SUMMARY
Various single nucleotide polymorphisms have been reported to be associated with a higher risk of hepatocellular carcinoma in alcoholic cirrhotic patients. Until now, only common variants conferring a small increase in liver cancer risk have been identified. These inherited factors are able to modulate several biological pathways involved in alcohol-induced hepatocarcinogenesis, such as ethanol metabolism, inflammation, oxidative stress, or iron and lipid homeostasis. How the combination of these variants might collectively define an individual genomic risk prediction is currently being investigated. The other challenge in clinical practice lies in defining how to integrate this genetic information with other clinical parameters so as to refine selection of alcoholic cirrhotic patients according to various classes of hepatocellular carcinoma risk.
KEYWORDS : alcoholic liver disease, cirrhosis, liver cancer, single nucleotide polymorphisms
Practice Points.
The study of genetic predisposition to alcohol-related liver cancer aims to identify among patients with alcoholic cirrhosis those particularly prone to develop hepatocellular carcinoma (HCC).
Although large cohorts of patients with alcoholic liver disease are still scarce, several variants modulating various biological pathways involved in alcoholic hepatocarcinogenesis have been reported as associated with a higher risk of HCC in these patients.
These genetic traits can affect ethanol metabolism, oxidative stress, inflammation process, and iron and lipid homeostasis.
The challenge is to understand how the combination of several variants might refine stratification of alcoholic cirrhotic patients according to different HCC risk classes.
These advances will enable implementation of personalized medicine in these patients, that is, the adaptation of screening and preventive measures, criteria of early diagnosis and the choice of therapeutic procedures based on inter-individual susceptibility.
Large prospective cohorts are urgently needed to validate and integrate predisposing genetic traits into individual HCC risk assessment models.
Context of genetic predisposition to liver cancer in patients with alcoholic liver disease
Although chronic infections by hepatitis B virus (HBV) and hepatitis C virus (HCV) represent the main causes of hepatocellular carcinoma worldwide, the percentage of cases attributable to excessive chronic alcohol consumption in Europe is high [1] and, as a whole, is increasing [2,3]. This trend will be confirmed with the decline in viral-induced hepatocellular carcinoma (HCC) expected in forthcoming years in Western countries on the heels of more effective viral eradication and control [4], and by the increase in alcohol consumption observed in developing countries [5]. It is therefore pivotal to identify, among millions of excessive drinkers, those susceptible to developing HCC, that is, primarily cirrhotic patients, and, among the latter, to select those especially prone to developing liver cancer [6].
• Particularities of HCC in patients with alcoholic cirrhosis
Longitudinal studies dealing with the natural history of alcoholic cirrhosis are scarce [7–9]. Moreover, major selection biases in studies conducted in decompensated patients or based on registry data hamper their interpretation in terms of HCC occurrence, as they mainly report the incidence of other liver-related complications that constitute risks of death competing with liver cancer occurrence in these populations [10,11]. In these patients, the annual incidence of HCC has been estimated between 2 and 4% [12,13], thus justifying periodic screening. Risk factors for HCC include classical features such as older age, components of metabolic syndrome and severity of underlying cirrhosis [8,12–13]. Interestingly, by combining these features, cirrhotic patients can be stratified into various HCC risk classes and thus define a specific phenotype of patients at risk [12,13]. However, all alcoholic cirrhotic patients are subjected to the same management, namely 6-month periodic HCC screening, with no preventive measures undertaken [14]. Genetic risk markers of HCC in patients with alcoholic cirrhosis have also been described and might at least partly explain the wide inter-individual susceptibility to HCC in these patients [15].
• Genetic predisposition to HCC in patients with alcoholic cirrhosis
Genetic heterogeneity is the result of a combination of single nucleotide polymorphisms (SNPs). These variations in a DNA sequence due to the change in a single nucleotide are scattered throughout the human genome, every 1000 pairs of bases, and define the phenotype of a given individual. Some of these variants are situated in coding regions and can modify gene product expression and/or function, which may result in biological consequences. If the latter are implicated in alcoholic liver disease (ALD) progression or liver carcinogenesis, then these modifications might affect individual susceptibility to HCC in the case of excessive alcohol consumption.
Various lines of evidence suggest a role for genetic traits in the emergence of liver cancer in patients with alcoholic cirrhosis. First, several variants are reported to be associated with a higher risk of evolution towards cirrhosis in individuals with chronic alcohol abuse [16], suggesting that these markers might influence progression through all steps of ALD up to cancer. Secondly, several SNPs have been linked to development of other alcohol-related cancers such as that of the upper aerodigestive tract, in particular, those modulating the formation of highly carcinogenic acetaldehyde [17]. The third and strongest argument is derived from the example of other types of chronic liver diseases: mainly conducted in Asia, results from large cohorts of HBV- or HCV-infected patients have established that the presence of HCC in first-degree relatives is associated with a higher incidence of this tumor [18], and have also identified various SNPs through multiple candidate gene or genome-wide association (GWA) approaches [15]. The extent to which these findings are reproducible in Caucasian patients with alcoholic cirrhosis and exposed to risk of HCC occurrence warrants clarification.
• Exploration of genetic risk markers of HCC in patients with ALD
Conversely to HCV- or HBV-induced HCC, no reports from GWAs conducted in patients with ALD and HCC have been yet published. Only candidate-gene approaches have been developed thus far to explore genetic susceptibility to liver cancer in heavy drinkers. While GWAs compare the genotypic distributions of millions of SNPs between HCC patients and ‘controls’ in order to discover previously unsuspected variants associated with HCC, candidate gene studies usually select one or several SNPs with known functional consequences in a biological pathway that is implicated in hepatocarcinogenesis. Table 1 describes the characteristics and specific biases of these methods. These studies propose comparing allelic or genotypic distributions of SNPs thought to influence liver carcinogenesis between 'cases' (ALD patients with HCC) and 'controls' (ALD patients without HCC). However, this case–control approach is subject to numerous biases. Indeed, most of the published studies were conducted in small samples from single centers and did not systematically take into account the stage of ALD in included patients. As a consequence, cirrhotic patients exposed to risk of liver cancer, a major prerequisite for the study of risk factors in HCC development, were not always included as controls: such selection biases do not allow the conclusion that a given variant is clearly implicated in alcoholic carcinogenesis or if it simply favors the progression of ALD towards end-stage liver disease. More importantly, the criteria of selection of these control populations is rarely precise. Some cohorts include or even mix outpatients with a long-standing uncomplicated liver disease or patients with decompensated cirrhosis; it has, however, been shown that these patients could bear a strikingly different genetic profile [19], another selection bias that can lead to major changes in genotypic distributions and risks of misinterpretation. Moreover, power calculation according to the expected prevalence of genotype distribution should always be performed prior to analyses: it has been indeed shown that less than one-third of reported associations between genetic variants and cancer diseases are actually statistically significant [20], due to high risk of false-positive report probability. Finally, in order to validate these associations, there is an urgent need for prospective cohorts of alcoholic cirrhotic patients with long follow-up, allowing the occurrence of numerous events, to allow multivariate analyses to be performed that, along with genetic traits, take into account competing risks of death as well as clinical and environmental factors known to influence HCC occurrence.
Table 1. . Methodological quality criteria for critical analysis of genetic associations studies in alcoholic hepatocarcinogenesis.
| Candidate-gene studies | GWAs | |
|---|---|---|
| Number of tested SNPs | Several, choice based on literature data | Millions, unsupervised |
| Type of studies | case–control or longitudinal | case–control |
| Number of published studies in the setting of alcohol-induced HCC | Several decades | None |
| Choice of control group for case–control studies | Patients with alcoholic cirrhosis and without HCC | Patients with alcoholic cirrhosis and without HCC |
| Selection of patients/controls | Context should be precise (compensated outpatients vs decompensated hospitalized patients) | Should precise the context (compensated outpatients vs decompensated hospitalized patients) |
| Statistical differences | According to Bonferroni correction | Usually below 10-8 |
| Power calculation and number of patients | • Depending on frequency of genetic variation, HCC incidence and length of follow-up in prospective cohorts | • Depending on frequency of genetic variation |
| • Usually several hundreds of individuals | • Usually several thousands of individuals | |
| Confirmation of identified SNPs in discovery cohort | • Replication in independent case–control cohorts | • Replication in independent case–control cohorts |
| • Validation in prospective cohorts | • Validation in prospective cohorts | |
| Main biases | • False-positive report probability | • Non selection of possible variants of interest due to prerequired high levels of p values for difference |
| • Selection biases of cirrhotic population | • Selection biases of cirrhotic population | |
| • Confounding factors degree of liver failure, comorbidities, competing risks of death | • Confounding factors degree of liver failure, comorbidities, competitive risks of death | |
GWA: Genome-wide association; HCC: Hepatocellular carcinoma; SNP: Single nucleotide polymorphism.
• Pathophysiology of alcohol-induced liver carcinogenesis
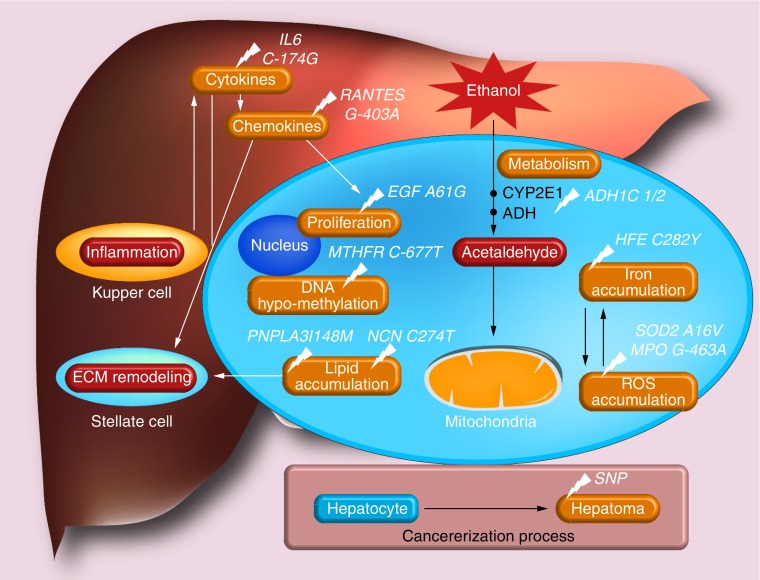
Figure 1 shows the major pathways thought to be implicated in alcohol-related hepatocarcinogenesis [21]. Ethanol is metabolized by alcohol dehydrogenase (ADH) into acetaldehyde, which acts as a carcinogen, by directly damaging DNA and proteins. In addition, ethanol is oxidized by CYP2E1, enhancing acetaldehyde, but also reactive oxygen species (ROS) formation. Oxidative stress plays a pivotal role in cancer promotion during ALD, in particular through mitochondrial dysfunction. ROS accumulation triggers lipid peroxidation and DNA damage through the formation of mutagenic adducts. The effects of iron and lipid metabolism also interfere with oxidative stress, creating a complex interplay that further influences the progression of the carcinogenic process during ALD. Furthermore, acetaldehyde alters methyl transfer, leading to DNA hypomethylation that is able to modify the expression of oncogenes and tumor suppressor genes. Finally, ethanol decreases the level of retinoic acid, leading to generation of toxic metabolites associated with changes in cell cycle regulation and cellular proliferation.
Figure 1. . Major biological pathways implicated in alcohol-induced hepatocarcinogenesis and their genetic heterogeneity, thought to influence liver cancer risk in patients with alcohol liver disease. The main variants reported as possible genetic markers of liver cancer in these patients are also depicted, according to their specific interaction with these various biological pathways.
ROS: Reactive oxygen species; SNP: Single nucleotide polymorphism.
• Challenges for research programs focusing on liver cancer susceptibility in ALD patients
Studying genetic predisposition to alcohol-induced HCC might provide the possibility for individualized management of patients according to specific genetic backgrounds. Various SNPs have been reported to be associated with a higher risk of HCC in alcoholic cirrhotic patients, but up until now, only common variants conferring a small increase in liver cancer risk (usually with a relative risk below 3) have been identified [15]. How the combination of several variants might collectively define a 'genomic risk prediction' for these individuals is a current challenge in this field. The other challenge in clinical practice lies in defining how to integrate this genetic information in order to refine stratification of alcoholic cirrhotic patients according to risk classes. The questions addressed in the case of HCC surveillance are the identification of individuals at risk, modalities and periodicity of HCC screening, and the ability to adapt preventive or curative management according to a specific genetic background [22]. Finally, these findings might change the decision-making process in clinical practice and could be translated into more global perspectives that would modify the approach to liver cancer at the population level [3].
This review provides an overview of genetic traits that modulate the risk of HCC development in alcoholic patients with cirrhosis and, when necessary, highlights the influence of these variants on other spectra of ALD progression and their role in the liver cancer process due to other etiologies.
Genetic variants associated with HCC risk in patients with alcoholic cirrhosis
Table 2 shows the most reliable and robust results based on previous methodological considerations. Far from being exhaustive, this selection highlights the quality of these studies and classifies them according to the specific biological pathways explored through their genetic modulation. Differences or similarities with viral hepatocarcinogenesis are highlighted, as well as limitations or quality criteria allowing a critical assessment of these studies.
Table 2. . Published single nucleotide polymorphisms associated with risk of hepatocellular carcinoma in patients with alcoholic liver disease.
| Biological pathway | Study (year) | SNP (rs number) | Other HCC etiologies | Progression of ALD | Type of study (n = number of included publications if meta-analysis) | Cases/controls or n if a prospective cohort of cirrhotic patients | OR or HR | Ethnicity | Limitations according to quality criteria exposed in Table 1 | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Ethanol metabolism enzymatic systems | Homann et al. (2006) | ADH1C 1/2 (rs283413) | No | Yes | Case–control | 86/217† | OR = 3.6 | Caucasian | • No replication • Low number of cases |
[23] |
| Sakamoto et al. (2006) | ALDH2 1/2 (rs 886205) | No | No | Case–control | 275/481 | OR = 4.5 | Asian | • No replication • Choice of controls • Selection of patients not precised |
[24] | |
| Munaka et al. (2003) | ALDH2 1/2 (rs 886205) | No | No | Case–control | 78/138 | OR = 2.53 | Asian | • No replication • Low number of cases and controls • Choice of controls • Selection of patients not precised |
[25] | |
| Oxidative stress/iron metabolism | Nahon et al. (2009) | SOD2 A16V (rs4880) | No | Controversial | Prospective | n = 190 | HR = 1.7 | Caucasian | • No replication | [26] |
| Nahon et al. (2009) | MPO G-463A (rs2333227) | HCV | No | Prospective | n = 190 | HR = 3.8 | Caucasian | • No replication | [26] | |
| Nahon et al. (2008) | HFE C282Y (rs1800562) | Controversial | Controversial | Prospective | n = 165 | HR = 2.7 | Caucasian | • No replication | [27] | |
| Jin et al. (2010) | HFE C282Y(rs1800562) | Controversial | Controversial | Meta-analysis (n = 9) | 1102/3766 | OR = 4.06 | Caucasian | • Limitations of meta-analyses • Not performed on individual participant data |
[28] | |
| Inflammation | Charni et al. (2011) | RANTES G-403A (rs2107538) | No | No | Prospective | n = 253 | HR = 2.7 | Caucasian | • No replication | [29] |
| Falleti et al. (2009) | IL6 C-174G(rs1800795) | HBV/HCV | Yes | Case–control | 66/153† | OR = 7.5 | Caucasian | • No replication • Low number of cases and controls |
[30] | |
| DNA synthesis and repair mechanisms | Saffroy et al. (2004) | MTHFR C-677T (rs1801133) | HCV/HBV | No | Case–control | 72/122 | OR = 2.03 | Caucasian | • No replication • Low number of cases and controls • Choice of controls |
[31] |
| Fabris et al. (2009) | MTHFR C-677T (rs1801133) | HCV/HBV | No | Case–control | 17/46 | OR = 4.47 | Caucasian | • No replication • Low number of cases and controls • Choice of controls • Selection of patients not precised |
[32] | |
| Growth factors | Tanabe et al. (2008) | EGF A61G(rs4444903) | HBV/HCV | No | Case–control | 44/121† | OR = 2.9 | Caucasian | • Low number of cases and controls (subgroup analyses) | [33] |
| Zhong et al. (2012) | EGF A61G (rs4444903) | HBV/HCV | No | Meta-analysis (n= 8) | 1304/2613 | OR = 1.79 | Mixed | • Limitations of meta-analyses • Not performed on individual participant data |
[34] | |
| Lipid metabolism | Falleti et al. (2011) | PNPLA3 I148M (rs738409) | HCV/NASH | Yes | Case–control | 66/132† | OR = 1.45 | Caucasian | • No replication • Low number of cases and controls |
[35] |
| Nischalke et. (2011) | PNPLA3 I148M(rs738409) | HCV/NASH | Yes | Case–control | 80/80† | OR = 2.8 | Caucasian | • No replication • Low number of cases and controls |
[36] | |
| Trépo et al. (2012) | PNPLA3 I148M(rs738409) | HCV/NASH | Yes | Case–control | 145/425† | OR = 4.7 | Caucasian | • No replication | [37] | |
| Guyot et al. (2013) | PNPLA3 I148M(rs738409) | HCV/NASH | Yes | Prospective | n = 279 | HR = 1.72 | Caucasian | • No replication | [38] | |
| Trépo et al. (2013) | PNPLA3 I148M (rs738409) | HCV/NASH | Yes | Meta-analysis (n = 5) | 442/932† | OR = 2.13 | Caucasian | • Limitations of meta-analyses | [39] | |
| Nischalke et al. (2014) | NCAN C274T (rs2228603) | No | No | Case–control (with replication) | 126/356† (83/146†) | OR = 1.84 | Caucasian | • None | [40] | |
Note: OR or HR takes into account the highest reported value according to allelic or genotypic expression of results and, when available, after multivariate analysis.
†Case–control studies conducted in alcoholic cirrhotic patients with or without HCC (i.e., appropriate control group).
ALD: Alcoholic liver disease; HBV: Hepatitis B virus; HCC: Hepatocellular carcinoma; HCV: Hepatitis C virus; HR: Hazard ratio; NASH: Non-alcoholic steato-hepatitis; OR: Odds ratio; SNP: Single nucleotide polymorphism.
• Inherited variations in alcohol-metabolizing enzymes: first steps in tumor promotion
As previously mentioned, acetaldehyde is generated and then metabolized by the successive effects of alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) families [41]. This complex enzymatic system is subjected to genetic heterogeneity, possibly providing an attractive explanation for individual susceptibility to alcohol-related malignancies. Consequently, genetic variants involved in alcohol-metabolizing enzymes that modulate subsequent amounts of carcinogenic acetaldehyde were explored as potential candidates, suggesting a possible interaction between genetic susceptibility and alcohol exposure in cancer diseases, including those of the liver [17].
The study of Caucasian patients with ALD highlighted functional polymorphism in the ADH1C isoenzyme as a possible inherited factor implicated in the emergence of various alcohol-related cancers, particularly HCC [23]. Indeed, individuals harboring ADH1C*1, thought to confer higher enzymatic activity, may be hypothetically exposed to stronger acetaldehyde formation and accumulation. By comparing genotype distribution in ALD patients with cirrhosis complicated or not by HCC, Homann et al. concluded that homozygosity for allele ADH1C*1 was a risk factor in liver cancer development. This association is further supported by the fact that excessive drinkers carrying the same genotype are more likely to develop cirrhosis [42], thus suggesting genetic modulation of this variant throughout the entire ALD spectrum. Furthermore, the same study reported a higher prevalence of alcohol-related malignancies of the upper gastrointestinal tract in the same individuals, a fact that strengthens the role of the carcinogenic effects of genetically influenced production of acetaldehyde in chronic alcohol abuse.
A large proportion of individuals of Asian descent are unable to sufficiently detoxify acetaldehyde in case of ethanol consumption due to frequent genetic variations in ALDH2 resulting in weak enzymatic activity [43]. This explains their poor tolerance for even small amounts of alcohol ingestion, resulting in nausea, vomiting and facial flushing. These observations led several teams from Japan to explore this genetic heterogeneity as a possible modifier of liver cancer risk [24–25]. As expected from the acetaldehyde accumulation hypothesis, individuals carrying the weakly active ALDH2*2-conferring allele were more prone to developing HCC in case of excessive alcohol intake. However, it is important to note that those studies, conducted in Asia, primarily included patients with liver disease of viral etiology in whom alcohol consumption was recorded, a consideration to take into account for interpreting these data as the same genotype could also lead patients to refrain from consuming large amounts of alcohol.
• Genetic modulation of oxidative stress & iron metabolism: pivotal role in alcohol-induced liver carcinogenesis
ROS promote damage to cellular macromolecules and participate in liver carcinogenesis progression [44]. The combination of DNA damage with continuous DNA replication causes gene mutations that will accumulate over the years, some of them may eventually hamper the control of the cell cycle and/or apoptosis, to produce a cancerous clone. Given the importance of oxidative stress (particularly inside mitochondria) in the pathogenesis of ALD, genetic variants affecting enzymes regulating the production and detoxification of ROS have been shown to modulate its outcome. In particular, MPO is expressed in neutrophils and Kupffer cells, both of which are involved in the pathogenesis of ALD, and catalyzes the reaction of H2O2 with Cl- to form the highly reactive hypochlorous acid (HOCl) and anion (OCl-) [45]. A G to A base exchange at position -463 affects its promoter [46]. The GG-MPO genotype is associated with higher protein expression and favors cirrhosis constitution in patients with hemochromatosis [47] as well as various malignancies [48].
In addition, manganese superoxide dismutase (SOD2) generates H2O2 within mitochondria [49] that can form hypochlorous acid in the presence of MPO or the hydroxyl radical in the presence of iron. A genetic dimorphism substitutes either alanine (Ala) or valine (Val) in the mitochondrial targeting sequence of SOD2 [50] and results in higher mitochondrial activity for the Ala-SOD2 variant. Indeed, in acute mitochondrial import and transfection experiments, the Ala-SOD2 variant achieved higher mitochondrial activity than the Val-SOD2 variant [50,51]. However, MnSOD is inducible by ROS, cytokines, and ethanol [52], and is inactivated by peroxynitrite [53] or by the misincorporation of iron instead of manganese into the MnSOD active site [54]. The transfection of Ala-SOD2 variant in cell lines is associate with higher hydrogen peroxide formation [55], a finding that may explain the multiple reports linking this variant to numerous cancers.
It has been shown, in large prospective cohorts of alcoholic cirrhotic patients, that carriage of two G-MPO alleles, or possession of at least one Ala-SOD2 allele alone, were independent risk factors for HCC occurrence [26]. Furthermore, the occurrence of HCC increased dramatically in individuals who cumulated these two genetic traits, suggesting a synergic effect of these SNPs on ROS-mediated liver carcinogenesis. Interestingly, these variants also favor liver iron overload, possibly through enhanced mitochondrial hydrogen peroxide production [55]. Liver iron overload, as well as HFE gene mutations have also been reported as associated with the risk of HCC in Caucasians with alcoholic cirrhosis [27–28] These findings suggest that hepatic iron accumulation might not only be a cause of enhanced oxidative stress in the course of ALD but also a consequence of mitochondrial dysfunction, thus establishing the basis of a vicious circle, an association that does not seem to be as relevant in HCV-related liver cancer development [27,56].
Interestingly, the comparison of these results with those obtained in patients with HCV-related cirrhosis suggests both similarities and differences in HCV- and alcohol-induced hepatocarcinogenesis. HCV-related liver cancer is also associated with increased ROS production and iron accumulation [57]. A possible explanation for the role of the Ala16Val-SOD2 and iron metabolism in alcohol-induced but not HCV-mediated carcinogenesis could reside in the fact that both ROS and iron downregulate HCV replication [58,59]. Thus any genetic trait increasing ROS formation and/or iron accumulation within HCV-infected hepatocytes could also decrease HCV replication in these cells. These differences might also explain why liver iron accumulation seems to act as a strong carcinogen in alcohol-induced carcinogenesis, while its effect on HCV carcinogenesis seems to be less relevant [27].
Since the benefits of using antioxidant treatments or implementing iron depletion to prevent cancer remain controversial [60], individual variations influenced by the cause of liver disease and by individual characteristics as well, point out the need for development of targeted antioxidant chemopreventive strategies.
• Inflammation, cytokine & chemokine systems
Chronic alcohol consumption induces activation of macrophages and Kupffer cells, in particular, through the IL-1 pathway, which leads to production of pro-inflammatory cytokines such as IL-6 and TNFα. IL-6/STAT3 and TNFα/NF-kB axes have been pointed out as being major biological pathways involved in hepatocarcinogenesis [61]. In the liver, NF-kB activation in nonparenchymal cells is necessary for HCC tumor promotion, but also stimulates production of growth factors and cytokines such as EGF and IL-6, leading to an increase in ROS production [62]. Although the mechanisms by which pro-inflammatory cytokines promote liver cancer are not clear, their signals regulate gene expression through transcription factors STAT3 and NF-kB. NF-kB is the intracellular signaling effector for many pro-inflammatory cytokines, including TNFα, IL-1, and toll-like receptors (TLRs). In the liver, NF-kB activation in nonparenchymal cells is necessary for HCC tumor promotion, but also stimulates production of growth factors and cytokines such as EGF and IL-6, leading to an increase in ROS production [62]. Interestingly, TNFα and IL-6 create a positive feedback loop and form a complex of regulatory networks that promote hepatocarcinogenesis. In this setting, downstream from TNFα and IL-1 production, IL-6 activates STAT3 and thereby enhances cell growth [63].
This pro-inflammatory cytokine signaling pathway can be further modulated by complex genetic heterogeneity. In this setting, variants influencing TNFα, IL1β, IL6 or EGF production were reported to be modifiers of liver cancer risk, particularly in patients infected by HBV or HCV [64]. These SNPs also seem to influence the course of ALD, in particular variants affecting TNFα and IL6 secretion, which have been shown to favor progression to cirrhosis in heavy drinkers [65,66]. In particular, the highly productive cytokine IL6–174G allele has been reported to be a possible genetic variation influencing the risk of liver cancer in Caucasians with alcoholic cirrhosis [30]. The influence of TNFα variants on alcoholic carcinogenesis, however, seems more controversial [67].
EGF promotes cancer growth and invasiveness [68] and triggers liver carcinogenesis in animal models [69]. This factor is subjected to functional polymorphism involving an A to G exchange in the 5′ untranslated region of the EGF gene, resulting in stronger transcription and development of malignancies in individuals harboring 2 G-alleles [70]. Tanabe et al. [33] genotyped two distinct case–control sample cohorts of cirrhotic Caucasian patients (alcohol- or HCV-related), and reported a higher frequency of the at-risk homozygote GG genotype in both cohorts. This observation, consistent with a ubiquitous role for this variant in carcinogenesis progression in both viral and alcoholic disease, is further supported by a recent meta-analysis [34]. Furthermore, the impact of this genetic trait on the occurrence of liver cancer was validated in patients included in the HALT-C trial [71], indicating that this SNP is an essential candidate gene driving hepatocarcinogenesis.
Chemokines are chemotactic cytokines involved in the attraction and activation of specific inflammatory cells to sites of inflammation in the course of chronic liver disease [72]. The role of chemokines and their receptors in promotion of tumor growth and subsequent invasion has been reported, via lymphocyte recruitment in the livers of patients with HCC [73]. Chronic ethanol consumption is characterized by an increase in the expression of a number of inflammatory cytokines and chemokines. In particular, TNFα induces the expression of RANTES, a T-cell chemoattractant and immunoregulatory molecule that may play a pivotal role in migration of inflammatory cells to the liver in patients with ALD. Indeed, RANTES/CCL5 is secreted either from tumoral or stromal cells, and enhances motility and invasion of cancer cells [74]. A RANTES promoter polymorphism (G-403A) results in increased RANTES expression for A-403 alleles (10, 11) and favors development of various malignancies (12–14). A prospective study conducted in Caucasian patients with cirrhosis revealed the influence of genetic heterogeneity modulating RANTES chemokine expression upon the risk of HCC. In particular, alcoholic cirrhotic patients bearing two G-403 alleles had higher incidences of HCC during follow-up. As described earlier, this association seemed to be specific to patients with alcohol-induced HCC and could not transposed to all patients with chronic liver diseases, as this observation was not replicated in HCV-infected individuals [29].
Taken together, these data highlight the importance of inflammation in initiation and progression of liver carcinogenesis, as well as its modulation by complex genetic heterogeneity, that might help to refine selection of alcoholic cirrhotic patients who may benefit from HCC chemoprevention. These approaches, by affecting anti-tumor-specific immune responses through antitumoral immunotherapy [75] or by blocking the EGF/EGFR pathway, for example [69], warrant further testing in the setting of specifically targeted therapies.
• Genetic modulation of folate metabolism
Variants in the MTHFR gene, leading to alterations in folate metabolism, an essential component of DNA synthesis and methylation, have been reported to be associated with HCC development [76], irrespective of the cause of liver disease or ethnic origin. The polymorphism of MTHFR, resulting in a substitution of C to T at nucleotide 677, which converts an alanine to a valine residue, seems to be a possible susceptibility locus for liver cancer in alcoholic patients [31–32]. Although based on the hypothesis that MTHFR activity and folate availability might affect gene expression (through DNA methylation) and genome integrity (through DNA synthesis and repair) [17], these studies nonetheless showed discrepancies, warranting further elucidation.
• Synergic effect of alcoholic consumption & genetic factors affecting risk of liver cancer in other chronic liver diseases
As mentioned earlier, most studies in the field of HCC predisposition are currently conducted in Asia, where the most prevalent causes of chronic liver disease are dominated by chronic viral infections. However, some reports have identified several interactions between various SNPs influencing liver cancer emergence and the amount of chronic ethanol consumption in these individuals. This is for example the case of genetic modifiers of cytochrome CYP2E1, for which a recent meta-analysis performed in nearly 4000 individuals confirmed that while CYP2E1 Pst I/Rsa polymorphism was not associated with HCC risk, the interaction between this variant and alcohol consumption increased the risk of liver cancer [77]. Other emerging data focused on genetic modulation of miRNA genes, leading to changes in expression of mature miRNAs have been identified as genetic modifiers of HCC risk in HBV- or HCV-infected individuals from Asia such as miR-146a, miR-122 and miR-378 [15]. In the same way, some of these SNPs such as rs4938723 in the promoter region of pri-miR-34b/c, seem to modify liver cancer risk in these patients through specific interaction with ethanol consumption [78]. The extent to which these observations might be translated in patients with alcoholic cirrhosis, and thus to ethanol-related hepatocarcinogenesis, remains to be clarified and should be specifically addressed in populations with ALD.
• PNPLA3 rs738409: a robust association with alcohol-induced liver cancer, but as yet no clear explanation
Several GWAs conducted in NASH and ALD patients identified a single nucleotide polymorphism (rs738409 C>G) which encodes for an isoleucine to methionine substitution at position 148 in the PNPLA3 protein sequence [79–81], as a promoter of intracellular triglyceride accumulation [82]. Subsequently, the rs738409 (G) allele was reported to be associated with more pronounced steatosis, advanced fibrosis and a higher incidence of cirrhosis in the most prevalent causes of chronic liver diseases affecting Caucasians, namely NASH [83,84], HCV infection [85,86] and ALD [87,88]. It was thus tempting to speculate that the same genetic trait that influences development of initial steps common to all chronic liver disease might also promote the emergence of liver cancer. In this context, case–control studies were subsequently conducted in alcoholic and/or HCV-infected patients from Europe with cirrhosis, complicated or not by HCC, in order to test this hypothesis. These initial reports identified an association between the rs738409 (GG) genotype and HCC in these patients [35–37,85,88–89]. However, results seemed concordant for ALD, but remained conflicting in HCV-infected patients. More recently, a prospective cohort study conducted in cirrhotic patients screened for HCC confirmed that the G-PNPLA3 allele was a risk factor for HCC in alcoholic patients, while the same variant did not influence the outcome of HCV-infected individuals [38].
All studies in this field were conducted in Europe and included a homogeneous population of more than 2500 Caucasian patients with cirrhosis, complicated or not by HCC. This led to a recent meta-analysis [39], which also had the advantage of using individual participant data. As a whole, this approach confirmed the influence of PNPLA3 genotypes on liver carcinogenesis in Caucasian cirrhotic patients, which was more pronounced in alcoholic patients, in whom a twofold HCC risk was observed for those bearing the G variant. A recent second meta-analysis confirmed this specific association [90].
The exact mechanisms by which PNPLA3 influences hepatocarcinogeneis in ALD patients remain to be determined. Indeed, the question of whether the rs738409 variant promotes liver carcinogenesis by creating a favorable microenvironment constituted by steatosis, inflammation and fibrosis, or by specifically triggering the carcinogenic process, is still unclear. In particular, these complex entities composing the tissue environment might interact and thereby drive cancer/host cross-talk, but the details of this interplay are not clear. However, it is noteworthy that the PNPLA3 variant seems to influence progression of chronic liver disease independently of the grade of steatosis [85–86,91], and that this histological feature has not been clearly associated with the risk of HCC occurrence in prospective cohorts of cirrhotic patients [92]. Mechanistic studies should unravel specific impaired biological pathways explaining the frequently reported associations between rs738409 and HCC in patients with ALD. Such results are currently emerging, with, for example, the recent report of an unsuspected interaction between PNPLA3 and retinoid metabolism [93]; indeed, these liposoluble micronutrients are known to influence cell proliferation and differentiation [94] and have been shown to be effective in liver cancer prevention trials [95].
Interestingly, other genetic modifiers of lipid metabolism seem to further influence HCC risk in ALD. A variant in extracellular matrix proteoglycan NCAN has been identified in a GWAS conducted in patients with NASH as a polymorphism affecting hepatic steatosis [96]. Very recently, a case–control study reported a higher prevalence of the same NCAN-T variant with HCC developed in patients with alcoholic cirrhosis [40]. This particular study presented the advantage of meeting several methodological quality criteria. First, it was conducted in a large case–control series including an appropriate control group of patients with alcoholic cirrhosis. Second, the authors concomitantly explored the influence of the genetic trait in other clinical settings such as ALD progression or HCV-related HCC. Third, a validation set confirmed this association. Fourth, the authors also took into account variations in PNPLA3, and were thus able to observe that the proportion of HCC patients among carriers of both NCAN-T and PNPLA3-G risk variants were increased as compared with carriers of only one or none of these traits (OR: 1.891 and OR: 4.575, respectively). Finally, genotype–phenotype correlations were undertaken in liver samples, highlighting a possible involvement for this ECM protein in ALD progression, although mechanisms of cancerization process remain to be clarified.
Future perspective
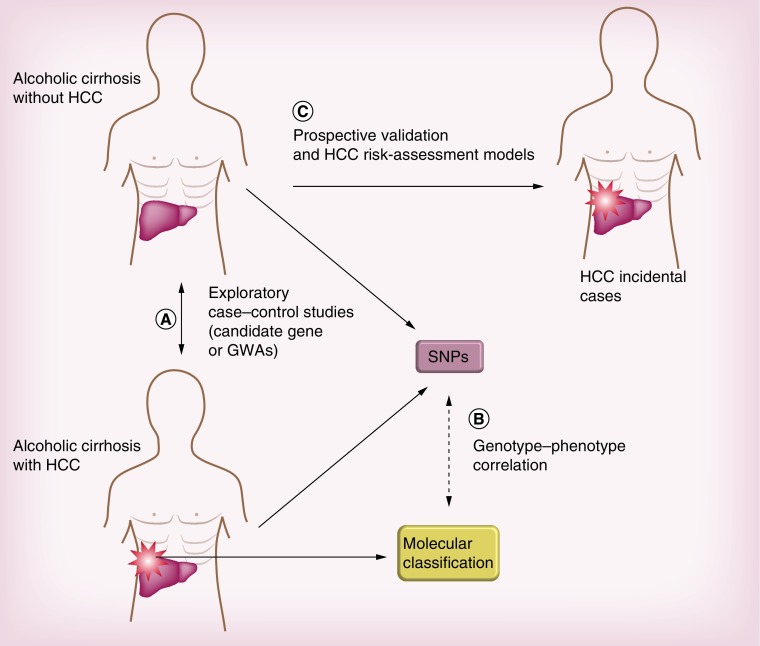
Except for the involvement of PNPLA3 variants in alcohol-related HCC, most associations still suffer from a lack of robustness or remain inconclusive. Numerous methodological limitations hamper validation of these candidate genes as definitive risk markers of liver cancer in ALD patients. Structuring of this field of research is urgently needed, as recently demonstrated by genetic studies conducted in the field of lipid metabolism genetic modulation, but will only be feasible by using large, well-defined populations in order to overcome these pitfalls. Figure 2 provides an example of optimal research programs in three steps that could lead to progress in discovering, exploring and then validating genetic traits associated with liver cancer risk in patients with alcoholic cirrhosis.
Figure 2. . Challenges and perspectives in research programs.
(A) Optimal design for case–control studies aimed at exploring HCC susceptibility through candidate gene or GWA approaches. (B) Translational studies should explore both germ-line and functional genetics of alcohol-induced liver cancer. (C) Longitudinal studies should validate and incorporate genetic data into complex HCC risk assessment models in order to refine selection of alcoholic cirrhotic patients according to prognosis.
GWA: Genome-wide association; HCC: Hepatocellular carcinoma; SNP: Single nucleotide polymorphism.
First, case–control studies should compare genotypic distribution between HCC patients and individuals exposed to liver cancer, namely cirrhotic patients. Whether performed in the setting of supervised exploration or GWASs, such a rigorous approach might limit selection bias. This is well illustrated by the converging results on PNPLA3 variants obtained in several homogeneous cohorts of patients with alcoholic cirrhosis complicated or not by HCC. In the absence of such an in-depth approach at both the genotypic and phenotypic level, the exact implications of these variants in liver cancer emergence cannot be correctly assessed, leading to a high risk of misinterpretation.
Second, the exploration of biological consequences of newly discovered variants should be assessed in liver samples in which exploration at the molecular level can be performed. The precise knowledge of effects of a given variant on tumor promotion is indeed a prerequisite for confirming its role in modulation of cancer risk, and may also pave the way for development of possible therapeutic targets. Furthermore, the question as to whether genetic factors at the individual level are associated with the occurrence of specific genomic subtypes of liver tumors remains to be elucidated. HCC is indeed a very heterogeneous tumor and can be classified into various transcriptomic groups [97], a fact that can impact treatment response [98] and also differ according to underlying liver disease [99]. The combination and integration of these complex genetic signatures at the individual and tumor level in clinical decision-making is another exciting challenge in forthcoming years.
Third, all newly discovered SNPs should be validated in cohorts of compensated alcoholic cirrhotic patients prospectively followed up and screened for HCC, so as to include all confounding factors. The ultimate challenge will be to successfully integrate genetic data into individual HCC risk assessment models. The incremental value of this genetic information must be assessed in order to refine the prognosis in ALD patients, that is, to measure its relative weight in various phenotypes of patients with alcoholic cirrhosis encountered in clinical practice. This approach is currently providing interesting results in both alcoholic- and HCV-induced cirrhotic cohorts [38,67,71]. For example, it has been shown that adding PNPLA3 genotypes to simple clinical scoring systems in prospectively followed-up patients with alcoholic cirrhosis led to a better prediction of liver cancer emergence at the individual level [38]. In this setting, numerous SNPs could be incorporated into complex HCC predictive scores combining genetic and clinical parameters, enabling selection of patients according to level of liver cancer risk. This integrative approach could distinguish specific sets of genes as new theranostic markers of HCC in alcoholic cirrhotic patients, and might change our perception of the individual risk level of patients in clinical situations where pivotal therapeutic decisions could be made.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special notes have been highlighted as: • of interest
- 1.Schoniger-Hekele M, Muller C, Kutilek M, Oesterreicher C, Ferenci P, Gangl A. Hepatocellular carcinoma in Austria: aetiological and clinical characteristics at presentation. Eur. J. Gastroenterol. Hepatol. 2000;12(8):941–948. doi: 10.1097/00042737-200012080-00015. [DOI] [PubMed] [Google Scholar]
- 2.Bosch Fx, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127(5 Suppl. 1):S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Bruix J, Gores Gj, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63(5):844–855. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis Gl, Alter Mj, El-Serag H, Poynard T, Jennings Lw. Aging of hepatitis C virus (HCV)-infected persons in the United States: a multiple cohort model of HCV prevalence and disease progression. Gastroenterology. 2010;138(2):513–521. doi: 10.1053/j.gastro.2009.09.067. 521.e511–e516. [DOI] [PubMed] [Google Scholar]
- 5.Lim SS, Vos T, Flaxman AD, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2224–2260. doi: 10.1016/S0140-6736(12)61766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology. 2004;127(5 Suppl. 1):S87–S96. doi: 10.1053/j.gastro.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Benvegnu L, Noventa F, Bernardinello E, Pontisso P, Gatta A, Alberti A. Evidence for an association between the aetiology of cirrhosis and pattern of hepatocellular carcinoma development. Gut. 2001;48(1):110–115. doi: 10.1136/gut.48.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mair RD, Valenzuela A, Ha NB, et al. Incidence of hepatocellular carcinoma among US patients with cirrhosis of viral or nonviral etiologies. Clin. Gastroenterol. Hepatol. 2012;10(12):1412–1417. doi: 10.1016/j.cgh.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toshikuni N, Izumi A, Nishino K, et al. Comparison of outcomes between patients with alcoholic cirrhosis and those with hepatitis C virus-related cirrhosis. J. Gastroenterol. Hepatol. 2009;24(7):1276–1283. doi: 10.1111/j.1440-1746.2009.05851.x. [DOI] [PubMed] [Google Scholar]
- 10.Bell H, Jahnsen J, Kittang E, Raknerud N, Sandvik L. Long-term prognosis of patients with alcoholic liver cirrhosis: a 15-year follow-up study of 100 Norwegian patients admitted to one unit. Scand. J. Gastroenterol. 2004;39(9):858–863. doi: 10.1080/00365520410006350. [DOI] [PubMed] [Google Scholar]
- 11.Jepsen P, Ott P, Andersen PK, Sorensen HT, Vilstrup H. Risk for hepatocellular carcinoma in patients with alcoholic cirrhosis: a Danish nationwide cohort study. Ann. Intern. Med. 2012;156(12):841–847. W295. doi: 10.7326/0003-4819-156-12-201206190-00004. [DOI] [PubMed] [Google Scholar]
- 12.Mancebo A, Gonzalez-Dieguez Ml, Cadahia V, et al. Annual incidence of hepatocellular carcinoma among patients with alcoholic cirrhosis and identification of risk groups. Clin. Gastroenterol. Hepatol. 2013;11(1):95–101. doi: 10.1016/j.cgh.2012.09.007. [DOI] [PubMed] [Google Scholar]; • Large cohort study of prospectively followed-up patients with alcoholic cirrhosis which assessed specific risk factors for hepatocellular carcinoma occurrence in this population.
- 13.N’kontchou G, Paries J, Htar MT, et al. Risk factors for hepatocellular carcinoma in patients with alcoholic or viral C cirrhosis. Clin. Gastroenterol. Hepatol. 2006;4(8):1062–1068. doi: 10.1016/j.cgh.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 14.Trinchet JC, Chaffaut C, Bourcier V, et al. Ultrasonographic surveillance of hepatocellular carcinoma in cirrhosis: A randomized trial comparing 3- and 6-month periodicities. Hepatology. 2011;54(6):1987–1997. doi: 10.1002/hep.24545. [DOI] [PubMed] [Google Scholar]
- 15.Nahon P, Zucman-Rossi J. Single nucleotide polymorphisms and risk of hepatocellular carcinoma in cirrhosis. J. Hepatol. 2012;57:663–674. doi: 10.1016/j.jhep.2012.02.035. [DOI] [PubMed] [Google Scholar]; • General review focusing on genetic predisposition to hepatocellular carcinoma in patients with cirrhosis.
- 16.Stickel F, Hampe J. Genetic determinants of alcoholic liver disease. Gut. 2011;61(1):150–159. doi: 10.1136/gutjnl-2011-301239. [DOI] [PubMed] [Google Scholar]; • General review focusing on genetic predisposition to alcoholic liver disease.
- 17.Druesne-Pecollo N, Tehard B, Mallet Y, et al. Alcohol and genetic polymorphisms: effect on risk of alcohol-related cancer. Lancet Oncol. 2009;10(2):173–180. doi: 10.1016/S1470-2045(09)70019-1. [DOI] [PubMed] [Google Scholar]
- 18.Lee MH, Yang HI, Liu J, et al. Prediction models of long-term cirrhosis and hepatocellular carcinoma risk in chronic hepatitis B patients: risk scores integrating host and virus profiles. Hepatology. 2013;58(2):546–554. doi: 10.1002/hep.26385. [DOI] [PubMed] [Google Scholar]
- 19.Nahon P, Sutton A, Pessayre D, et al. Genetic dimorphism in superoxide dismutase and susceptibility to alcoholic cirrhosis, hepatocellular carcinoma, and death. Clin. Gastroenterol. Hepatol. 2005;3(3):292–298. doi: 10.1016/s1542-3565(04)00718-9. [DOI] [PubMed] [Google Scholar]
- 20.Dong LM, Potter JD, White E, Ulrich CM, Cardon LR, Peters U. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. JAMA. 2008;299(20):2423–2436. doi: 10.1001/jama.299.20.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat. Rev. Cancer. 2007;7(8):599–612. doi: 10.1038/nrc2191. [DOI] [PubMed] [Google Scholar]
- 22.Della Corte C, Aghemo A, Colombo M. Individualized hepatocellular carcinoma risk: the challenges for designing successful chemoprevention strategies. World J. Gastroenterol. 2013;19(9):1359–1371. doi: 10.3748/wjg.v19.i9.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homann N, Stickel F, Konig IR, et al. Alcohol dehydrogenase 1C*1 allele is a genetic marker for alcohol-associated cancer in heavy drinkers. Int. J. Cancer. 2006;118(8):1998–2002. doi: 10.1002/ijc.21583. [DOI] [PubMed] [Google Scholar]
- 24.Sakamoto T, Hara M, Higaki Y, et al. Influence of alcohol consumption and gene polymorphisms of ADH2 and ALDH2 on hepatocellular carcinoma in a Japanese population. Int. J. Cancer. 2006;118(6):1501–1507. doi: 10.1002/ijc.21505. [DOI] [PubMed] [Google Scholar]
- 25.Munaka M, Kohshi K, Kawamoto T, et al. Genetic polymorphisms of tobacco- and alcohol-related metabolizing enzymes and the risk of hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2003;129(6):355–360. doi: 10.1007/s00432-003-0439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nahon P, Sutton A, Rufat P, et al. Myeloperoxidase and superoxide dismutase 2 polymorphisms comodulate the risk of hepatocellular carcinoma and death in alcoholic cirrhosis. Hepatology. 2009;50(5):1484–1493. doi: 10.1002/hep.23187. [DOI] [PubMed] [Google Scholar]; • Example of prospective study of the influence of a panel of genetic variants modulating oxidative stress on the risk of hepatocellular carcinoma emergence in a longitudinal cohort of patients with alcoholic cirrhosis.
- 27.Nahon P, Sutton A, Rufat P, et al. Liver iron, HFE gene mutations, and hepatocellular carcinoma occurrence in patients with cirrhosis. Gastroenterology. 2008;134(1):102–110. doi: 10.1053/j.gastro.2007.10.038. [DOI] [PubMed] [Google Scholar]
- 28.Jin F, Qu LS, Shen XZ. Association between C282Y and H63D mutations of the HFE gene with hepatocellular carcinoma in European populations: a meta-analysis. J. Exp. Clin. Cancer Res. 2010;29:18. doi: 10.1186/1756-9966-29-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charni F, Sutton A, Rufat P, et al. Chemokine RANTES Promoter Dimorphisms and Hepatocellular Carcinoma Occurrence in Patients with Alcoholic or Hepatitis C Virus-Related Cirrhosis. Cancer Epidemiol. Biomarkers Prev. 2011;20(7):1439–1446. doi: 10.1158/1055-9965.EPI-11-0341. [DOI] [PubMed] [Google Scholar]
- 30.Falleti E, Fabris C, Toniutto P, et al. Interleukin-6 polymorphisms and gender: relationship with the occurrence of hepatocellular carcinoma in patients with end-stage liver disease. Oncology. 2009;77(5):304–313. doi: 10.1159/000260057. [DOI] [PubMed] [Google Scholar]
- 31.Saffroy R, Pham P, Chiappini F, et al. The MTHFR 677C > T polymorphism is associated with an increased risk of hepatocellular carcinoma in patients with alcoholic cirrhosis. Carcinogenesis. 2004;25(8):1443–1448. doi: 10.1093/carcin/bgh147. [DOI] [PubMed] [Google Scholar]
- 32.Fabris C, Toniutto P, Falleti E, et al. MTHFR C677T polymorphism and risk of HCC in patients with liver cirrhosis: role of male gender and alcohol consumption. Alcohol Clin. Exp. Res. 2009;33(1):102–107. doi: 10.1111/j.1530-0277.2008.00816.x. [DOI] [PubMed] [Google Scholar]
- 33.Tanabe KK, Lemoine A, Finkelstein DM, et al. Epidermal growth factor gene functional polymorphism and the risk of hepatocellular carcinoma in patients with cirrhosis. JAMA. 2008;299(1):53–60. doi: 10.1001/jama.2007.65. [DOI] [PubMed] [Google Scholar]; • Report of the association between EGF variant and hepatocellular carcinoma in patients with alcohol- or hepatitis-C-related cirrhosis
- 34.Zhong JH, You XM, Gong WF, et al. Epidermal growth factor gene polymorphism and risk of hepatocellular carcinoma: a meta-analysis. PLoS ONE. 2012;7(3):e32159. doi: 10.1371/journal.pone.0032159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falleti E, Fabris C, Cmet S, et al. PNPLA3 rs738409C/G polymorphism in cirrhosis: relationship with the aetiology of liver disease and hepatocellular carcinoma occurrence. Liver Int. 2011;31(8):1137–1143. doi: 10.1111/j.1478-3231.2011.02534.x. [DOI] [PubMed] [Google Scholar]
- 36.Nischalke HD, Berger C, Luda C, et al. The PNPLA3 rs738409 148M/M genotype is a risk factor for liver cancer in alcoholic cirrhosis but shows no or weak association in hepatitis C cirrhosis. PLoS ONE. 2011;6(11):e27087. doi: 10.1371/journal.pone.0027087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trepo E, Guyot E, Ganne-Carrie N, et al. PNPLA3 (rs738409 C>G) is a common risk variant associated with hepatocellular carcinoma in alcoholic cirrhosis. Hepatology. 2012;55(4):1307–1308. doi: 10.1002/hep.25518. [DOI] [PubMed] [Google Scholar]
- 38.Guyot E, Sutton A, Rufat P, et al. PNPLA3 rs738409, hepatocellular carcinoma occurrence and risk model prediction in patients with cirrhosis. J. Hepatol. 2013;58(2):312–318. doi: 10.1016/j.jhep.2012.09.036. [DOI] [PubMed] [Google Scholar]; • Example of integration of PNPLA3 genotypes into hepatocellular carcinoma risk assessment models enabling to refine the selection of patients with alcoholic cirrhosis according to prognosis
- 39.Trepo E, Nahon P, Bontempi G, et al. Association between the PNPLA3 (rs738409 C>G) variant and hepatocellular carcinoma: evidence from a meta-analysis of individual participant data. Hepatology. 2013;59(6):2170–2177. doi: 10.1002/hep.26767. [DOI] [PubMed] [Google Scholar]; • Meta-analysis of the association between PNPLA3 variants and hepatocellular carcinoma in nearly 2500 Caucasian patients from Europe.
- 40.Nischalke HD, Lutz P, Kramer B, et al. A common polymorphism in the NCAN gene is associated with hepatocellular carcinoma in alcoholic liver disease. J. Hepatol. 2014;16:S0168–S8278. doi: 10.1016/j.jhep.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Duester G, Farres J, Felder MR, et al. Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem. Pharmacol. 1999;58(3):389–395. doi: 10.1016/s0006-2952(99)00065-9. [DOI] [PubMed] [Google Scholar]
- 42.Bosron WF, Ehrig T, Li TK. Genetic factors in alcohol metabolism and alcoholism. Semin. Liver Dis. 1993;13(2):126–135. doi: 10.1055/s-2007-1007344. [DOI] [PubMed] [Google Scholar]
- 43.Crabb DW, Edenberg HJ, Bosron WF, Li TK. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. J. Clin. Invest. 1989;83:314–316. doi: 10.1172/JCI113875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarsour EH, Kumar MG, Chaudhuri L, Kalen AL, Goswami PC. Redox control of the cell cycle in health and disease. Antioxid. Redox Signal. 2009;11(12):2985–3011. doi: 10.1089/ars.2009.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klebanoff SJ. Myeloperoxidase: friend and foe. J. Leukoc. Biol. 2005;77(5):598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- 46.Piedrafita FJ, Molander RB, Vansant G, Orlova EA, Pfahl M, Reynolds WF. An Alu element in the myeloperoxidase promoter contains a composite SP1-thyroid hormone-retinoic acid response element. J. Biol. Chem. 1996;271(24):14412–14420. doi: 10.1074/jbc.271.24.14412. [DOI] [PubMed] [Google Scholar]
- 47.Osterreicher CH, Datz C, Stickel F, et al. Association of myeloperoxidase promotor polymorphism with cirrhosis in patients with hereditary hemochromatosis. J. Hepatol. 2005;42(6):914–919. doi: 10.1016/j.jhep.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 48.Ahn J, Gammon MD, Santella RM, et al. Myeloperoxidase genotype, fruit and vegetable consumption, and breast cancer risk. Cancer Res. 2004;64(20):7634–7639. doi: 10.1158/0008-5472.CAN-04-1843. [DOI] [PubMed] [Google Scholar]
- 49.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283(5407):1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 50.Sutton A, Khoury H, Prip-Buus C, Cepanec C, Pessayre D, Degoul F. The Ala16Val genetic dimorphism modulates the import of human manganese superoxide dismutase into rat liver mitochondria. Pharmacogenetics. 2003;13(3):145–157. doi: 10.1097/01.fpc.0000054067.64000.8f. [DOI] [PubMed] [Google Scholar]
- 51.Sutton A, Imbert A, Igoudjil A, et al. The manganese superoxide dismutase Ala16Val dimorphism modulates both mitochondrial import and mRNA stability. Pharmacogenet. Genomics. 2005;15(5):311–319. doi: 10.1097/01213011-200505000-00006. [DOI] [PubMed] [Google Scholar]
- 52.Perera CS, St Clair DK, Mcclain CJ. Differential regulation of manganese superoxide dismutase activity by alcohol and TNF in human hepatoma cells. Arch. Biochem. Biophys. 1995;323(2):471–476. doi: 10.1006/abbi.1995.0069. [DOI] [PubMed] [Google Scholar]
- 53.Macmillan-Crow LA, Crow JP, Thompson JA. Peroxynitrite-mediated inactivation of manganese superoxide dismutase involves nitration and oxidation of critical tyrosine residues. Biochemistry. 1998;37(6):1613–1622. doi: 10.1021/bi971894b. [DOI] [PubMed] [Google Scholar]
- 54.Yang M, Cobine PA, Molik S, et al. The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J. 2006;25(8):1775–1783. doi: 10.1038/sj.emboj.7601064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nahon P, Charnaux N, Friand V, et al. The manganese superoxide dismutase Ala16Val dimorphism modulates iron accumulation in human hepatoma cells. Free Radic. Biol. Med. 2008;45(9):1308–1317. doi: 10.1016/j.freeradbiomed.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 56.Lambrecht RW, Sterling RK, Naishadham D, et al. Iron levels in hepatocytes and portal tract cells predict progression and outcomes of patients with advanced chronic hepatitis C. Gastroenterology. 2011;140(5):1490–1500. e1493. doi: 10.1053/j.gastro.2011.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korenaga M, Wang T, Li Y, et al. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J. Biol. Chem. 2005;280(45):37481–37488. doi: 10.1074/jbc.M506412200. [DOI] [PubMed] [Google Scholar]
- 58.Choi J, Lee KJ, Zheng Y, Yamaga AK, Lai MM, Ou JH. Reactive oxygen species suppress hepatitis C virus RNA replication in human hepatoma cells. Hepatology. 2004;39(1):81–89. doi: 10.1002/hep.20001. [DOI] [PubMed] [Google Scholar]
- 59.Fillebeen C, Pantopoulos K. Iron inhibits replication of infectious hepatitis C virus in permissive Huh7.5.1 cells. J. Hepatol. 2010;53(6):995–999. doi: 10.1016/j.jhep.2010.04.044. [DOI] [PubMed] [Google Scholar]
- 60.Bjelakovic G, Nikolova D, Gluud LL, Simonetti RG, Gluud C. Mortality in randomized trials of antioxidant supplements for primary and secondary prevention: systematic review and meta-analysis. JAMA. 2007;297(8):842–857. doi: 10.1001/jama.297.8.842. [DOI] [PubMed] [Google Scholar]
- 61.Park EJ, Lee JH, Yu GY, et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140(2):197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stauffer JK, Scarzello AJ, Jiang Q, Wiltrout RH. Chronic inflammation, immune escape, and oncogenesis in the liver: a unique neighborhood for novel intersections. Hepatology. 2012;56(4):1567–1574. doi: 10.1002/hep.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chiba T, Marusawa H, Ushijima T. Inflammation-associated cancer development in digestive organs: mechanisms and roles for genetic and epigenetic modulation. Gastroenterology. 2012;143(3):550–563. doi: 10.1053/j.gastro.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 64.Yang Y, Luo C, Feng R, Bi S. The TNF-alpha, IL-1B and IL-10 polymorphisms and risk for hepatocellular carcinoma: a meta-analysis. J. Cancer Res. Clin. Oncol. 2011;137(6):947–952. doi: 10.1007/s00432-010-0959-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gleeson D, Bradley MP, Jones J, et al. Cytokine gene polymorphisms in heavy drinkers with and without decompensated liver disease: a case–control study. Am. J. Gastroenterol. 2008;103(12):3039–3046. doi: 10.1111/j.1572-0241.2008.02150.x. [DOI] [PubMed] [Google Scholar]
- 66.Marcos M, Gomez-Munuera M, Pastor I, Gonzalez-Sarmiento R, Laso FJ. Tumor necrosis factor polymorphisms and alcoholic liver disease: a HuGE review and meta-analysis. Am. J. Epidemiol. 2009;170(8):948–956. doi: 10.1093/aje/kwp236. [DOI] [PubMed] [Google Scholar]
- 67.Tarhuni A, Guyot E, Rufat P, et al. Impact of cytokine gene variants on the prediction and prognosis of hepatocellular carcinoma in patients with cirrhosis. J. Hepatol. 2014;61(2):342–350. doi: 10.1016/j.jhep.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 68.Blanc P, Etienne H, Daujat M, et al. Mitotic responsiveness of cultured adult human hepatocytes to epidermal growth factor, transforming growth factor alpha, and human serum. Gastroenterology. 1992;102(4 Pt 1):1340–1350. [PubMed] [Google Scholar]
- 69.Schiffer E, Housset C, Cacheux W, et al. Gefitinib, an EGFR inhibitor, prevents hepatocellular carcinoma development in the rat liver with cirrhosis. Hepatology. 2005;41(2):307–314. doi: 10.1002/hep.20538. [DOI] [PubMed] [Google Scholar]
- 70.Shahbazi M, Pravica V, Nasreen N, et al. Association between functional polymorphism in EGF gene and malignant melanoma. Lancet. 2002;359(9304):397–401. doi: 10.1016/S0140-6736(02)07600-6. [DOI] [PubMed] [Google Scholar]
- 71.Abu Dayyeh BK, Yang M, Fuchs BC, et al. A functional polymorphism in the epidermal growth factor gene is associated with risk for hepatocellular carcinoma. Gastroenterology. 2011;141(1):141–149. doi: 10.1053/j.gastro.2011.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boisvert J, Kunkel EJ, Campbell JJ, Keeffe EB, Butcher EC, Greenberg HB. Liver-infiltrating lymphocytes in end-stage hepatitis C virus: subsets, activation status, and chemokine receptor phenotypes. J. Hepatol. 2003;38(1):67–75. doi: 10.1016/s0168-8278(02)00328-8. [DOI] [PubMed] [Google Scholar]
- 73.Yoong KF, Afford SC, Jones R, et al. Expression and function of CXC and CC chemokines in human malignant liver tumors: a role for human monokine induced by gamma-interferon in lymphocyte recruitment to hepatocellular carcinoma. Hepatology. 1999;30(1):100–111. doi: 10.1002/hep.510300147. [DOI] [PubMed] [Google Scholar]
- 74.Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449(7162):557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 75.Iida N, Nakamoto Y, Baba T, et al. Antitumor effect after radiofrequency ablation of murine hepatoma is augmented by an active variant of CC Chemokine ligand 3/macrophage inflammatory protein-1alpha. Cancer Res. 2010;70(16):6556–6565. doi: 10.1158/0008-5472.CAN-10-0096. [DOI] [PubMed] [Google Scholar]
- 76.Yuan JM, Lu SC, Van Den Berg D, et al. Genetic polymorphisms in the methylenetetrahydrofolate reductase and thymidylate synthase genes and risk of hepatocellular carcinoma. Hepatology. 2007;46(3):749–758. doi: 10.1002/hep.21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu C, Wang H, Pan C, Shen J, Liang Y. CYP2E1 PstI/RsaI polymorphism and interaction with alcohol consumption in hepatocellular carcinoma susceptibility: evidence from 1,661 cases and 2,317 controls. Tumour Biol. 2012;33(4):979–984. doi: 10.1007/s13277-012-0326-2. [DOI] [PubMed] [Google Scholar]
- 78.Xu Y, Liu L, Liu J, et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int. J. Cancer. 2011;128(2):412–417. doi: 10.1002/ijc.25342. [DOI] [PubMed] [Google Scholar]
- 79.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tian C, Stokowski Rp, Kershenobich D, Ballinger Dg, Hinds Da. Variant in PNPLA3 is associated with alcoholic liver disease. Nat. Genet. 2010;42(1):21–23. doi: 10.1038/ng.488. [DOI] [PubMed] [Google Scholar]
- 81.Yuan X, Waterworth D, Perry JR, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am. J. Hum. Genet. 2008;83(4):520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He S, Mcphaul C, Li JZ, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J. Biol. Chem. 2010;285(9):6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sookoian S, Pirola Cj. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53(6):1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 84.Speliotes EK, Butler JL, Palmer CD, Voight BF, Hirschhorn JN. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52(3):904–912. doi: 10.1002/hep.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valenti L, Rumi M, Galmozzi E, et al. Patatin-like phospholipase domain-containing 3 I148M polymorphism, steatosis, and liver damage in chronic hepatitis C. Hepatology. 2011;53(3):791–799. doi: 10.1002/hep.24123. [DOI] [PubMed] [Google Scholar]
- 86.Trepo E, Pradat P, Potthoff A, et al. Impact of patatin-like phospholipase-3 (rs738409 C>G) polymorphism on fibrosis progression and steatosis in chronic hepatitis C. Hepatology. 2011;54(1):60–69. doi: 10.1002/hep.24350. [DOI] [PubMed] [Google Scholar]
- 87.Stickel F, Buch S, Lau K, et al. Genetic variation in the PNPLA3 gene is associated with alcoholic liver injury in caucasians. Hepatology. 2011;53(1):86–95. doi: 10.1002/hep.24017. [DOI] [PubMed] [Google Scholar]
- 88.Trepo E, Gustot T, Degre D, et al. Common polymorphism in the PNPLA3/adiponutrin gene confers higher risk of cirrhosis and liver damage in alcoholic liver disease. J. Hepatol. 2011;55(4):906–912. doi: 10.1016/j.jhep.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 89.Corradini SG, Burza MA, Molinaro A, Romeo S. Patatin-like phospholipase domain containing 3 sequence variant and hepatocellular carcinoma. Hepatology. 2011;53(5):1776. doi: 10.1002/hep.24244. author reply 1777. [DOI] [PubMed] [Google Scholar]
- 90.Singal AG, Manjunath H, Yopp AC, et al. The effect of PNPLA3 on fibrosis progression and development of hepatocellular carcinoma: a meta-analysis. Am. J. Gastroenterol. 2014;109(3):325–334. doi: 10.1038/ajg.2013.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Valenti L, Al-Serri A, Daly AK, et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51(4):1209–1217. doi: 10.1002/hep.23622. [DOI] [PubMed] [Google Scholar]
- 92.Nkontchou G, Bastard JP, Ziol M, et al. Insulin resistance, serum leptin, and adiponectin levels and outcomes of viral hepatitis C cirrhosis. J. Hepatol. 2011;53(5):827–833. doi: 10.1016/j.jhep.2010.04.035. [DOI] [PubMed] [Google Scholar]
- 93.Pirazzi C, Valenti L, Motta BM, et al. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum. Mol. Genet. 2014;23(15):4077–4085. doi: 10.1093/hmg/ddu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fields AL, Soprano DR, Soprano KJ. Retinoids in biological control and cancer. J. Cell Biochem. 2007;102(4):886–898. doi: 10.1002/jcb.21530. [DOI] [PubMed] [Google Scholar]
- 95.Muto Y, Moriwaki H, Saito A. Prevention of second primary tumors by an acyclic retinoid in patients with hepatocellular carcinoma. N. Engl. J. Med. 1999;340(13):1046–1047. doi: 10.1056/NEJM199904013401315. [DOI] [PubMed] [Google Scholar]
- 96.Speliotes EK, Yerges-Armstrong LM, Wu J, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7(3):e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Boyault S, Rickman DS, De Reynies A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45(1):42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 98.Nault JC, De Reynies A, Villanueva A, et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145(1):176–187. doi: 10.1053/j.gastro.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 99.Derambure C, Coulouarn C, Caillot F, et al. Genome-wide differences in hepatitis C- vs alcoholism-associated hepatocellular carcinoma. World J. Gastroenterol. 2008;14(11):1749–1758. doi: 10.3748/wjg.14.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]