Abstract
Immunotherapies (IT) require induction, expansion, and maintenance of specific changes to a patient’s immune cell repertoire which yield a therapeutic benefit. Recently, mechanistic understanding of these changes at the cellular level has revealed that IT results in complex phenotypic transitions in target cells, and that therapeutic effectiveness may be predicted by monitoring these transitions during therapy. However, monitoring will require unique tools that enable capture, manipulation, and profiling of rare immune cell populations. In this study, we introduce a method of automated and unsupervised separation and processing of rare immune cells, using high-force and multidimensional magnetic ratcheting (MR). We demonstrate capture of target immune cells using samples with up to 1:10,000 target cell to background cell ratios from input volumes as small as 25 microliters (i.e. a low volume and low cell frequency sample sparing assay interface). Cell capture is shown to achieve up to 90% capture efficiency and purity, and captured cell analysis is shown using both on-chip culture/activity assays and off-chip ejection and nucleic acid analysis. These results demonstrate that multi-directional magnetic ratcheting offers a unique separation system for dealing with blood cell samples that contain either rare cells or significantly small volumes, and the “sample sparing” capability leads to an expanded spectrum of parameters that can be measured. These tools will be paramount to advancing techniques for immune monitoring under conditions in which both the sample volume and number of antigen-specific target cells are often exceedingly small, including during IT and treatment of allergy, asthma, autoimmunity, immunodeficiency, cell based therapy, transplantation, and infection.
Introduction
Immunotherapy (IT) requires the induction, expansion, and maintenance of specific changes to a patient’s immune system which yield a therapeutic benefit.1–4 Often these changes are a result of a complex progression of interactions between an exogenously introduced agent (e.g. cell, antigen, biologic), a target cell (or tissue), and several supporting cells that modulate the target’s response.4 These complex interactions that give rise to the therapeutic benefit have driven a major clinical research emphasis on defining the underlying immunological mechanism of action in IT; a strategy which is apparent across disease applications. Recently, investigation of mechanisms at the cellular level has revealed that IT results in complex phenotypic transitions in the “therapeutic” cells, and that the effectiveness of treatment may be predicted by monitoring these phenotypic transitions during therapy.
Some examples of IT cell states that require monitoring include the activation state of therapeutically modified T cells (which has been shown to indicate sustained therapeutic activity in chimeric antigen receptor (CAR) T applications).5 In food allergy ITs, investigation of the mode-of-action has revealed that immune changes accompanying therapy may not persist, as allergen specific immune cells may transition into transient “non-allergic” versus more permanent tolergenic phenotypes.4,6,7 In infectious disease applications, screening of antigen-specific T cells has been applied to examine whether seasonal influenza vaccinations induce response to novel re-assorted strains.8–10 And, in studies of human autoimmune disease, such as multiple sclerosis, similar approaches have been used to demonstrate that there are specific phehnotypic changes in the frequency of autoreactive T cells that secrete pro-inflammatory cytokines, while in control patients autoreactive T cells predominately secrete suppressive cytokines.11–13
However, these specific, disease associated immune cells are rare and often only available from precious and low volume clinical samples (see S1 for more information on the need for sample sparing assays in immunotherapy diagnostics). In human autoimmune disease applications, the target antigen specific immune cells can exist at frequencies of 1:100,000 in peripheral blood.14 And, the cost, complexity, and skill associated with capturing and analysing these cells has limited the total number of clinical and mechanistic studies on antigen-specific cell populations. Even within the studies that exist, sample sizes of participants are small and, to ensure significance, researchers depend instead on comparison to many different control settings.4 A primary reason for this remains the expense and complexity of multivariate rare cell assays. To date, steps to capture and analyse these cells have included capture using antigen-specific multimers, FACS sorting in multiwell plates for single cell nucleic acid isolation and preparation, and multivariate gene expression analysis on microfluidic PCR devices.4,15,16 While seemingly trivial, the method used to capture the low-abundant single cells and prime them for down-stream phenotyping remains one of the main hurdles.
Fluorescence activated cell sorting (FACS) remains the primary separation technology used in clinical research. Yet, FACS instruments have limited success with extremely rare cell populations, and require a dedicated flow cytometrist to operate (with substantial maintenance costs to reproducibly enrich cells for single-cell assays).17 Other automated optical systems are often limited in throughput of cells per run, and are not suited for rare or low-abundant cell analysis (i.e. >1000 target cells are required for each step), or have a prohibitive cost per sample.4 Alternative magnetic activated cell sorting (MACS) systems provide only binary separations (separate particle bound versus unbound cells) and can yield low purity extractions for rare cells due to non-specific binding or non-specific uptake of magnetic particles.18
Within the last two years new classes of cell sequencing technologies have emerged that rely on random priming of individual cells with molecular barcodes for post-sequencing indexing of transcripts to its cell of origin.19,20 The studies have been performed using both microfluidic droplets and microwells for partitioning and barcoding each cell’s contents, prior to post-capture single-tube library preparation protocols and sequencing. These new platforms have increased the cell throughput of multivariate (genome-wide) expression profiling. However, the primary application of these tools remains biological discovery, as direct head-to-head comparisons between non-barcoded (SMART-seq) and the high-throughput bar-coded sequencing protocols have shown up to a 50% decrease in the number of genes detected per cell.21 These technical challenges may prohibit the use of these high-throughput single cell sequencing methods in clinical settings where quantitative assessment of sub-sets of genes (including those with low transcript number) is the primary goal.
In this study, we introduce a method of automated and unsupervised separation and processing of rare antigen-specific immune cells from blood cell samples. The method utilizes a magnetic separation technology22 that employs arrays of ferromagnetically soft micropillars to separate and isolate low-abundant immune cell populations (labelled with magnetic beads) in a quantitative fashion (based on the quantity of attached beads) using high-force magnetic ratcheting (MR). Unlike the current FACS or microfluidic rare cell handling platforms used today, the MR system requires no fluidic handling system. Instead, the target cells are automatically extracted from clinical samples and processed using discrete magnetically driven displacements across the device. Furthermore, unlike the highly supervised FACS separations that currently dominate cell analysis methods (requiring monitoring, analysis, and gating based on multiple light scatter and fluorescence detection lines), the MR system uses an unsupervised mechatronic device that generates a cycling magnetic field capable of deterministic displacement, separation, and processing of individual cells. Using the system, automated capture of low-abundant target immune cells was demonstrated from up to 1: 10,000 target cell:PBMC ratios. Target cells were localized to specific points on the device by ratcheting cells across an increasing gradient of micropillar traps. This technique was shown effective in automatically gating magnetic cells based on surface expression of several cell surface markers. Captured rare cell populations (from ~50 to 4000 total target cells within millions of background PBMCs) were then deterministically loaded into wells on the MR device using multi-directional MR. A macro-scale valve enabled isolation of the chamber containing the captured cells, injection of reagents for cell processing or ejection of the cells for use in other down-stream analytical systems. The magnetic ratcheting technique is shown compatible with several post-capture analytical methods, including imaging cytometry, culture and amplification of antigen-specific cells, protein secretion assays, and RNA sequencing analysis. These results suggest that unsupervised immune cell handling platforms (such as the magnetic ratcheting system) are a viable means to replace the complex and costly FACS and microfluidic systems currently limiting isolation of rare cells in clinical studies
Materials and methods
Magnetic Ratcheting Device and System
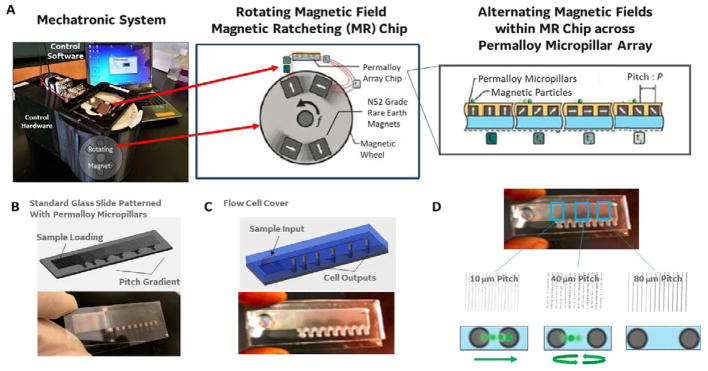
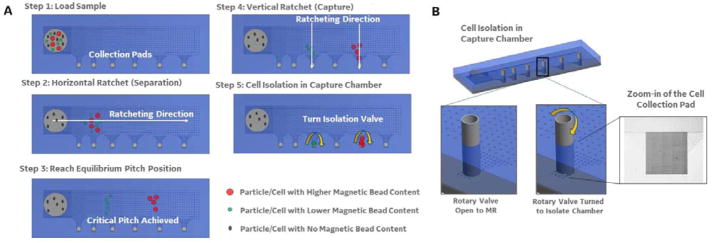
We have developed a multi-directional magnetic ratcheting platform, which digitizes magnetic bead-based cell capture and processing. Figure 1 shows the simple control system (1A) and device (1A-D) used to perform highly parallel magnetic ratcheting of cells from clinical samples. The device (1B) consists of a standard glass microscope slide patterned with a two-dimensional array of magnetically soft micro-pillars (fabrication described previously22). The micro-pillars magnetize in alignment to larger applied magnetic fields (1A, right), and application of a directionally cycled magnetic field creates a dynamic array of potential energy “wells” that transit across the device. The simple mechatronic control system contains a rotating magnetic wheel and functions to continuously cycle the potential energy wells across the surface of the device (1A, left). As shown in the zoom-in image, superparamagnetic particles (or cells labelled with particles) added to the top of the device follow the potential wells and ratchet from pillar to pillar with each passing rotation of the magnetic wheel. The micro-pillars are fabricated with a defined inter-pillar distance (or pitch) which increases across the chip, and the frequency of the rotating magnetic field can be adjusted until there is no longer a sufficient time averaged magnetic force for the particle to traverse to the next pillar (i.e. the critical pitch). Therefore, particle-labelled cells equilibrate at a location with a critical pitch that depends on the quantity of magnetic particles bound to the cell.
Figure 1.
A. (left) The mechatronic system used to control cell ratcheting across the device. The hardware consists of permanent magnets enclosed in a wheel, and a motor controller to rotate the wheel at defined frequencies. (middle) A stage holds the magnetic ratcheting chip at a fixed distance above the rotating magnetic field, allowing a time varying magnetic field to be applied to the chip. (right) As the magnetic field rotates, the permalloy paramagnetic pillars are magnetized in sequence (t0–t4), thus magnetic particles (or magnetic bead bound cells) experience a time varying magnetic force that pushes the particle toward the neighbouring pillar.22 B. The paramagnetic pillars are patterned on a glass slide in an arrangement with increasing distance between pillars (from left to right on the slide).22 At a certain point across the chip, the rotating magnetic field fails to provide enough magnetic force to push a particle (or cell with a specific number of attached magnetic beads) to the next pitch region. C. The changing pillar pitch across the slide creates critical pitch “capture regions”, where the termination points at the bottom of each pitch region become collection points (i.e. areas where cells achieving equivalent critical pitch are concentrated and collected). In addition, the sample input has a constant pitch of very closely packed pillars, so that all magnetic particles (or labelled cells) are ratcheted away from the sample input and toward the varying pitch separation region. D. The images show an example of 1 micron magnetic beads after introduction to the sample input, and ratcheting for 15 minutes at 15 Hz. The 1 micron particles moved across the ratchet, and achieved a “critical pitch” at 40 micron pillar pitch (as shown).
The device (1C) is covered with a simple fluid chamber that contains three key features: designed to accept clinical samples (sample input), separate out target immune cells based on cell-surface proteins labelled by beads (coated with a recognition element), and localize those cells in open wells for further processing (sample outputs). The sample loading region consists of a patch of seven-micron pitch micro-pillars covered by a simple open well (in which the clinical sample can be injected using a standard pipette). The tight inter-pillar distance within this region is designed to be less than the critical pitch and ensures that any particle-bound cell will be ratcheted away from this loading region. Upon actuation of the rotating magnetic wheel, labelled cells begin translating or ratcheting into the neighbouring separation region at an average particle velocity of Pitch (microns) x Frequency.22 There is no other fluid motion within the device, and the cells traversing the separation region experience only the force applied by the rotating magnetic field. Within this region, the cells move across a gradient of micro-pillar patches (1D), each fabricated with an increasing pitch. The equations governing ratcheting (previously characterized by the balance of the time averaged magnetic versus drag force22) provide a means to estimate both particle velocity (i.e. separation speed) and critical pitch (i.e. equilibrium position of separated cells across the pillar array).
The magnetic particles utilized to label and actuate cells across the micro-pillar array are labelled with specific antibodies or tetramers with high affinity to cell surface proteins. Therefore, the total number of magnetic particles on each cell depends on the surface expression of that target protein. Cells labelled with only a few non-specifically bound magnetic particles will reach a critical pitch very early and remain on the left side of the device, cells with low surface expression of the target or cell-specific protein will be bound to a few more magnetic particles and traverse further to a larger critical pitch, while cells with high surface expression will traverse across the length of the device. This quantization of a magnetic separation process results in a cell population distribution across the device that depends on the surface expression of the target protein. However, unlike flow cytometry that requires significant supervision (determination of specific light scatter and fluorescent “gates”) to achieve cell separation, magnetic ratcheting provides completely unsupervised separation that is based entirely on underlying physical properties. As shown in figure 1D (using 1 micron magnetic particles with a critical pitch of 40 micron pillar spacing at magnetic rotation of 5 Hz), each particle or cell traverses the micro-pillar array to its critical pitch, once that particle does not have sufficient migration time to reach the next pillar it will oscillate and become trapped indefinitely at that device position.
System Operation and Multi-Direction Ratcheting
The final challenge to enabling rare cell analysis is to enable simple collection of the separated cells for processing. To accomplish this, we designed collection pads at the bottom of each pitch region (2A and B). After sample loading (step 1), the magnetic wheel rotates horizontally (parallel with the separation region, step 2). However, once a critical pitch is achieved (step 3) and during cell collection, the magnetic wheel’s direction of rotation is turned 90° driving the separated cells toward the bottom of the device (step 4). The collection pads consist of a grouping of termination points for each vertical aligned set of micropillars. The vertical inter-pillar pitch is held constant across the device so that, although the cells are separating into each pitch region horizontally, all cells across the device will ratchet vertically at the same speed. The zoom-in of the collection pad (2B; left) shows that a series of diagonal micro-pillars connect all the vertical micro-pillar columns from each pitch region into a single collection pad (effectively concentrating cells from each pitch region to a corresponding 2 mm × 2 mm spot). In addition, each collection pad is covered by an open well that contains a rotary valve (2B). An opening in the rotary valve is aligned to face the micropillar separation array during processing (to allow cell entrance into the collection pad). The rotary valve is then manually turned, shutting off the connection between the collection pad and the separation array. There is a 200 micron space between the rotary valve opening and the ratcheting surface when open, allowing cells to enter the collection pad. After ratcheting is complete and the valve is manually shut (i.e. rotated to the side that does not contain an opening), the separated cells can be imaged in place, cultured in place (through the addition of fresh culture media), or ejected through simple pipetting without disturbing the cells/liquid in the surrounding areas of the device, which would reduce the sample purity and potentially dilute secreted compounds.
Cell Samples and Magnetic Bead Reagents
Peripheral blood mononuclear cells (PBMCs) were obtained from either the UCLA CFAR Virology Core or the GE Cell Therapy Group core and were thawed using RPMI complete media (10% FBS and 1% Pen-Strep) and counted using a hemocytometer. For spiking experiments separate PBMC samples were stained with either Calcein AM green to represent target cells or Calcein AM red to represent background cells. Both samples were then centrifuged and washed three times to prevent cross staining when mixed together. To simulate rare antigen-specific cell populations, between 100–1000 target cells were injected into a background of 1 million background PBMCs then magnetically labelled using either 500nm, 1μm, or 4μm magnetic beads which were functionalized to target either CD11b, CD4/8, or CD69. To create functionalized beads, commercially available streptavidin coated magnetic particles (Micromod GmbH or Invitrogen) were incubated with a molar excess of biotinylated antibody for CD11b, CD4/8 or CD69. The beads were washed to elute unbound antibody and added to cell samples at particle to target cell ratios of 100:1, 10:1, and 1:1, corresponding to the 500nm, 1μm, and 4μm particle diameters. After addition of magnetic particles, cells were incubated (for up to 30 minutes) at room temperature under a constant stirring rotation of 20 RPM. Samples were then added to the ratcheting chips and separated.
Flow cytometry was also performed to confirm the presence of surface antigen on target cell populations. Target cell populations were stained with DRAQ5 nuclear stain (Thermo Fisher) and labelled with phycoerythrin (PE) fluorescent antibody for either CD11b (BioLegend 101207 [M1/70]), CD4/8 (Miltenyi Biotec, 130-110-676, 130-098-543), CD154 (Miltenyi Biotec, 130-092-289) or CD69 (BioLegend [FN50]). Samples were then analysed on a BD FACS Canto II flow cytometer.
CD154 Stimulation and Spiking
Cryopreserved Peripheral Blood Mononuclear Cells (PBMCs) were purchased from ALLCELLs (Alameda, CA, USA; Cat No. PB003F) and stored at −180°C in vapor phase liquid nitrogen until ready for experimental use. Cells were thawed by placing cryovial in 37°C water bath for 2 minutes. Vial content (1mL) was resuspended in 9mL of serum containing culture media and centrifuged at 300 × g for 10 minutes. Supernatant was discarded and cell pellet was resuspended in serum containing culture media at a density of 1×107 PBMCs/mL. Serum containing culture media: 5% HSA (Human Serum Albumin; Off the clot, Cat No. HS1017; Valley Biomedical Service Inc, Winchester, VA, USA), ImmunoCult™ XF T Cell Expansion Medium (Cat No. 10981, STEMCELL Technologies Inc, Cambridge, MA, USA), 1% GlutaMAX Supplement (Cat No. 35050061, ThermoFisher Scientific, Waltham, MA USA), 1% Penicillin-Streptomycin (5,000 U/mL) (Cat No. 15070063, ThermoFisher Scientific). 1% N-Acetyl-L-cysteine (Cat No. A9165-25G, Sigma Aldrich, USA).
T cells were activated using CytoStim (Cat No. 130-092-172, Miltenyi Biotec, Bergisch Gladbach, Germany). PBMCs in serum containing culture media at a cell density of 1.0×107 cells/mL were plated in tissue culture plates (Corning, Tewsksbury, MA, USA) at a density of 5×106 cells/cm2. Using a 12 well plate design, total volume used was 1mL with 2.0×107 cells per well. CytoStim was added at 20μL/mL and CD40 pure-functional grade at 1μg/mL (Cat No. 130-094-133, Miltenyi Biotec) to the cell suspension. Cell suspension was mixed carefully and incubated at 37°C, 5% CO2 for 4 hours.
After stimulation of PBMCs with CytoStim, CD154+ T cells were labelled with magnetic beads. For magnetic bead labelling isolation buffer was prepared by diluting MACS BSA Stock Solution (Cat No. 130-091-376, Miltenyi Biotec) 1:20 with autoMACS Rinsing Solution (Cat No. 130-091-222, Miltenyi Biotec). Stimulated cells were carefully collected from tissue culture plate by pipetting up and down, and rinsing the tissue culture plate with cold isolation buffer. During magnetic labelling cells were kept cold, using pre-cooled solutions. Cell number was determined using an automated cell nucleo-coutner NC200 (Chemometec). Cells were centrifuged at 300 × g for 10 minutes, supernatant was aspirated and cells were resuspended in 40 uL of isolation buffer per 1×107 total cells. Dynabead MyOne Streptavidin T1 (Thermofisher) beads were labelled with CD154-Biotin (Miltenyi Biotec) per manufacturer’s instructions, and resuspended to a final volume that allowed incubation of 1×107 cell/mL at a particle to total cell ratio of 10:1.
The ratio of CD154+ T cells within each sample was established using a CytoFLEX (Beckman Coulter) volumetric flow cytometer, and a number of additional and confirmatory markers including CD8+ FITC (Cat No. 557696, BD Biosciences, San Jose, CA, USA), CD4+ V500 (Cat No. 560768), CD25+ PE (Cat No. 555432), and CD3+ PerCpCy5.5 (Cat No. 560835). CD4-APC (Miltenyi, 130-092-374) and Anti-Biotin-PE (Miltenyi, 130-090-756) were used to check the level of expression for CD154 in the positive and negative fractions. A specific volume of sample was then chosen (based on the number of desired input CD154+ cells within each experiment) and spiked into fresh non-labelled PBMCs at the desired ratio.
Tetramer Staining and Spiking
DRB1*0401 soluble class II molecules were generated as previously described.14 MOG97-109 was used to load the DRB1*0401 molecule to generate the DRB1*0401/MOG97-109 tetramers. Phycoerythrin (PE, BioSource International, Camarillo, CA) conjugated streptavidin was used for cross-linking of peptide-MHC II monomers. PBMC from HLA-DR4+ HLA-DR4+ MS patients were isolated from venous peripheral blood (as described above), cryopreserved and kept in liquid N2 until used. The cells were thawed, washed with PBS, and then stained with 10 μg/ml DR4 tetramers (MOG97-109 tetramers), anti-CD4, and live/dead dye (Life technologies) at 37 °C for 30 minutes in RPMI 1640 medium with 2% human AB serum. After washing with PBS, the percentage of live D4+tetramer+ and/or CD4+tetramer− T cells was determined by flow cytometry using a FACS Aria.14 For the isolation of CD4+tetramer+ cells using the magnetic ratcheting device. 5×106 PBMCs were labeled with tetramers as for flow cytometry. After washing the excess of unbound tetramer, the cells were incubated with 1/100 anti-PE biotin (13–4120, ebioscience) for 15 minutes at room temperature, washed once, then labeled for 15 minutes with a 3:1 paramagnetic particles to total cells ratio (65601, Dynabeads). The sample was diluted to 5×106 PBMCs/ml and loaded onto the magnetic ratcheting system, magnetic field rotation was set at 5 Hz for 10 minutes and cell separation was checked at the end of the run under the microscope.
Optical Characterization and Validation
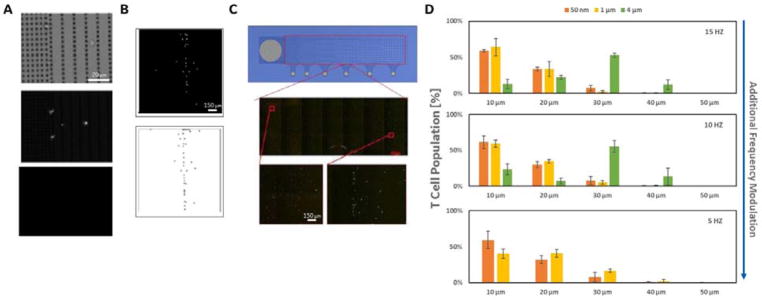
Figure 3 A–D shows the process for optical characterization of the MR separation using immune cells labelled with 500 nm anti-CD11b (myeloid cell marker) labelled paramagnetic particles spiked to a known concentration within a background of unlabelled PBMCs. The magnetic field rotation speed for this example was set at 5 Hz. Figure 3A shows one microscopic field of view taken from the interface between the sample loading region (7 micron pitch) and the separation region (10 micron pillar pitch) after ratcheting. Each field of view is characterized using three optical channels (3A), bright field (showing an image of the cells captured), a target channel (using a fluorescently labelled antibody specific for a target cell specific surface marker), and a non-specific channel (using a fluorescently labelled antibody that binds to a non-target cell specific surface marker). For the CD11b experiments, the magnetic bead labelled cells were further labelled with fluorescent antibodies against anti-CD11b-FITC (target channel) and anti-CD3-PR (non-specific channel) markers. After background subtraction, a particle counting script was applied to each image, with each image object tested using both size (>1 and < 20 microns) and specificity filters (i.e. positive particles in both the bright field and target channel, but not the non-specific channels). Figure 3A shows an example 20X magnification image of captured CD11b labelled cells in the bright field (top), FITC (middle), and PE (bottom) channels. Figure 3B shows an additional 10X magnification example of captured CD11b labelled cells in the FITC channel before (top) and after (bottom) background subtraction and application of the size and specificity filters (showing 37 specific CD11b labelled cells within this example field of view). The total number of cells for each separation run was calculated using this method, and scanning across the full width of the MR device using an automated microscope stage.
Figure 3.
A. Images (20X magnification) showing the bright field (top), the target channel (middle; using a fluorescent label anti-CD11b-FITC), and the non-specific control channel (bottom; using a fluorescent label anti-CD3-PR) for ratcheted 500 nm anti-cd11b bead labelled cells (separated from background PBMCs at 5 Hz magnetic field rotation). The images for these 500 nm bead labelled cells are taken at the interface between the sample loading region (7 micron pillar pitch) and the separation region (10 micron pillar pitch). B. Additional 10X magnification images of the captured CD11b labelled cells in the FITC channel after use of the particle counting scripts described (top) and background subtraction (bottom). C. The middle picture shows an example of a whole device image, taken by stitching together 21 separate images taken across the device with an automated microscope/stage. In these images, equal mixtures of cells labelled with anti-CD3 magnetic particles of 500 nm, 1 micron, and 4 micron sizes were loaded. Prior to ratcheting (15 Hz for 15 minutes), cells labelled with 500 nm and 4 micron particles were incubated in fluorescent dye cell tracker® green and red respectively. The zoom-in images of the 500 nm bead labelled cells (left) and 4 micron bead labelled cells, and demonstrates use of differential ratcheting of multiple cell population to different positions on the device (i.e. multi-marker/multi-cell separation). D. The graphs show quantitative data on the multi-marker separation capability of the ratcheting system. Each graph shows the distribution of three separate ratcheting runs, performed using the three CD3-labelled magnetic particle sizes at three different magnetic field rotation speeds. The slower magnetic field rotations (e.g. 5 Hz; bottom) produce the largest ratcheting forces; in this case ratcheting the 4 micron bead labelled cell populations entirely off of the device (i.e. no critical pitch achieved). At the faster magnetic field rotation speeds, the 4 micron bead labelled cells reach a critical pitch that places them at the right-side (i.e. larger pitch regions) of the device compared to the 500 nm and 1 micron bead labelled populations.
Figure 3C shows an example of 21 separate images taken and stitched together to create a map of all cells captured across the separation region. In this image, equal mixtures of cells labelled with anti-CD3 magnetic particles of 500 nm, 1 micron, and 4 micron sizes were loaded. Prior to ratcheting, the cells labelled with the 500 nm and 4 micron particles were incubated in the fluorescent dye cell tracker® green and cell tracker® red, respectively. The image shows the entire device after ratcheting at 15 Hz for 15 minutes, with both cell tracker green and red labelled cells visible at distinct points (or critical pitches). As shown, the cells labelled with larger particles (cell tracker red labelled) ratcheted farther across the device achieving a larger critical pitch. Figure 3D shows that this differential ratcheting can be used to perform multi-marker (i.e. multi-cell) separation and processing on the device. Each graph shows the distribution of three separate ratcheting runs performed with the three CD3-labelled magnetic particle sizes at three different magnetic field rotation frequencies. As the frequency of rotation decreases the time averaged magnetic force of the particles increases, resulting in larger critical pitches. The 5 Hz graph shows the extreme result of this phenomenon, as the cells labelled with the largest particle are ratcheted entirely through the separation region and off the device. Those cells may be collected, and the frequency can then be further lowered to achieve additional ratcheting of a cell population labelled with a smaller particle.
Post-Ratcheting Cell Culture and Cytokine Secretion
As described above, after separation, the magnetic field direction was rotated 90 degrees and the captured cells at each pitch region were ratcheted into their respective collection pads. For cell culture and incubation, the rotary valves were shut isolating the captured cells from the remaining sample. Once captured, the entire device was removed from the mechatronic system and moved to a standard mammalian cell incubator (37°C, 5% CO2 with water tray). During incubation, a series of reagents were added to the well above the collection pad that enabled monitoring of IL2 secretion from the separated cells (based on a commercial Miltenyi IL2 secretion kit). First, an Il-2 catch reagent was added to the captured cells (which consists of an IL2 antibody conjugated to a cell surface specific monoclonal antibody), this primed the cells for capture of secreted IL2. A permanent magnet was placed on the device during this time to provide a constant magnetic field trapping force within the collected pad. The entire device was placed in an incubator for 45 minutes to allow cytokine secretion, capture, and fluorescent labelling on the trapped cells. The device was then removed from the incubator and a buffer wash was performed by adding a phosphate buffer saline (PBS) solution to the wells above the collection pads. After wash, a secondary IL2 detection antibody (labelled with APC) was added to the well and allowed to incubate with the cells for 10 minutes, a second wash with PBS was performed and the cells were then imaged again (to test for secreted IL2).
Post-Ratcheting Ejection and Cell Line Expansion
Cell collection was performed after switching the magnetic wheel’s direction of rotation by 90° driving the separated cells toward the bottom of the device. Cells were removed from the collection pad and transferred to 96 well culture plates for expansion. Wells contained X-vivo 15 medium supplemented with 20 units/mL of IL-2. Cell lines thus obtained were expanded for 28 days by stimulation with irradiated allogenic PBMCs in the presence of soluble anti-CD3 (1 ug/mL), anti-CD28 (1 ug/mL) and IL-2 (10 U/mL). Cell lines were tested by restimulating with autologous monocytes either unpulsed or pulsed with MOG97-109 to check the specificity by CFSE dilution (proliferation assay). The proliferation assay was performed as follows, cell lines were rested for 3 days without IL-2, washed twice with PBS, then incubated with 1 uM CFSE (Invitrogen) for 7 minutes. After quenching and washing out the excess of CFSE with PBS containing 2% BSA, the cells were resuspended in X-vivo 15 medium at 1×106 cells/mL and co-cultured with irradiated autologous monocytes in the presence or without 1 ug/mL MOG97-109. After 5 days of co-culture, the cells were washed and labelled with anti-CD4 APC, anti-CD8 APC-H7 and live-dead viability dye. Proliferation was assessed by flow cytometry as a sequential halving of CFSE fluorescence signal while gating on CD4 cells.23
Post-Ratcheting Ejection and RNA Sequencing
RNA extraction and RNA sequencing was performed at the Roswell Park Cancer Institute, (Genomics Shared Resource24, Buffalo, New York, USA) on grouped sample, collected from collection outlets on multiple ratcheting runs. The purification of total and small RNAs was prepared using the miRNeasy micro kit (Qiagen). Cells are pelleted at 300g for 10 minutes at 4°C. Supernatant is removed and cells are immediately lysed and resuspended with the addition of 700 ul of QIAzol Lysis Reagent. The lysate was then left at room temperature for 30 minutes to allow for adequate cell lysis. After addition of chloroform, the homogenate is then separated into aqueous and organic phases by centrifugation. RNA partitions to the upper, aqueous phase, while DNA partitions to the interphase and proteins to the lower, organic phase or the interphase. The upper, aqueous phase is extracted, and ethanol is added to provide appropriate binding conditions for all RNA molecules from 18 nucleotides upwards. The sample is then applied to the RNeasy Mini spin column, where the total RNA binds to the membrane and phenol and other contaminants are efficiently washed away. On-column DNAse digestion is also performed to remove any residual genomic DNA contamination followed by additional washes. High quality RNA is then eluted in 20 ul of RNase-free water. Quantitative assessment of the purified total RNA is then accomplished by using a Qubit High Sense RNA kit (Thermofisher). The RNA is further evaluated qualitatively by a 2100 Bioanalyzer Pico Chip (Agilent technologies).
The sequencing libraries are prepared with the TruSeq Stranded mRNA kit (Illumina Inc), from 300ng total RNA. Following manufacturer’s instructions, the first step selects for PolyA RNA. After PolyA selection, the remaining RNA is purified, fragmented and primed for cDNA synthesis. Fragmented RNA is then reverse transcribed into first strand cDNA using random primers. The next step removes the RNA template and synthesizes a replacement strand, incorporating dUTP in place of dTTP to generate ds cDNA. AMPure XP beads (Beckman Coulter) are used to separate the ds cDNA from the second strand reaction mix resulting in blunt-ended cDNA. A single ‘A’ nucleotide is then added to the 3′ ends of the blunt fragments. Multiple indexing adapters, containing a single ‘T’ nucleotide on the 3′ end of the adapter, are ligated to the ends of the ds cDNA, preparing them for hybridization onto a flow cell. Adapter ligated libraries are amplified by PCR, purified using Ampure XP beads, and validated for appropriate size on a 4200 TapeStation D1000 Screentape (Agilent Technologies, Inc.). The DNA libraries are quantitated using KAPA Biosystems qPCR kit, and are pooled together in an equimolar fashion, following experimental design criteria. Each pool is denatured and diluted to 2.4pM with 1% PhiX control library added. The resulting pool is then loaded into the NextSeq 75 cycle high output Reagent cartridge, and sequenced on a NextSeq500 following the manufacturer’s recommended protocol (Illumina Inc.).
Sequencing output from the Illumina NextSeq 500 platform consisted of 75 base pair single ended reads with an average of 30 million reads per sample. Base quality control was checked using FastQC v0.10.1 from Babraham Bioinformatics.25 Sequencing reads were mapped to the human reference genome GRCh38.p12 (Ensembl release 92) using STAR_2.5.3a aligner26 (Ran STAR with parameter quantMode set to GeneCounts). To allow comparison across samples, gene read counts for each sample were converted into units of TPM (Transcripts Per Kilobase Million) after first filtering out non-protein coding transcripts. Genes that were compared across ratcheted versus bulk stimulated samples included CD69, CD25 (IL2RA), TNF, CD3D, CD28, IL4, IL13).
Results
MR Capture for Multiple Cell Types
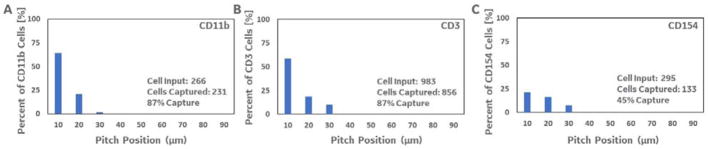
We first worked with the immune cells labelled with 500 nanometer anti-cd11b (myeloid cell marker) labelled paramagnetic particles and spiked a known number of pre-separated target cells into unlabelled PBMCs. The cells were again further labelled with fluorescent antibodies against CD3, to detect and quantify non-specific cells (as described above). Figure 4A shows the full post-separation distribution of specific (CD11b positive) cells across the device after loading, based on the cell counting algorithm. As shown, the 500 nanometer beads result in a relatively low critical pitch (i.e. most of the ratcheted cells achieve an equilibrium at the first two, 10 and 20 micron, pitch regions). The estimated target cell input for this experiment was 266 labelled CD11b cells (based on both quantitative cell counting and flow cytometry data), spiked into unlabelled PBMCs at a 1:8,000 ratio. After ratcheting, 231 target CD11b cells were recovered across the device with only 15 non-specific CD3+ cells, resulting in an estimated capture efficiency of 87%.
Figure 4.
Three graphs quantifying the total number of target cells captured on the ratcheting device for three different 500 nm bead labelled populations (anti-CD11b, anti-CD3, and anti-CD154). A. 231 out of an estimated 266 target CD11b cells were captured across the ratcheting device. B. 956 out of an estimated 983 CD3 target CD3 cells were captured across the ratcheting device. C. 133 out of 295 target CD154 cells were captured across the ratcheting device. Each device/experimental run was loaded with 100 microliters of sample containing bead-labelled target cells in an excess of back-ground (non-labelled) PBMCs as described above.
We next tested the cell processor using cells labelled with a mixture of 500 nanometer anti-CD4/anti-CD8 (CD3; lymphoid cell markers) labelled paramagnetic particles, and spiked a known number of pre-separated target cells into unlabeled PBMCs. Figure 4B shows the histogram of target CD3 (anti-CD4/anti-CD8-FITC labelled; using non-specific anti-CD14-PE marker) cell counts across the device after ratcheting. The estimated target cell input for this experiment was 983 labelled CD4/CD8 cells, spiked into unlabelled PBMCs at a 1:2000 ratio. As shown, 856 specific cells were counted across the device after ratcheting compared to 44 non-specific cells (yielding an estimated capture efficiency of 88%). Again, most cells labelled with the 500 nm particles ratcheted to the first few pitch regions (yielding a critical pitch of 10 – 30 microns). S2 contains additional data that compares the sample sparing ratcheting run (using the same CD3 labelled samples) with detection and quantification using a top-end volumetric flow cytometer. When using the same sample sparing inputs (i.e. 20–50 microliters of sample) and analysis times the flow cytometer did not perform as well as the magnetic ratcheting device.
Next, the MR was loaded with Cytostim® activated cells labelled with 500 nm anti-CD154 (activated T cell marker) conjugated paramagnetic particles spiked into unlabelled PBMCs at a ratio of 1:6500. CD154 is only transiently upregulated on activated CD4+ T cells (as a costimulatory molecule in T cell/antigen-presenting cell interactions through ligation of CD40. However, use of a CD40 blocking-antibody during stimulation has been shown to prevent down-regulation of CD154 expression (induced by CD40 interactions), and enable use as a robust cell surface marker of T cell activation. We used a commercial T cell stimulation and capture kit (Miltenyi CD154 microbead kit) containing this CD40 antibody and CytoStim to stimulate and label activated T cells (as described above). The magnetic field rotation for this experiment was again set at 5 Hz, and Figure 4C shows the distribution of the activated T cells across the device after separation. In this run, 295 CD154 particle labelled cells were added to the device, and 133 were counted across the separation region (yielding an estimated capture efficiency of 45%). Lower surface expression of CD154 (compared to other surface markers such as CD11b and CD4/8 used above) is a well-known challenge, and the 45% efficiency compares favourably to reports of FACS or MACS based sorting. Importantly, the capture purity for CD154 isolation can also be increased significantly compared to MACS (95±1.4% vs.2.8±0.27%), based on the quantitative separation capability that reduces the isolation of cells with only a few bound beads (see S2 for data and additional discussion).
Altering MR Particles for Increased Purity
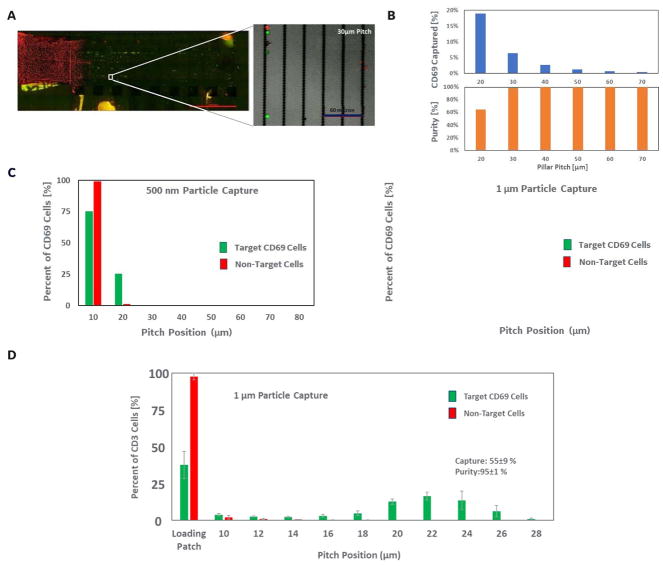
To maximize separation performance of target cells from background we evaluated the separation profiles of cells tagged with 500nm, 1μm, and 4μm diameter magnetic particles (specific for the CD69 surface marker) on the ratcheting chip (Figure 5A–C). Figure 5A shows fluorescent images of the entire ratcheting device. The non-specific cells (red channel) are observed to remain behind in the input well, while the specific cells (green channel) are ratcheted across the device by a distance dependent on the magnetic force applied (partially controlled by the particle label size). This full view of the device enables monitoring of several separation parameters (including collection efficiency, as described above), but also target cell purity at each separation or pitch region. As shown in figure 5B, as the target cells are ratcheted further away from the input well the purity of the separated cells in a pitch band increases, and very high cell purities (i.e. greater than 95%) are achievable at one to two pitch regions away from the input well (i.e. the 30 or 40 micron pitch regions on this device). Cells with increasing volume of magnetic particles have stronger magnetic forces applied and therefore will equilibrate at higher pillar pitches within the chip, enabling differentiation between specific binding due to association with target antigens and non-specific binding of a few magnetic particles. Figure 5C shows the profiles of cells tagged with particles of two diameters where the 500nm labelled populations equilibrated between the 10–30μm pitch and the 1μm labelled population peaked at the 30μm pitch. The larger particle labels spread the captured cell population further down chip, as the increased magnetic force on the cells allowed pillar to pillar cell movement over the magnetic field rotation time that enables traversing to the next pillar pitch region.
Figure 5.
A. An additional full ratcheting device image of an experiment performed using 1 micro bead anti-CD69 labelled cells (spiked into excess PBMCs as described above). Target cells were again pre-labelled with green fluorescent tracking dye and non-target cells were labelled with the red dye (false coloured images of the combined fluorescent channels used for visualization). The image shows the separation of the non-target (red) cells from the target/ratcheted green populations. B. Results from the anti-CD69 bead labelled ratcheting experiment showing the percent of total target cells captured in each pitch region on the device (top), and the purity of target versus non-target or background cells in each pitch region (bottom). C. Results compared anti-CD69 bead labelled experimental runs using either the 500 nm (left) or 1 micron (right) capture beads.
A follow-up anti-CD69 bead labelled ratcheting experiment performed using ratcheting devices containing a narrower distribution of “pitch regions” (i.e. 10–28 micron pitch spacings across the device versus 20–70 micron pitch distribution above). The distribution of cells across the device shows the capability of using tight pitch distribution to drive a more complete separation of target versus non-target cells.
This behaviour draws a parallel to flow cytometry where fluorophores of differing excitation and emission are used to quantify biomarkers by tuning the voltage of a photomultiplier tube (PMT). The fluorophore excitation/emission and the PMT voltage can be tuned to achieve an optimal signal-to-noise ratio to resolve the desired cell sub-populations. In a similar fashion, the ratcheting system uses immunomagnetic particles with differing magnetic contents which can be tuned by modulating the ratcheting frequency to maximize separation. In evaluating these different particle types, it was determined that the 1μm particle exhibited the optimal separation performance because it achieved a larger dynamic range, or pillar pitch range, than the 500nm particles (due to the higher magnetic content). Additionally, the smaller size (compared to 4μm particles) enabled better resolution in discriminating between target and off target labelled cells as well as increased binding efficiency due to higher particle diffusivity (data not shown).
Altering MR Geometry and Particle Labelling for Increased Purity
We also evaluated the effect of having a finer increment of pillar pitch to determine the resolving power of the ratcheting cell separation platform. We constructed a ratcheting chip which consisted of a 7μm pitch loading patch leading into a gradient pitch array which had an initial pitch of 10μm and incremented by 2μm. Cells were labelled with 1μm anti-CD3 particles in these experiments and separated on the chip under a 5Hz ratchet (as above) in a background of CD3(-) cells. As shown in Figure 5D, the CD3+ target population can be more easily separated away from the input well using the finer pitch increment, achieving a purity of 95±1% across the entire separation region. This fine gradient enabled clear separability between the populations which can be critical in distinguishing weakly tagged off target cells from the higher expressing target cell type.
On-Chip Cell Culture and Cytokine Secretion Assays
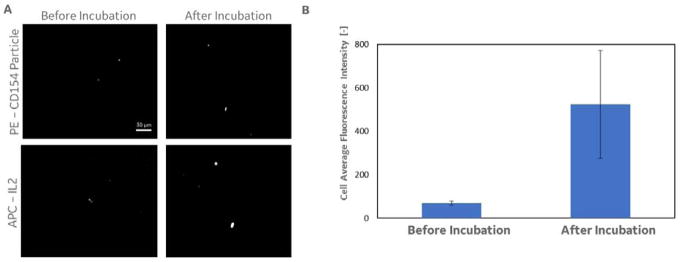
We next tested whether cells captured within the separation region (at each critical pitch) could be further processed, cultured in-place, and analysed. Upon completion of the separation run, the magnetic field direction was rotated 90 degrees and the captured cells at each pitch region were ratcheted into their respective collection pads (as outlined in Figure 2A). Once captured, the cells were incubated with IL-2 secretion capture reagents which were serially added to the collection chamber, as described above. Figure 6A shows representative images of capture cells before and after incubation with the IL-2 capture reagents (PE signal from anti-biotin-PE label for the CD154 capture particles, and APC signal from anti-IL-2-APC detection antibody). After incubation, the cells showed an increase in average APC intensity as the cells secreted IL-2, which was captured using the IL-2 cell surface capture reagent. Figure 6B shows the average APC signal for each cell captured before and after culture and incubation, for all cells that were captured (n = 6; see culture time and protocol above) and concentrated from the 20 micron critical pitch region.
Figure 2.
A. A step-by-step schematic of the multi-directional magnetic ratcheting technique used to separate out rare immune cells from blood samples. Step 1: A previously labelled cell sample (i.e. sample incubated with magnetic particles specific for cell surface target molecules) is loaded into the cell input chamber with underlying densely packed paramagnetic pillars. Multiple labels can be used to separate non-labelled cells (black) from cells labelled with smaller magnetic particles (i.e. 50 – 500 nanometers; green) and cells labelled with large magnetic particles (i.e. 1 – 4 microns; red). Step 2: Ratcheting begins when the rotating magnetic wheel in the mechatronic control hardware is enabled to spin at a pre-defined frequency. This initiates movement of all magnetically labelled cells across the ratcheting chip at an average cell/particle velocity of Pitch (microns) x Freqency.22 Step 3: Ratcheting (i.e. magnetic wheel rotation) is continued until all particles are given enough time to traverse the device. Cells with differing numbers of bound magnetic particles (i.e. differing magnetic content) will stop ratcheting at different critical pitch. This is the pitch at which the rotating magnetic field does not produce enough magnetic force to propel that specific cell to the next pillar (of increasing interpillar distance or pitch). Step 4: The magnetic wheel is then rotated 90 degrees and all cells that terminated in each pitch region are moved to the bottom of the device (which contains an open collection reservoir above the collection pads). Step 5: A rotating isolation valve in each collection chamber enable closing of each chamber from the larger ratcheting device, and collection of only those cells that achieved that particular critical pitch. B. A CAD drawing and close-up of the rotating isolation valve is shown; these cylindrical valve components were fabricated using stereolithography (Fineline Prototyping; high resolution watershed material build) and contained a small opening that could be turned toward or away from the ratcheting pillar array. The image also shows a microscope close-up of the collection pad (with densely packed 5 micron inter-pillar distances) and the termination points for all the column of pillars within that pitch region.
Figure 6.
Results from a post-ratchet analysis of cytokine secreting cells captured within the device. A. The images show cells captured using the anti-CD154 beads (as described above) before and after incubation with reagents designed to capture secreted cytokines on the cell surface. B. Incubation of the captured cells with reagents designed to capture secreted cytokines (in this case IL2) on the cells surface resulted in increased fluorescence intensity over incubation time (data shown is for a 45 minute incubation and cytokine capture).
Capture and Expansion of Myelin-Specific T Cells
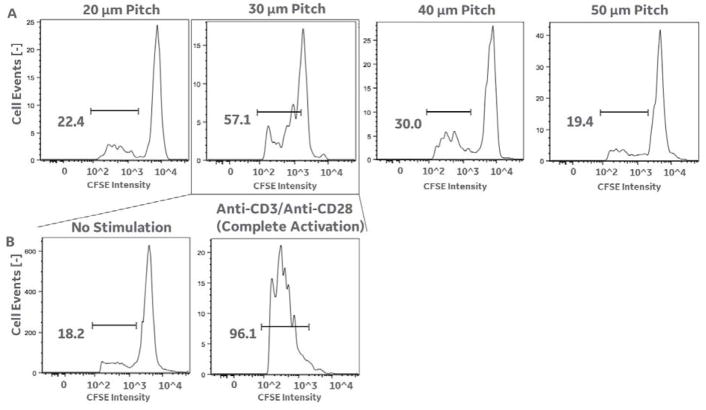
In a separate assay, we tested the specificity of captured cells for their proliferation upon challenge with the myelin antigen peptide MOG97-109.11,13,14 PBMCs from HLA-DR4+ blood samples were labelled with tetramer and isolated using the magnetic ratcheting system as described in materials and methods. Using flow cytometry, a blood sample was tested as positive for containing MOG specific T cells,11,13,14 and that sample was utilized in the ratcheting test described below. Tetramer/bead captured cells were ratcheted and collected from the wells over collection pads at 20, 30, 40 and 50 micron pitches (Fig. 7A) and expanded for 4 weeks. At the end of the four weeks the cell lines were allowed to rest for 3 days without IL-2, were restimulated with DR4-restricted myelin peptides (MOG97-109) using irradiated autologous monocytes as antigen presenting cells.11,13,14 As shown, cell lines that exhibited the highest scores for proliferation (i.e. the largest decrease in CFSE intensity due to antigen stimulated proliferation) were collected within the 30 micron pitch (the expected center of cell distribution based on magnetic bead size used, and the previous results above). The specificity of this cell line to MOG97-109 was compared to no stimulation (negative control) and to multiple anti-CD3+antiCD28 (positive control) (Fig. 7B). The positive control demonstrated >92% proliferative response from the cell line generated from the 30 micron pitch sample, showing that the cells collected produced viable immune cell lines. And, the negative controls showed no proliferative response, suggesting that proliferation within the captured sample is specific to the applied antigen.
Figure 7.
Results showing capture of antigen specific T cells and cell line expansion. A. The flow cytometry data show the results of a CFSE cell proliferation assay for all cells captured from the 20 micron, 30 micron, 40 micron, and 50 micron pitch regions. Cells in this experiment were captured using 1 micron beads attached to DRB1*0401/MOG97-109 tetramers (as described above), and based on our previous results (figure 6C; right) should produce target cell separation into the 30 micron pitch region. The flow cytometry results do demonstrate the largest proliferative response (i.e. antigen specific cell expansion in response to stimulation with the myelin antigen peptide MOG97-109) in the 30 micron pitch region (i.e. 57.1% of cells showing a lower fluorescent CFSE intensity due to proliferation, compared to 22.4% in the 20 micron pitch region, 30% in the 40 micron pitch region, and 19.4 percent in the 50 micron pitch region respectively). B. Further analysis of the cells captured within the 30 micron pitch revealed an 18.2% cell count within the proliferative intensity window in unstimulated cells (providing a background level of zero proliferative response for the other conditions), and a 96.1% cell count within the proliferative intensity window in broadly stimulated cells (i.e. stimulated using anti-CD3/anti-CD28 instead of an antigen specific peptide). These results indicate a significant proliferative response to the antigen specific peptide (in 7A) in 30 and 40 micron pitch regions (centered on the 30 micron pitch region), in agreement with figure 6C.
Cell Ejection and Gene Expression
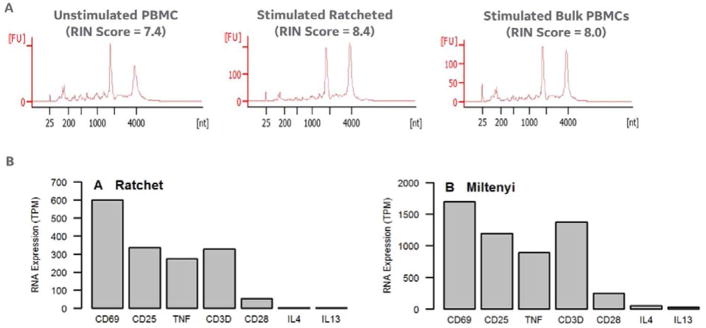
The cell viability of CD154+ cells ratcheted and then pipetted off the device (using the collection pads as described above) averaged 87% (data not shown; NC-200 cytometer). In addition, processing of the captured cells for nucleic acids produced nucleic acid with adequate purity for down-stream processing. Figure 8A shows that cells processed on the magnetic ratchet (i.e. Cytostim activated and then ratcheted using CD154+ beads) produced the highest RIN score, producing RNA with higher quality than the unprocessed/unstimulated PBMCS (left) and bulk stimulated/miltenyi column processed CD154+ cells (middle). Full transcriptome sequencing was performed as described above and Figure 8B shows a comparison of RNA levels from cells captured after ratcheting (i.e. Cytostim activated and then ratcheted using CD154+ beads) or stimulated in bulk. The relative distribution of genes in the ratcheted samples agrees well with the “bulk stimulated” cell samples. (including those markers for activated immune cells (CD69, CD25), T cell specific markers (CD3, CD28), and secreted cytokines (TNF, IL4, IL13)).
Figure 8.
A. Agilent bioanalyzer electrophoresis results on RNA extracted from unstimulated (or bulk) PBMCs (left), cytostim stimulated PBMCs labelled with anti-CD154 beads and separated on the ratcheting device (middle), and cytostim stimulated and separated PBMCs using the milytenyi anti-CD154 column-based separation kit (right). All samples (including the ratchet separated cells) produced acceptable RIN scores (RIN score ~8.0 is a typical desired value for down-stream gene expression analysis). B. The RNA extracted from the ratcheted cell sample supported whole transcriptome sequencing (as described above), and the gene read counts for CD69, CD25, TNF, CD3d, IL4, and IL13 showed a similar distribution between the ratcheted and bulk separated cell samples.
Discussion
In this report, we present application of multi-directional magnetic ratcheting for handling and analysing rare populations of immune cells. As discussed above, recent success in immunotherapies (across several major clinical disciplines, including oncology, allergy, and autoimmune disease)1–4 have increased the need to capture and profile specific sets of immune cells that are involved in the IT response1–14. However, cumbersome and costly FACS and microfluidic separation technologies remain the primary means to select, capture, and analyse specific immune cells from a background of non-specific cells (such as, antigen specific lymphocytes).15,16 These technologies have limited the number of mechanistic studies that seek to identify the specific immune system changes that underlie successful IT, and screen for those changes to improve therapies.4
We provide a method for completely unsupervised (i.e. dependent on only physical properties of the separation without use of complex FACS gates or microfluidic fluid manipulations) separation, capture, and isolation of rare immune cells through the introduction of multi-directional and multi-parameter magnetic ratcheting. In this method, discrete single-cell magnetic manipulations are utilized to 1) select for specific and rare cell types within complex clinical samples (Figure 3), 2) drive near complete separation of those selected cells from the non-specific cell types within the sample (Figure 4 and 5), and 3) move cells to specific sites within the device for down-stream analysis and profiling (Figure 6–8). We show the capability to perform multiple follow-on assays that remain important in immunology and IT research including live-cell cytokine secretion assays, cell capture and antigen specific stimulation and expansion for cell line creation, and post-capture ejection and nucleic acid purification for RNA sequencing. Our future work with this platform will include modification to enable separation and isolation of single cells (i.e. rare cell output into a well-plate like single cell isolation), and testing across a panel of clinical samples in the autoimmune and allergy application areas.
Conclusions
Our goal was to demonstrate a device that rapidly separates and profiles rare and low-abundance immune cells from clinical samples within a simple-to-use instrument. Using a unique magnetic ratcheting strategy, we first demonstrated unsupervised capture and collection of several different immune cell types from PBMC samples (CD11b, CD3 or CD4/CD8, CD69, and CD154 expressing cells) and automated post-extraction culture and processing. We demonstrated additional on-device processing by using magnetic ratcheting to collect cells onto specific locations within the device (without external handling or fluidic manipulations), culturing those cells in-place, and adding reagents to perform cell subpopulation cytokine secretion analysis. We further demonstrated separation of particle-labelled antigen specific T cells from clinical samples at extremely low collection frequencies (1:20,000), ejected, and expanded these cells for post-capture analysis. Finally, we demonstrated the capability to eject the captured cells and perform nucleic acid purification and RNA sequencing analysis. To our knowledge, this is the first application of a magnetic trapping array to completely automate capture and process rare immune cells in an unsupervised process, in which there are no requirements for optical characterization and gating (as in standard FACS protocols) or complex fluidic manipulations (as with microfluidic based cell sorters). The clinical samples used to test the device were chosen to demonstrate its capability in supporting mechanistic studies on rare immune cell populations which would be important for clinical immunotherapy trials. Current dependence on FACS as the primary cell selection tool within clinical research has limited researchers’ ability to access data on specific single cell phenotypes. Within this report, we show that extremely low-abundant myelin specific T cells are rapidly separated within the device, and made available within a small pipettable well for expansion or analysis. This separation is done in a completely unsupervised manner, and to the clinician or researcher functions by simply pipetting in a magnetic bead labelled clinical sample on one side of the device and pulling out rare cell populations from the other. These results suggest immediate utility of the device to replace the expensive and bulky methods currently being used to identify both transitory and long-lived antigen-specific cell phenotypes within current immunotherapy trials. In addition, the extremely low sample input requirements offer a unique solution for small volume processing and maximal data collection during longitudinal monitoring.
Supplementary Material
Acknowledgments
The work was supported by NIH NIAID (1U24AI118667-01). However, any opinions, findings, conclusions or other recommendations expressed herein are those of the authors and do not necessarily reflect the views of the NIH.
The authors would like to thank Prashant Singh at the Roswell Park Cancer Institute for sequencing advice and processing.
Footnotes
Electronic Supplementary Information (ESI) available: S1: Description and Citations on Sample Sparing Assays and the Challenges Associated with Antigen-Specific Immune Cell Monitoring; S2: Comparison of Magnetic Ratcheting Separation to Standard Flow Cytometry and MACS based techniques.
References
- 1.Zou W. Mechanistic Insights into Cancer Immunity and Immunotherapy. Cell Mol Immunol. 2018 doi: 10.1038/s41423-018-0011-5. Epub Ahead of Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfaar O, Bonini S, Cardona V, et al. Perspectives in allergen immunotherapy: 2017 and beyond. Allergy. 2018;73(104):5–23. doi: 10.1111/all.13355. [DOI] [PubMed] [Google Scholar]
- 3.Jones AP, Kermode AG, Lucas RM, et al. Circulating immune cells in multiple sclerosis. Clin Exp Immunol. 2016;187(2):193–203. doi: 10.1111/cei.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryan JF, Hoyde R, Glanville J, et al. Successful immunotherapy induces previously unidentified allergen-specific CD4+ T-cell subsets. PNAS. 2016;113(9):E1286–E1295. doi: 10.1073/pnas.1520180113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porter DL, Hwang WT, Frey NV, et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci Transl Med. 2015;7(303):303ra139. doi: 10.1126/scitranslmed.aac5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renand A, Shami MH, Harris KM, et al. Synchronous immune alterations mirror clinical response during allergen immunotherapy. J Allergy Clin Immunol. 2017;S0091–6749(17):31735–9. doi: 10.1016/j.jaci.2017.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wambre E, Bajzik V, DeLong JH, et al. A phenotypically and functionally distinct human Th2 cell subpopulation is associated with allergic disorders. Sci Transl Med. 2017;9(401):eaam9171. doi: 10.1126/scitranslmed.aam9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James EA, LaFond RE, Gates TJ, et al. Yellow fever vaccination elicits broad functional CD4+ T cell responses that recognize structural and nonstructural proteins. J Virol. 2013;87(23):12794–804. doi: 10.1128/JVI.01160-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, James E, Gates TJ, et al. CD4+ T cells recognize unique and conserved 2009 H1N1 influenza hemagglutinin epitopes after natural infection and vaccination. Int Immunol. 2013;25(8):447–57. doi: 10.1093/intimm/dxt005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge X, Tan V, Bollyky PL, et al. Assessement of Seasonal Influenza A Virus-Specific CD4 T-cell responses to 2009 pandemic H1N1 Swine-origin influenza A Virus, 2010. J Virology. 84(7):3312–3319. doi: 10.1128/JVI.02226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y, Goods BA, Raddassi K, et al. Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis. Sci Tranl Med. 2015;7(287):287ra74. doi: 10.1126/scitranslmed.aaa8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerosaletti K, Barahmand-Pour-Whitman F, Yang J, et al. Single-Cell RNA Sequencing Reveals Expanded Clones of Islet Antigen-Reactive CD4+ T Cells in Peripheral Blood of Subjects with Type 1 Diabetes. J Immunol. 2017;199(1):323–335. doi: 10.4049/jimmunol.1700172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T helper type-1-like Fox3+ regulatory T cells in human autoimmune disease. Nature Medicine. 17(6):673–675. doi: 10.1038/nm.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radassi K, Kent SC, Yang J, et al. Increased Frequencies of MOG/MHC Class II binding CD4 cells in patients with Multiple Sclerosis. 2011. J Immunol. 187(2):1039–1046. doi: 10.4049/jimmunol.1001543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seah YFS, Hu H, Merten CA. Microfluidic single-cell technology in immunology and antibody screening. Mol Aspects Med. 2018;58:47–61. doi: 10.1016/j.mam.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18(1):35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 17.Swennenhuis JF, Terstappen L. Sample Preparation Methods Following CellSearch Approach Compatible with Single-Cell Whole Genome Amplification: An Overview. Methods Mol Biol. 2015;1347:57–67. doi: 10.1007/978-1-4939-2990-0_4. [DOI] [PubMed] [Google Scholar]
- 18.Bacher P, Schink C, Teutschbein J, et al. Antigen-reactive T cell enrichment for direct, high-resolution analysis of the human naïve and memory Th Cell repertoire. J Immunol. 2013;190(8):3967–76. doi: 10.4049/jimmunol.1202221. [DOI] [PubMed] [Google Scholar]
- 19.Macosko EZ, Basu A, Satija R, et al. Highly Parellel Genome-wide Expression Profiling of Individual Cells using Nanoliter Droplets. Cell. 161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gierahn TM, Wadsworth MH, Hughes TK, et al. Seqwell: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017 doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziegenhain C, Vieth B, Parekh S, et al. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell. 2017;65(4):631–643. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 22.Murray C, Pao E, Tseng P, et al. Quantitative Magnetic separation of Particles and Cells using Gradient Magnetic Ratcheting. Small. 2016 doi: 10.1002/smll.201502120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyons AB, Parish CR. Determination of lymphocyte division by flow cytometry. J Immunol Methods. 1994;171:131. doi: 10.1016/0022-1759(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 24.https://www.roswellpark.edu/shared-resources/genomics; a shared resource funded by NCI P30CA16056
- 25.http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 26.Dobin A, Gingeras TR. Mapping RNA-seq Reads with STAR. Current protocols in bioinformatics/editoral board, Andreas D Baxevanis [et al] 2015;51:11.4.1–4.9. doi: 10.1002/0471250953.bi1114s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.