Abstract
Ca2+ is an important second messenger, and it is involved in many cellular processes such as cell death and proliferation. The rise in intracellular Ca2+ levels can be due to the generation of inositol 1,4,5-trisphosphate (InsP3), which is a product of phosphatidylinositol 4,5-bisphosphate (PIP2) hydrolysis by phospholipases C (PLCs), that leads to Ca2+ release from endoplasmic reticulum by InsP3 receptors (InsP3R). Ca2+ signaling patterns can vary in different regions of the cell and increases in nuclear Ca2+ levels have specific biological effects that differ from those of Ca2+ increase in the cytoplasm. There are PLCs in the cytoplasm and nucleus, but little is known about the functions of nuclear PLCs. This work aimed to characterize phenotypically the human PLCδ4 (hPLCδ4) in mesenchymal stem cells. This nuclear isoform of PLC is present in different cell types and has a possible role in proliferative processes. In this work, hPLCδ4 was found to be mainly nuclear in human adipose-derived mesenchymal stem cells (hASC). PLCδ4 knockdown demonstrated that it is essential for hASC proliferation, without inducing cell death. An increase of cells in G1, and a reduction of cells on interphase and G2/M in knockdown cells were seen. Furthermore, PLCδ4 knockdown increased the percentage of senescent cells, p16INK4A+ and p21Cip1 mRNAs expression, which could explain the impaired cell proliferation. The results show that hPLCδ4 is in involved in cellular proliferation and senescence in hASC.
Keywords: human PLCδ4, nuclear calcium signaling, calcium signaling, cellular proliferation, cellular senescence
1. Introduction
Ca2+ is a versatile second messenger, and it is involved in many cellular processes, such as muscular contraction, cell death and proliferation, egg fertilization, and secretion [1–4]. Although it is not fully established how Ca2+ coordinates such diverse functions, there is evidence that they may be regulated by the spatial patterns of Ca2+ distribution [1]. The rise in intracellular Ca2+ levels can occur through the generation of inositol 1,4,5-trisphosphate (InsP3), which is a product of phosphatidylinositol 4,5-bisphosphate (PIP2) hydrolysis by phospholipases C (PLCs), leading to Ca2+ release by InsP3 receptors (InsP3R) present in the membrane of the endoplasmic reticulum [5, 6]. In turn, Ca2+ activates sensor molecules, like calmodulin, which modulate cellular activity [7]. This pathway is activated by G protein-coupled receptors (GPCR), or by tyrosine kinase receptors (RTKs), via PLCs.
Some events regulated by Ca2+ occur specifically in the cell nucleus, and several pieces of evidence indicate that cytosolic and nuclear Ca2+ can regulate different cellular processes [4, 8–11]. Therefore, the nucleus must contain Ca2+ stores and its signaling machinery. Ryanodine and InsP3 receptors can be found in the nucleoplasmic reticulum [12–15]. In addition, the nucleus contains enzymes and substrates necessary for the production of InsP3 [16]. Previous studies demonstrated the detection of PIP2 in the nuclear envelope and nucleoplasm [17, 18], as well as the presence of nuclear isoforms of PLCs, such as γ1, β1, δ1 and δ4 [19]. These enzymes could be involved in primary nuclear Ca2+ signaling since activation of InsP3 receptors is the most implicated mechanism in the release of Ca2+ from the nucleus [20]. In mammals, phosphoinositide-specific phospholipases C (PI-PLCs; 1-phosphatidyl-1D-myoinositol-4,5-bisphosphate inositoltrisphosphohydrolases) consist of a protein family containing 13 isozymes, divided into 6 classes: β (1–4), γ (1–2), δ (1,3,4), ε (1), ζ (1) e η (1,2), according to their structures [21]. All PI-PLCs isozymes catalyze PIP2 cleavage, generating InsP3 and DAG, but each isozyme possesses unique physiological functions [21]. Some PLC isozymes have been described within the nucleus in specific cell types, such as PLCβ1, PLCγ1, PLCδ1, PLCδ4 and PLCζ [19, 22]. The activity of these nuclear PLCs has been related to the regulation of several cellular processes, including proliferation and differentiation, and diseases, such as myelodysplastic syndromes (PLCβ1), neurological diseases (PLCγ1) and infertility (PLCζ and PLCδ1–4) [23], indicating the importance of the study and characterization of these isozymes.
PLCδ4 is a nuclear-localized phospholipase C isozyme that may be involved in proliferative processes [22, 24, 25]. It was first purified from regenerating rat liver protein extracts [26], and its gene was cloned by Liu and colleagues in 1996 [27] from a regenerating rat liver cDNA library, indicating a possible role of PLCδ4 in cell proliferation. Furthermore, this enzyme is abundantly expressed in the brain and testes of rats [24, 25]. Human PLCδ4 gene (hPLCδ4) was cloned from an oligodendroglioma cDNA library, and it is located on the long arm of chromosome 2 in region 35 (2q35; [28]). Despite its well-described chromosomal location, studies of hPLCδ4 protein are scarce.
Given the importance of phosphoinositide/Ca2+ signaling in the cell nucleus, we aimed to study PLCδ4 function in standard cell culture condition. To do so, human adipose-derived mesenchymal stem cells (hASC) were used as an experimental model [29]. These cells were chosen for their proliferation capacity, being this cellular process probably involved in PLC signaling. PLCδ4 showed to be mainly nuclear in hASC. Knockdown of PLCδ4 induced cell cycle arrest, and increased senescence, with increased p16INK4A+ and p21Cip1 mRNAs expression. Our results indicate that PLCδ4 is a nuclear protein that is involved in hASC proliferation and senescence.
2. Materials and Methods
2.1. hASC isolation and culture
Human adipose-derived mesenchymal stem cells (hASC) were obtained from the subcutaneous adipose tissue removed during liposuction surgeries or abdominoplasties. The samples were donated freely by lipoplasty surgery patients according to the regulations approved by the “Universidade Federal de Minas Gerais’s Research Ethics Committee” (CAAE: 55698116.2.0000.5149). Cell extraction was carried out as described by Zuk and colleagues (2001) [29]. The tissue was washed twice using a phosphate-buffered saline solution (PBS, pH 7.4) to remove residual blood, centrifuged at 300 × g, 7 minutes, at 25°C and then digested using 0.1% (w/v) type I collagenase (Sigma-Aldrich, St. Louis, MO, USA) for 60 minutes at 37°C. The solution was centrifuged at 300 × g for 10 minutes at 25°C, the precipitate was resuspended in complete Dulbecco´s Modified Eagle´s Medium (DMEM medium, Sigma-Aldrich) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA) and 1% penicillin/streptomycin (PS; Sigma-Aldrich) and transferred to T25 culture flasks (Sarstedt, Nümbrecht, Germany), kept in a humidified atmosphere at 37°C and 5% CO2. Cell media was replaced every 3 days. Passages 3 and 4 were used for performing the assays.
2.2. Immunofluorescence
Stem cells were plated at a concentration of 2 × 105 cells per plate, in 6-well plates containing coverslips. After a 24 hour cultivation, cells were fixed, incubated with rabbit polyclonal primary antibody anti-PLCδ4 antibody (1:200, Santa Cruz Biotechnology, Dallas, TX, USA), and anti-rabbit IgG secondary antibody conjugated to Alexa Fluor® 488 (1:500, Thermo-Fisher Scientific). Coverslips were assembled using SlowFade Gold Antifade with DAPI (Thermo-Fisher Scientific). Fluorescence images were obtained using confocal microscope Zeiss 5 LIVE (Carl Zeiss, Jena, Germany), at “Centro de Aquisição e Processamento de Imagens do ICB/UFMG” (CAPI), using 405 and 488 nm lasers for DAPI and Alexa Fluor® 488 excitations, respectively. The 63× immersion objective lenses, 1.4 numerical aperture was used, and the pinhole was kept near 1 Airy unit. Images were analyzed by Image Browser or ZEN software (Carl Zeiss). At least three assays were performed.
2.3. siRNA transfection
The knockdown of human PLCδ4 was achieved using siRNA ON-TARGETplus SMARTpool (84812), siRNA J-005065-01 (Dharmacon/Thermo Scientific; target sequences: CAAGAAGUUCAGCGGUUAU, GCUCAAUCCCAUACCGACA, GACCAAUGGCUGAGCGAUU, CAACAAGGUUACCGCCACA). Scrambled siRNA siGENOME Non-targeting siRNA pool #2 (Dharmacon/Thermo Scientific; target sequences: UAAGGCUAUGAAGAGAUAC, AUGUAUUGGCCUGUAUUAG, AUGAACGUGAAUUGCUCAA, UGGUUUACAUGUCGACUAA) was used as a control. Cells were transfected using Lipofectamine RNAiMAX (Thermo-Fisher Scientific). Lipofectamine alone, or 25 nM siRNA were diluted in the minimal medium Opti-MEM (Thermo-Fisher Scientific), incubated for 5 minutes, then combined 1:1 and incubated for 20 minutes. The lipofectamine-siRNA complex was added to resuspended cells (the number of cells and the medium volume varied according to each assay). The cells were kept at 37°C, 5% CO2, in Opti-MEM for 5 hours for cell adhesion after which the medium was replaced by complete DMEM, and the cells maintained at 37°C, 5% CO2, for the duration for each assay.
2.4. Quantitative Real-Time PCR (qPCR)
RNA was extracted with TRIzol reagent (Thermo-Fisher Scientific), according to the manufacturer recommendations. RNA samples were treated using DNAse RQ1 (Promega, Madison, WI, USA) and reverse transcription reactions were performed using High-capacity cDNA Reverse Transcription Kit (Thermo-Fisher Scientific) with random primers. Primers were designed for PLCδ4 (accession number on GenBank: NM_032726; hPLCd4 forward (F): 5’-AGGTGGATGTATGGGATGGACC-3’; hPLCd4 reverse (R): 5’-GGGTAGTCTGATGTCTGGAAGG-3’), RPL13A (GenBank accession number: AB082924; RPL13A F: 5’-TATGAGTGAAAGGGAGCC-3’; RPL13A R: 5’-ATGACCAGGTGGAAAGTC-3’), p21 (GenBank accession number: NM_000389.4; p21 CDKN1A (1) Fw: 5’-CTGTCTTGTACCCTTGTGCCT-3’; p21 CDKN1A (1) Rv: 5’-AATCTGTCATGCTGGTCTGCC-3’) and p16 (GenBank accession number: NM_000077.4; p16 CDKN2A (1) Fw: 5’-GAGCAGCATGGAGCCTTCG-3’; p16 CDKN2A (1) Rv: 5’-CGTAACTATTCGGTGCGTTGG-3’) genes, using Primer-BLAST [30]. Primers for GAPDH (GenBank accession number: NM_001256799) were designed by Thibeaux and colleagues (2014) [31]. Primers were chemically synthesized by Integrated DNA Technologies (IDT, Coralville, IA, USA). For qPCR, primers were added to cDNAs and to GoTaq® qPCR Master Mix (Promega), which contained the fluorescent intercalating agent DNA BRYT Green®, according to the manufacturer instructions. Reactions were read using CFX96 Touch™ RT-PCR (Bio-Rad). CFX Manager 3.0 software (Bio-Rad) was used to analyze the results. Relative PLCδ4 expression was calculated using REST-MCS® software, version 2 [32, 33]. At least three assays were performed.
2.5. Western Blot
Total protein extracts were obtained from cells using sterile cell scraper and NETN buffer (150 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl pH 8.0, 0.5% Nonidet P-40) and 1% of protease and phosphatase inhibitors cocktail (Sigma-Aldrich). After extraction, protein concentration was quantified by Bradford assay (Sigma-Aldrich), according to the manufacturer recommendations. SDS-PAGE was performed with 30–50 µg of protein, followed by transference to a polyvinylidene fluoride membrane (PVDF; Bio-Rad, Hercules, CA, USA) using the semi-dry transfer system (Bio-Rad) for 90 minutes. Immunoblotting was performed by standard methods. Primary antibodies against human PLCδ4 (Santa Cruz Biotech; 1:200) and α-tubulin (Sigma-Aldrich; 1:2000) were used. Protein bands were revealed by chemiluminescence using ECL Plus (Thermo Scientific) and BioMax® MR (Carestream Dental, Atlanta, GA, USA) films. The films were scanned and quantitative analyses were performed using ImageJ software [34]. Relative protein expression was calculated using α-tubulin as a loading control. At least three assays were performed.
2.6. Growth curve
Mesenchymal stem cells were plated on 24-well plates at 1 × 104 cells/well and transfected using siRNA. Cells were trypsinized every 2 days, resuspended in 1 mL DMEM medium, and counted in a Neubauer chamber using Trypan blue for viability exclusion, in triplicate. Results were plotted in time (hours) versus cell number (×104). The assay was repeated three times.
2.7. Cell death assay
To evaluate cell viability, Dead Cell Apoptosis Kit (Thermo Fisher Scientific) was used as manufacturer instructions. Briefly, cells were transfected as previously described (item 2.4) in a 96-well plate (Nunc), 3 wells per group, using 1 × 104 cells per plate, for the 2 days post-transfection assays, and 2.5 × 103 cells per plate, for the 7 days post-transfection assays. Cells were allowed to grow for 2 or 7 days. Then, the cells were washed in PBS 1× and incubated with Annexin-V Alexa Fluor-488 (1:50) and propidium iodide (PI, 0.1 µg) for 15 minutes, washed and incubated with PBS 1×. Fluorescence intensity was measured using Cytation 5 (BioTek Instruments, Winooski, VT, USA) fluorimeter with filters sets for Annexin-V (excitation 485/20, emission 528/20) and PI (excitation 535/20, emission 617/20). The assay was repeated twice.
2.8. Cell cycle
Stem cells were plated on 6-well plates, 2 × 105 cells/well, 3 wells per group, and transfected using siPLCδ4 or siSCR. After 48 hours, cells were enzymatically removed and 1.5 × 105 cells were used for DNA extraction [35]. A solution containing 0.25 µM TO-PRO3® (Thermo-Fisher Scientific) and 0.2 mg.mL−1 RNase A (Thermo-Fisher Scientific) was used to stain the cells. The analysis was performed using a Guava Easycyte 6-2L flow cytometer (Millipore, Temecula, CA, USA). The control group (not transfected) was used for adjusting the forward and side scatter parameters to determine the population of interest on a logarithmic scale. Samples were excited (Ex) at 633 nm and observed with 661/19 emission (Em) filter. The cytometer assessed 5000 events, and the data were analyzed using FlowJo software version 7.2.5 (LLC, Ashland, OR, USA). The results were plotted as a percentage of cells in G1, S or G2/M cell cycle phases, relative to the total cell count. At least three assays were performed.
2.9. 5-ethinyl-2’-deoxyuridine (EdU) incorporation assay
EdU (5-ethinyl-2’-deoxyuridine), a thymidine analog, was used to quantify proliferating cells [36]. hASC were plated on 96-well plates at a density of 1 × 104 cells per plate, for the 2 days post-transfection assays, and 2.5 × 103 cells per plate, for the 7 days post-transfection assays, 3 wells per group in each assay, using the transfection procedures previously described. For marking and quantifying EdU-positive cells, Click-iT EdU Kit (Thermo-Fisher Scientific) was used. EdU detection solution, containing Azida-Alexa 555, and nuclear staining solution HCS Nuclear Mask Blue (1:2000) was used. Cells were photo-documented using 4× magnification objective lens in an Olympus IX70 microscope, QIClick camera, and Image Pro Plus 7.01. Filters used for excitation and emission were: Ex 350/50, Em 455/50 for HCS Nuclear Mask Blue and Ex 555/25, Em 605/52 for Azida-Alexa 555. The assays were replicated three times. The total number of cells per field (HCS Nuclear Mask Blue nuclear labeling) and EdU-positive number of cells (Azida-Alexa 555) were counted, processed and presented as the percentage of EdU-positive cells. At least three assays were performed.
2.10. Mitotic index
Phosphorylated H3 histone at serine 10 is a mitosis marker [37]. The phosphorylated protein was assessed by immunofluorescence assay to quantify cells at this cell cycle phase. Stem cells were plated on 96-well plates, at a concentration of 1 × 104 cells per well for the 2 days post-transfection assays, and 2.5 × 103 cells per well for the 7 days post-transfection assays, 3 wells per group in each assay, and transfected. Cells were incubated with the rabbit polyclonal anti-pH3 (phosphorylated histone H3, S10p) primary antibody (1:500, Millipore), secondary antibody conjugated to Alexa Fluor® 488 IgG anti-rabbit (1:1000, Thermo-Fisher Scientific) and the nuclear marker Hoechst 33258 (1 µg.mL−1, Invitrogen). The cells were photographed using an Olympus IX70 microscope at 4× magnification with QIClick camera and the software Image Pro Plus 7.01. For detecting fluorophore excitation and emission, the filters Ex 350/50 and Em 455/50 were used for Hoescht, and Ex 490/20 and Em 525/36 were used for Alexa Fluor® 488. The assays were replicated three times. The total number of cells per field (nuclear marker, Hoescht) and the number of pH3-positive cells (Alexa Fluor® 488 marking) were counted and expressed as the percentage of pH3-positive cells compared to total cell number. At least three assays were performed.
2.11. Senescence assay
Mesenchymal stem cells were plated in 24-well plates at a density of 1 × 104 cells per well and transfected, 2 wells per group. 2 or 7 days after transfection, the cells were stained for detection of senescence-associated β-galactosidase activity (SA-β-Gal; [38]), using Senescence Cells Histochemical Staining Kit (Sigma-Aldrich). Cells were photo-documented using an Olympus IX70 microscope at 4× magnification, QIClick camera, and Image Pro Plus 7.01 software. Assays were performed in triplicate. The total number of cells per field and the number of senescent cells (perinuclear blue staining) were counted, and expressed as a percentage of senescent cells, relative to total cell number. At least three assays were performed.
2.12. Reactive oxygen species assay
The Fluorometric Intracellular reactive oxygen species (ROS) kit (Sigma-Aldrich) was used for hASC ROS detection following manufacturer instructions. First, cells were transfected as described in item 2.3, in a 96-well white opaque plate (Costar), 3 wells per group using 1 × 104 cells or 2.5 × 103 per well for the 2 or 7 days post-transfection assays, respectively. Then, the cells were allowed to grow for 2 or 7 days, followed by incubation with ROS reaction mix for one hour at 37°C, 5% CO2. The plate was then read using Cytation 5 (BioTek) fluorimeter with the filter 490/20 for excitation and 525/20 for emission. The assay was repeated twice.
2.13. Statistical analyses
At least three independent assays were performed for each experiment. Statistical analysis was performed using GraphPad Prism 5 software (La Jolla, CA, USA). Results were expressed as a mean ± standard error (SEM) and compared through variance analysis (One-way or Two-way ANOVA), Bonferroni’s multiple comparison tests, Student’s t-test (for parametric samples), Kruskal-Wallis and Dunn’s multiple comparison tests, or Mann-Whitney test (for non-parametric samples). Statistical significance was defined for p<0.05.
3. Results
3.1. hPLCδ4 is mainly a nuclear protein
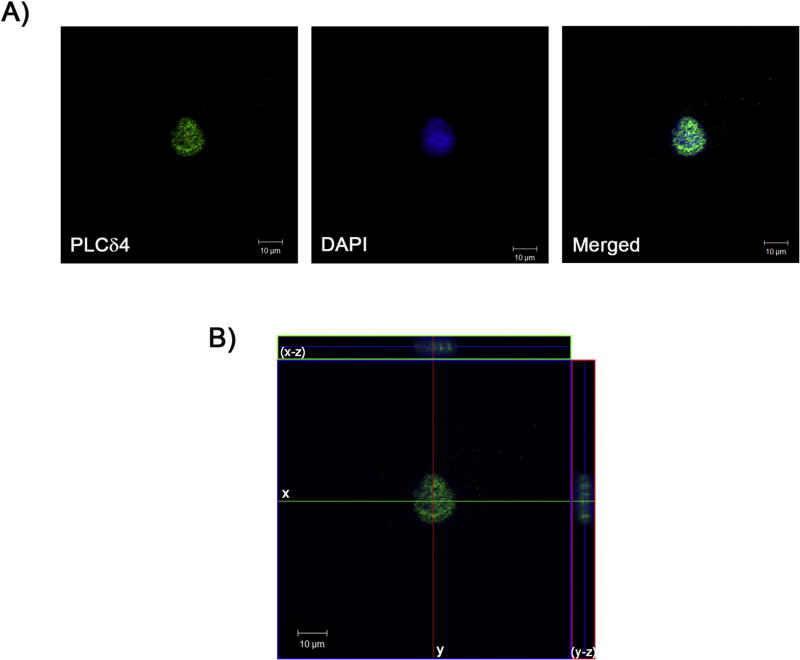
In silico analysis of the hPLCδ4 protein sequence revealed the presence of a Nuclear Localization Signal (NLS). NucPred software [39] showed that PLCδ4 has a 86% probability of being nuclear, and SeqNLS [40] suggested that there is an 86–89% chance that PLCδ4 contains a genuine NLS. Nuclear localization of PLCδ4 in hASC was confirmed by confocal microscopy. As seen in Figure 1A (merged images, right panel), the PLCδ4 antibody conjugated with Alexa 488 (green; left panel) colocalized with DAPI stained nuclei (blue; central panel). To further demonstrate the nuclear compartmentalization of PLCδ4, serial images were collected, and used to build a tridimensional reconstruction (Figure 1B). In the medial plan, PLCδ4 and nuclear labeling colocalized.
Figure 1. PLCδ4 was primarily nuclear localized in hASC.
hASC were incubated with primary antibody anti-PLCδ4 and a secondary antibody conjugated with Alexa 488 (green). The nucleus was stained with DAPI (blue). A) PLCδ4 colocalizes with nuclear staining in hASC, indicated by the merged image. B) Tridimensional reconstruction. Central panel: medial plane inside the nucleus. Upper panel: 3D reconstruction of the x-z plane. Right panel: 3D reconstruction of the y-z plane. Scale bar: 10 µm. Representative image of what was observed in at least samples of three patients.
3.2. siPLCδ4 impairs cell growth but does not increase cell death
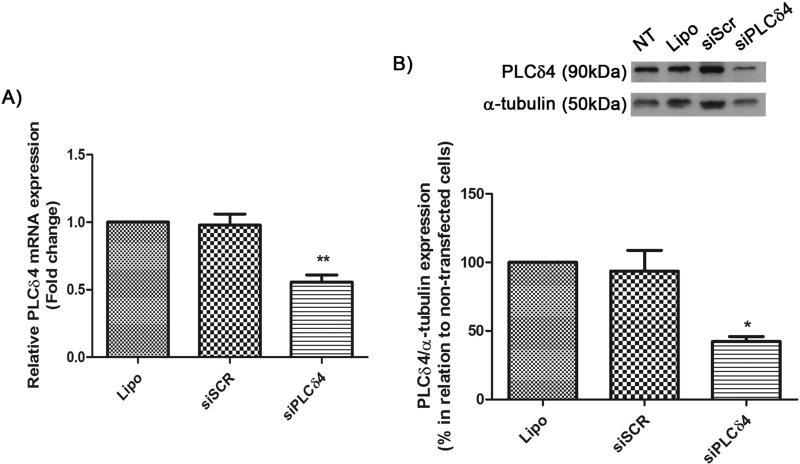
To study PLCδ4 function, siRNA knockdown was performed. Silencing efficiency was assessed by qPCR and Western Blotting (Figure 2). 48 hours after siRNA transfection, total RNA and protein extraction was performed. Three groups were analyzed: Lipo (stem cells treated only with the transfection reagent, Lipofectamine), siSCR (cells transfected with scrambled siRNA), and siPLCδ4 (cells transfected with siRNA for siPLCδ4). In qPCR assays and Western Blot analysis, PLCδ4 gene expression was reduced by almost half, compared to control groups (Figure 2A and B). PLCδ4 expression reduced almost by half, going from 0.98 ± 0.08 SEM in siSCR to 0.56 ± 0.05 SEM in siPLCδ4. In Western Blot analysis, siPLCδ4 cells also showed a similar reduction in protein expression to the one observed for PLCδ4 mRNA, there was about 50% reduction in PLCδ4 expression (Figure 3B). The specificity of PLCδ4 siRNA was tested against the expression level of PLCγ1, and any nonspecific knockdown for PLCγ1 was observed (data not shown). These data validate the efficiency of the siPLCδ4 sequence in reducing PLCδ4 levels in hASC.
Figure 2. siPLCδ4 sequence reduces the expression of PLCδ4 in hASC.
A) Relative PLCδ4 mRNA expression measured by Real-Time qPCR. Cells were transfected with 25 nM of scrambled siRNA (siSCR) or siRNA for PLCδ4 (siPLCδ4). 48 hours post transfection, total RNA was extracted from cells and was used for cDNA synthesis. PLCδ4 expression was normalized with GAPDH and RPL13A, and samples treated only with lipofectamine (non-transfected cells) were used as calibrators. There was a reduction in PLCδ4 expression in siPLCδ4 cells. B) Western Blot of total protein extracts 48 hours posttransfection. α-tubulin was used as a loading protein control, and protein expression levels were normalized with lipofectamine samples values (in percentage). The reduction of PLCδ4 protein expression can be observed for siPLCδ4 treated cells when compared to controls. Lipo = cells treated with lipofectamine (transfection reagent); siSCR = cells transfected with scrambled siRNA; siPLCδ4 = cells transfected with siRNA for PLCδ4. One-way ANOVA with Bonferroni’s multiple comparison tests (* p<0.05; ** p<0.01, n=3).
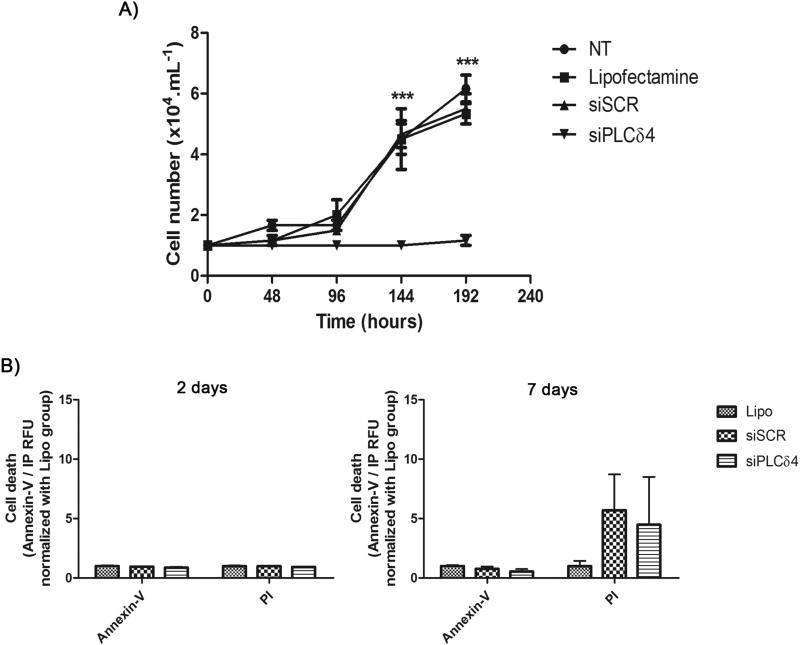
Figure 3. PLCδ4 knockdown abolishes cell proliferation in hASC, without a decrease in cell viability.
A) Growth curve of transfected cells. The curve was started with 1 × 104 cells. Cells were counted every two days, in a Neubauer chamber, using Trypan Blue for viability exclusion. From 0 to 192 hours the number of cells in siPLCδ4 were stable. The time points of 144 hours and 192 hours in siPLCδ4 are different from the other groups. B) Cell death assay, using Annexin-V/PI. 2 or 7 days post-transfection cells were stained Annexin-V/PI and their fluorescence were measured. There was no difference between the groups. NT = non-treated cells; Lipo = cells treated with lipofectamine (transfection reagent); siSCR = cells transfected with scrambled siRNA; siPLCδ4 = cells transfected with siRNA for PLCδ4. RFU = relative fluorescence unit. Two-way ANOVA with Bonferroni’s multiple comparison tests (*** p<0.001, n=3 for growth curve and n=2 for Annexin-V/PI).
Once the efficiency of PLCδ4 silencing was confirmed, functional analyses were conducted with PLCδ4 knockdown cells. Cell growth assays revealed a pattern for the control groups (NT, non-treated cells; Lipo; and siSCR, Figure 3A). In contrast, cells treated with siPLCδ4 exhibited no proliferation, compared to controls. The number of cells was stable for 8 days of culture (192 hours), suggesting that knockdown cells were not proliferating. Despite the impaired cell growth, transfected cells did not present a reduction in cell viability. Annexin-V/PI stained cells was not different between the analyzed groups (Figure 3B). These findings suggest that PLCδ4 knockdown induce cell cycle arrest without affect cell death.
3.3. siPLCδ4 causes cell cycle arrest
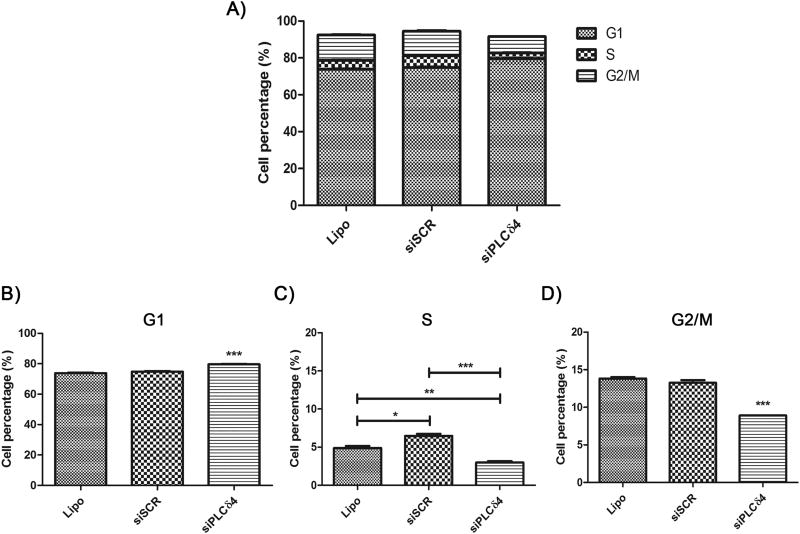
Next, the effect of siPLCδ4 on cell cycle profile was assessed. After 48 hours of transfection, stem cells were incubated with To-Pro®3 nuclear marker for evaluation of cell cycle by flow cytometry. The percentages of cells in each phase were analyzed. In Figure 4, it is possible to note a distinct distribution profile of cell cycle phases between siPLCδ4 cells and control cells. An increase in hASC siPLCδ4 cells in G1 phase (Figure 4B; 74.75 ± 0.25 in siSCR to 79.63 ± 0.18 in siPLCδ4), and a decrease in the percentages of these cells in S and G2/M (Figure 4C and D; S phase: 6.47 ± 0.27 in siSCR to 2.97 ± 0.19 in siPLCδ4; G2/M: 13.25 ± 0.35 in siSCR to 8.91 ± 0.01 in siPLCδ4) was observed.
Figure 4. Reduction of PLCδ4 expression in hASC leads to cell cycle arrest.
48 hours after transfection, cells were resuspended with trypsin, incubated with the nuclear probe To-Pro® 3 and analyzed by flow cytometry. A) Graphic showing cell cycle phases distribution among the different cell groups. B) Percentage of cells, in relation to total cell number, on G1 cell cycle phase. C) S phase. D) G2/M phases. Lipo = cells treated with lipofectamine (transfection reagent); siSCR = cells transfected with scrambled siRNA; siPLCδ4 = cells transfected with siRNA for PLCδ4. One-way ANOVA with Bonferroni’s multiple comparison tests (* p<0.05; ** p<0.01; *** p<0.001, n≥3).
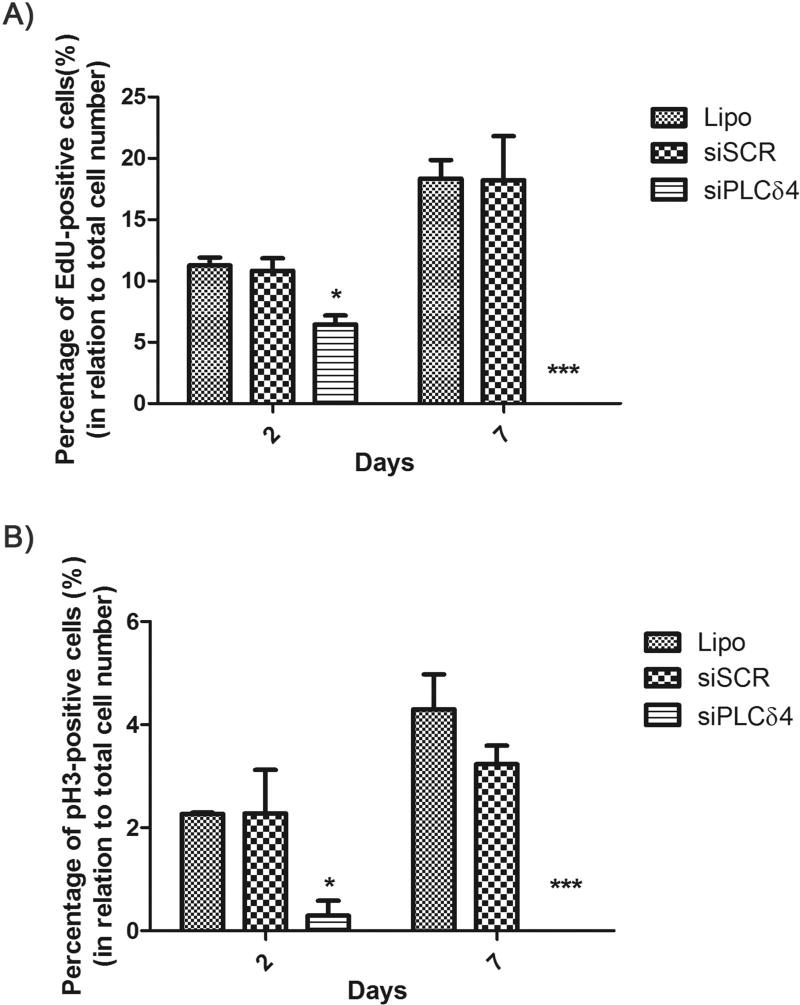
To confirm the results obtained from cell cycle profile analysis, we used assays with specific probes for S and M cell cycle phases, 5-ethynyl-2'-deoxyuridine (EdU) and phosphorylated histone H3 (pH3), respectively. 2 and 7 days post-transfection, cells were labeled with the probes and photo documented. Hoescht probe was used as a nuclear marker. For both EdU (S phase) and pH3 (M phase), the number of positive-labeled cells for siPLCδ4 group is lower than controls (Figure 5). The data indicated that PLCδ4 knockdown promoted cell cycle arrest, which continued for at least 7 days post-transfection.
Figure 5. PLCδ4 knockdown reduces the number of EdU-positive and pH3-positive cells.
2 or 7 days after transfection, hASC were incubated with Hoescht (nuclear staining) and EdU (a probe for dividing cells) or pH3 (a marker for mitosis). The figures show the percentage of cells marked simultaneously with Hoechst and EdU or pH3, in relation to total cell number (stained only with Hoescht). A) Percentage of EdU-positive cells. B) Percentage of pH3-positive cells. For both probes and for the two-time points analyzed, there was a reduction of positive-labeled cells for the siPLCδ4 group, when compared to control groups. One-way ANOVA with Bonferroni’s multiple comparison tests (*p<0.05; *** p<0.001, n=3).
3.4. PLCδ4 silencing induces cellular senescence
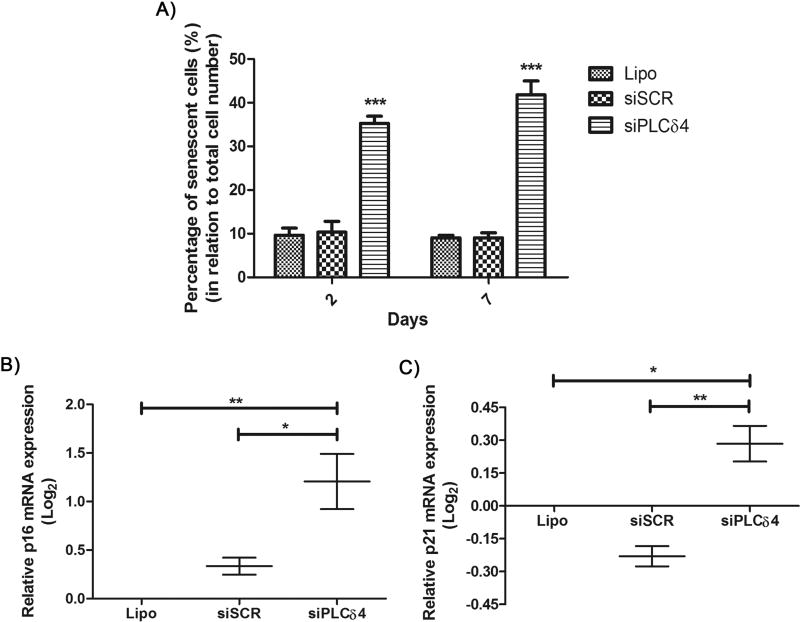
There are several causes of cell cycle arrest, one being cellular senescence, which can be measured by the senescence-associated β-galactosidase assay (SA-β-Gal) [38]. At pH 6, senescent cells express β-galactosidase, and its activity can be visualized and measured by addition of X-Gal to cells. If X-Gal is hydrolyzed by β-galactosidase, a blue color in the perinuclear region is generated. Transfected stem cells were subjected to senescence assays 2 and 7 days post-transfection. The cells were imaged, and, for each field, total and senescent cell counts were performed. For both time points (2 and 7 days), the number of senescent cells in the group transfected with siPLCδ4 was higher than for control cells (Figure 6A; 2 days: 10.36 ± 2.44 in siSCR to 35.26 ± 1.64 in siPLCδ4, 7 days: 9.07 ± 1.12 in siSCR to 41.81 ± 3.19 in siPLCδ4). To further characterize the senescent state of siPLCδ4 cells, mRNA expression levels of p16INK4A+ and p21Cip1, two biomarkers commonly associated with senescence [41], were measured (Figure 6 B and C). There was an increase in the expression of both mRNAs for cells treated with siPLCδ4, which corroborate the SA-β-Gal assay result. This finding may explain the reduction of siPLCδ4 cell proliferation and cell cycle arrest (Figures 3–5).
Figure 6. PLCδ4 silencing increases the percentage of senescent cells.
A) 2 or 7 days after transfection, hASC were assayed for SA-β-Gal activity. The figure shows the percentage of cells stained for SA-β-Gal activity, in relation to total cell number. For the two-time points analyzed, there was a reduction of stained cells for the siPLCδ4 group, when compared to control groups as determined by one-way ANOVA with Bonferroni’s multiple comparison tests (***p<0.001, n=3). B and C) Relative p16INK4A+ and p21Cip1 mRNA expression measured by Real-Time qPCR. 48 hours post transfection, total RNA was extracted from cells and was used for cDNA synthesis. p16 and p21 expression was normalized with RPL13A, and Lipo group was used as calibrator. There was an increase in p16INK4A+ and p21Cip1 expression in siPLCδ4 cells. One-way ANOVA with Bonferroni’s multiple comparison tests (* p<0.05; ** p<0.01, n=2).
4. Discussion
PLCδ4 has been most exhaustively characterized in murine models, such as rat and mice. In these models, it was observed that PLCδ4 participates in proliferative processes and that it is mainly located in the nucleus [27, 42]. In this work, hPLCδ4 also showed preferential nuclear localization in hASC (Figure 1). Therefore, PLCδ4 subcellular localization in hASC is similar to this protein’s distribution described for other cell types, like rat hepatic cells [27], mouse cells [42], and also corroborate with in silico protein sequence analysis. Several aspects of PLCδ4 function, such as activation and signaling, remain unknown. Moreover, human PLCδ4 has not been well characterized and could be associated with pathologies, as described for other PLC isoforms, which may have roles in the development of cancer, neurodegenerative diseases, and neoplasia [21, 23].
The nucleus contains molecules and machinery for Ca2+ signaling, such as PIP2 and InsP3, among others [43]. This would suggest that nuclear PLCs are active, once their substrate and products are present in the nucleus. PLCs are classified as either primary or secondary, depending on their activation mechanisms. Primary PLCs are activated directly by receptors that bind to extracellular modulators, while secondary PLCs are activated by intracellular signals [44]. PLCβ subtypes are activated by G protein-coupled receptors (GPCR) by several mechanisms, and PLCγ is activated by tyrosine kinases receptors (RTK) [45]. PLCε is activated by proteins recruited by GPCRs and RTKs signaling, whereas evidence indicates that the δ and η classes are regulated by Ca2+, through a positive feedback mechanism of the activity of the other PLCs [46, 47]. Although the activation of cytoplasmic PLCs is well understood, the mechanism of nuclear PLC activation is different. PLCγ1 in hepatic cell nuclei of mice is activated by nuclear epidermal growth factor (EGFR) receptor [48]. It was proposed by Xu and colleagues in 2001 [49] that nuclear PLCβ1 from rats would be phosphorylated by extracellular signal-regulated kinase (ERK) after stimulation with insulin-like growth factor-1 (IGF-1), pointing to a possible mechanism of nuclear PLC activation through RTKs. Interestingly, the murine PLCδ4 promoter region is activated by growth factors [50].
To further characterize PLCδ4 functions in mesenchymal stem cells, siRNA knockdown was performed to understand the function of this enzyme in standard cell culture conditions. Silencing efficiency was assessed by qPCR and Western Blot (Figure 2). Growth curves of hASC cells silenced for PLCδ4 indicated impaired proliferation of these cells, compared to control cells (Figure 3A). Nuclear Ca2+ has been described as an important messenger for cell proliferation [4]. When hepatocytes had their nuclear Ca2+ chelated by nuclear-directed parvalbumin, their proliferation was significantly reduced, demonstrating a connection between nuclear Ca2+ signaling and cell proliferation. The reduction in proliferation of hASC cells silenced for PLCδ4 could be explained by an increase in cell death, which would reduce the number of viable cells, or by cell cycle arrest. The first hypothesis was discarded because no increase in cell death was observed for hASC PLCδ4 knockdown (Annexin-V/PI labeling, Figure 3B). However, cell cycle arrest for cells silenced for PLCδ4 was observed (Figures 4 and 5). Arrest probably occurred due to the entry of the cells in a senescent stage (Figure 6), in which cells are metabolically active, but unable to divide [51].
The relation between nuclear PLCs and cell cycle regulation is described in the literature [23, 52–54]. In 2003, Irvine [55] highlighted the connection between nuclear lipid oscillations and cell cycle progression. Cell cycle can be defined as a series of steps that leads to cell division. In each of these steps, there are molecules responsible for the regulation of cell cycle progression, such as cyclins, CDKs (cyclin-dependent kinases), pRB (retinoblastoma protein) and Cip/Kip inhibitors (p16, p21, and p27) [56]. Further investigation is necessary to determine if PLCs interfere with the cell cycle through interaction or signaling with regulatory molecules. Expression of the hPLCβ1 nuclear isoform, hPLCβ1b, peaks at G1/S and G2/M transitions [54]. One of the mediators of PLCβ1b cell cycle modulation is diacylglycerol (DAG), generated as a product of the enzymatic activity of PLCβ1b over PIP2. DAG, at G1/S transition, activates the cyclin D3-CDK4 complex, but in G2/M progression DAG leads to the translocation of PKCα and cyclin B1 to the nucleus. In rat fibroblasts, unlike PLCβ1, PLCγ1 overexpression promotes DNA synthesis independently of its lipase activity [57]. For rat PLCδ1, Kaproth-Joslin and colleagues [52], described the reduction of Rat-1 and NIH 3T3 cell proliferation following PLCδ1 silencing with shRNA. They observed no increase in cell death, but altered G1/S transition, due to the reduction of cyclin E-CDK2 complex activity, without alteration of cyclin D-CDK4 complex. The authors, however, did not observe an increase in senescence, as seen in our work (Figure 6). A better characterization of the molecules involved in PLCδ4 knockdown cell arrest is necessary to determine more precisely at which point in the cell cycle these cells are accumulated.
Cell cycle studies in rat cells and human tumor cells show that PLCδ4 expression increases during the G1/S transition, and throughout M phase, but when cells enter a new G1 phase, the expression of PLCδ4 practically disappears [27]. In regenerative rat livers, which underwent partial hepatectomy, an increase in PLCδ4 and PKCs α and ε activities were observed in PLCδ4+/+ animals during G1/S transition, but not for PLCδ4−/− rats [58]. Our cell cycle results corroborate with these findings as a reduction of PLCδ4 levels interrupted G1 to S progression (Figures 4 and 5). The reduction of EdU (S phase) and pH3 (mitosis) labeling (Figure 5) for siPLCδ4 cells indicated that the inhibition of cycle progression started in G1 phase. Moreover, this inhibitory response remained stable, for at least 7 days after the transfection.
Defects in the signaling of any PI-PLC could potentially lead to the development of tumors as these enzymes participate in cell cycle control. Leung and colleagues [22] analyzed matched samples of human cDNAs from tumor and healthy cells, and observed that hPLCδ4 was overexpressed in tissues derived from breast and testicular cancers. They also reported that PLCδ4 overexpression led to a faster proliferation of MCF-7 cells in serum-free medium, indicating a possible role of hPLCδ4 in oncogenesis.
Cell senescence is a state in which cells stop dividing, leading to a profile of apoptosis resistance, often with increased protein synthesis, increased glycolytic metabolism, reduced fatty acid oxidation, increased ROS, increased p16INK4A+ and p21Cip1 expression, and increased secretion of factors associated with senescence (SASP; [59]). The increase in senescent cells in hASC PLCδ4 knockdown (Figure 6) has not been described in the literature. However, in human breast cancer cells, BT474 and MCF-7, it was observed that PIP4K silencing (PI4P-phosphorylating kinase, which generates PIP2) led to the accumulation of ROS in the cells, directing them to senescence [60]. Despite not showing an increase in ROS levels (Supplementary Figure 1), siPLCδ4 cells exhibited cell morphology modification, as cells treated with siPLCδ4 appear to be more vacuolated and flattened (Supplementary Figure 2), a morphology trait commonly associated with senescence [41], increased SA-β-Gal staining, p16INK4A+ and p21Cip1 expression (Figure 6). While the p53/p21 pathway leads cells to growth arrest, the pRB/p16 pathway inhibits cell cycle progression, taking cells to irreversible senescence [41]. Senescence of siPLCδ4 cells, observed from 2 days after transfection, could explain the non-transient effects seen in the reduction of cell growth (Figure 3) and in the reduction of EdU and pH3 labeling (Figure 5), which remained reduced in comparison to controls, even seven days post-transfection. Kawano and colleges showed that human mesenchymal stem cells have spontaneous Ca2+ oscillations [61]. It has been speculated that increased Ca2+ levels activate PLCδ, that may amplify Ca2+ elevation to sufficient levels to induce downstream signaling [21]. We speculate that these spontaneous Ca2+ oscillations could activate the PLCδ4 that is important to amplify Ca2+ levels to induce cell proliferation.
Our data show that human PLCδ4 is in involved in cellular proliferation, which is a process possibly mediated by nuclear Ca2+. Further characterization of PLCδ4 signalization is needed, to depict how PLCδ4 is activated and which cell signaling pathways this PLC activates.
5. Conclusions
In summary, this work provides evidence that hPLCδ4 is essential for cell proliferation in human mesenchymal stem cells and contribute to understand better the proliferation processes in stem cells, which could be used for tissue engineering purposes.
Supplementary Material
Supplementary Figure 1: ROS levels in transfected cells. 2 or 7 days after transfection, cells were assayed for ROS levels using Fluorometric Intracellular ROS kit (Sigma-Aldrich). There was no difference in ROS levels between the groups. Lipo = cells treated with lipofectamine (transfection reagent); siSCR = cells transfected with scrambled siRNA; siPLCδ4 = cells transfected with siRNA for PLCδ4. RFU = relative fluorescence unit. n=2.
Supplementary Figure 2: Cell morphology of transfected hASC. 2 days after transfection, cells were photographed, using the 10× objective lens on an Olympus IX70 microscope, using a QIClick camera, and Image Pro Plus 7.01 software. Lipofectamine = cells treated with lipofectamine (transfection reagent); siSCR = cells transfected with scrambled siRNA; siPLCδ4 = cells transfected with siRNA for PLCδ4. At least three independent experiments were performed. Scale bar: 200 µm.
Highlights.
Little is known about the functions of nuclear PLCs.
PLCδ4 is a nuclear protein
PLCδ4 is essential for mesenchymal stem cells proliferation
The percentage of senescent cells is increased after PLCδ4 knockdown
Acknowledgments
This work was supported by the NIH grant 1R03TW008709 and by grants from the “INCT-Regenera”, “Rede Mineira de Engenharia de Tecidos e Terapia Celular (REMETTEC, RED-00570-16),” FAPEMIG, CAPES, and CNPq. The authors thank “Pró-Reitoria de Pesquisa da Universidade Federal de Minas Gerais” for its financial sponsorship. The authors also thank “Centro de Aquisição e Processamento de Imagens do ICB/UFMG” (CAPI) for technical support and Professor Elio Anthony Cino for language revision.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that they have no competing interests.
References
- 1.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nature reviews. Molecular cell biology. 2000;1(1):11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Mendes CC, Gomes DA, Thompson M, Souto NC, Goes TS, Goes AM, Rodrigues MA, Gomez MV, Nathanson MH, Leite MF. The type III inositol 1,4,5-trisphosphate receptor preferentially transmits apoptotic Ca2+ signals into mitochondria. The Journal of biological chemistry. 2005;280(49):40892–900. doi: 10.1074/jbc.M506623200. [DOI] [PubMed] [Google Scholar]
- 3.Minagawa N, Nagata J, Shibao K, Masyuk AI, Gomes DA, Rodrigues MA, Lesage G, Akiba Y, Kaunitz JD, Ehrlich BE, Larusso NF, Nathanson MH. Cyclic AMP regulates bicarbonate secretion in cholangiocytes through release of ATP into bile. Gastroenterology. 2007;133(5):1592–602. doi: 10.1053/j.gastro.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigues MA, Gomes DA, Leite MF, Grant W, Zhang L, Lam W, Cheng YC, Bennett AM, Nathanson MH. Nucleoplasmic calcium is required for cell proliferation. The Journal of biological chemistry. 2007;282(23):17061–8. doi: 10.1074/jbc.M700490200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikoshiba K. The InsP3 receptor and intracellular Ca2+ signaling. Current opinion in neurobiology. 1997;7(3):339–45. doi: 10.1016/s0959-4388(97)80061-x. [DOI] [PubMed] [Google Scholar]
- 6.Fedorenko OA, Popugaeva E, Enomoto M, Stathopulos PB, Ikura M, Bezprozvanny I. Intracellular calcium channels: inositol-1,4,5-trisphosphate receptors. European journal of pharmacology. 2014;739:39–48. doi: 10.1016/j.ejphar.2013.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nature reviews. Molecular cell biology. 2003;4(7):517–29. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 8.Hardingham GE, Chawla S, Johnson CM, Bading H. Distinct functions of nuclear and cytoplasmic calcium in the control of gene expression. Nature. 1997;385(6613):260–5. doi: 10.1038/385260a0. [DOI] [PubMed] [Google Scholar]
- 9.Malviya AN, Rogue PJ. "Tell me where is calcium bred": clarifying the roles of nuclear calcium. Cell. 1998;92(1):17–23. doi: 10.1016/s0092-8674(00)80895-8. [DOI] [PubMed] [Google Scholar]
- 10.Pusl T, Wu JJ, Zimmerman TL, Zhang L, Ehrlich BE, Berchtold MW, Hoek JB, Karpen SJ, Nathanson MH, Bennett AM. Epidermal growth factor-mediated activation of the ETS domain transcription factor Elk-1 requires nuclear calcium. The Journal of biological chemistry. 2002;277(30):27517–27. doi: 10.1074/jbc.M203002200. [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R. The C terminus of the L-type voltage-gated calcium channel Ca(V)1.2 encodes a transcription factor. Cell. 2006;127(3):591–606. doi: 10.1016/j.cell.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tasker PN, Taylor CW, Nixon GF. Expression and distribution of InsP(3) receptor subtypes in proliferating vascular smooth muscle cells. Biochemical and biophysical research communications. 2000;273(3):907–12. doi: 10.1006/bbrc.2000.3036. [DOI] [PubMed] [Google Scholar]
- 13.Echevarria W, Leite MF, Guerra MT, Zipfel WR, Nathanson MH. Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nature cell biology. 2003;5(5):440–6. doi: 10.1038/ncb980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leite MF, Thrower EC, Echevarria W, Koulen P, Hirata K, Bennett AM, Ehrlich BE, Nathanson MH. Nuclear and cytosolic calcium are regulated independently. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(5):2975–80. doi: 10.1073/pnas.0536590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marius P, Guerra MT, Nathanson MH, Ehrlich BE, Leite MF. Calcium release from ryanodine receptors in the nucleoplasmic reticulum. Cell calcium. 2006;39(1):65–73. doi: 10.1016/j.ceca.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Gomes DA, Rodrigues MA, Leite MF, Gomez MV, Varnai P, Balla T, Bennett AM, Nathanson MH. c-Met must translocate to the nucleus to initiate calcium signals. The Journal of biological chemistry. 2008;283(7):4344–51. doi: 10.1074/jbc.M706550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payrastre B, Nievers M, Boonstra J, Breton M, Verkleij AJ, Van Bergen en Henegouwen PM. A differential location of phosphoinositide kinases, diacylglycerol kinase, and phospholipase C in the nuclear matrix. The Journal of biological chemistry. 1992;267(8):5078–84. [PubMed] [Google Scholar]
- 18.Vann LR, Wooding FB, Irvine RF, Divecha N. Metabolism and possible compartmentalization of inositol lipids in isolated rat-liver nuclei. The Biochemical journal. 1997;327(Pt 2):569–76. doi: 10.1042/bj3270569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faenza I, Fiume R, Piazzi M, Colantoni A, Cocco L. Nuclear inositide specific phospholipase C signalling - interactions and activity. The FEBS journal. 2013;280(24):6311–21. doi: 10.1111/febs.12450. [DOI] [PubMed] [Google Scholar]
- 20.Bootman MD, Fearnley C, Smyrnias I, MacDonald F, Roderick HL. An update on nuclear calcium signalling. Journal of cell science. 2009;122(Pt 14):2337–50. doi: 10.1242/jcs.028100. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura Y, Fukami K. Regulation and physiological functions of mammalian phospholipase C. Journal of biochemistry. 2017;161(4):315–321. doi: 10.1093/jb/mvw094. [DOI] [PubMed] [Google Scholar]
- 22.Leung DW, Tompkins C, Brewer J, Ball A, Coon M, Morris V, Waggoner D, Singer JW. Phospholipase C delta-4 overexpression upregulates ErbB1/2 expression, Erk signaling pathway, and proliferation in MCF-7 cells. Molecular cancer. 2004;3:15. doi: 10.1186/1476-4598-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramazzotti G, Faenza I, Fiume R, Billi AM, Manzoli L, Mongiorgi S, Ratti S, McCubrey JA, Suh PG, Cocco L, Follo MY. PLC-beta1 and cell differentiation: An insight into myogenesis and osteogenesis. Advances in biological regulation. 2017;63:1–5. doi: 10.1016/j.jbior.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Lee SB, Rhee SG. Molecular cloning, splice variants, expression, and purification of phospholipase C-delta 4. The Journal of biological chemistry. 1996;271(1):25–31. doi: 10.1074/jbc.271.1.25. [DOI] [PubMed] [Google Scholar]
- 25.Nagano K, Fukami K, Minagawa T, Watanabe Y, Ozaki C, Takenawa T. A novel phospholipase C delta4 (PLCdelta4) splice variant as a negative regulator of PLC. The Journal of biological chemistry. 1999;274(5):2872–9. doi: 10.1074/jbc.274.5.2872. [DOI] [PubMed] [Google Scholar]
- 26.Asano M, Tamiya-Koizumi K, Homma Y, Takenawa T, Nimura Y, Kojima K, Yoshida S. Purification and characterization of nuclear phospholipase C specific for phosphoinositides. The Journal of biological chemistry. 1994;269(16):12360–6. [PubMed] [Google Scholar]
- 27.Liu N, Fukami K, Yu H, Takenawa T. A new phospholipase C delta 4 is induced at S-phase of the cell cycle and appears in the nucleus. The Journal of biological chemistry. 1996;271(1):355–60. doi: 10.1074/jbc.271.1.355. [DOI] [PubMed] [Google Scholar]
- 28.Kim H, Suh PG, Ryu SH, Park SH. Assignment of the human PLC delta4 gene (PLCD4) to human chromosome band 2q35 by fluorescence in situ hybridization. Cytogenetics and cell genetics. 1999;87(3–4):254–5. doi: 10.1159/000015437. [DOI] [PubMed] [Google Scholar]
- 29.Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue engineering. 2001;7(2):211–28. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 30.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thibeaux R, Ave P, Bernier M, Morcelet M, Frileux P, Guillen N, Labruyere E. The parasite Entamoeba histolytica exploits the activities of human matrix metalloproteinases to invade colonic tissue. Nature communications. 2014;5:5142. doi: 10.1038/ncomms6142. [DOI] [PubMed] [Google Scholar]
- 32.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic acids research. 2002;30(9):e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9(7):671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nature protocols. 2006;1(3):1458–61. doi: 10.1038/nprot.2006.238. [DOI] [PubMed] [Google Scholar]
- 36.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(7):2415–20. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(13):7480–4. doi: 10.1073/pnas.95.13.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(20):9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brameier M, Krings A, MacCallum RM. NucPred--predicting nuclear localization of proteins. Bioinformatics. 2007;23(9):1159–60. doi: 10.1093/bioinformatics/btm066. [DOI] [PubMed] [Google Scholar]
- 40.Lin JR, Hu J. SeqNLS: nuclear localization signal prediction based on frequent pattern mining and linear motif scoring. PloS one. 2013;8(10):e76864. doi: 10.1371/journal.pone.0076864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pole A, Dimri M, Dimri GP. Oxidative stress, cellular senescence and ageing. AIMS Molecular Science. 2016;3(3):300–324. [Google Scholar]
- 42.Fiume R, Ramazzotti G, Faenza I, Piazzi M, Bavelloni A, Billi AM, Cocco L. Nuclear PLCs affect insulin secretion by targeting PPARgamma in pancreatic beta cells. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26(1):203–10. doi: 10.1096/fj.11-186510. [DOI] [PubMed] [Google Scholar]
- 43.Gomes DA, Leite MF, Bennett AM, Nathanson MH. Calcium signaling in the nucleus. Canadian journal of physiology and pharmacology. 2006;84(3–4):325–32. doi: 10.1139/y05-117. [DOI] [PubMed] [Google Scholar]
- 44.Yang YR, Follo MY, Cocco L, Suh PG. The physiological roles of primary phospholipase C. Advances in biological regulation. 2013;53(3):232–41. doi: 10.1016/j.jbior.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annual review of biochemistry. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim YH, Park TJ, Lee YH, Baek KJ, Suh PG, Ryu SH, Kim KT. Phospholipase C-delta1 is activated by capacitative calcium entry that follows phospholipase C-beta activation upon bradykinin stimulation. The Journal of biological chemistry. 1999;274(37):26127–34. doi: 10.1074/jbc.274.37.26127. [DOI] [PubMed] [Google Scholar]
- 47.Kim JK, Choi JW, Lim S, Kwon O, Seo JK, Ryu SH, Suh PG. Phospholipase C-eta1 is activated by intracellular Ca(2+) mobilization and enhances GPCRs/PLC/Ca(2+) signaling. Cellular signalling. 2011;23(6):1022–9. doi: 10.1016/j.cellsig.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 48.Klein C, Gensburger C, Freyermuth S, Nair BC, Labourdette G, Malviya AN. A 120 kDa nuclear phospholipase Cgamma1 protein fragment is stimulated in vivo by EGF signal phosphorylating nuclear membrane EGFR. Biochemistry. 2004;43(50):15873–83. doi: 10.1021/bi048604t. [DOI] [PubMed] [Google Scholar]
- 49.Xu A, Suh PG, Marmy-Conus N, Pearson RB, Seok OY, Cocco L, Gilmour RS. Phosphorylation of nuclear phospholipase C beta1 by extracellular signal-regulated kinase mediates the mitogenic action of insulin-like growth factor I. Molecular and cellular biology. 2001;21(9):2981–90. doi: 10.1128/MCB.21.9.2981-2990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fukami K, Takenaka K, Nagano K, Takenawa T. Growth factor-induced promoter activation of murine phospholipase C delta4 gene. European journal of biochemistry. 2000;267(1):28–36. doi: 10.1046/j.1432-1327.2000.00943.x. [DOI] [PubMed] [Google Scholar]
- 51.Althubiti M, Lezina L, Carrera S, Jukes-Jones R, Giblett SM, Antonov A, Barlev N, Saldanha GS, Pritchard CA, Cain K, Macip S. Characterization of novel markers of senescence and their prognostic potential in cancer. Cell death & disease. 2014;5:e1528. doi: 10.1038/cddis.2014.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaproth-Joslin KA, Li X, Reks SE, Kelley GG. Phospholipase C delta 1 regulates cell proliferation and cell-cycle progression from G1- to S-phase by control of cyclin E-CDK2 activity. The Biochemical journal. 2008;415(3):439–48. doi: 10.1042/BJ20080233. [DOI] [PubMed] [Google Scholar]
- 53.Poli A, Billi AM, Mongiorgi S, Ratti S, McCubrey JA, Suh PG, Cocco L, Ramazzotti G. Nuclear Phosphatidylinositol Signaling: Focus on Phosphatidylinositol Phosphate Kinases and Phospholipases C. Journal of cellular physiology. 2016;231(8):1645–55. doi: 10.1002/jcp.25273. [DOI] [PubMed] [Google Scholar]
- 54.Ratti S, Ramazzotti G, Faenza I, Fiume R, Mongiorgi S, Billi AM, McCubrey JA, Suh PG, Manzoli L, Cocco L, Follo MY. Nuclear inositide signaling and cell cycle. Advances in biological regulation. 2017 doi: 10.1016/j.jbior.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 55.Irvine RF. Nuclear lipid signalling. Nature reviews. Molecular cell biology. 2003;4(5):349–60. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- 56.Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nature reviews. Molecular cell biology. 2007;8(2):149–60. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- 57.Smith MR, Liu YL, Matthews NT, Rhee SG, Sung WK, Kung HF. Phospholipase C-gamma 1 can induce DNA synthesis by a mechanism independent of its lipase activity. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(14):6554–8. doi: 10.1073/pnas.91.14.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akutagawa A, Fukami K, Banno Y, Takenawa T, Kannagi R, Yokoyama Y, Oda K, Nagino M, Nimura Y, Yoshida S, Tamiya-Koizumi K. Disruption of phospholipase Cdelta4 gene modulates the liver regeneration in cooperation with nuclear protein kinase C. Journal of biochemistry. 2006;140(5):619–25. doi: 10.1093/jb/mvj194. [DOI] [PubMed] [Google Scholar]
- 59.Kirkland JL, Tchkonia T. Cellular Senescence: A Translational Perspective. EBioMedicine. 2017;21:21–28. doi: 10.1016/j.ebiom.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emerling BM, Hurov JB, Poulogiannis G, Tsukazawa KS, Choo-Wing R, Wulf GM, Bell EL, Shim HS, Lamia KA, Rameh LE, Bellinger G, Sasaki AT, Asara JM, Yuan X, Bullock A, Denicola GM, Song J, Brown V, Signoretti S, Cantley LC. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell. 2013;155(4):844–57. doi: 10.1016/j.cell.2013.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kawano S, Shoji S, Ichinose S, Yamagata K, Tagami M, Hiraoka M. Characterization of Ca(2+) signaling pathways in human mesenchymal stem cells. Cell Calcium. 2002 Oct;32(4):165–74. doi: 10.1016/s0143416002001240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: ROS levels in transfected cells. 2 or 7 days after transfection, cells were assayed for ROS levels using Fluorometric Intracellular ROS kit (Sigma-Aldrich). There was no difference in ROS levels between the groups. Lipo = cells treated with lipofectamine (transfection reagent); siSCR = cells transfected with scrambled siRNA; siPLCδ4 = cells transfected with siRNA for PLCδ4. RFU = relative fluorescence unit. n=2.
Supplementary Figure 2: Cell morphology of transfected hASC. 2 days after transfection, cells were photographed, using the 10× objective lens on an Olympus IX70 microscope, using a QIClick camera, and Image Pro Plus 7.01 software. Lipofectamine = cells treated with lipofectamine (transfection reagent); siSCR = cells transfected with scrambled siRNA; siPLCδ4 = cells transfected with siRNA for PLCδ4. At least three independent experiments were performed. Scale bar: 200 µm.