Abstract
Cholangiocarcinoma (CCA) is a devastating malignancy with high mortality, in part due to the combination of late presentation, significant diagnostic challenges and limited effective treatment options. Late presentation and diagnosis contribute to the high mortality in CCA and there is an urgent unmet need for diagnostic and prognostic biomarkers to facilitate early diagnosis and treatment stratification to improve clinical outcomes. MiRs are small ncRNA molecules that regulate gene expression and modulate both tumor suppressive and oncogenic pathways. They have a well-defined role in carcinogenesis, including CCA. In this review, we outline the evidence for MiRs in the pathogenesis of CCA and their potential utility as diagnostic and prognostic biomarkers to guide clinical management.
KEYWORDS : biomarkers, carcinogenesis, cell growth, hepatobiliary malignancy, liver cancer, oncogenesis, prognosis markers, signaling pathways
Practice points.
MiRs directly influence the translation of thousands of genes involved in cellular proliferation, differentiation, migration and invasiveness.
MiR expression can be altered via various mechanisms in cancer, including MiR gene mutation and methylation, altered MiR gene expression through transcription factors and post-translational events, such as MiR cleavage and transport.
MiR alteration contributes to cholangiocarcinogenesis via increased cell proliferation, reduced cell apoptosis, increased inflammation-mediated oncogenesis and enhanced cell migration and invasiveness.
MiRs may also play a role in tumor sensitivity and resistance to chemotherapy.
Microarray studies suggest the strongest evidence for upregulation of MiR21, MiR223 and MiR625 and downregulation of MiR145, MiR200c, MiR222, MiR338 and let-7a in human cholangiocarcinoma tissues compared with healthy biliary tissue.
While theoretically MiRs are attractive biomarkers for diagnosis and prognosis in cholangiocarcinoma, current evidence for their clinical utility at the bedside is limited and further validation studies are warranted.
Background
Cholangiocarcinoma (CCA) is a devastating malignancy with high mortality, in part due to the combination of late presentation, significant diagnostic challenges and limited effective treatment options [1,2]. CCA develops from the ductular biliary epithelium either within the liver (intrahepatic) or extrahepatic biliary ducts (extrahepatic). It is the second most common primary liver cancer, accounting for 3% of all gastrointestinal malignancies [3,4]. Of concern, the incidence and mortality of intrahepatic CCA appears to be increasing, despite a reduction in extrahepatic CCA globally [2,4–6].
CCA often presents a diagnostic dilemma as the clinical picture and radiological appearance may mimic other common conditions, such as benign biliary strictures, autoimmune cholangiopathy, primary sclerosing cholangitis, gallbladder cancer and hepatocellular carcinoma [2,7]. The intense desmoplastic reaction in perihilar and distal ductal CCA and the location of these cancers makes obtaining biopsies of malignant tissue challenging, especially in patients with background primary sclerosing cholangitis [7]. Moreover, brush tissue specimens only have a sensitivity of 20–40% for CCA diagnosis [3] and current blood biomarkers, such as CA 19-9 and carcinoembryonic antigen, have limited diagnostic accuracy for CCA [8,9]. Late presentation and diagnosis contribute to the high mortality in CCA and there is an urgent unmet need for diagnostic and prognostic biomarkers to facilitate early diagnosis and treatment stratification.
Within this context, there has been considerable research interest in MiRs as potential biomarkers in CCA. MiRs are small ncRNA molecules that regulate gene expression through direct binding to mRNA gene transcripts, preventing translation [10]. They operate through either tumor suppressive or oncogenic pathways and have a well-defined role in carcinogenesis, with considerable evidence for their role in many human malignancies, including lung, breast, prostate and GI tract cancers [10,11]. Emerging evidence also suggests an important role for MiRs in CCA [12,13].
In this review, we outline the evidence for MiRs in the pathogenesis of CCA, defining their central role in key oncogenic pathways and the effects of individual MiRs commonly altered in CCA. The potential utility of MiRs as diagnostic and prognostic biomarkers to guide clinical management is also evaluated.
The role of miRNAs in the pathogenesis of malignancy
MiRs are small, ncRNA molecules that bind to the 3′ untranslated region of specific mRNAs, targeting them for degradation and therefore preventing gene translation [10]. Thirty percent of MiRs are derived from processed introns of protein-coding genes, with the remainder encoded by specific genes [10]. A primary MiR transcript (pri-MiR) is transcribed by RNA polymerase II in the nucleus, then undergoes further modification and cleavage by microprocessor to produce one or more MiRs [14,15]. Each MiR is regulated by multiple other MiRs, with each MiR in turn inhibiting translation of thousands of mRNA transcripts [10,16]. Functionally, MiRs exert modest effects on multiple targets along a given signaling pathway, allowing refined and orchestrated control of cell processes such as cell proliferation and differentiation, survival and metastatic capacity [10,17–19].
Since the first description of MiR-15 and MiR-16 deletion in B cell lymphocytic leukemia [20], a myriad of publications have defined the key role of MiRs in many tumor types [10,20–22]. Although MiRs may function as either tumor suppressors (such as let-7 class inhibition of c-Myc) [23] or oncogenes (such as MiR-17-92 cluster activation of c-Myc) [24], MiRs have a predominantly tumor-suppressor role in cancer [10,25–26].
Mechanisms of altered MiR biogenesis in cancer
Several mechanisms exist that alter MiR expression in cancer. MiR genes are frequently located in fragile genomic regions with high mutation rates in malignancy [10,27]. Hypermethylation of CpG islands located in MiR genes also suppress MiR transcription in some cancers [28,29]. Cell singling pathways, such as c-Myc, modulate MiR expression [30–35]. Other steps in the biosynthesis and regulation of MiRs, such as pri-MiR processing, cleavage and export from the cell nucleus and hypoxia-induced stress responses in rapidly growing tumors mediated via EGFR signaling also orchestrate suppression of MiR transcription in human cancers [10,15,36–37].
Role of MiRs in the pathogenesis of CCA
• MiR expression profiles in CCA
Several studies describe MiR gene expression profiles in CCA using targeted PCR or untargeted MiR microarray techniques [38–43]. A summary of these studies is outlined in Table 1. While the findings reported in these studies diverge, there are nine MiRs differentially expressed in CCA compared with controls that were identified in more than one study. Mir21 was upregulated in both a cell line and 2 human CCA tissue studies; MiR223 and MiR625 were upregulated in two human CCA tissue studies; whereas MiR145, MiR200c, MiR222, MiR338, MiR451 and let-7a were all downregulated in two human CCA studies.
Table 1. . Summary of miRNAs expression studies in cholangiocarcinoma.
| Study (year) | MiRs upregulated in CCA compared to controls | MiRs downregulated in CCA | Study CCA model | Ref. |
|---|---|---|---|---|
| Kawahigashi et al. (2009) | 22, 125a, 127, 199a, 214, 376a, 424 | CCA cell line (TFK, HuCCT1, MEC) | [40] | |
| Meng et al. (2006) | 21†, 200b, 141 | CCA cell line (KMCH-1, Mz-ChA-1, TFK) | [41] | |
| Yang et al. (2014) | 29b-1, 7, 34b, 146a, 192, 194, 203, 215, 221, 361-3p, 375, 582-5p, 625†, 892b | 135b, 144, 200a/b/c†, 204, 429, 451†, 486-5p, 495, 513a-5p, 1826 | Human CCA tissue (three CCA, three matched normal) | [42] |
| Zhang et al. (2015) | 92b-5p, 188-5p, 331-3p 423-5p, 491-5p, 566, 612, 625-3p†, 675-5p, 765 | 19b-3p, 24-3p, 26a-5p, 29a, 29c-3p, 99a-5p, 100-5p, 101-3p, 103a-3p, 130a-3p, 141-3p, 143-3p, 338-3p†, 451a†, 497-5p, let-7a†, let-7d, let-7e, let-7f | Human CCA tissue (66 CCA, nine matched normal) | [39] |
| Karakatsanis et al. (2013) | 21†, 31, 223† | 122, 145†, 200c†, 221, 222† | Human CCA tissue (21 CCA, 60 HCC, 98 controls) | [38] |
| Chen et al. (2009) | 15a, 15b, 17-5p, 17-3p, 19a, 20, 21†, 25, 103, 106a, 130b, 142-3p, 193, 223†, 224, 331, 324-5p, 374 | 98, 145†, 184, 185, 197, 198, 200c†, 204, 214, 222†, 302b, 302d, 320, 328, 337, 338†, 371, 373, let-7a†, let-7b | Human CCA tissue (27 CCA, 18 controls) | [43] |
†miRNA with evidence for a role in CCA from more than one study.
CCA: Cholangiocarcinoma; HCC: Hepatocellular carcinoma.
The largest study by Zhang et al. [39] included 66 CCA tissue samples and nine matched adjacent nonmalignant biliary tissue controls, however, the small number of controls compared with cases may have reduced power to detect differences in expression profile between cases and controls. Moreover, qPCR confirmation was only performed for three MiRs combined as a prognostic score for survival, not to confirm all 29 differentially expressed MiRs identified by microarray [39]. Chen et al. [43] identified 38 differentially expressed MiRs in CCA compared with controls in a study, which included 27 CCA tissue samples and 18 biliary tissue samples from healthy controls, allowing premalignant or ‘at-risk’ MiR expression changes associated with underlying disease to be excluded. Functional assays also confirmed the significance of miR204 and 320 modulation in CCA cell models. Karakatsanis et al. [38] evaluated expression of nine MiRs by targeted PCR and included biliary tissue samples from 21 CCA, 60 HCC and 98 healthy controls to allow comparison of MiR expression between HCC and intrahepatic CCA, which often poses a difficult clinical diagnostic scenario. Notably, none of these studies included subpopulations commonly presenting as a differential diagnosis, such as PSC or biliary obstruction.
Involvement of individual MiRs in CCA carcinogenesis pathways
Further studies have focused on the functional roles of individual MiRs in CCA. An outline of available evidence from human studies for the roles of specific MiRs in CCA is presented in Table 2 and Figure 1 outlines the main MiRs associated with key processes of carcinogenesis in CCA. It is important to note that for many MiRs there is only one study that has reported their significance in CCA. Overall, there were ten MiRs upregulated (MiR21, 25, 29, 31, 126, 199, 210, 223, 331-3p and 625) and 18 MiRs downregulated (MiR26a, 34a, 101-3p, 103a-3p, 127, 130a, 141, 145, 451, 200b and c, 204, 221, 222, 338-3p, 370, 373, 494 and let-7a) in CCA for which there were published evidence from more than one study.
Table 2. . Summary of evidence for specific miRNA expression changes in cholangiocarcinoma that has been demonstrated in human studies.
| MiR | Expression in CCA | MiR target | Clinical association | Sample size | Study model | Ref. |
|---|---|---|---|---|---|---|
| 15 (a & b) | Increased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 16 | Increased | Diagnostic utility | 46 CCA; 50 controls | Human CCA | [44] | |
| 17 (3p & 5p) | Increased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 19a, b | 19a increased (39) 19b decreased (39) | 66 CCA; nine controls | Human CCA | [39] | ||
| 19b | Decreased | 66 CCA; nine controls | Human CCA | [39] | ||
| 20 | Increased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 21 | Increased | PTEN, PDGH | Intrahepatic CCA (87); survival (70, 87) | 18–27 CCA; 18–98 controls | CCA cell lines; murine models; human CCA | [38,41,43,45–51] |
| 23b | Increased | High CA19-9 | 27 CCA; 18 controls | Human CCA | [43] | |
| 24-3p | Decreased | 66 CCA; nine controls | Human CCA | [39] | ||
| 25 | Increased | TRAIL-mediated apoptosis/DR4 | 27 CCA; 18 controls | CCA cell lines; human CCA | [43,52] | |
| 26a | Decreased (39); Increased (52) | 21–66 CCA; nine controls | CCA cell lines; human CCA | [39,53] | ||
| 29 | Increased (74); decreased 29b (74), 29c (39, 46) | Mcl-1 | 66 CCA; nine controls | CCA cell lines; human CCA | [39,54–55] | |
| 30c | Increased | High CA19-9 | 27 CCA; 18 controls | Human CCA | [43] | |
| 31 | Increased | RASA1 | 21 CCA; 60 HCC; 98 controls | CCA cell lines; human CCA | [38,56] | |
| 34a | Decreased | c-Myc | Murine; hamster models; cell lines | [57–60] | ||
| 92b-5p | Increased | 66 CCA; nine controls | Human CCA | [39] | ||
| 98 | Decreased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 99a-5p | Decreased | 66 CCA; nine controls | Human CCA | [39] | ||
| 100-5p | Decreased | 66 CCA; nine controls | Human CCA | [39] | ||
| 101-3p | Decreased | VEGF | 46–66 CCA; nine controls | Human CCA | [39,61] | |
| 103a-3p | Decreased | 27–66 CCA; 9–18 controls | Human CCA | [39,43] | ||
| 106a | Increased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 122 | Decreased | 21 CCA; 60 HCC; 98 controls | Human CCA | [38] | ||
| 126 | Increased | Survival; high CA19-9 (43) | 27–32 CCA; 18–32 controls | Human CCA | [43,62] | |
| 127 | Decreased | 127-4b diagnosis (84) | 46 CCA; 50 controls | CCA cell line; human CCA | [40,44] | |
| 130a, b | 130a decreased (39); 130b increased (43) | 27–66 CCA; 9–18 controls | Human CCA | [39,43] | ||
| 135a, b | Decreased | High CA19-9 | 27 CCA; 18 controls | Human CCA | [43] | |
| 138 | Decreased | Rho C, p-ERK, MMP-2/9 | Metastases | CCA cell lines; human CCA | [63] | |
| 141 | Increased (41); decreased (39, 82) | CLOCK | High CA19-9 (43); progenitor-cluster CCA (82) | 27–91 CCA; 18 controls | CCA cell lines; human CCA | [39,41,43,64] |
| 142-3p | Increased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 143-3p | Decreased | 66 CCA; nine controls | Human CCA | [39] | ||
| 144 | Decreased | Three CCA; three controls | Human CCA | [42] | ||
| 145 | Decreased | 21–27 CCA; 18–98 controls | Human CCA | [38,43] | ||
| 150 | Increased | Diagnostic utility | 53 CCA | CCA cell lines; human CCA | [49] | |
| 151-3p | Decreased | Survival | 32 CCA; 32 controls | Human CCA | [62] | |
| 182 | Decreased | High CA19-9 | 27 CCA; 18 controls | Human CCA | [43] | |
| 184 | Decreased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 185 | Decreased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 188-5p | Increased | 66 CCA; nine controls | Human CCA | [39] | ||
| 191 | Increased | Diagnostic utility | 46 CCA; 50 controls | Human CCA | [44] | |
| 192 | Increased | Opisthorcis infection; survival | 11 CCA; nine controls | Human CCA | [65] | |
| 193 | Increased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 195 | Increased | High CA19-9 | 27 CCA; 18 controls | Human CCA | [43] | |
| 197 | Decreased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 198 | Decreased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 199a, b | Increased; 199a decreased (40) | High CA19-9 | CCA Cell line; human CCA | [40,43] | ||
| 200b, c | 200b increased (41); 200c decreased (38, 43, 60, 61, 82) | ZEB1/2; SUZ12/ROCK2 | Progenitor cluster CCA (82); high CA19-9 (43) | 14–27 CCA; 18–98 controls | CCA cell lines; human CCA | [38,41,43,64,66–67] |
| 204 | Decreased (43, 91) | Bcl-1 | High CA19-9 (43) | 27–30 CCA; 18 controls | Human CCA | [43,68] |
| 210 | Increased (50); decreased (43) | Mnt | High CA19-9 (43) | 27 CCA; 18 controls | Human CCA | [43,60] |
| 214 | Decreased | Twist | Metastases | 27 CCA; 18 controls | CCA cell line; human CCA | [40,43,69] |
| 221 | Decreased | PIK3R1 | Sensitivity to gemcitabine | 21 CCA; 60 HCC; 98 controls | CCA cell line; human CCA | [38,55] |
| 222 | Decreased | 21–27 CCA; 18–98 controls | Human CCA | [38,43] | ||
| 223 | Increased (38, 43) | High CA19-9 (43) | 21–27 CCA; 18–98 controls | Human CCA | [38,43] | |
| 224 | Increased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 302b,d | Decreased | High CA19-9 | 27 CCA; 18 controls | Human CCA | [43] | |
| 320 | Decreased | Mcl-1 | 27 CCA; 18 controls | Human CCA | [43] | |
| 324-5p | Increased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 328 | Decreased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 331-3p | Increased | 27–66 CCA; 9–18 controls | Human CCA | [39,43] | ||
| 337 | Decreased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 338-3p | Decreased | 27–66 CCA, 9–18 controls | Human CCA | [39,43] | ||
| 370 | Decreased | 34 CCA; 24 controls | CCA cell lines; human CCA | [70,71] | ||
| 371 | Decreased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 373 | Decreased | RASSF1A; MBD2 | Survival | 27 CCA; 18 controls | CCA cell lines; human CCA | [43,72] |
| 374 | Increased | 27 CCA; 18 controls | Human CCA | [43] | ||
| 388-3p | Decreased | Survival | 66 CCA; nine controls | Human CCA | [39] | |
| 423-5p | Increased | 66 CCA; nine controls | Human CCA | [39] | ||
| 429 | Decreased | 14 CCA | Human CCA | [66] | ||
| 451a | Decreased | 66 CCA; nine controls | Human CCA | [39] | ||
| 484 | Decreased | Diagnostic utility | 46 CCA; 50 controls | Human CCA | [44] | |
| 486-3p | Increased | Diagnostic utility | 46 CCA; 50 controls | Human CCA | [44] | |
| 491-5p | Increased | 66 CCA; nine controls | Human CCA | [39] | ||
| 494 | Decreased | Cell cycle arrest (G2) via CCNB1, CDC20, TOP2A | 43 CCA; 30 controls | CCA cell lines; human CCA | [73,74] | |
| 566 | Increased | 66 CCA; nine controls | Human CCA | [39] | ||
| 612 | Increased | 66 CCA; nine controls | Human CCA | [39] | ||
| 765 | Increased | 66 CCA; nine controls | Human CCA | [39] | ||
| 497-5p | Decreased | 66 CCA; nine controls | Human CCA | [39] | ||
| 625-3p | Increased | 66 CCA; nine controls | Human CCA | [39] | ||
| 652-3p | Decreased | Survival | 66 CCA; nine controls | Human CCA | [39] | |
| 675-5p | Increased | Survival | 66 CCA; nine controls | Human CCA | [39] | |
| 886 | Decreased | PKR | 16 CCA | CCA cell lines; human CCA | [75] | |
| let-7a-5p, 7d-5p, 7e-5p, 7f-5p | Decreased | c-Myc; NF2 | 27–66 CCA; 9–18 controls | Human CCA | [39,43,57–60] | |
CCA: Cholangiocarcinoma; HCC: Hepatocellular carcinoma.
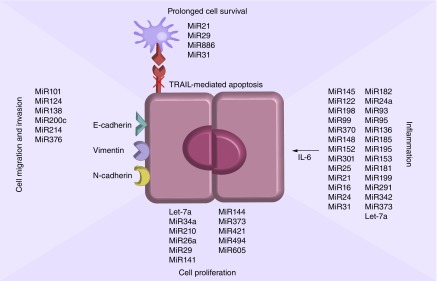
Figure 1. . MiRs associated with cell proliferation, invasiveness and migratory capacity, inflammation-based signaling pathways and reduced cellular apoptosis in cholangiocarcinoma cells.
• Cell proliferation
The c-Myc pathway plays a central role in cholangiocarcinogenesis. c-Myc is a basic helix-loop leucine zipper transcription factor that activates transcription of many genes involved in cell proliferation [76] and also drives cyclin D1 signaling and cell de-differentiation in CCA [76,77]. c-Myc forms a heterodimer with to activate gene transcription, though Max also complexes with the c-Myc inhibitor Mnt to antagonize c-Myc transcription [76]. Additionally, c-Myc signaling, along with Toll-like receptor and hedgehog pathway signaling, appears to protect CCA cells from TRAIL-mediated apoptosis [54].
MiR34a and the let-7 family inhibit c-Myc transcription and are both downregulated in CCA compared with healthy controls [57,58], leading to elevated levels of c-Myc [59,60]. Murine DEN and BDL-induced CCA models suggest MiR-34a is the key driver of enhanced c-Myc signaling, with later contributions by let-7 and MiR210 [60]. Similarly, MiR210 is also upregulated in CCA and leads to downregulation of Mnt and, therefore, enhanced c-Myc gene transcription and signaling [60]. HIF-2α induced by tissue hypoxia in CCA appears to drive MiR210-mediated Mnt inhibition [60,78].
MiR26a upregulation also leads to cell proliferation in both CCA cell lines and human CCA tissue samples, via GSK-3β and activation of the β-catenin signaling cascade, including c-Myc, cyclin D1 and ρPAR-γ canonical pathways [53]. Moreover, depletion of β-catenin leads to a reduction in MiR26a-mediated cell proliferation and colony formation [53]. MiR29 expression is also controlled by c-Myc, hedgehog and NF-κB inflammatory signaling pathways and appears downregulated in CCA tissue samples in association with enhanced cell proliferation [54].
During cell division, MiR494 halts progression from G2 to M phase through modulation of PLK1, PTTG1, CCNB1, CDK6, CDC2, CDC20 and TOP2A [73,74].
A study by Olaru et al. [74] identified downregulation of MiR494 in five CCA tissue specimens compared to five normal controls using microarray and confirmatory qPCR, then validated this in a larger set of 43 CCA tissue specimens and 30 normal biliary tissue controls. MiR494 also inhibited CCA cell growth in vivo in murine xenograft models [74]. Others have confirmed downregulation of MiR494 in CCA [73,74].
Additional studies support roles for MiRs in cell proliferation in CCA. MiR141 reduces cell growth via PTEN inhibition [41], and a single study found MiR144 inhibits cell growth via LIS1 and both are downregulated in murine and cell line models of CCA. However, data on MiR141 and 144 from human tissue studies are currently lacking. MiR605 was downregulated in CCA in one study by PSMD10 overexpression and increases cell proliferation [79]. MiR421 was also overexpressed in human CCA in one study compared with normal biliary tissue and inversely correlates with farsenoid X receptor expression, leading to increased cell growth, migration and colony formation [80], whereas MiR421 downregulation leads to cell cycle arrest in CCA cell lines [80]. MiR373 also modulates cell growth and is inactivated in CCA cell lines by MBD2 hypermethylation, being located in a CpG-rich region of the genome [72].
• Cell migration & invasiveness
Cell migration and invasiveness are modified by MiRs and determine metastatic potential of cancer cells. The MiR200 family target ZEB1 and 2, therefore reducing E-cadherin-mediated EMT, cell proliferation, migration and invasion [81,82]. An elegant study by Peng et al. [66] evaluated MiR gene expression in 14 CCA tissue samples using microarray. They found that miR200a/b/c and MiR429 were downregulated in CCA tissue samples compared to healthy controls. Moreover, this group [66] determined that miR200b/c expression increases metastatic potential of cells, by inhibiting cell migration and invasion via interactions with SUZ12 and rho-kinase 2 both in vitro and in vivo, suggesting MiR200b/c are important checkpoints against metastases in CCA [66]. MiR200b/c downregulation drives tumor initiation, sphere formation and drug resistance in CCA [66]; however, the association between metastases and MiR200a/b/c has not been confirmed in clinical studies. Other groups have also confirmed the importance of the MiR200 family in CCA development [38]. MiR200c is downregulated in CCA by the SPRR2-α complex, ZEB1 and CtPB, thereby modulating cell growth and remodeling, particularly in cholangiocytes undergoing wound healing and regeneration [67].
MiR376c is downregulated in CCA cell line HuCCT1 via promotor methylation [83]. MiR376c inhibits transcription of GRB2 and subsequently EGF-dependent cell migration [83]. Downregulation of miR376c may therefore be a mechanism of enhanced metastatic potential in CCA, however, this requires confirmation in human clinical studies.
MiR124 was downregulated in HCV-related intrahepatic CCA in one study. Evidence from human CCA tissue samples suggests that the HCV core protein inhibits DNMT-1 and low levels of MiR124 lead to increased cholangiocyte migration and invasiveness through upregulated SMYD-3 [84].
MiR214 was also downregulated in patients with metastatic CCA compared with nonmetastatic CCA in one study [69]. This leads to increased Twist expression, which in turn reduces E-cadherin expression and enhances EMT, a crucial step in tumors developing metastatic potential [69]. MiR138 was also downregulated in metastatic CCA compared to nonmetastatic disease in a single study, leading to increased proliferation and invasiveness through upregulation of Rho C, p-ERK and MMP-2 and -9 [63].
By contrast, MiR101 inhibits angiogenesis in cholangiocytes, also a key step in metastasis, via VEGF and COX2 transcription inhibition and is downregulated in CCA cell lines and human CCA tissue samples compared with controls [61]. MiR101 downregulation also increases cell proliferation in CCA [61].
• Cellular apoptosis
Another essential step in carcinogenesis is induction of immortality in malignant cells and various MiRs have been shown modulate apoptosis in CCA. Several studies have identified MiR21 upregulation in human CCA tissue samples [41,45–47]. Ars2 is also overexpressed in CCA and has been shown to reduce MiR21 expression in murine knockout models [45]. Overexpression of MiR21 leads to enhanced PTEN-mediated cell proliferation and reduces TIMP3 and PCD4-mediated cell apoptosis [46–48], which is countered by Ars2 [45]. However, in this small study, a direct relationship between Ars2, MiR21 and PTEN and PCD4 expression could not be demonstrated and it may be that Ars2 contributes to, but is not the sole driver of, MiR21-mediated cell proliferation [45]. MiR21 also appears necessary for liver progenitor cell survival, particularly in intrahepatic CCA [48]. Additionally, MiR21 inhibits transcription of 15-PDGH, promoting carcinogenesis through increased prostaglandin E2 (PGE2) signaling [47,49]. PGE2 and COX2, both increased in inflammation, work synergistically as positive feedback stimulants of MiR21 expression [49]. Importantly, MiR21 is also associated with metastatic CCA migration and invasion through reductions in E-cadherin and upregulation of vimentin and N-cadherin, key drivers of EMT [50,85].
MiR29 is strongly expressed in cholangiocytes and binds to Mcl-1, an immortality member of the Bcl-2 family, directly reducing Mcl-1 expression and sensitizing cells to TRAIL-mediated apoptosis [55]. MiR29b is downregulated in human CCA tissue samples compared with nonmalignant biliary tissue, with an inversely correlated upregulation of Mcl-1 and prolonged CCA cell survival [55].
Other routes of cellular immortality through miR modulation in cancer are described. Pri-MiR886 completely suppresses the canonical PKR/elf2-α apoptosis pathway and MiR886 downregulation prolonged cholangiocyte survival through NF-κB signaling in human CCA cells in one study [75]. MiR31 is upregulated in human CCA (cell lines and tissue), reduces cellular apoptosis and increases cell proliferation via inhibition of RASSF1A and subsequent enhanced RAS-MAPK signaling [56].
• MiR expression & inflammation in CCA carcinogenesis
Inflammation is a key driver in oncogenesis, evident in the relationship between chronic inflammatory conditions of the biliary tree, such as primary sclerosing cholangitis, liver fluke infection and CCA development [2,7]. Inflammatory IL-6-driven cell proliferation is well described in CCA [70,71]. Work by Meng et al. [70] demonstrated that overexpression of IL-6 in cholangiocyte cell lines downregulates seven specific MiRs in CCA compared to normal biliary controls: MiR182, MiR 291-5p, MiR145, MiR122a, MiR198, MiR99a and MiR370 [70]. Specifically, MiR370 lies within a CpG island and is also subject to silencing through hypermethylation [70]. MiR370 normally suppresses MAP3K8, therefore, Il-6 mediated suppression of MiR370 increases MAP3K8 signaling in CCA [70]. IL-6 also increases expression of DNMT1, which epigenetically modifies gene expression through hypermethylation of CpG islands, including MiR genes, such as MiR370 [71,86]. In a large study by An et al. [71], MiR370 was silenced through IL-6-dependent gene methylation. Interestingly, in males the MiR370 gene is already silenced, however, in females the miR370 gene is also silenced in CCA compared to healthy controls [71]. This is an important demonstration of tumor suppressor MiR gene imprinting in relation to carcinogenesis [71].
DNMT-1 appears to be a target of MiR 148a, 152 and 301 in CCA, suggesting modification of gene methylation by these MiRs, all of which are downregulated in human CCA tissues compared with normal biliary tissues, CCA cell lines and murine xenograft CCA models [86]. MiR148a and MiR152 also increase tumor suppressor RASA1 and PI6INK4 expression by reducing inflammatory-driven DNMT1-mediated gene methylation [86].
Inflammation-induced hedgehog signaling also stimulates MiR25 expression in CCA cell lines and higher expression of MiR25 is seen in human CCA tissue samples compared to noncancerous biliary tissue samples [52]. Importantly, MiR25 appears to protect CCA cells from TRAIL-mediated apoptosis [52], similar to MiR21-related and MiR210-enhanced c-Myc signaling, thereby reducing TRAIL-mediated apoptosis of cancerous cells. Additionally, TRAIL-DR4 appears to also be a target for MiR25 inihibition, providing further escape for cholangiocytes from TRAIL-mediated destruction [52].
In Clonorchis sinensis infection, where chronic biliary obstruction and inflammation leads to CCA development, in vitro experiments using CCA cell lines have demonstrated that the clonorchis excretory-secretory products upregulate 13 MiRs (MiR16-2, MiR24, MiR31, MiR93, MiR95, MiR136, MiR153, MiR181d, MiR185, MiR195, MiR199a-3p, MiR342-5p and MiR373) and downregulate three MiRs (let-7a, let-7i and MiR124a) compared to controls [87]. These MiRs were involved with cellular proliferation, DNA methylation, inflammation and cell migration [87]. Importantly, these data suggest specific carcinogenic pathways are triggered by infection rather than generic inflammatory pathways [87]. Moreover, downregulation of let-7i also leads to upregulation of TLR4 gene transcription, further augmenting inflammation-mediated cell damage in clonorchis infection [87].
Clinical utility of miRNAs in CCA
• Diagnosis & disease classification
Differential expression of MiRs in CCA compared to healthy controls has encouraged considerable research interest in MiRs as diagnostic biomarkers. In a study by Oishi et al. [64], nanostring transcriptome profiling of 16 intrahepatic CCA specimens and seven combination hepatocellular CCA specimens from Asian patients with early-stage resectable disease were compared to MiR expression profiles of seven matched nontumor biliary specimens and seven benign liver lesion samples (two adenomas, five focal nodular hypeplasias). Among 623 MiR genes differentially expressed between CCA and controls, two distinct miR expression signatures corresponded to two tumor phenotypes [64]. One MiR cluster shared attributes with a previously described progenitor cell hepatocellular carcinoma phenotype [64]. Both MiR141 and MiR200c were downregulated in the stem cell cluster signature and these cells had undergone EMT, whereas CCA cells that had normal MiR200c levels still retained an epithelial phenotype [64]. Interestingly, both miR141 and MiR200c share a gene transcript [64], suggesting a common regulator in CCA, which may be c-Myc or TGF-β given their upstream binding sites.
In the previously described study by Chen et al. [43], an MiR signature was developed that predicted CA19-9 level, itself a negative prognostic marker in CCA [43]. Upregulated MiR126, MiR195, MiR199a and b, MiR 204, MiR23b and MiR30c and downregulation of MiR135a and b, MiR141, MiR182, MiR200c, MiR210, MiR223 and MIR302b were associated with high CA19-9 levels in CCA patients [43]. However, this was not correlated with clinical outcomes.
Few studies have evaluated the diagnostic utility of MiRs. MiR21 was shown to have a sensitivity of 95% and specificity of 100% for distinguishing CCA from normal biliary epithelium in one validation study of 18 CCA samples and 12 controls [46]. The work by Zhang et al. [39] included development of a 30 MiR diagnostic signature with 100% accuracy for determining CCA from normal biliary epithelium [39]. However, in this study there was no validation cohort, the control group was small (nine patients) and overfitting of the diagnostic model is possible [39]. A single small study in 11 opisthorcis-related CCA patients found serum MiR-192 was higher in CCA patients compared to healthy controls, with sensitivity of 74% and specificity of 72% for CCA diagnosis (AUC: 0.803). A further small study of 15 CCA patients determined high expression of MiR150 in both plasma and CCA tissue specimens compared to healthy controls and found the addition of MiR150 expression to CA19-9 levels improved the diagnostic sensitivity and specificity of CA19-9 measurement alone in CCA. However, all these studies had small sample sizes and, therefore, inadequate power to reliably assess diagnostic utility. Furthermore, they lacked validation cohorts [49].
Pancreatic cancer and CCA sometimes pose a diagnostic dilemma as both are associated with extrahepatic stricturing and associated mass lesion, as well as obstructive jaundice and elevated CA19-9 levels [2]. Collins et al. [88], therefore, explored whether a diagnostic MiR signature could be developed to reliably distinguish between pancreatic cancer and CCA. They conducted a small study comparing MiR expression in 31 matched pairs of malignant CCA tissue and adjacent normal biliary tissue from CCA patients, compared to nine matched pairs of pancreatic carcinoma and adjacent noncancerous pancreatic tissue, using the nCounter® gene expression approach (NanoString, WA, USA) [88]. They found 41 MiRs differentially expressed between CCA and normal biliary tissue and 52 differentially expressed MiRs between pancreatic cancer and normal pancreatic tissue [88]. Seven MiRs had opposite patterns of expression between CCA and pancreatic cancer: MiR100, MiR145, MiR125b and MiR127-3p were all downregulated in CCA and upregulated in pancreatic cancer, while MiR30a, MiR96 and MiR30b were all upregulated in CCA but downreglated in pancreatic cancer [88]. These data suggest MiR profiling may be useful for distinguishing pancreatic cancer and CCA, however, this pilot study was small and diagnostic accuracy was not adequately assessed.
One important question regarding the future clinical use of MiRs is where they should be measured. MiRs are present in various biofluids, therefore offering a minimally invasive method to measure CCA behavior compared with tissue biopsy. Unfortunately, biliary RNA degrades very quickly at room temperature, complicating their measurement in bile [44]. Within this context, a very interesting paper by Li et al. [44] offers a potential solultion by demonstrating stable MiR expression within extracellular biliary vesicles in human bile by microarray. They found MiR191, MiR486-3p, MiR127-4b, MiR16 and MiR 484 had sensitivity of 67% and specificity of 96% for distinguishing human CCA from normal controls in a cohort including 13 PSC patients, 16 patients with biliary obstruction and three patients with bile leak syndromes [44]. This study had a high NPV for CCA diagnosis even in a patient sample that included important differential diagnoses for CCA, therefore, these data are very promising and further validation is required.
• MiRs as prognostic & treatment stratification biomarkers in CCA
A handful of studies have assessed the prognostic utility of MiRs in CCA. The strongest survival data come from a study by Oishi et al. [64], where a stem cell/progenitor cell-like MiR profile in CCA including downregulation of MiR200c was associated with reduced survival in both their original 23-Asian test cohort and a validation cohort of 68 Caucasian CCA subjects [64]. NCAM1 levels also correlated with survival in CCA and inversely correlated with MiR200c levels, being a direct target of miR200c [64]. These findings are logical given NCAM1 is highly expressed in hepatic progenitor cells and is essential for EMT [89].
Zhang et al. [39] found the combination of upregulated MiR675-5p and downregulated MiR652-3p and MiR388-3p independently predicted overall survival in CCA with moderate accuracy (AUC: 0.747) [39]. McNally et al. [62] also used the NanoString gene expression method in 32 matched pairs of CCA and adjacent normal biliary tissue specimens and found upregulation of MiR151-3p and MiR126 in CCA cells were both independently associated with prolonged survival [62]. However, no functional role for these MiRs has been confirmed in CCA and there was no control group in the second study [39,62]. MiR21 upregulation is associated with reduced survival and lymph node metastases in CCA [47–48,51]. Though sample size in all three studies was small, functional studies provide a likely mechanism to support a prognostic role for MiR21 [46–48].
More specifically, in hilar CCA, one group reported MiR373 downregulation is associated with reduced overall survival, advanced tumor clinical stage and de-differentiation of CCA cells to a more primitive and aggressive cell type [72,90]. MiR192 overexpression was also associated with reduced survival and lymph node metastases in one small study of 11 patients with opisthorcis-related CCA [65].
With respect to treatment response, an interesting study by Okamoto et al. [91] determined that MiR29b, MiR205 and MiR221 expression was higher in HuCCT1 cells, which are sensitive to gemcitabine compared to HuH28 cells. Furthermore, ectopic expression of MiR29b, MiR205 and MiR221 restored gemcitabine sensitivity in HuH28 cells [91]. A further study found MiR21 and MiR200b expression enhances sensitivity of CCA cell lines to gemcitabine via PTEN and importantly that gemcitabine exposure of CCA cell lines in vitro alters MiR expression and drug-induced cell death [41]. These studies describe potential mechanisms of drug sensitivity and resistance in CCA and further validation studies in human CCA tissue and normal biliary tissue specimens are warranted.
Therapeutic potential of miRNAs in CCA
While miRNAs have shown considerable potential as biomarkers in CCA, the ability to delicately control cell processes through miRNA manipulation also has obvious therapeutic appeal, particularly for miRNAs with a tumor-suppressor role. There is preliminary evidence of the therapeutic benefit of miRNAs in some cardiovascular diseases, including atherosclerosis and lipid metabolism [92], however to date they have not been trialed in human cancer patients. miRNAs with specific inhibitory roles in oncogenesis and proven low levels in CCA could have therapeutic potential through restoration of intrinsic levels. For example, MiR34a and the let-7 family inhibit c-Myc transcription [57,58], thereby providing a potential way to inhibit CCA growth. An alternative use would be to restore chemotherapy responsiveness, for example by using therapeutic MiR21 and MiR200b, the expression of which is associated with greater response to gemcitabine-based chemotherapy in CCA cells [41]. However, the therapeutic role of miRNAs in CCA has not been evaluated and further studies exploring this important arena are urgently needed.
Limitations in the current literature & future directions
While the data presented in this review shed light on potential signaling pathways in CCA modulated by MiRs, the majority of studies have small samples sizes and are likely to be underpowered, particularly for investigating the clinical significance of MiRs in CCA, and the data require validation in larger human tissue sample studies. Similarly, inadequate clinical information prevents evaluation of bias due to important confounding variables, such as age, race and other carcinogens, such as tobacco. Contrasting MiR expression in CCA patients with different disease etiologies would be useful to glean a detailed understanding of carcinogenesis in different disease contexts, such as PSC, toxin-related or liver fluke-associated CCA compared to sporadic. Evaluation of potential diagnostic and prognostic biomarkers in large mixed-race and mixed-etiology validation cohorts is needed before these findings can be incorporated into current CCA management algorithms.
Another important limitation is the lack of a uniform baseline comparator for assessment of MiR relative expression. The use in recent studies of a known quantity of control MiR spiked into clinical samples is a more robust method for determining relative MiR expression in clinical studies [10]. Finally, detailed functional studies of MiRs identified in microarray expression studies are still required to delineate the pathogenic role of some MiRs in CCA.
Conclusion & future perspective
Available data support an important role for MiRs in the pathogenesis of CCA and certain MiRs have shown some promise as diagnostic and prognostic biomarkers in patients with CCA. In particular, there is good evidence from several studies of upregulation of MiR21, 625 and 223 and downregulation of MiR451, 200b/c, 338-3p, let-7a, 145 and 222 in CCA, with supportive evidence for a plausible functional role in cholangiocarcinogenesis shown by mechanistic studies. These MiRs are worthy of further investigation as both biomarkers and therapeutic targets in CCA.
However, current clinical association data are preliminary and further validation studies are needed to confirm the pathogenic role of MiRs with altered expression in CCA and to determine the clinical utility of MiR-based biomarkers when incorporated into CCA clinical management guidelines.
Footnotes
Financial & competing interests disclosure
JA Howell is supported by a National Health and Medical Research Council (Australia) Early Career Fellowship grant. The authors are grateful to the NIHR Biomedical Research Centre Funding scheme at Imperial College London for support. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest
- 1.Bridgewater J, Galle PR, Khan SA, et al. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J. Hepatol. 2014;60(6):1268–1289. doi: 10.1016/j.jhep.2014.01.021. [DOI] [PubMed] [Google Scholar]; • An excellent evidence-based consensus guideline for cholangiocarcinoma (CCA) management.
- 2.Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD. Cholangiocarcinoma. Lancet. 2005;366(9493):1303–1314. doi: 10.1016/S0140-6736(05)67530-7. [DOI] [PubMed] [Google Scholar]; • A detailed overview of CCA epidemiology, pathophysiology, diagnosis and management.
- 3.Blechacz B, Komuta M, Roskams T, Gores GJ. Clinical diagnosis and staging of cholangiocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 2011;8(9):512–522. doi: 10.1038/nrgastro.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel T. Worldwide trends in mortality from biliary tract malignancies. BMC Cancer. 2002;2:10. doi: 10.1186/1471-2407-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaib YH, Davila JA, McGlynn K, El-Serag HB. Rising incidence of intrahepatic cholangiocarcinoma in the United States: a true increase? J. Hepatol. 2004;40(3):472–477. doi: 10.1016/j.jhep.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 6.Khan SA, Emadossadaty S, Ladep NG, et al. Rising trends in cholangiocarcinoma: is the ICD classification system misleading us? J. Hepatol. 2012;56(4):848–854. doi: 10.1016/j.jhep.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 7.Patel T. Cholangiocarcinoma – controversies and challenges. Nat. Rev. Gastroenterol. Hepatol. 2011;8(4):189–200. doi: 10.1038/nrgastro.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nehls O, Gregor M, Klump B. Serum and bile markers for cholangiocarcinoma. Semin. Liver Dis. 2004;24(2):139–154. doi: 10.1055/s-2004-828891. [DOI] [PubMed] [Google Scholar]
- 9.Patel AH, Harnois DM, Klee GG, LaRusso NF, Gores GJ. The utility of CA19-9 in the diagnoses of cholangiocarcinoma in patients without primary sclerosing cholangitis. Am. J. Gastroenterol. 2000;95(1):204–207. doi: 10.1111/j.1572-0241.2000.01685.x. [DOI] [PubMed] [Google Scholar]
- 10.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]; • An excellent review of the mechanisms by which MiRs can be altered in cancer.
- 11.Kohlhapp FJ, Mitra AK, Lengyel E, Peter ME. MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene. 2015;34(48):5857–5868. doi: 10.1038/onc.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A detailed overview of the different pathways by which alterations in MiR function lead to carcinogenesis.
- 12.Haga H, Yan I, Takahashi K, Wood J, Patel T. Emerging insights into the role of microRNAs in the pathogenesis of cholangiocarcinoma. Gene Expr. 2014;16(2):93–99. doi: 10.3727/105221614X13919976902174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piontek K, Selaru FM. MicroRNAs in the biology and diagnosis of cholangiocarcinoma. Semin. Liver Dis. 2015;35(1):55–62. doi: 10.1055/s-0034-1397349. [DOI] [PubMed] [Google Scholar]
- 14.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory RI, Yan KP, Amuthan G, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowarsch A, Marr C, Schmidl D, Ruepp A, Theis FJ. Tissue-specific target analysis of disease-associated microRNAs in human signaling pathways. PLoS ONE. 2010;5(6):e11154. doi: 10.1371/journal.pone.0011154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010;11(4):252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 20.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol. Med. 2010;16(6):257–267. doi: 10.1016/j.molmed.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23(15):1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 26.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39(5):673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Huang J, Yang N, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl Acad Sci. USA. 2006;103(24):9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl Acad. Sci. USA. 2008;105(36):13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guil S, Esteller M. DNA methylomes histone codes and miRNAs: tying it all together. Int. J. Biochem. Cell Biol. 2009;41(1):87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454(7200):56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori M, Triboulet R, Mohseni M, et al. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156(5):893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Heldin CH, Landstrom M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr. Opin. Cell Biol. 2009;21(2):166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 35.Pardali K, Moustakas A. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim. Biophys. Acta. 2007;1775(1):21–62. doi: 10.1016/j.bbcan.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Rupaimoole R, Wu SY, Pradeep S, et al. Hypoxia-mediated downregulation of miRNA biogenesis promotes tumor progression. Nat. Commun. 2014;5:5202. doi: 10.1038/ncomms6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melo SA, Moutinho C, Ropero S, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18(4):303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol. Carcinog. 2013;52(4):297–303. doi: 10.1002/mc.21864. [DOI] [PubMed] [Google Scholar]
- 39.Zhang MY, Li SH, Huang GL, et al. Identification of a novel microRNA signature associated with intrahepatic cholangiocarcinoma (ICC) patient prognosis. BMC Cancer. 2015;15:64. doi: 10.1186/s12885-015-1067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawahigashi Y, Mishima T, Mizuguchi Y, et al. MicroRNA profiling of human intrahepatic cholangiocarcinoma cell lines reveals biliary epithelial cell-specific microRNAs. J. Nippon. Med. Sch. 2009;76(4):188–197. doi: 10.1272/jnms.76.188. [DOI] [PubMed] [Google Scholar]
- 41.Meng F, Henson R, Lang M, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130(7):2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]; • An important paper outlining potential mechanisms of drug resistence in cholangiocarcinoma via MiR expression.
- 42.Yang R, Chen Y, Tang C, et al. MicroRNA-144 suppresses cholangiocarcinoma cell proliferation and invasion through targeting platelet activating factor acetylhydrolase isoform 1b. BMC Cancer. 2014;14:917. doi: 10.1186/1471-2407-14-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Yan HX, Yang W, et al. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J. Hepatol. 2009;50(2):358–369. doi: 10.1016/j.jhep.2008.09.015. [DOI] [PubMed] [Google Scholar]; • An important paper outlining MiR expression profiles in human cholangiocarcinoma samples compared to healthy biliary sample controls.
- 44.Li L, Masica D, Ishida M, et al. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology. 2014;60(3):896–907. doi: 10.1002/hep.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He Q, Cai L, Shuai L, et al. Ars2 is overexpressed in human cholangiocarcinomas and its depletion increases PTEN and PDCD4 by decreasing microRNA-21. Mol. Carcinog. 2013;52(4):286–296. doi: 10.1002/mc.21859. [DOI] [PubMed] [Google Scholar]
- 46.Selaru FM, Olaru AV, Kan T, et al. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology. 2009;49(5):1595–1601. doi: 10.1002/hep.22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chusorn P, Namwat N, Loilome W, et al. Overexpression of microRNA-21 regulating PDCD4 during tumorigenesis of liver fluke-associated cholangiocarcinoma contributes to tumor growth and metastasis. Tumour Biol. 2013;34(3):1579–1588. doi: 10.1007/s13277-013-0688-0. [DOI] [PubMed] [Google Scholar]
- 48.Wang LJ, He CC, Sui X, et al. MiR-21 promotes intrahepatic cholangiocarcinoma proliferation and growth in vitro and in vivo by targeting PTPN14 and PTEN. Oncotarget. 2015;6(8):5932–5946. doi: 10.18632/oncotarget.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu L, Byrnes K, Han C, Wang Y, Wu T. miR-21 targets 15-PGDH and promotes cholangiocarcinoma growth. Mol. Cancer Res. 2014;12(6):890–900. doi: 10.1158/1541-7786.MCR-13-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z, Jin ZY, Liu CH, Xie F, Lin XS, Huang Q. MicroRNA-21 regulates biological behavior by inducing EMT in human cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 2015;8(5):4684–4694. [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, Jiao J, Cermelli S, et al. miR-21 inhibition reduces liver fibrosis and prevents tumor development by inducing apoptosis of CD24+ progenitor cells. Cancer Res. 2015;75(9):1859–1867. doi: 10.1158/0008-5472.CAN-14-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Razumilava N, Bronk SF, Smoot RL, et al. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55(2):465–475. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Han C, Wu T. MicroRNA-26a promotes cholangiocarcinoma growth by activating beta-catenin. Gastroenterology. 2012;143(1):246–256. e8. doi: 10.1053/j.gastro.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mott JL, Kurita S, Cazanave SC, Bronk SF, Werneburg NW, Fernandez-Zapico ME. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J. Cell. Biochem. 2010;110(5):1155–1164. doi: 10.1002/jcb.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mott JL, Kobayashi S, Bronk SF, Gores GJ. MirR-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26(42):6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu C, Huang F, Deng G, Nie W, Huang W, Zeng X. miR-31 promotes oncogenesis in intrahepatic cholangiocarcinoma cells via the direct suppression of RASA1. Exp. Ther. Med. 2013;6(5):1265–1270. doi: 10.3892/etm.2013.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christoffersen NR, Shalgi R, Frankel LB, et al. p53-independent upregulation of miR-34a during oncogene-induced senescence represses Myc. Cell Death Differ. 2010;17(2):236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 58.Mendell JT. Tumors line up for a letdown. Nat. Genet. 2009;41(7):768–769. doi: 10.1038/ng0709-768. [DOI] [PubMed] [Google Scholar]
- 59.Prakobwong S, Khoontawad J, Yongvanit P, et al. Curcumin decreases cholangiocarcinogenesis in hamsters by suppressing inflammation-mediated molecular events related to multistep carcinogenesis. Int. J. Cancer. 2011;129(1):88–100. doi: 10.1002/ijc.25656. [DOI] [PubMed] [Google Scholar]
- 60.Yang H, Li TW, Peng J, et al. A mouse model of cholestasis-associated cholangiocarcinoma and transcription factors involved in progression. Gastroenterology. 2011;141(1):378–388. doi: 10.1053/j.gastro.2011.03.044. 388.e1–e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, Han C, Zhu H, Song K, Wu T. miR-101 inhibits cholangiocarcinoma angiogenesis through targeting vascular endothelial growth factor (VEGF) Am. J. Pathol. 2013;182(5):1629–1639. doi: 10.1016/j.ajpath.2013.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McNally ME, Collins A, Wojcik SE, et al. Concomitant dysregulation of microRNAs miR-151-3p and miR-126 correlates with improved survival in resected cholangiocarcinoma. HPB (Oxford) 2013;15(4):260–264. doi: 10.1111/j.1477-2574.2012.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Q, Tang H, Yin S, Dong C. Downregulation of microRNA-138 enhances the proliferation, migration and invasion of cholangiocarcinoma cells through the upregulation of RhoC/p-ERK/MMP-2/MMP-9. Oncol. Rep. 2013;29(5):2046–2052. doi: 10.3892/or.2013.2304. [DOI] [PubMed] [Google Scholar]
- 64.Oishi N, Kumar MR, Roessler S, et al. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology. 2012;56(5):1792–1803. doi: 10.1002/hep.25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silakit R, Loilome W, Yongvanit P, et al. Circulating miR-192 in liver fluke-associated cholangiocarcinoma patients: a prospective prognostic indicator. J. Hepatobiliary Pancreat. Sci. 2014;21(12):864–872. doi: 10.1002/jhbp.145. [DOI] [PubMed] [Google Scholar]
- 66.Peng F, Jiang J, Yu Y, et al. Direct targeting of SUZ12/ROCK2 by miR-200b/c inhibits cholangiocarcinoma tumourigenesis and metastasis. Br. J. Cancer. 2013;109(12):3092–3104. doi: 10.1038/bjc.2013.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mizuguchi Y, Isse K, Specht S, et al. Small proline rich protein 2a in benign and malignant liver disease. Hepatology. 2014;59(3):1130–1143. doi: 10.1002/hep.26889. [DOI] [PubMed] [Google Scholar]
- 68.Qiu YH, Wei YP, Shen NJ, et al. miR-204 inhibits epithelial to mesenchymal transition by targeting slug in intrahepatic cholangiocarcinoma cells. Cell Physiol. Biochem. 2013;32(5):1331–1341. doi: 10.1159/000354531. [DOI] [PubMed] [Google Scholar]
- 69.Li B, Han Q, Zhu Y, Yu Y, Wang J, Jiang X. Down-regulation of miR-214 contributes to intrahepatic cholangiocarcinoma metastasis by targeting Twist. FEBS J. 2012;279(13):2393–2398. doi: 10.1111/j.1742-4658.2012.08618.x. [DOI] [PubMed] [Google Scholar]
- 70.Meng F, Wehbe-Janek H, Henson R, Smith H, Patel T. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene. 2008;27(3):378–386. doi: 10.1038/sj.onc.1210648. [DOI] [PubMed] [Google Scholar]
- 71.An F, Yamanaka S, Allen S, et al. Silencing of miR-370 in human cholangiocarcinoma by allelic loss and interleukin-6 induced maternal to paternal epigenotype switch. PLoS ONE. 2012;7(10):e45606. doi: 10.1371/journal.pone.0045606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen Y, Gao W, Luo J, Tian R, Sun H, Zou S. Methyl-CpG binding protein MBD2 is implicated in methylation-mediated suppression of miR-373 in hilar cholangiocarcinoma. Oncol. Rep. 2011;25(2):443–451. doi: 10.3892/or.2010.1089. [DOI] [PubMed] [Google Scholar]
- 73.Yamanaka S, Campbell NR, An F, et al. Coordinated effects of microRNA-494 induce G(2)/M arrest in human cholangiocarcinoma. Cell Cycle. 2012;11(14):2729–2738. doi: 10.4161/cc.21105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olaru AV, Ghiaur G, Yamanaka S, et al. MicroRNA down-regulated in human cholangiocarcinoma control cell cycle through multiple targets involved in the G1/S checkpoint. Hepatology. 2011;54(6):2089–2098. doi: 10.1002/hep.24591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kunkeaw N, Jeon SH, Lee K, et al. Cell death/proliferation roles for nc886, a non-coding RNA, in the protein kinase R pathway in cholangiocarcinoma. Oncogene. 2013;32(32):3722–3731. doi: 10.1038/onc.2012.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang H, Li TW, Ko KS, Xia M, Lu SC. Switch from Mnt-Max to Myc-Max induces p53 and cyclin D1 expression and apoptosis during cholestasis in mouse and human hepatocytes. Hepatology. 2009;49(3):860–870. doi: 10.1002/hep.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sugimachi K, Aishima S, Taguchi K, et al. The role of overexpression and gene amplification of cyclin D1 in intrahepatic cholangiocarcinoma. J. Hepatol. 2001;35(1):74–79. doi: 10.1016/s0168-8278(01)00079-4. [DOI] [PubMed] [Google Scholar]
- 78.Menrad H, Werno C, Schmid T, et al. Roles of hypoxia-inducible factor-1alpha (HIF-1alpha) versus HIF-2alpha in the survival of hepatocellular tumor spheroids. Hepatology. 2010;51(6):2183–2192. doi: 10.1002/hep.23597. [DOI] [PubMed] [Google Scholar]
- 79.Li J, Tian F, Li D, et al. MiR-605 represses PSMD10/Gankyrin and inhibits intrahepatic cholangiocarcinoma cell progression. FEBS Lett. 2014;588(18):3491–3500. doi: 10.1016/j.febslet.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 80.Zhong XY, Yu JH, Zhang WG, et al. MicroRNA-421 functions as an oncogenic miRNA in biliary tract cancer through down-regulating farnesoid X receptor expression. Gene. 2012;493(1):44–51. doi: 10.1016/j.gene.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 81.Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iwaki J, Kikuchi K, Mizuguchi Y, et al. MiR-376c down-regulation accelerates EGF-dependent migration by targeting GRB2 in the HuCCT1 human intrahepatic cholangiocarcinoma cell line. PLoS ONE. 2013;8(7):e69496. doi: 10.1371/journal.pone.0069496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zeng B, Li Z, Chen R, et al. Epigenetic regulation of miR-124 by hepatitis C virus core protein promotes migration and invasion of intrahepatic cholangiocarcinoma cells by targeting SMYD3. FEBS Lett. 2012;586(19):3271–3278. doi: 10.1016/j.febslet.2012.06.049. [DOI] [PubMed] [Google Scholar]
- 85.Huang Q, Liu L, Liu CH, et al. MicroRNA-21 regulates the invasion and metastasis in cholangiocarcinoma and may be a potential biomarker for cancer prognosis. Asian Pac. J. Cancer Prev. 2013;14(2):829–834. doi: 10.7314/apjcp.2013.14.2.829. [DOI] [PubMed] [Google Scholar]
- 86.Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51(3):881–890. doi: 10.1002/hep.23381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pak JH, Kim IK, Kim SM, et al. Induction of cancer-related microRNA expression profiling using excretory-secretory products of Clonorchis sinensis . Parasitol. Res. 2014;113(12):4447–4455. doi: 10.1007/s00436-014-4127-y. [DOI] [PubMed] [Google Scholar]
- 88.Collins AL, Wojcik S, Liu J, et al. A differential microRNA profile distinguishes cholangiocarcinoma from pancreatic adenocarcinoma. Ann. Surg. Oncol. 2014;21(1):133–138. doi: 10.1245/s10434-013-3240-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Turner R, Lozoya O, Wang Y, et al. Human hepatic stem cell and maturational liver lineage biology. Hepatology. 2011;53(3):1035–1045. doi: 10.1002/hep.24157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen YJ, Luo J, Yang GY, Yang K, Wen SQ, Zou SQ. Mutual regulation between microRNA-373 and methyl-CpG-binding domain protein 2 in hilar cholangiocarcinoma. World J. Gastroenterol. 2012;18(29):3849–3861. doi: 10.3748/wjg.v18.i29.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Okamoto K, Miyoshi K, Murawaki Y. miR-29b, miR-205 and miR-221 enhance chemosensitivity to gemcitabine in HuH28 human cholangiocarcinoma cells. PLoS ONE. 2013;8(10):e77623. doi: 10.1371/journal.pone.0077623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ono K. Functions of microRNA-33a/b and microRNA therapeutics. J. Cardiol. 2016;67(1):28–33. doi: 10.1016/j.jjcc.2015.10.017. [DOI] [PubMed] [Google Scholar]