Abstract
The objective of this study was to determine bacterial community profiles of the equine cecum in response to abrupt inclusion of varying levels of dietary starch. Seven cecally cannulated Quarter Horse geldings (497 to 580 kg) were used in a crossover design with two 28-d periods and a 28-d washout between each. Horses were randomly assigned to dietary treatments consisting of a commercial concentrate offered as fed at either 0.6 (low starch [LS]) or 1.2% BW (high starch [HS]) daily that was divided into 2 meals at 12-h intervals. Prior to the start of each period, horses were allowed ad libitum access to coastal bermudagrass (Cynodon dactylon) hay. Concentrate was fed on d 1 with no adaptation. Cecal fluid was collected on d 1 at h 0 and at 3, 6, 9, and 12 h relative to the initial concentrate meal on d 1. Additional samples were collected 6 h after feeding on d 2, 3, and 7 of each period. Cecal contents were used to determine pH and VFA concentrations and extract microbial DNA. The V4 through V6 region of 16S rRNA gene was amplified using PCR and sequenced on the Roche 454 FLX platform. Sequence analysis was performed with QIIME, and data were analyzed using the MIXED procedure of SAS. Cecal pH tended to decrease (P = 0.09) in horses fed HS in the first 12 h after the first concentrate meal and remained lower (P ≤ 0.05) the following 7 d. Total VFA were greater (P ≤ 0.05) in horses fed HS in the initial 12 h and 7 d after addition of concentrate. Species richness determined using the Chao1 index was unchanged (P > 0.20) over the initial 12 h and decreased (P = 0.01) over 7 d for both treatments. Community diversity determined using the Shannon index tended to decrease (P = 0.06) over the 7 d. Relative abundances of Paraprevotellaceae were greater (P ≤ 0.05) in HS in the first 12 h. Over 7 d, relative abundances of Paraprevotellaceae, Veillonellaceae, and Succinivibrionaceae were greater (P ≤ 0.05) in HS compared with LS. Abrupt and short-term exposure to dietary starch does alter cecal fermentation and microbial community structure in horses, but the impact on horse health is unknown.
Keywords: cecal pH, cecum, equine, microbiome, starch, volatile fatty acid
INTRODUCTION
Increased concentrate in equine rations affects cecal fermentation dynamics by altering the community structure of resident microflora and subsequent fermentation end products. Abrupt changes in the cecal environment can negatively affect digestive health and increase the incidence of gastrointestinal upset (Argenzio et al., 1974; Willard et al., 1977; de Fombelle et al., 2001). Performance horses are often subjected to abrupt nutritional changes in diet with little adaption to meet increased nutrient and energy demands. Typical concentrate diets fed to horses in light and moderate work contain higher levels of nonstructural carbohydrates and can range from 0.5 to 1.5% BW per day. Response to nutritional changes are adequately described in other species in vivo; however, the majority of equine microbial research to date has heavily relied on in vitro culture–based methods for quantifying viable counts of bacteria after collecting gastrointestinal contents from horses following euthanasia (Lin and Stahl, 1995; Daly et al., 2001).
Culture-independent methods have revealed a more efficient and inclusive means to identify microbiota by extracting their genetic material and characterizing the taxonomy based on the 16S rRNA gene. However, these methods have characterized only the equine microbiome in gastrointestinal contents of euthanized horses (Dougal et al., 2012; Moreau et al., 2014; Costa et al., 2015), in a laminitis or colic model (Respondek et al., 2008; Milinovich et al., 2010), or in fecal samples (Costa et al., 2012; Steelman et al., 2012; Weese et al., 2015). Few studies have used next-generation sequencing techniques to identify the cecal microbiota’s response to nutritional changes in living animals (Hansen et al., 2015). Therefore, the objective of this trial was to investigate the response of the microbial community structure in the equine cecum to diets containing varying levels of starch.
MATERIALS AND METHODS
All procedures and handling of horses were approved by the Texas A&M University Institutional Animal Care and Use Committee Permit (2011-24).
Experimental Design
Seven previously cecally cannulated Quarter Horse geldings (10 to 23 yr and 497 to 580 kg) were used in a crossover design with two 28-d treatment periods separated by a 28-d washout period between. Prior to the start of the study, horses were stratified by BW and randomly assigned to 1 of 2 dietary treatments by pair. Horses received either 0.6 (low starch [LS]; 0.9 g nonstructural carbohydrate/kg BW, as-fed basis) or 1.2% BW (high starch [HS]; 1.8 g nonstructural carbohydrate/kg BW, as-fed basis) of a commercial concentrate (Vitality Perform 14 Horse Feed; Cargill Animal Nutrition, Elk River, MN) divided into 2 meals and individually fed twice daily at 0630 and 1830 h (Table 1). Horses were assigned to HS (n = 4) or LS (n = 3) in period 1 and HS (n = 3) or LS (n = 4) in period 2. The remainder of the diet consisted of ad libitum access to coastal bermudagrass hay (Cynodon dactylon) round bales group-fed in a drylot pen. The same hay was fed for 28 d prior to the start of period 1 and again during the washout period. On d 0 of each period, the BW was measured and the amount of concentrate offered was determined for the period. Horses were allowed 1 h to consume concentrate, and refusals were recorded.
Table 1.
Nutrient components of concentrate and forage (DM basis) fed to mature Quarter Horse geldings
| Item | Concentrate1 | Forage2 |
|---|---|---|
| DM, % | 89.9 | 91.9 |
| CP, % | 14.0 | 9.6 |
| NDF, % | 14.3 | 70.1 |
| ADF, % | 5.9 | 36.2 |
| NSC,3 % | 30.0 | - |
| Ca, % | 0.9 | 0.4 |
| P, % | 0.6 | 0.2 |
Diets consisted of 0.06% BW (low starch) or 1.2% BW (high starch; as-fed basis) per day of a commercially pelleted concentrate (Vitality Perform 14 Horse Feed; Cargill Animal Nutrition, Elk River, MN).
Hay consisted of coastal bermudagrass (Cynodon dactylon) offered ad libitum.
NSC = nonstructural carbohydrate.
Sample Collection
After fed solely hay during the 28 d washout period, all horses began treatments on d 1 of each period with no prior adaptation to grain to determine the immediate (12 h) and short-term (7 d) response to dietary concentrate. Cecal samples were collected at 0 h, prior to the morning meal; at 3, 6, 9, and 12 h after the morning meal on d 1 to evaluate the immediate response; and at 6 h after the morning meal on d 2, 3, and 7 of each period to evaluate the short-term response. Cecal cannulas were opened, and contents were collected into insulated containers. The pH was immediately measured in the insulated containers using a handheld pH meter (Thermo Fischer Scientific, West Chester, PA). Approximately 30 mL of cecal contents was frozen at −20°C for later isolation of DNA, and 10 mL was strained and frozen at −20°C for later analysis of VFA. After freezing, samples were thawed and 8 mL of cecal fluid was combined with 2 mL of 25% m-phosphoric acid for VFA analysis. All samples were then analyzed at Kansas State University (Manhattan, KS) using the procedure described by Vanzant and Cochran (1994).
Deoxyribonucleic Acid Extraction
Deoxyribonucleic acid was extracted using the QIAamp DNA stool mini kit (Qiagen, Valencia, CA) protocol with previously described modifications (McCann et al., 2014a). Specifically, 3 mL of liquid or a similar volume of solid sample were mixed with 3 mL Buffer ASL (QIAamp DNA Stool Kit; Qiagen) and vortexed for 15 min. After centrifugation at 500 × g for 1 min at room temperature, 500 μL of supernatant was transferred to tubes with 0.15-mm garnet beads (Mo Bio Laboratories, Inc., Carlsbad, CA) and 1,000 μL phenol chloroform. Vortexing for 5 min completed bacterial lysis, and the suspension was incubated at 95°C for 5 min. Particulate matter was pelleted using centrifugation at 13,000 × g for 1 min at room temperature; 400 μL of the supernatant was added to 1,000 μL Buffer ASL and 1 InhibitEX tablet (QIAamp DNA Stool Kit; Qiagen) to incubate for 1 min at room temperature. Samples were centrifuged (16,000 × g) for 7 min at room temperature, and 535 μL supernatant was added to 25 μL Proteinase K followed by 535 μL Buffer AL (QIAamp DNA Stool Kit; Qiagen). The samples were subsequently incubated at 70°C for 20 min. After addition of 535 μL ethanol, the mixture was added to a DNA spin column, and QIAamp DNA stool mini kit recovery protocols were used to finish extraction. The concentration of extracted DNA was estimated using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, LLC, Wilmington, DE), and integrity was determined using electrophoresis on a 1% agarose gel stained with ethidium bromide. Ethanol precipitation was performed on all samples possessing a 260:280 nm ratio below 1.7. Extracted DNA was normalized to 20 ng/μL concentration for downstream amplification and pyrosequencing.
16S rRNA Amplification and Pyrosequencing
The V4 throughV6 segment of the 16s rRNA gene was amplified using the universal eubacterial primers 530F (5′-GTGCCAGCMGCNGCGG-3′) and 1100R (5′-GGGTTN CGNTCGTTG-3′) as previously described (Dowd et al., 2008). A single-step 30-cycle PCR using a HotStarTaq Plus Master Mix Kit (Qiagen) was used under the following conditions: 1) denaturing at 94°C for 3 min, 2) 28 cycles of 94°C for 30 s, 3) annealing at 53°C for 40 s, 4) extension at 72°C for 1 min, and 5) a final elongation step at 72°C for 5 min. Following PCR, all amplicon products from different samples were uniquely barcoded and mixed in equal concentrations and purified using Agencourt AMPure X beads (Agencourt Bioscience Corp., Beverly, MA). Pyrosequencing was performed with a Genome Sequencer FLX System with Titanium chemistry (Roche, USA, Branford, CT) at the MR DNA Molecular Research Lab (MR DNA [Molecular Research LP], Shallowater, TX).
Sequence Analysis
Prior to further analysis, barcodes, primers, short reads < 200 bp, and reads with homopolymer runs exceeding 6 bp were removed. A total of 890,204 sequences were denoised and chimera checked using a proprietary analysis pipeline (MR DNA [Molecular Research LP]) prior to further analysis yielding a total of 672,371 sequence reads. Reads were processed using QIIME (Caporaso et al., 2010b) and were quality trimmed to 325 bp to reduce sequencing errors. One sample was removed from analysis due to lack of sequences. Operational taxonomic units (OTU) were classified based on a 97% sequence identity threshold using the UCLUST software package (Edgar et al, 2010). A representative sequence was selected for each OTU based on abundance. Reads were aligned using PyNast (Caporaso et al., 2010a), and taxonomy was assigned using basic local alignment search tool (BLAST) against the 12_10 Greengenes database (http://greengenes.secondgenome.com/downloads/database/12_10; accessed 1 April 2013; McDonald et al., 2012). Operational taxonomic units were considered part of the core microbiome if they were present in 100% of the samples in each treatment.
Bacterial Diversity and Statistical Analysis
Sequencing depth per sample ranged from 1,615 to 28,783 reads, with a mean value of 6,057. Samples were rarefied to 4,700 reads for α-diversity and richness estimations using the Shannon, Simpson, Chao1, and observed OTU indices. The rarefaction level provided equal coverage for more than 90% of samples. Bray–Curtis dissimilarity (Beals, 1984) matrices were used in a principal coordinate analysis to generate 2-dimensional plots in PRIMER version 6 software (Clarke and Gorley, 2006). Permutational multivariate ANOVA was implemented to test differences in β-diversity for the 2 statistical models described below. Cecal pH, VFA, bacterial α-diversity, and relative abundance data were analyzed using the MIXED procedure of SAS 9.3 (SAS Inst. Inc., Cary, NC) with the repeated measures statement for hour or day. Terms in the model for immediate adaptation data included treatment, hour, treatment × hour, and period as fixed effects and horse as a random effect. The model for short-term adaptation data included treatment, day, treatment × day, and period as fixed effects and horse as a random effect. Treatment means were calculated using the LSMEANS statement. If necessary, data were transformed to ensure normality of residual using logit or Box–Cox transformation. Treatment means have been back-transformed to original units for consistency and interpretation. Significance was declared at P ≤ 0.05 and tendencies are discussed at 0.05 < P ≤ 0.10.
RESULTS
Immediate Effect on Cecal pH and VFA over 12 h
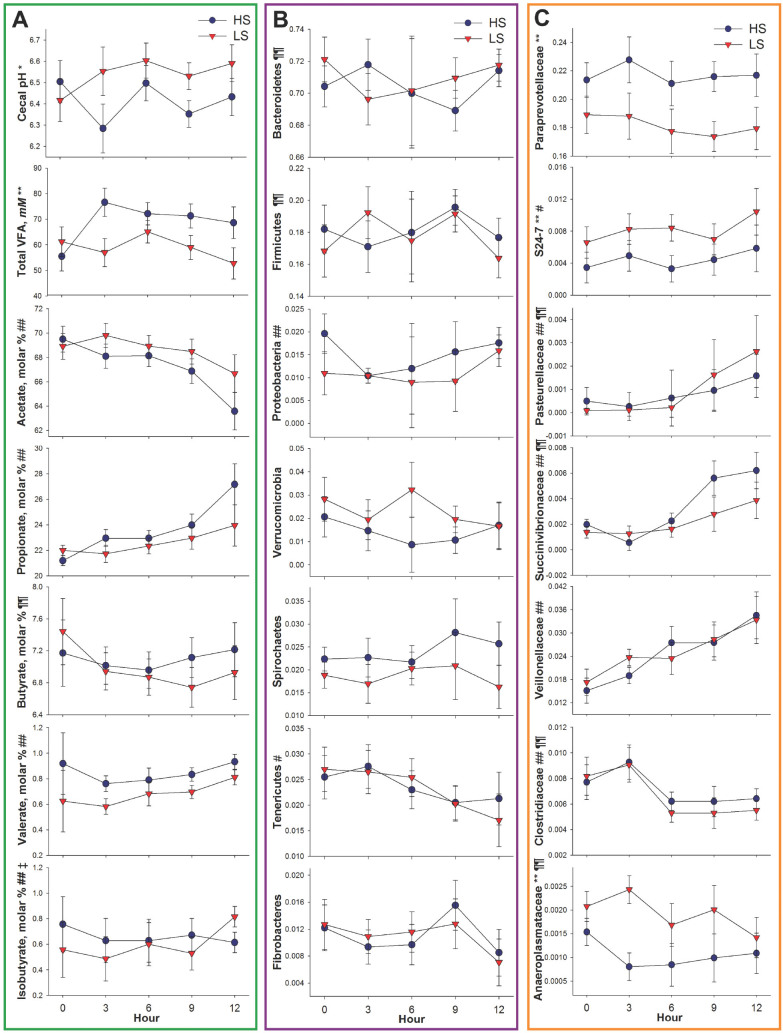
Cecal pH tended to decrease (P = 0.09; Fig. 1A) in horses fed HS for the first 12 h after the first concentrate meal when compared with horses fed LS; however, there was no effect of time or treatment × time interaction (P ≥ 0.29). During this first 12 h after abrupt concentrate exposure, the cecal pH of most horses did not reach levels indicative of subclinical acidosis. However, 2 horses on the HS diet in period 1 did have at least 1 time point with cecal pH < 6.0 during the initial 3 d after the diet change. Horses fed HS had greater (P = 0.02) concentrations of total VFA. Molar proportions of acetate were greater (P ≤ 0.05) in horses fed LS and decreased (P ≤ 0.05) over 12 h, whereas propionate increased (P ≤ 0.05) in the first 12 h after abrupt concentrate meal administration.
Figure 1.
Influence of short-term concentrate diet adaptation (low starch [LS] = 0.6% BW/d; high starch [HS] = 1.2% BW/d, as-fed basis) on equine cecum over 12 h. (A) Cecal fermentation is described by pH, total VFA concentration (mM), and molar proportion of VFA. (B) Microbiome response at the phyla level is described by the relative abundance of bacterial phyla present at greater than 1% of 16S rRNA sequences. (C) Microbiome response at the family level is described by relative abundance of bacterial families present at greater than 0.1% of 16S rRNA sequences. Symbols on the y-axis in Fig. 1A, 1B, and 1C denote the following effects treatment (**P < 0.05; *0.1 > P > 0.05), hour (##P < 0.05; #0.1 > P > 0.05), treatment × hour (‡0.1 > P > 0.05), and period (¶¶P < 0.05; ¶0.1 > P > 0.05). Error bars represent a pooled SEM.
Immediate Effect on Cecal Microbiome over 12 h
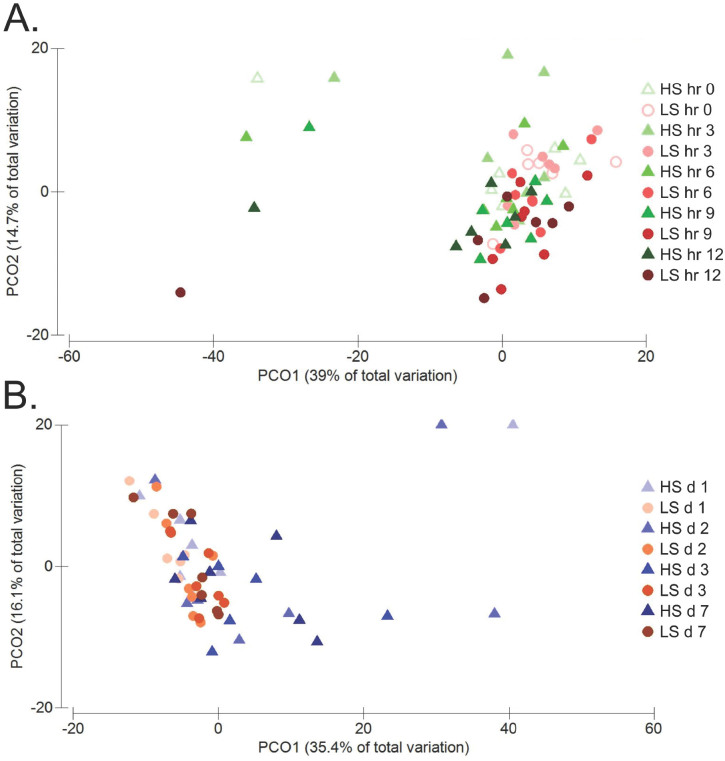
Although no treatment effects (P ≥ 0.10) were observed for Shannon, Simpson, and Chao1 indices in the first 12 h (Table 2), the α-diversity measured using the Simpson index was affected (P = 0.02) over the initial 12 h. Observed OTU tended to decrease (P = 0.10) after the abrupt concentrate feeding. All 4 α-diversity and richness indices were not influenced by treatment in the first 12 h. The Bray–Curtis dissimilarity index, used to measure β-diversity, or the dissimilarity between samples, indicated an effect of treatment (P = 0.002), hour (P = 0.001), period (P = 0.0002), and horse (P = 0.0001; Fig. 2A).
Table 2.
Alpha-diversity over first 12 h after an abrupt concentrate meal in the cecum of horses
| Hour | P-value1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Item | 0 | 3 | 6 | 9 | 12 | SEM | Trt | Hour | Trt ×hour |
| Shannon index | |||||||||
| LS2 | 7.703 | 7.708 | 7.672 | 7.650 | 7.307 | 0.135 | 0.99 | 0.14 | 0.17 |
| HS2 | 7.741 | 7.553 | 7.570 | 7.601 | 7.564 | ||||
| Simpson index3 | |||||||||
| LS | 0.987 | 0.986 | 0.985 | 0.987 | 0.981 | 0.002 | 0.92 | 0.02 | 0.38 |
| HS | 0.988 | 0.982 | 0.986 | 0.987 | 0.986 | ||||
| Chaol index | |||||||||
| LS | 1,133 | 1,122 | 1,153 | 1,112 | 987 | 39 | 0.84 | 0.28 | 0.54 |
| HS | 1,119 | 1,102 | 1,102 | 1,051 | 1,077 | ||||
| Observed OTU4 index | |||||||||
| LS | 701 | 702 | 695 | 665 | 614 | 26 | 0.92 | 0.10 | 0.65 |
| HS | 712 | 692 | 676 | 661 | 655 | ||||
Trt = effect of dietary treatment (high starch or low starch). Hour represents the effect of time at 0, 3, 6, 9, and 12 h. Trt × hour represents the interaction.
LS = low starch; HS = high starch. Diets consisted of a commercial concentrate (Vitality Perform 14 Horse Feed; Cargill Animal Nutrition, Elk River, MN) fed at 0.6 (LS) or 1.2% BW/d (HS; as-fed basis) and horses had ad libitium access to coastal bermudagrass hay (Cynodon dactylon).
Period effect P ˂ 0.05.
OTU = operational taxonomic units.
Figure 2.
Influence of concentrate diet adaptation (low starch [LS] = 0.6% BW/d; high starch [HS] = 1.2% BW/d, as-fed basis) on equine cecum microbiome. (A) Effects on the bacterial community during the initial 12 h using principal coordinate analysis (PCoA) of β-diversity with Bray–Curtis dissimilarity. Permutational multivariate ANOVA analysis revealed an effect of treatment (P = 0.002), hour (P = 0.001), period (P = 0.0002), and horse (P = 0.0001) PCO1 =39% of total variation; PCO2 = 14.7% of total variation. (B) Effects on the bacterial community during adaptation over 7 d using PCoA of β-diversity with Bray–Curtis dissimilarity. Permutational multivariate ANOVA analysis revealed an effect of treatment (P = 0.0001), period (P = 0.0002), and horse (P = 0.0001) with a trend (P = 0.06) for a day effect
Taxonomic classification of cecal fluid identified 18 phyla, 35 classes, 70 orders, 120 families, and 208 genera. A total of 5,531 OTU were observed with a minimum of 309 OTU/sample and a maximum of 1,169 OTU/sample. Of the 18 observed phyla, 7 phyla were present at greater than 1% relative abundance including Bacteroidetes (69.82%), Firmicutes (19.58%), Proteobacteria (2.56%), Verrucomicrobia (2.27%), Tenericutes (1.14%), Spirochaetes (1.12%), and Fibrobacteres (1.18%). Phyla accounting for less than 1% of sequences included Cyanobacteria, Fusobacteria, Synergistetes, TM7, WPS2, Planctomycetes, Elusimicrobia, Actinobcteria, Chloroflexi, Deferribacteres, and GN02.
The core microbiome was determined by identifying OTU that were present in all samples including the control sample, which was taken before morning meal administration on 0 h, when horses consumed solely forage for 28 d before, and LS and HS samples. The core community across all samples consisted of 10 OTU belonging to the Bacteroidetes and Firmicutes phyla (Table 3).
Table 3.
Core microbiome in 0-h baseline sample and low starch (LS) and high starch (HS) samples over 7 d in the cecum of horses1
| Taxonomic classification | ||||||
|---|---|---|---|---|---|---|
| No.OTU2 | Kingdom | Phylum | Class | Order | Family | Genus |
| 4,960 | Bacteria | Bacteroidetes | Bacteroidia | Bacteroidales | ||
| 20 | Bacteria | Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae | Prevotella |
| 3,261 | Bacteria | Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae | Prevotella |
| 2,631 | Bacteria | Bacteroidetes | Bacteroidia | Bacteroidales | Paraprevotellaceae | |
| 79 | Bacteria | Bacteroidetes | Bacteroidia | Bacteroidales | Paraprevotellaceae | YRC22 |
| 7,076 | Bacteria | Bacteroidetes | Bacteroidia | Bacteroidales | Paraprevotellaceae | |
| 4,186 | Bacteria | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | |
| 1,693 | Bacteria | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Roseburia |
| 5,899 | Bacteria | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Roseburia |
| 237 | Bacteria | Firmicutes | Clostridia | Clostridiales | Veillonellaceae | |
Ad libitum access to coastal bermudagrass hay (Cynodon dactylon).
OTU = operational taxonomic units.
The most abundant bacterial phyla, Bacteroidetes and Firmicutes, were not affected during the first 12 h after the initial concentrate meal (Fig. 1B). However, relative abundances of Paraprevotellaceae were greater (P ≤ 0.05) in horses fed HS (Fig. 1C) and Ruminococcaceae decreased (P ≤ 0.05) in the first 12 h (Supplemental Table S1). Additionally, genus YRC22 was greater (P = 0.01) in horses fed HS whereas genus Prevotella tended to increase (P = 0.09) over time (Supplemental Table S2; see the online version of the article at http://journalofanimalscience.org). Relative abundances of S24-7 were greater (P ≤ 0.05) in horses fed LS and tended to gradually increase (P ≤ 0.10) over the first 12 h, regardless of diet (Fig. 1C). A period effect was observed in the immediate response to dietary starch as Bacteroidetes increased (P < 0.01) in period 2 compared with the first period.
Greater changes were observed in Firmicutes taxa immediately after concentrate inclusion (Table 1C; Supplemental Tables S1 and S2 [see the online version of the article at http://journalofanimalscience.org]). Veillonellaceae and its genus Anaerovibrio increased (P ≤ 0.05) over time in the first 12 h. Relative abundances of Lactobacillaceae steadily increased (P = 0.04). Clostridiaceae and genus Clostridium decreased (P = 0.002 and P = 0.04, respectively) over time regardless of treatment in the first 12 h (Fig. 1C; Supplemental Table S2 [see the online version of the article at http://journalofanimalscience.org]). Ruminococcaceae decreased (P ≤ 0.05) whereas genus Oscillospira increased (P = 0.04) over 12 h (Supplemental Tables S1 and S2; see the online version of the article at http://journalofanimalscience.org). Unassigned reads within orders Coriobacteriales and Clostridiales steadily decreased (P ≤ 0.01 and P = 0.02, respectively) in the first 12 h, whereas Erysipelotrichaceae and Coprobacillaceae tended to decrease (P = 0.07 and P = 0.07, respectively; Supplemental Table S1 [see the online version of the article at http://journalofanimalscience.org]). Streptococcaceae tended to be greater (P ≤ 0.10), with HS horses having higher relative abundances than LS horses (Supplemental Table S1; see the online version of the article at http://journalofanimalscience.org). Firmicutes decreased (P < 0.01) in period 2 compared with the first period.
Relative abundances of Proteobacteria, Succinivibrionaceae, and Pasteurellaceae increased (P ≤ 0.05) by 12 h regardless of treatment. Tenericutes tended to decrease (P = 0.08) over 12 h, and taxon Anaeroplasmataceae were greater (P ≤ 0.05) in horses fed LS. Relative abundances of an unknown family belonging to the order RF39 and to the Tenericutes phylum tended to decrease (P ≤ 0.10) immediately after concentrate inclusion (Supplemental Table S1; see the online version of the article at http://journalofanimalscience.org).
Short-Term Effect on Cecal pH and VFA
Horses fed HS had lower (P ≤ 0.05; Fig. 3A) cecal pH at 6 h after the morning meal compared with horses fed LS over the 7 d after transitioning to concentrate feed. Regardless of treatment, cecal pH decreased (P ≤ 0.01) on d 2 and failed to return to baseline by d 7. Although average pH measurements did not fall below 6 when measured 6 h after feeding, 2 individual HS horses did display symptoms of lactic acidosis, with body temperatures exceeding 38.8°C on d 2 and 3, with pH values at or below 6. Although a treatment × day interaction was not observed (P = 0.66), on d 7, pH in horses fed LS was similar to those recorded on d 1 (P = 0.25) but horses fed HS had decreased (P = 0.01) cecal pH values on d 7 compared with d 1. Similar to the initial 12 h, greater (P < 0.05) concentrations of total VFA were observed in horses fed HS. During the short-term effect of concentrate feeding over 7 d, propionate was greater (P ≤ 0.05) in horses fed HS. Propionate increased (P ≤ 0.01) on d 2, which coincided with the lowest (P ≤ 0.05) molar proportion of acetate and isovalerate. Prior to concentrate feeding at 0 h, calculated means of the acetate:propionate decreased from 3.54 to either 3.07 (LS) or 2.60 (HS) in the first 12 h.
Figure 3.
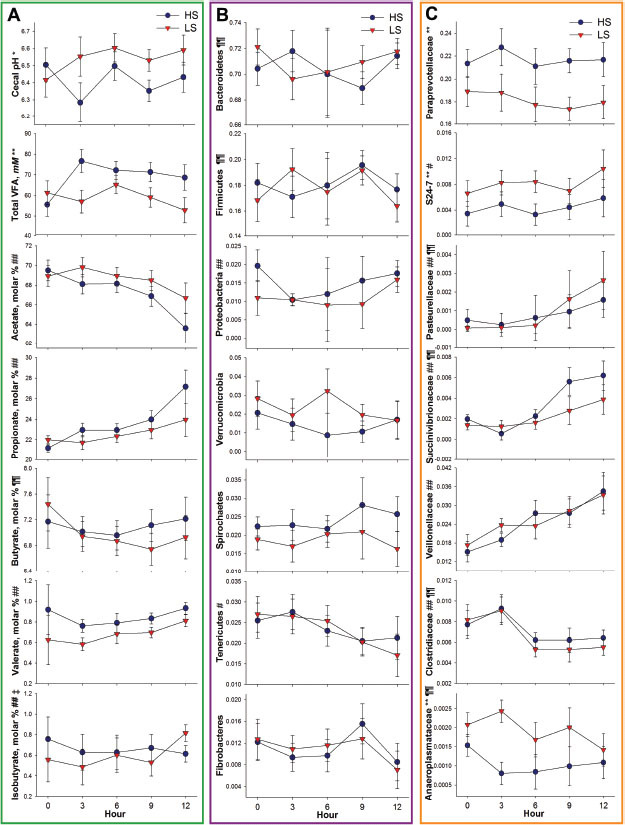
Influence of short-term concentrate diet adaptation (low starch [LS] = 0.6% BW/d; high starch [HS] = 1.2% BW/d, as-fed basis) on equine cecum over 7 d. (A) Cecal fermentation is described by pH, total VFA concentration (mM), and molar proportion of VFA. (B) Microbiome response at the phyla level is described by the relative abundance of bacterial phyla present at greater than 1% of 16S rRNA sequences. (C) Microbiome response at the family level is described by relative abundance of bacterial families present at greater than 0.1% of 16S rRNA sequences. Symbols on the y-axis in Fig. 2A, 2B, and 2C denote the following effects treatment (**P < 0.05; *0.1 > P > 0.05), day (##P < 0.05; #0.1 > P > 0.05), treatment × day (‡0.1 > P > 0.05), and period (¶¶P < 0.05; ¶0.1 > P > 0.05). Error bars represent a pooled SEM.
Short-Term Effect on Cecal Microbiome over 7 d
Species richness estimated with the Chao1 and observed OTU indices decreased (P = 0.01) over the 7 d after concentrate feeding (Table 4). The Shannon index revealed a tendency for a day and a treatment × day interaction (P = 0.07 and P = 0.09, respectively) caused by a greater decrease in α-diversity in horses fed HS. Bray–Curtis dissimilarity revealed an effect (P < 0.01) of treatment (P = 0.0001), period (P = 0.0002), and horse (P = 0.0001) and a trend (P = 0.06) for a day effect (Fig. 2B).
Table 4.
Alpha-diversity over 7 d after an abrupt concentrate meal in the cecum of horses
| Day | P-value1 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Item | 1 | 2 | 3 | 7 | SEM | Trt | Day | Trt × day | |||||||
| Shannon index | |||||||||||||||
| LS2 | 7.677 | 7.662 | 7.774 | 7.676 | 0.208 | 0.15 | 0.07 | 0.09 | |||||||
| HS2 | 7.552 | 7.359 | 6.884 | 7.145 | |||||||||||
| Simpson index | |||||||||||||||
| LS | 0.985 | 0.987 | 0.991 | 0.988 | 0.008 | 0.28 | 0.69 | 0.11 | |||||||
| HS | 0.986 | 0.983 | 0.946 | 0.981 | |||||||||||
| Chao1 index | |||||||||||||||
| LS | 1,147 | 1,058 | 1,070 | 1,034 | 48 | 0.14 | 0.01 | 0.62 | |||||||
| HS | 1,102 | 935 | 917 | 924 | |||||||||||
| Observed OTU3 index | |||||||||||||||
| LS | 694 | 663 | 685 | 673 | 33 | 0.14 | 0.01 | 0.10 | |||||||
| HS | 673 | 609 | 561 | 574 | |||||||||||
Trt = effect of dietary treatment (high starch or low starch). Day represents the effect of time at 1, 2, 3, and 7 d. Trt × day represents the interaction.
LS = low starch; HS = high starch. Diets consisted of a commercial concentrate (Vitality Perform 14 Horse Feed; Cargill Animal Nutrition, Elk River, MN) fed at 0.6 (LS) or 1.2% BW/d (HS; as-fed basis) and horses had ad lbitum access to coastal bermudagrass hay (Cynodon dactylon).
OTU = operational taxonomic units.
More than 88% of reads from samples collected over 7 d were assigned to Bacteroidetes and Firmicutes (Fig. 1B). Bacteroidetes taxa, including family Paraprevotellaceae and genus YRC22, were more abundant (P < 0.05) in horses fed HS. Relative abundances of Paludibacter were greater (P = 0.01) in horses fed LS, whereas S24-7 tended to be greater (P = 0.06) in horses fed LS over 7 d (Table 2C; Supplemental Tables S3 and S4 [see the online version of the article at http://journalofanimalscience.org]). Period effects were also observed for Bacteroidetes (P ≤ 0.01), with increased relative abundances observed in period 2 compared with period 1.
Relative abundances of Firmicutes steadily increased (P ≤ 0.01) over 7 d (Fig. 3B). Firmicutes taxa Veillonellaceae and Phascolarctobacterium were greater (P ≤ 0.05) in horses fed HS and increased (P ≤ 0.05) regardless of treatment over 7 d (Fig. 2C; Supplemental Table S4 [see the online version of the article at http://journalofanimalscience.org]). Streptococcaceae tended to be higher (P = 0.09) in horses fed HS (Supplemental Table S3; see the online version of the article at http://journalofanimalscience.org). An unknown genus belonging to Lachnospiraceae tended to be higher (P = 0.08) in horses fed LS. Firmicutes (P ≤ 0.01) was also influenced by period, with decreased relative abundances observed in period 2 compared with period 1.
Proteobacteria was greater (P ≤ 0.01) in horses fed HS compared with horses fed LS and tended to increase (P = 0.09) over 7 d. Relative abundances of Succinivibrionaceae and Pasteurellaceae increased (P < 0.01) during the short-term period. Pasteurellaceae and an unknown genus belonging to this family were greater (P ≤ 0.05) in horses fed HS. The family Desulfovibrionaceae and the genus Desulfovibrio, which belong to the Proteobacteria phylum, tended to decrease (P < 0.08) over 7 d (Supplemental Tables S3 and S4; see the online version of the article at http://journalofanimalscience.org).
Other phyla were influenced during the short-term adaption period over 7 d. Tenericutes were greater (P = 0.02) in horses fed LS horses compared with horses fed HS and decreased (P < 0.01) regardless of treatment. Specifically, genus RF39 decreased (P < 0.01) over time and tended to be more prevalent (P = 0.06) in horses fed LS. Spirochaetes tended to be influenced by treatment (P = 0.07), with horses fed LS having higher relative abundances than horses fed HS. Spirochaetaceae tended to be higher (P = 0.09) whereas Spirochaetes tended to decrease (P < 0.10) over 7 d. Verrucomicrobia tended to be more abundant (P = 0.06) in horses fed LS compared with horses fed HS.
The Fibrobacteres phylum decreased (P ≤ 0.01) over 7 d regardless of treatment. Similarly, Fibrobacteraceae and Fibrobacter decreased (P ≤ 0.01) and tended to decrease (P = 0.08), respectively, over 7 d regardless of treatment. Fibrobacter tended to be greater (P = 0.06) in horses fed LS compared with horses fed HS. Period effects were also observed for Fibrobacteres (P = 0.04), with increased relative abundances observed in period 2 compared with period 1.
DISCUSSION
The cecum is an integral part of the equine digestive system responsible for microbial fermentation of fiber and undigested nutrients from the small intestine. Modern feeding regimes involve feeding increased concentrate meals, which allow horses to meet elevated energy and nutrient requirements of work and reproduction. However, feeding regimens used in the performance horse industry often do not provide suficient time for adaptation of the microbial community to the change in substrate. In this study, both LS and HS diets affected cecal microflora and the resulting physiology measured by changes in relative abundance of bacterial taxa, VFA concentration, and pH levels both immediately and after 7 d of adaptation to the diet.
Cecal pH and VFA
Cecal pH decreases as greater amounts of fermentable substrate are incorporated into the diet (Julliand et al., 2001) and changes can be detected 4 to 6 h after the meal when digesta enters the cecum from the ileum (Willard et al., 1977). This was also observed in the present study, with the lowest pH values reported at 6 h after the meal on d 2, 3, and 7 in horses fed HS. Julliand et al. (2001) reported decreases in cecal pH values from 6.74 to 6.41 to 6.26 as horses were transitioned from an all-forage diet to a diet with 30 and 50% barley, respectively. Similarly, Willard et al. (1977) reported changes in cecal pH from 7.14 to 6.12 6 h after concentrate was administered to an all-forage diet. In the present study, cecal pH values dropped 4 to 6 h after feeding and were lowest in horses fed the HS diet, which contained higher amounts of nonstructural carbohydrates.
Volatile fatty acids are metabolic end products from bacterial fermentation of substrates present and have been studied to determine large-scale changes in fermentation; however, specific VFA production of many taxa identified in this study has not been evaluated. Volatile fatty acids have been quantified in the horse consuming an all-forage diet (Hintz et al., 1971) and when varying levels of concentrate were added to the ration (Wilson, 2009). Molar percentages in cecal fluid from forage-only diets can range from approximately 74 to 77% acetate, 15.7 to 17% propionate, 5.3 to 6% butyrate, and 2 to 4% isobutyrate, valerate, and isovalerate, depending on the type of forage offered (Hintz et al., 1971; Coverdale et al., 2004). Molar percentages in samples taken at 0 h in the present study were approximately 69% acetate, 22% propionate, 7% butyrate, and 2% isobutyrate, valerate, and isovalerate.
Feeding a highly fermentable substrate alters VFA proportions in the cecum of cannulated horses (Coverdale et al., 2004). Although soy hulls were fed instead of a commercial concentrate, Coverdale et al. (2004) reported that cecal molar percentages of propionate linearly increased (15.7, 18.0, 16.6, and 21.9 mol/100 mol total VFA) as 0, 25, 50, and 75% soybean hulls, respectively, were incorporated into the diets. Molar percentages of propionate in the present study at 6 h after the meal are similar to those reported in the 75% soybean hull diet (Coverdale et al., 2004). Proportions of butyrate decreased as more fermentable substrate was added into the diet, as reported by Coverdale et al. (2004) and in the present study at 7 d. Coverdale et al. (2004) reported that the acetate:propionate ratio decreased, with means of 4.9, 4.2, 4.9, and 3.3, when soy hulls were added to the diet. A similar decrease was also seen in the present study when horses were fed only hay prior to concentrate feeding (3.54), LS (3.07), and HS (2.60) in the first 12 h. According to Leek (1993), cellulolytic bacteria survivability and acetate production decline below a pH of 6.2, and 2 horses in the current study fell below this threshold.
Levels of D/L-lactate were not measured in the current study. A previous study conducted with cannulated ponies determined concentrations of lactate-utilizing bacteria were not influenced in the cecum or colon at 5 and 29 h following an abrupt change in diet from hay only to 50% hay + 50% barley (de Fombelle et al., 2001). When horses were adapted over 14 d from the 70:30 to the 50:50 diet), the concentration of lactate-utilizing bacteria, lactobacilli, and streptococci increased in the colon, although it did not significantly interfere with these populations in the cecum. As lactate-utilizing bacteria diminish, the concentrations of lactate will tend to accumulate in the colon rather than in the cecum.
Bacterial Diversity and Richness
Although the core microbiome may provide insights for the development of nutritional and therapeutic strategies for disease prevention, criteria used to identify taxa in the core community structure have not been thoroughly defined. The current standard requires every OTU to be present in every sample across animals but does not account for the abundance of those particular taxa. Only 1 study has evaluated and characterized the core microbial community in different segments of the gastrointestinal tract when horses were fed a forage-only diet (Dougal et al., 2012). Dougal et al. (2012) reported that 31 out of 2,263 (1.4%) total OTU make up the core microbiome in the ceca of 5 horses and 5 ponies. Results suggest that a horse’s hindgut core microbiome comprises many low abundant OTU, which could explain the lack of stability of the core community and subsequent onset of digestive disturbances (Dougal et al., 2012). Our study is the first to characterize the cecal core microbial community across different diets in living animals. The core community across all samples consisted of 10 OTU (0.18%), belonging to either the Bacteroidetes or Firmicutes phylum, out of 5,531 total OTU identified from the trial. Of these phyla, the greatest number of OTU belonged to either the Prevotellaceae or Lachnospiraceae families. Several studies investigating the rumen microbiome have identified Prevotellaceae and its genera as dominant and genetically diverse members in the microbial community (Jami and Mizrahi, 2012; McCann et al., 2014b). However, the core microbiome in the cow rumen is much larger and phylogenetically related, although a large number of OTU differ between samples (Jami and Mizrahi, 2012). It is important to note that one of the predominant cellulolytic genera, Fibrobacter, belonging to the Fibrobacteres phylum, was not represented in the cecal core microbiome in this trial, likely due to sequencing depth. Metagenomic analysis of the rumen microbiome has suggested that the abundance of these taxa varies considerably across cows and diets and they may not be represented due to low abundance below the threshold for detection using pyrosequencing techniques (Jami and Mizrahi, 2012).
Ecological estimators (e.g., α-diversity and richness) are commonly used in microbiome studies to describe changes in community composition (Lehman et al., 2000; McCann et al., 2014b). Ruminants fed a high-concentrate diet had lower diversity and richness measures compared with those animals fed solely hay (Fernando et al., 2010). Higher Chao1 and abundance-based coverage estimator (ACE) estimates of richness were reported in animals consuming solely forage-based diets compared with animals consuming high-grain diets (Fernando et al., 2010). Pitta et al. (2010) also observed greater diversity and richness in the rumen of steers fed coastal bermudagrass compared with steers fed a wheat diet.
Several studies have investigated richness and diversity of microbial communities in equine fecal samples with low animal numbers (Shepherd et al., 2012; Steelman et al., 2012) or in the gastrointestinal tract of horses following euthanasia (Dougal et al., 2012; Costa et al., 2015). The fecal microbiome has been thoroughly categorized and has been used as an option to model the cecal microbial community profile (Steelman et al., 2012). However, microbial communities can differ throughout the gastrointestinal tract of animals (Malmuthuge et al., 2014; Costa et al., 2015). In 3-wk-old preweaned calves, mucosal and digesta microbial communities differed within each particular segment of the gastrointestinal tract and also differed among each distinct region of the gastrointestinal tract. The distinct differences between content and epithelial-specific bacterial genera further suggest that fecal samples do not adequately represent the intricacy of the microbiome (Malmuthuge et al., 2014).
The diversity estimates of the equine microbiome in response to high nutrient inclusion have been described in a live animal model in only 1 study (Hansen et al., 2015). Estimates of diversity and temporal stability of cecal microbiota were lower in horses fed a high nutrient available diet (Hansen et al., 2015). In the present study, lower values measured using the Simpson index were reported, indicating lower bacterial diversity of the cecal microbiome as starch was added to the diet in the first 12 h. Similarly, Pitta et al. (2010) reported higher α-diversity and richness in ruminants fed hay compared with ruminants fed wheat, indicating a that greater species richness may be required to break down cellulose in forage diets. Although no treatment effects were observed between horses fed HS and horses fed LS in the present study, diversity and richness decreased in the first 12 h after concentrate feeding and over the short-term 7-d adaption period, which is consistent with studies performed in ruminants and cannulated horses (Pitta et al., 2010; Fernando et al., 2010; Hansen et al., 2015). The increased retention time of digesta in horses fed solely forage compared with horses fed concentrate may facilitate the growth of a more complex and rich bacterial community.
Cecal Microbiome
Taxonomic classification of equine cecal bacteria in living animals fed controlled diets has been evaluated in few studies (Hansen et al., 2015). Bacteroidetes has been reported as the dominant phylum in the rumen of cattle (Jami and Mizrahi, 2012; McCann et al., 2014b), whereas other trials have reported Firmicutes as the dominant phylum in the rumen of dairy cattle (Mao et al., 2013) or in the cecum of horses (Costa et al., 2015). Bacteroidetes (69.82%) and Firmicutes (19.58%) were the dominant phyla in the cecum, and this is consistent with results from Hansen et al. (2015) and Dougal et al. (2012), who reported Bacteroidetes as the dominant phylum. Although Costa et al. (2015) observed more Firmicutes in the cecum of horses, comparisons of overall taxa abundance across microbiome experiments are difficult due to biases existing in DNA extractions and 16S PCR primers. Similarities exist between the rumen microbial communities in 3-wk-old preweaned calves and the equine cecum in the present study. Malmuthuge et al. (2014) compared luminal contents and mucosal tissue-associated microbial communities along different segments in the gastrointestinal tract. The majority of digesta-associated Bacteroidetes (54.8%) in calves were Bacteroides (15.8%), Prevotella (15.1%), and Paludibacter (4.1%); however, equine cecal communities in the first 12 h comprised Prevotella (14.2%), Paludibacter (2.41%), and Bacteroides (0.13%). The large-intestinal mucosa-associated bacterial communities in these calves primarily comprised Bacteroidetes (cecal tissue; 56.8%) and Firmicutes (cecal tissue; 21.1%). However, the rumen tissue-associated bacterial community in beef steers fed a concentrate diet and in beef heifers fed hay- or grain-based diets consisted of more Firmicutes than of Bacteroidetes (Chen et al., 2011; Li et al., 2012; Jami et al., 2013).
Despite no observed effect in the relative abundances of Bacteroidetes or Firmicutes in the first 12 h, multiple taxa were affected by diet. Relative abundances of the genus YRC22, which belongs to the Bacteroidetes phylum, were greater in horses fed HS in both the immediate and short-term period, whereas relative abundances of Paludibacter and S24-7 were greater in horses fed LS in the short-term period, indicating that members of this phylum do play a role in carbohydrate fermentation. Increases in relative abundances of Prevotella have also been reported in the rumen of cows transitioned to a diet containing 50% dried distiller’s grains (Callaway et al., 2010) and have been shown to increase propionate (Strobel, 1992). In the present study, Prevotella only tended to increase after immediate incorporation of grain and no differences were observed in the short-term adaptation.
Cereal grains are introduced into calf diets during weaning and influence the ability of the developing rumen to adapt to these new nutritional changes. Meale et al. (2016) documented changes in the microbial community structure in pre- and postweaned dairy calves. The 3 most abundant phyla in the preweaned calf rumen were Bacteroidetes (66.1%), Firmicutes (18.6%) and Proteobacteria (10.5%) and changed to Bacteroidetes (42.2%), Firmicutes (34.4%), and Proteobacteria (20.3%) after weaning, indicating the ability of Firmicutes and Proteobacteria to ferment diets with higher nutrient availability (Meale et al., 2016). Higher abundances of Firmicutes were found to be directly proportionate to obesity levels in humans and rats and are positively correlated to caloric intake (Bervoets et al., 2013; Hartstra et al., 2015). A long-term trial investigating the role of members of Firmicutes in the gut microbiome of horses introduced to concentrate rations may uncover their role in the development of equine metabolic and digestive disorders.
Relative abundances of the genera Anaerovibrio and Oscillospira from the phyla Firmicutes increased immediately after grain inclusion. Anaerovibrio, a member of the Veillonellaceae family, produces propionate, acetate, and succinate as end products, whereas other species are characteristically lipolytic (Bergey et al., 1974). Decreases in cecal pH can be attributed to increases in certain bacterial taxa with an affinity to metabolize nonstructural carbohydrates (Al Jassim et al.,2005). Members of the Veillonellaceae family, which belongs to the Firmicutes phylum, ferment starch to produce lactate or ferment lactate to product propionate and acetate (Biddle et al., 2013). These observations support the need for more sensitive and accurate detection methods for analysis of microbial changes in genera across treatments and over time. Greater changes were observed in the Firmicutes phylum and the entire bacterial community over 7 d compared with the initial 12 h. The changes observed at the phylum level for Firmicutes were primarily driven by increases in Streptococcus, Anaerostipes, and Phascolarctobacterium. However, Ruminococcaceae, Clostridiaceae, Coriobacteriaceae, and Clostridiales decreased over time in horses fed LS and HS diets, indicating that these families’ fermentation capabilities were negatively affected by the addition of concentrate into equine diets. These findings differ from those of Fernando et al. (2010), who detected significantly higher numbers of organisms belonging to the families Clostridiaceae in ruminants fed a high-concentrate diet.
Over the first 12 h after abrupt inclusion of the first concentrate meal, 2 phyla were significantly affected by grain feeding. Tenericutes tended to decrease in the first 12 h, which was correlated with the decreased of the genus Anaeroplasma. These findings are similar to those found in Hansen et al. (2015), who reported greater Tenericutes in horses fed the hay-only diet. Relative abundances of Proteobacteria increased over time immediately and over the short-term adaptation period and were greatest in horses fed HS, indicating Proteobacteria’s role in fermenting greater levels of protein and starch entering the equine cecum. Pitta et al. (2016) observed an increase in Proteobacteria in the rumen of steers affected with frothy bloat, whereas Petri et al. (2013) observed increases in Proteobacteria in the first 12 h after the acidotic challenge. Members of this phylum represent a diverse group of commensal and pathogenic bacteria (Naushad et al., 2015).
Fibrobacteres’ relative abundance was negatively affected as more starch was incorporated into the diet. These changes were more apparent over the 7-d adaption period when the relative abundances decreased by approximately 50%. Fibrobacteres is a fiber-fermenting inhabitant of the equine cecum. Shifts in the microbiota toward starch-fermenting species can lower cecal pH and has detrimental effects on cellulolytic bacteria, resulting in decreased fiber fermentation rates (Milinovich et al., 2010). The decrease in Fibrobacteres abundances was expected when dietary starch was incorporated into the diet. Verrucomicrobia relative abundances were greater in horses fed LS compared with horses fed HS over the 7-d period, and Verrucomicrobia has been identified as an inhabitant of the equine hindgut (Costa et al., 2015; Hansen, 2015); however, functional characteristics of this organism still need to be determined. A similar trend was observed in Spirochaetes, which thrived better under LS conditions. Spirochaetes ferment polymers, such as xylan, pectin, and arabinogalactan, in plant materials (Paster and Canale-Parola, 1982).
Many taxa at the phylum, family, and genus level were affected by period within the first 12 h and over the next 7 d of continued concentrate feeding. This data set was part of a large-scale study in which samples were taken until d 28, with a 28-d washout between both periods, indicating that this washout period may be insufficient. Other factors that may contribute to period effects are variations between hay, as round bales were fed over the 4-mo duration of this study. Increasing temperatures in summer months may also have contributed to period effects observed in the present study. A more thorough evaluation of the duration of a washout period needs to be investigated, as some microbial effects may not be reversible. Although dietary treatments were fed for a longer period of time, 28 d vs. 7 d in the current study, similar trends were observed. A complete adaptation period needs to be thoroughly investigated because acetate, propionate, and butyrate were influenced by period, indicating an insufficient washout period between treatment periods.
Conclusion
This study reports the cecal fermentation and bacterial responses to meal feeding of performance horses. Results from this study can be used to further investigate the role of specific microbes that were affected by the addition of dietary starch and their contribution to the equine microbiome in response to immediate and short-term adaptation to concentrate diets. Grain feeding modifies the cecal environment by decreasing pH and altering VFA concentrations and the bacterial community. Alpha-diversity analysis revealed a less diverse microflora with the addition of dietary starch. This was further supported by core microbiome analysis, which revealed only 10 shared OTU among horses, representing 0.18% of the total observed microbial community. Significant changes were reported in the phylum, family, and genus level community structure, as many taxa were affected by treatment and time in the first 12 h and over the next 7 d. Although our diets were fed below the threshold needed to induce starch overload or subclinical acidosis, diets in the present study with less than 1.8 g nonstructural carbohydrate/kg BW per meal do modify cecal environment and fermentation.
Supplementary Material
Footnotes
Funding for this project was provided by Morris Animal Foundation Grant number D13EQ-040 to Josie Coverdale and Jan Janecka at Texas A&M University.
The authors would like to acknowledge Josie Coverdale for her many contributions to the animal industry, discoveries in equine research, and her commitment to student development. We would also like to thank Josh McCann for his expertise in bioinformatics and microbiome data interpretation.
LITERATURE CITED
- Al Jassim R. A. M., Scott P. T., Trebbin A. L., D. Trott, and Pollitt C. C.. 2005. The genetic diversity of lactic acid producing bacteria in the equine gastrointestinal tract. FEMS Microbiol. Lett. 248: 75–81. doi:10.1016/j.femsle.2005.05.023 [DOI] [PubMed] [Google Scholar]
- Argenzio. R, Southworth M., and Stevens C. E.. 1974. Sites of organic acid production and absorption in the equine gastrointestinal tract. Am. J. Physiol. 226: 1043–1050. [DOI] [PubMed] [Google Scholar]
- Bergey D., Buchanan R. E., and Gibbons N. E.. 1974. Bergey’s manual of determinative bacteriology. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- Bervoets L., K. Van Hoorenbeeck, Kortleven I., C. Van Noten, Hens N., C. Vael, Goossens H., Desager K. N., and V. Vankerckhoven. 2013. Differences in gut microbiota composition between obese and lean children: A cross-sectional study. Gut Pathog. 5:10. doi:10.1186/1757-4749-5-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beals E. W. 1984. Bray-Curtis ordination: An effective strategy for analysis of multivariate ecological data. Adv. Ecol. Res. 14: 1–55. doi:10.1016/S0065-2504(08)60168-3 [Google Scholar]
- Biddle A. S., Black S. J., and Blanchard J. L.. 2013. An in vitro model of the horse gut microbiome enables identification of lactateutilizing bacteria that differentially respond to starch induction. PLoS One 8(10):e77599. doi:10.1371/journal.pone.0077599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway T. R., Dowd S. E., and Edrington T. S.. 2010. Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J. Anim. Sci. 88(12):3977-3983. doi:10.2527/jas.2010-2900 [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Bittinger K., Bushman F. D., DeSantis T. Z., Andersen G. L., and Knight R.. 2010a. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. doi:10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Pena A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., J. Reeder, Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., T. Yatsunenko, Zaneveld J., and Knight R.. 2010b. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7: 335–336. doi:10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Penner G. B., Li M., M. Oba, and Guan L. L.. 2011. Changes in bacterial diversity associated with epithelial tissue in the beef cow rumen during the transition to a high-grain diet. Appl. Environ. Microbiol. 77: 5770–5781. doi:10.1128/AEM.00375-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K. R., and Gorley R. N.. 2006. PRIMER v6: User manual PRIMER-E. Plymouth. 192 pp. Plymouth, UK. [Google Scholar]
- Costa M. C., Arroyo L. G., E. Allen-Vercoe, Stampfli H. R., Kim P. T., Sturgeon A., and Weese J. S.. 2012. Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3-V5 region of the 16S rRNA gene. PLoS One 7:e41484. doi:10.1371/journal.pone.0041484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M. C., Silva G., Ramons R. V., Staempfli H. R., Arroyo L. G., Kim P., and Weese J. S.. 2015. Characterization and comparison of the bacterial microbiota in different gastrointestinal tract compartments in horses. Vet. J. 205: 74–80. doi:10.1016/j.tvjl.2015.03.018 [DOI] [PubMed] [Google Scholar]
- Coverdale J. A., Tyler H. D., Quigley J. D., and Brumm J. A.. 2004. Effect of various levels of forage and form of diet on rumen development and growth in calves. J. Dairy Sci. 87: 2554–2562. doi:10.3168/jds.S0022-0302(04)73380-9 [DOI] [PubMed] [Google Scholar]
- Daly K., Stewart C. S., Flint H. J., and Shirazi S. P.-Beechey. 2001. Bacterial diversity within the equine large intestine as revealed by molecular analysis of cloned 16S rRNA genes. FEMS Microbiol. Ecol. 38: 141–151. doi:10.1111/j.1574-6941.2001.tb00892.x [Google Scholar]
- de Fombelle A., Julliand V., C. Drogoul, and E. Jacotot. 2001. Feeding and microbial disorders in horses: 1-Effects of an abrupt incorporation of two levels of barley in a hay diet on microbial profile and activities. J. Equine Vet. Sci. 21: 439–445. doi:10.1016/S0737-0806(01)70018-4 [Google Scholar]
- Dougal K., Harris P. A., Edwards A., Pachebat J. A., Blackmore T. M., Worgan W. J., and Newbold C. J.. 2012. A comparison of the microbiome and the metabolome of different regions of the equine hindgut. FEMS Microbiol. Ecol. 82(3):642-652. doi:10.1111/j.1574-6941.2012.01441.x [DOI] [PubMed] [Google Scholar]
- Dowd S. E., Sun Y., Wolcott R. D., Domingo A., and Carroll J. A.. 2008. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: Bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog. Dis. 5: 459–472. doi:10.1089/fpd.2008.0107 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., C. Quince, and R. Knight. 2010. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. doi:10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando S. C., Purvis H. T., and Najar F. Z.. 2010. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 76: 7482–7490. doi:10.1128/AEM.00388-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen N. C., Avershina E., Mydland L. T., N J. A.æsset, D. Austbø, Moen B., I. Måge, and K. Rudi. 2015. High nutrient availability reduces the diversity and stability of the equine caecal microbiota. Microb. Ecol. Health Dis. 26:27216. doi:10.3402/mehd.v26.27216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartstra A. V., Bouter K. E., F. Bäckhed, and M. Nieuwdorp. 2015. Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes Care 38: 159–165. doi:10.2337/dc14-0769 [DOI] [PubMed] [Google Scholar]
- Hintz H. F., Argenzio R. A., and Schryver H. F.. 1971. Digestion coefficients, blood glucose levels and molar percentage of volatile acids in intestinal fluid of ponies fed varying forage-grain ratios. J. Anim. Sci. 33: 992–995. doi:10.2527/jas1971.335992x [DOI] [PubMed] [Google Scholar]
- Jami E., Israel A., A. Kotser, and I. Mizrahi. 2013. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 7: 1069–1079. doi:10.1038/ismej.2013.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jami E., and Mizrahi I.. 2012. Composition and similarity of bovine rumen microbiota across individual animals. PLoS One 7:e33306. doi:10.1371/journal.pone.0033306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julliand V., de Fombelle A., Drogoul C., and E. Jacotot. 2001. Feeding and microbial disorders in horses: 3 - Effects of three hay: Grain ratios on microbial profile and activities. J. Equine Vet. Sci. 21: 543–546. doi:10.1016/S0737-0806(01)70159-1 [Google Scholar]
- Leek B. F. 1993. Digestion in the ruminant stomach. In: Swenson M. J., editor, Dukes’ physiology of domestic animals. Comstock Publ. Assoc., Ithaca, NY. p. 438-474. [Google Scholar]
- Lehman C., Tilman D., and Gaines S. D.. 2000. Biodiversity, stability, and productivity in competitive communities. Am. Nat. 156(5):534-552. doi:10.1086/303402 [DOI] [PubMed] [Google Scholar]
- Li M., Zhou M., E. Adamowicz, Basarab J. A., and Guan L. L.. 2012. Characterization of rRNA sequencing, PCR-DGGE and qRT-PCR analysis. Vet. Microbiol. 155: 72–80. doi:10.1016/j.vetmic.2011.08.007 [DOI] [PubMed] [Google Scholar]
- Lin C., and Stahl D. A.. 1995. Taxon-specific probes for the cellulolytic genus Fibrobacter reveal abundant and novel equine-associated populations. Appl. Environ. Microbiol. 4: 1348–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmuthuge N., Griebel P. J., and Guan L. L.. 2014. Taxonomic identification of commensal bacteria associated with the mucosa and digesta throughout the gastrointestinal tracts of preweaned calves. Appl. Environ. Microbiol. 80: 2021–2028. doi:10.1128/AEM.03864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S. Y., Zhang R. Y., Wang D. S., and Zhu W. Y.. 2013. Impact of subacute ruminal acidosis (SARA) adaptation on rumen microbiota in dairy cattle using pyrosequencing. Anaerobe 24: 12–19. doi:10.1016/j.anaerobe.2013.08.003 [DOI] [PubMed] [Google Scholar]
- McCann J. C., Drewery M. L., Sawyer J. E., Pinchak W. E., and Wickersham T. A.. 2014a. Effect of postextraction algal residue supplementation on the ruminal microbiome of steers consuming low-quality forage. J. Anim. Sci. 92: 5063–5075. doi:10.2527/jas.2014-7811 [DOI] [PubMed] [Google Scholar]
- McCann J. C., Wiley L. M., Forbes T. D., Rouquette F. M. Jr., and Tedeschi L. O.. 2014b. Relationship between the rumen microbiome and residual feed intake-eficiency of Brahman bulls stocked on bermudagrass pastures. PLoS One 9(3):e91864. doi:10.1371/journal.pone.0091864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Price M. N., Goodrich J., Nawrocki E. P., DeSantis T. Z., Probst A., Andersen G. L., Knight R., and P. Hugenholtz. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meale S. J., Li S., P. Azevedo, Derakhshani H., Plaizier J. C., Khafipour E., and Steele M. A.. 2016. Development of ruminal and fecal microbiomes are affected by weaning but not weaning strategy in dairy calves. Front. Microbiol. 7:582. doi:10.3389/fmicb.2016.00582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milinovich G. J., Klieve A. V., Pollitt C. C., and Trott D. J.. 2010. Microbial events in the hindgut during carbohydrate-induced equine laminitis. Vet. Clin. North Am. Equine Pract. 26: 79–94. doi:10.1016/j.cveq.2010.01.007 [DOI] [PubMed] [Google Scholar]
- Moreau M. M., Eades S. C., Reinemeyer C. R., Fugaro M. N., and Onishi J. C.. 2014. Illumina sequencing of the V4 hypervariable region 16S rRNA gene reveals extensive changes in bacterial communities in the cecum following carbohydrate oral infusion and development of early-stage acute laminitis in the horse. Vet. Microbiol. 168: 436–441. doi:10.1016/j.vetmic.2013.11.017 [DOI] [PubMed] [Google Scholar]
- Naushad S., Adeolu M., N. Goel, Khadka B., A. Al-Dahwi, and Gupta R. S.. 2015. Phylogenomic and molecular demarcation of the core members of the polyphyletic Pasteurellaceae genera Actinobacillus, Haemophilus, and Pasteurella. Int. J. Genomics 2015:198560. doi:10.1155/2015/198560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster B. J., and Canale-Parola E.. 1982. Physiological diversity of rumen spirochetes. Appl. Environ. Microbiol. 43(3):686-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri R. M., Schwaiger T., Penner G. B., Beauchemin K. A., Forster R. J., McKinnon J. J., and McAllister T. A.. 2013. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS ONE, 8(12). 10.1371/journal.pone.0083424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitta D. W., Pinchak W. E., and Dowd S. E.. 2010. Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets. Microb. Ecol. 59(3):511-522. doi:10.1007/s00248-009-9609-6 [DOI] [PubMed] [Google Scholar]
- Pitta D. W., Pinchak W. E., Indugu N., B. Vecchiarelli, Sinha R., and Fulford J. D.. 2016. Metagenomic analysis of the rumen microbiome of steers with wheat-induced frothy bloat. Front. Microbiol. 7:689. doi:10.3389/fmicb.2016.00689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Respondek F., Goachet A. G., and V. Julliand. 2008. Effects of dietary short-chain fructooligosaccharides on the intestinal microflora of horses subjected to a sudden change in diet. J. Anim. Sci. 86: 316–323. doi:10.2527/jas.2006-782 [DOI] [PubMed] [Google Scholar]
- Shepherd M. L., Swecker W. S., Jensen R. V., and Ponder M. A.. 2012. Characterization of the fecal bacteria communities of forage-fed horses by pyrosequencing of 16S rRNA V4 gene amplicons. FEMS Microbiol. Lett. 326: 62–68. doi:10.1111/j.15746968.2011.02434.x [DOI] [PubMed] [Google Scholar]
- Steelman S. M., Chowdhary B. P., Dowd S., J. Suchodolski, and Jane J. E.čka. 2012. Pyrosequencing of 16S rRNA genes in fecal samples reveals high diversity of hindgut microflora in horses and potential links to chronic laminitis. BMC Vet. Res. 8:231. doi:10.1186/1746-6148-8-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel H. J. 1992. Vitamin B 12-dependent propionate production by the ruminal bacterium Prevotella ruminicola 23. Appl. Environ. Microbiol. 58: 2331–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzant E. S., and Cochran R. C.. 1994. Performance and forage utilization by beef cattle receiving increasing amounts of alfalfa hay as a supplement to low-quality, tallgrass-prairie forage. J. Anim. Sci. 72: 1059–1067. doi:10.2527/1994.7241059x [DOI] [PubMed] [Google Scholar]
- Weese J. S., Holcombe S. J., Embertson R. M., Kurtz K. A., Roessner H. A., Jalali M., and Wismer S. E.. 2015. Changes in the faecal microbiota of mares precede the development of post partum colic. Equine Vet. J. 47: 641–649. doi:10.1111/evj.12361 [DOI] [PubMed] [Google Scholar]
- Willard J. G., Willard J. C., Wolfram S. A., and Baker J. P.. 1977. Effect of diet on cecal pH and feeding behavior of horses. J. Anim. Sci. 45: 87–93. doi:10.2527/jas1977.45187x [DOI] [PubMed] [Google Scholar]
- Wilson K. L. 2009. The effect of graded levels of dietary starch. MS Thesis, Texas A&M University, College Station, TX. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.