiol 71, 45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhekadur PK, Das SK, Gredler R, Chen D, Srivastava J, Robertson C, Baldwin AS Jr., Fisher PB, and Sarkar D (2012). Multifunction protein staphylococcal nuclease domain containing 1 (SND1) promotes tumor angiogenesis in human hepatocellular carcinoma through novel pathway that involves nuclear factor kB and miR-221. J. Biol. Chem 287, 13952–13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, and Sasaki E (2018). Genetic engineering in nonhuman primates for human disease modeling. J. Hum. Genet 63, 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayeg MK, Weinberg BH, Cha SS, Goodloe M, Wong WW, and Han X (2015). Rationally Designed MicroRNA-Based Genetic Classifiers Target Specific Neurons in the Brain. ACS Synth. Biol 4, 788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Zhou YD, Zhang G, Jin Z, Stoppel DC, and Anderson MP (2011). Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci. Transl. Med 3, 103ra97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, and Kaneko T (2003). Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J. Comp. Neurol 467, 60–79. [DOI] [PubMed] [Google Scholar]
- Tan CL, Plotkin JL, Vene MT, von Schimmelmann M, Feinberg P, Mann S, Handler A, Kjems J, Surmeier DJ, O’Carroll D, et al. (2013). MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science 342, 1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. (2011). A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci 19, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay R, Lee S, and Rudy B (2016). GABAergic Interneurons in the Neocortex: From Cellular Properties to Circuits. Neuron 91, 260–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt D, Hunt RF, Mandal S, Sandberg M, Silberberg SN, Nagasawa T, Yang Z, Baraban SC, and Rubenstein JL (2014). Lhx6 directly regulates Arx and CXCR7 to determine cortical interneuron fate and laminar position. Neuron 82, 350–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Ohtsuka M, Kinoshita M, Takaji M, Isa K, Mizukami H, Ozawa K, Isa T, and Yamamori T (2015). Comparative analyses of adeno-associated viral vector serotypes 1, 2, 5, 8 and 9 in marmoset, mouse, and macaque cerebral cortex. Neurosci. Res 93, 144–157. [DOI] [PubMed] [Google Scholar]
- Xu R, Janson CG, Mastakov M, Lawlor P, Young D, Mouravlev A, Fitzsimons H, Choi KL, Ma H, Dragunow M, et al. (2001). Quantitative comparison of expression with adeno-associated virus(AAV-2) brain-specific gene cassettes. Gene Ther. 8, 1323–1332. [DOI] [PubMed] [Google Scholar]
- Xu X, Roby KD, and Callaway EM (2006). Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. J. Comp. Neurol 499, 144–160. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. (2015). Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


