Abstract
Little is known about the role of the gut mucosal microbiota and microbe–host signaling in the variation of pig’s feed efficiency (FE). This study therefore aimed to investigate the FE-related differences in the metabolically active mucosal bacterial microbiota and expression of genes for innate immune response, barrier function, nutrient uptake, and incretins in the cecum of finishing pigs. Pigs (n = 72) were ranked for their residual feed intake (RFI; metric for FE) between days 42 and 91 postweaning and were stratified within litter and sex into high (HRFI; n = 8) and low RFI (LRFI; n = 8). Cecal mucosa and digesta were collected on day 137–141 of life. After isolating total RNA from the mucosa, the RNA was transcribed into cDNA which was used for gene expression analysis, total bacterial quantification, and high-throughput sequencing (Illumina MiSeq) of the hypervariable V3-V4 region of the 16S rRNA gene. The RFI differed by 2.1 kg between low RFI (LRFI; good FE) and high RFI (HRFI; poor FE) pigs (P < 0.001). The cecal mucosa was mainly colonized by Helicobacteraceae, Campylobacteraceae, Veillonellaceae, Lachnospiraceae, and Prevotellaceae. Despite the lack of differences in microbial diversity and absolute abundance, RFI-associated compositional differences were found. The predominant genus Campylobacter tended (P < 0.10) to be 0.4-fold more abundant in LRFI pigs, whereas low abundant Escherichia/Shigella (P < 0.05), Ruminobacter (P < 0.05), and Veillonella (P < 0.10) were 3.4-, 6.6-, and 4.4-fold less abundant at the cecal mucosa of LRFI compared to HRFI pigs. Moreover, mucin 2 and zona occludens-1 were less expressed (P < 0.05) in the cecal mucosa of LRFI compared to HRFI pigs. Cecal mucosal expression of monocarboxylate transporter-1, glucagon-like peptide-1, and peptide YY further tended (P < 0.10) to be downregulated in LRFI compared to HRFI pigs, indicating an enhanced VFA uptake and signaling in HRFI pigs. Sparse partial least square regression and relevance networking support the hypothesis that certain mucosal bacteria and luminal microbial metabolites were more associated than others with differences in RFI and cecal gene expression. However, present results do not allow the determination of whether mucosal bacterial changes contributed to variation in FE or were rather a consequence of FE-related changes in the pig’s physiology or feeding behavior.
Keywords: cecum, gene expression, microbe, host interactions, mucosal microbiota, pig, residual feed intake
INTRODUCTION
The porcine gut microbiota contributes to nutrient processing and harvesting of the ingested energy and therefore may be a target to improve feed efficiency (FE) in pigs (McCormack et al., 2017). To date, few studies have evaluated differences in the bacterial community of the gastrointestinal content in pigs of diverging FE (McCormack et al., 2017; Tan et al., 2017; Yang et al., 2017), whereas there is no information on the contribution of the gut mucosal microbiota to variation in FE. The close proximity between the host and bacteria provides the mucosal community with a great potential to exert effects on the host, triggering immune activation and gut–brain communication (Leser and Mølback, 2009; Mayer et al., 2015), which may modify pigs’ FE. The host mucosa recognizes microbial activity via different routes, such as microbial metabolite sensing or receptor recognition of molecular patterns on the microbial cell surface (Kaiko and Stappenbeck, 2014).
So far, FE-associated bacteria in the porcine gut were assessed by targeting DNA (McCormack et al., 2017; Tan et al., 2017; Yang et al., 2017). However, DNA may not be a reliable marker for metabolically active bacteria because extracted DNA may originate from dead or dormant bacteria (de la Cruz-Leyva et al., 2011). Gut microbial populations can be more reliably quantified using RNA to correlate better with microbial growth rate and metabolic activity (Lettat et al., 2013).
The main objective was to investigate the differences in the metabolically active mucosal bacterial community and gene expression for innate immunity, barrier function, nutrient uptake, and incretins in the cecum of pigs phenotypically divergent for residual feed intake (RFI), a metric for FE. We hypothesized that FE-related differences in mucosal bacterial profiles are reflected in changes of the mucosal gene expression in the cecum of pigs due to alterations in microbe–host signaling with consequences for pig’s FE.
MATERIALS AND METHODS
Animals, Housing, and Selection for FE
Cecal mucosa and digesta samples used in this study originated from the Austrian pigs of the experiment reported earlier (Metzler-Zebeli et al., 2017). All animal experimentation procedures were approved by the institutional ethics committee of the University of Veterinary Medicine (Vienna, Austria) and the national authority according to paragraph 26 of Law for Animal Experiments, Tierversuchsgesetz 2012 – TVG 2012 (GZ 68.205/0058-WF/II/3b/2014).
The intact litters of 6 sows were used in this experiment (Metzler-Zebeli et al., 2017). Sows (Landrace) were inseminated with semen from 4 boars (Hermitage Genetics, Kilkenny, Ireland) which had a high estimated breeding value for feed conversion efficiency. Pigs were weaned with 28 d of age. During the test period between days 42 and 91 postweaning (pw), siblings were group-housed (n = 12) in pens and their feed intake was recorded via Feed Intake Recording Equipment (FIRE) feeders (Schauer Agrotonic, Wels, Austria). Water and feed were freely available throughout the nursery and fattening phases. All pigs were subjected to the same management, environmental, and nutritional conditions. The finisher diet was based on wheat and barley and contained 17.8% CP, 2.7% fat, and 14.16 MJ DE/kg. The detailed dietary ingredient and chemical composition is published in Metzler-Zebeli et al. (2017). During the test period, individual feed intake of pigs was recorded daily, while BW, back fat, and muscle depth were recorded weekly (Metzler-Zebeli et al., 2017). Back fat and muscle depth were measured between the third and fourth last lumbar vertebrae. Following this test period (day 91 pw), pigs were retrospectively ranked according to their RFI. To minimize the variability in FE due to external factors, pigs with extreme RFI were selected within litter and sex. Using the PROC REG procedure in SAS (version 9.4; SAS Inst. Inc., Cary, NC), a least squares regression model of ADFI on average daily weight gain, metabolic live weight, sex, and all relevant 2-way interactions, as well as the effects of back fat and muscle depth was used to estimate pig RFI as the residuals over the 49-d experimental period (Metzler-Zebeli et al., 2017). Extreme animals of the same sex within a litter were selected whose RFI values were greater than 2 SD distant from the mean of the 2 RFI ranks. Using this method, 8 low RFI (LRFI) and 8 high RFI (HRFI) pigs were selected.
Collection of Cecal Digesta and Mucosa
At a live weight of about 110 kg, between days 109 and 113 pw (day 137–141 of life), pigs were sacrificed by captive bold stunning followed by exsanguination (Metzler-Zebeli et al., 2017). Immediately thereafter, the abdominal cavity was opened and the visceral organs including the entire gastrointestinal tract were removed. The cecum was carefully emptied of digesta, cleaned with PBS (pH = 7.4) and, by taking care not to remove the mucus layer, carefully blotted dry with paper. The mucosa was scraped from the whole cecum, snap-frozen in liquid nitrogen, and stored at −80 °C. The cecal digesta was thoroughly homogenized and stored on ice until storage at −20 °C.
RNA Isolation
Total RNA was isolated from 20 mg cecal mucosal scrapings which were combined with lysis buffer (RNeasy Mini QIAcube kit, Qiagen, Hilden, Germany) and autoclaved ceramic beads (0.6 g; 1.4 mm; VWR). Mucosal samples were homogenized using the FastPrep-24 instrument (MP Biomedicals, Santa Ana, CA). The remainder of the RNA isolation protocol was completed according to the manufacturer’s instructions using the automated QIAcube robotic workstation (Qiagen, Hilden, Germany). The RNA isolates were treated with the Turbo DNA kit (Life Technologies Limited, Vienna, Austria) to remove genomic DNA. The RNA was quantified using the Qubit HS RNA Assay kit on the Qubit 2.0 Fluorometer (Life Technologies), and the quality of isolated RNA was evaluated with the Agilent Bioanalyzer 2100 (Agilent RNA 6000 Nano Assay, Agilent Technologies, Waghaeusel-Wiesental, Germany). Samples had RNA integrity numbers (RIN) from 8.1 to 9.4; except for 3 samples with RIN of 7.8. The cDNA was synthesized from 4 µg of RNA using the High Capacity cDNA RT kit (Life Technologies Limited, Vienna, Austria) after adding 1 µL of RNase inhibitor (Biozym, Hessisch Oldendorf, Germany). One aliquot of cDNA was used for 16S rRNA sequencing and bacterial quantification, whereas a second aliquot was employed in the gene expression experiment.
Complementary DNA Sequencing
Complementary DNA aliquots were sent to Microsynth (Balgach, Switzerland) for 16S rRNA gene PCRs, library preparation, and sequencing using the Illumina MiSeq sequencing platform (Illumina Inc., San Diego, CA). The V3-V4 hypervariable regions of bacterial 16S rRNA were amplified using the primers 341F-ill (5′-CCTACGGGNGGCWGCAG-3′) and 802R-ill (5′-GACTACHVGGGTATCTAATCC-3′) to produce an amplicon of approximately 460 bp. The 16S rRNA gene PCRs were performed using the KAPA HiFi HotStart PCR Kit (Roche, Baden, Switzerland). Libraries were constructed by ligating sequencing adapters and indices onto purified PCR products using the Nextera XT sample preparation kit (Illumina Inc.). Equimolar quantities of each library were pooled and sequenced on an Illumina MiSeq sequencing v2 platform using a 250 bp read length paired-end protocol. After sequencing, the overlapping paired-end reads were demultiplexed, trimmed of Illumina adaptor residuals using cutadapt (code.google.com/p/cutadapt/), and stitched using USEARCH (drive5/com) by Microsynth.
Stitched reads were processed using the software package Quantitative Insights into Microbial Ecology (QIIME, v1.9.4; Caporaso et al., 2010). Following quality filtering, through which low quality sequences were removed (q < 20), and assembly, chimeric sequences were filtered out with the UCHIME method using USEARCH8.1 and gold.fa database (Edgar, 2010; Edgar et al., 2011). Sequences were clustered into operational taxonomic units (OTUs) using a 16S rRNA distance of 0.03 and the USEARCH algorithm (Edgar et al., 2011) with a minimum cluster size of 10. The most abundant sequence was selected from each OTU cluster and taxonomically assigned using the Ribosomal Database Project (RDP) classifier tool (Cole et al., 2009) with the most recent version of the Greengenes database (13.8). Microbial richness and diversity were calculated using the nonparametric species estimator Chao1 as well as the Shannon and Simpson diversity indices in QIIME and were based on a rarefaction depth of 145,000 reads per sample. To determine the degree of similarity between samples, weighted and unweighted Unifrac distance matrices were calculated. Individual high abundance OTUs that correlated with RFI were additionally classified by using the Greengenes 16S rRNA gene database (http://greengenes.lbl.gov/). The raw sequence reads were uploaded to the NCBI Bioproject databank under the Bioproject number: PRJNA419713.
Microbial Metagenome Prediction
Microbial metagenome prediction for each cecal sample based on 16S rRNA gene sequencing data was determined using Pylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt; Langille et al., 2013). For this, closed-reference OTU picking was performed at 97% similarity level against the Greengenes database (downloaded from http://greengenes.secondgenome.com/downloads/database/13_5), and processed in the online Galaxy PICRUSt interface (http://galaxyproject.org/) with a workflow described by the developers (http://picrust.github.com/picrust/tutorials/quickstart.html#quickstartguide). The sequences were categorized by function based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in PICRUSt. Non-bacteria related KEGG orthology functions and functions <0.01% relative abundance were dismissed.
Absolute Bacterial Abundance
Quantification of 16S rRNA copies of total bacteria in mucosal cDNA samples was performed on a Stratagene Mx3000P QPCR System (Agilent Technologies, Santa Clara, CA) using the Fast Plus EvaGreen Master Mix with low ROX (Biotium, Hayward, CA) and the primer set 341-357F and 518-534R (Muyzer et al., 1993) in 20 µL reaction mixtures (Metzler-Zebeli et al., 2016). Each standard and sample reaction contained 10 µL of master mix, forward and reverse primers (62.5 pmol) and 1 ng of DNA template. The amplification program included an initial denaturation step at 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s, primer annealing at 60 °C for 30 s and elongation at 72 °C for 30 s. Fluorescence was measured at the last step of each cycle. The dissociation of PCR products was monitored to determine the specificity of the amplification. The standard was prepared from the purified and quantified PCR products generated by standard PCR using DNA from the cecal content of the present experiment and the total bacterial primer set (Metzler-Zebeli et al., 2016). Ten-fold standard serial dilutions (107 to 103 molecules/µL) were run on each 96-well plate, with an amplification efficiency (E = −1 + 10(−1/slope)) of 1.94 and R2 = 1.00.
Mucosal Gene Expression
Primers utilized for quantitative PCR are listed in Supplementary Table A1. Primers were newly designed and, similar to the previously published primers, verified using PrimerBLAST (www.ncbi.nlm.nih.gov/tools/primer-blast/) as well as tested for efficiencies and specificity using dissociation curve analysis. Amplifications were performed on the Stratagene Mx3000P QPCR System using the following conditions: 95 °C for 5 min, followed by 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s for 40 cycles, followed by the generation of dissociation curves. Negative controls as well as reverse transcription controls (RT minus) were included in order to control for residual DNA contamination. Each 20 µL reaction consisted of 50 ng cDNA, 10 µL Fast Plus EvaGreen Master Mix with low ROX (Biotium, Hayward, CA), 100 nM each of forward and reverse primers, and DEPC-treated water in a 96-well plate (VWR, Vienna, Austria). All reactions were run in duplicate.
Of the 5 housekeeping genes (HKG; Supplementary Table A1), ACTB and GAPDH were most stably expressed, which was analyzed using NormFinder (Andersen et al. 2004) and BestKeeper (Pfaffl et al. 2004). The geometric mean expression level of ACTB and GAPDH was used for normalization of target gene expression levels. For this, the mean raw gene expression data, obtained as Cq values, of the identified HKG were subtracted from the Cq of the target genes to determine ΔCq values. Relative gene expression was calculated relative to the pig with the lowest expression of the respective gene using the 2−ΔΔCq method.
Chemical Analysis
The VFA (acetate, propionate, butyrate, isobutyrate, valerate, and isovalerate) were analyzed after aliquots of 1 g cecal content were thawed on ice and mixed with 0.2 mL of 25% metaphosphoric acid, 1 mL of double-distilled water, and 200 μL of internal standard (4-methyl-valeric acid; Sigma-Aldrich, Vienna, Austria). The mixture was centrifuged at 3,148 × g for 10 min. The supernatant was again centrifuged at 15,000 × g for 25 min, and the clear supernatant was analyzed as previously described (Deckardt et al., 2016). Concentrations of cell-free lipopolysaccharides (LPS) were determined using the pyrochrome limulus amebocyte lysate (LAL) assay (Associates of Cape Cod Inc., East Falmouth, MA) as previously described (Metzler-Zebeli et al., 2013). Samples were diluted and deproteinized by heating, the supernatants were used to measure the optical density of samples at 405 nm against calibration curves using Pyros EQS software (Associates of Cape Cod Inc.). Reactions were run in duplicate and the intra-assay CV was <10 %.
Statistical Analysis
A power test analysis estimated according to Kononoff and Hanford (2006) and based on data with pigs divergent for FE or pig lines of divergent FE (Mani et al., 2013; Montagne et al., 2014) using the SAS software (version 9.3; SAS Inst. Inc., Cary, NC) was performed to identify the minimum number of observations needed. The power test analysis indicated that a statistical power of more than 90% for a sample size of n = 8 and α = 0.05 can be expected so that sufficient power will be available to reject the null-hypothesis (H0) if this H0 is false (p = 1 − β).
The Shapiro–Wilk test was used to test for normality of data distribution for all variables using the PROC UNIVARIATE procedure in SAS (version 9.4; SAS Inst. Inc., Cary, NC). Additionally, the Cook’s distance (Cook’s D) test was used to determine any potential influential observation on the model. Data of RFI, ADFI, and ADG (Metzler-Zebeli et al., 2017) were re-analyzed for the Austrian pigs.
All variables were normally distributed and analyzed by ANOVA using the MIXED procedure in SAS. Fixed effects included in the model were sex and RFI rank. The sow was included as random effect and pig was the experimental unit. Degrees of freedom were approximated by the method of Kenward–Roger and the covariance structure was compound symmetry. The Tukey correction for multiple testing was used for pairwise comparisons between least squares means. Least squares means were computed and significance declared at P ≤ 0.05. A trend was considered at 0.05 < P ≤ 0.10. Boxplots were generated using the package “ggplot2” (Wickham, 2009) in R Studio (version 1.0.136).
In order to identify relevant relationships of the mucosal bacterial genera and microbial metabolites with pig’s individual RFI and gene expression, sparse partial least squares (sPLS) regression and relevance network analysis (Lê Cao et al., 2009a, 2009b) were performed using the “mixOmics” package in R studio. The “network” function was used to generate the images from sPLS, whereby the “network” function calculated a similarity measure between X and Y variables in a pairwise manner (Mach et al., 2015). In the graph, each X and Y variable corresponds to a node, whereas the edges display the associations (Pearson’s correlations) between the nodes.
RESULTS
Feed Efficiency and Microbial Metabolites
The RFI values of LRFI pigs were 2.1 kg lower than those of HRFI pigs (P < 0.001; Table 1; Metzler-Zebeli et al., 2017). Moreover, the LRFI pigs ate 360 g/d less (P < 0.05) but gained similar weight compared to HRFI pigs between days 42 and 91 pw. Concentrations of total and individual VFA and LPS in cecal digesta were similar between LRFI and HRFI pigs (Table 2).
Table 1.
Residual feed intake, ADFI, ADG, and BW at slaughter of finishing pigs ranked on RFIa–c
| Item | LRFI | HRFI | SEM | P-valuesd |
|---|---|---|---|---|
| RFI (kg) | −1.07 | 1.06 | 0.259 | <0.001 |
| ADFI (kg) | 2.35 | 2.71 | 0.141 | 0.079 |
| ADG (kg) | 1.17 | 1.16 | 0.054 | 0.972 |
| BW at slaughter (kg) | 106 | 110 | 4.3 | 0.510 |
aValues are least squares means ± SEM.
bADFI, average daily feed intake; ADG, average daily weight gain; HRFI, high RFI; LRFI, low RFI.
cRFI, ADFI, and ADG were determined between days 42 and 91 pw (n = 8/RFI rank).
dMain effect of RFI rank.
Table 2.
Concentrations of VFA and lipopolysaccharides in cecal content of finishing pigs ranked on RFIa–c
| Itemd | LRFI | HRFI | SEM | P-valuese |
|---|---|---|---|---|
| Total VFA (µmmol/kg) | 185.1 | 174.4 | 8.711 | 0.368 |
| Acetate (µmol/g) | 111.2 | 105.5 | 4.896 | 0.393 |
| Propionate (µmol/g) | 49.8 | 46.7 | 2.760 | 0.408 |
| Butyrate (µmol/g) | 19.1 | 17.4 | 1.594 | 0.431 |
| Isobutyrate (µmol/g) | 1.92 | 1.91 | 0.199 | 0.969 |
| Valerate (µmol/g) | 2.44 | 2.24 | 0.407 | 0.716 |
| Isovalerate (µmol/g) | 0.52 | 0.54 | 0.149 | 0.922 |
| Caproate (µmol/kg) | 0.10 | 0.10 | 0.036 | 0.921 |
| Lipopolysaccharides (log10 EU/g) | 6.25 | 6.13 | 0.071 | 0.207 |
aValues are least squares means ± SEM.
bHRFI, high RFI; LRFI, low RFI.
cRFI was determined between days 42 and 91 pw (n = 8/RFI rank).
dResults are presented on fresh matter basis.
eMain effect of RFI rank.
Mucosal Metabolically Active Bacteria
The absolute abundance of metabolically active mucosal bacteria amounted to 9.8 log10 gene copies/g mucosa (Table 3). Illumina MiSeq sequencing of the V3-V4 region of the 16S rRNA generated a total of 9,273,830 for the 16 samples, with an average sequencing depth of 572,721 reads per sample and a mean read length of 409 bp. Using a dissimilarity threshold of 3% sequence divergence to classify OTUs and a relative abundance threshold of 0.002%, we identified 1,110 distinct OTUs residing at the cecal mucosa of LRFI and HRFI pigs.
Table 3.
Absolute abundance (log10 gene copies/g mucosa) and estimators for species richness and diversity of the metabolically active bacterial microbiota at the cecal mucosa of finishing pigs ranked on RFIa–c
| LRFI | HRFI | SEM | P-valuesd | |
|---|---|---|---|---|
| Bacterial abundance | 9.8 | 9.8 | 0.03 | 0.418 |
| Chao1 | 3,044 | 3,246 | 200.7 | 0.457 |
| Shannon | 4.59 | 5.01 | 0.385 | 0.424 |
| Simpson | 0.80 | 0.83 | 0.038 | 0.513 |
aValues are least squares means ± SEM.
bHRFI, high RFI; LRFI, low RFI.
cResidual feed intake was determined between days 42 and 91 pw (n = 8/RFI rank).
dMain effect of RFI rank.
Phylum-level analysis of the metabolically active mucosal microbiota among all pigs revealed that Proteobacteria and Firmicutes dominated, representing more than 50 and 26% of all sequences, respectively, followed by Bacteroides (6.4%), Spirochaetes (2.4%), and Cyanobacteria (1.7%; Table 4). Phylogenetic classification of the mucosal microbiota identified 44 families with a mean abundance greater than 0.01% of all reads (Table 5). The Proteobacteria mainly comprised the families Helicobacteriaceae (32.4% of all reads) and Campylobacteraceae (22.9% of all reads), whereas the Firmicutes community was dominated by the families Veillonellaceae (12.1% of all reads) and Lachnospiraceae (7.1% of all reads). The predominant Bacteroidetes family was Prevotellaceae (5.4% of all reads). Correspondingly, the most abundant mucosal genera were Helicobacter, Campylobacter, Anaerovibrio, an unclassified Lachnospiraceae genus, Prevotella, Flexispira, and Lactobacillus (Supplementary Table A2). These predominant genera were generally represented by 1 to 3 OTUs, which accounted for the majority of OTUs within that particular genus. The most predominant mucosal OTU with an average relative abundance of 24.7% of all reads showed 97.8% sequence identity with Helicobacter trogontum. The second most abundant OTU, accounting for 14.2% of all reads, was closely related with 97% sequence identity to Campylobacter hyointestinalis.
Table 4.
Relative abundance (% of total reads) of bacterial phyla at the cecal mucosa of finishing pigs ranked on RFIa–c
| Item | LRFI | HRFI | SEM | P-valuesd |
|---|---|---|---|---|
| Eubacteria | ||||
| Proteobacteria | 52.95 | 63.71 | 8.636 | 0.360 |
| Firmicutes | 33.90 | 26.06 | 5.344 | 0.285 |
| Bacteroidetes | 8.19 | 4.59 | 1.647 | 0.123 |
| Spirochaetes | 1.27 | 3.59 | 2.389 | 0.471 |
| Cyanobacteria | 2.59 | 0.74 | 0.807 | 0.107 |
| Verrucomicrobia | 0.047 | 0.244 | 0.1629 | 0.373 |
| Actinobacteria | 0.071 | 0.037 | 0.0151 | 0.114 |
| Deferribacteres | 0.053 | 0.093 | 0.0391 | 0.444 |
| Elusimicrobia | 0.064 | 0.047 | 0.0276 | 0.650 |
| Fibrobacteres | 0.011 | 0.023 | 0.0161 | 0.567 |
| Fusobacteria | 0.026 | 0.010 | 0.0085 | 0.194 |
| Tenericutes | 0.025 | 0.011 | 0.0067 | 0.140 |
| Synergistetes | 0.016 | 0.012 | 0.0058 | 0.624 |
| Archaea | ||||
| Euryarchaeota | 0.110 | 0.056 | 0.0829 | 0.631 |
aValues are least squares means ± SEM.
bHRFI, high RFI; LRFI, RFI.
cResidual feed intake was determined between days 42 and 91 pw (n = 8/RFI rank).
dMain effect of RFI rank.
Table 5.
Relative abundance (% of total reads) of the most abundant bacterial families at the cecal mucosa of finishing pigs ranked on RFIa–c
| Familyd | LRFI | HRFI | SEM | P-valuee |
|---|---|---|---|---|
| Helicobacteraceae | 33.28 | 31.49 | 6.322 | 0.832 |
| Campylobacteraceae | 27.57 | 18.16 | 4.681 | 0.057 |
| Veillonellaceae | 10.02 | 14.19 | 2.910 | 0.296 |
| Lachnospiraceae | 6.34 | 7.95 | 1.519 | 0.433 |
| Prevotellaceae | 5.04 | 5.82 | 1.400 | 0.677 |
| Lactobacillaceae | 2.31 | 4.05 | 0.939 | 0.184 |
| Ruminococcaceae | 3.40 | 3.44 | 0.607 | 0.957 |
| Spirochaetaceae | 1.73 | 3.74 | 2.202 | 0.498 |
| Unclassified Clostridiales | 1.53 | 1.85 | 0.522 | 0.643 |
| YS2 | 2.19 | 1.72 | 0.884 | 0.693 |
| Clostridiaceae | 1.24 | 1.59 | 0.635 | 0.686 |
| [Paraprevotellaceae] | 0.39 | 0.057 | 0.799 | 0.819 |
| Streptococcaceae | 0.45 | 0.60 | 0.224 | 0.616 |
| Succinivibrionaceae | 0.49 | 0.52 | 0.161 | 0.894 |
| Alcaligenaceae | 0.31 | 0.45 | 0.126 | 0.407 |
| Unclassified Bacteroidales | 0.31 | 0.29 | 0.140 | 0.914 |
| Unclassified Tremblayales | 0.50 | 0.26 | 0.233 | 0.438 |
| Unclassified RF32 | 0.17 | 0.23 | 0.084 | 0.606 |
| Erysipelotrichaceae | 0.14 | 0.19 | 0.049 | 0.467 |
| Turicibacteraceae | 0.15 | 0.19 | 0.054 | 0.631 |
| [Methanomassiliicoccaceae] | 0.128 | 0.069 | 0.076 | 0.563 |
| S24-7 | 0.074 | 0.102 | 0.032 | 0.520 |
| RF16 | 0.063 | 0.092 | 0.061 | 0.730 |
| Peptostreptococcaceae | 0.079 | 0.075 | 0.026 | 0.906 |
| Verrucomicrobiaceae | 0.009 | 0.113 | 0.117 | 0.512 |
| RFP12 | 0.045 | 0.061 | 0.048 | 0.801 |
| Coriobacteriaceae | 0.054 | 0.069 | 0.019 | 0.530 |
| Bacteroidaceae | 0.010 | 0.063 | 0.031 | 0.216 |
| Enterobacteriaceae | 0.019 | 0.081 | 0.017 | 0.017 |
| Peptococcaceae | 0.028 | 0.048 | 0.011 | 0.186 |
| Deferribacteraceae | 0.099 | 0.032 | 0.038 | 0.203 |
| Elusimicrobiaceae | 0.036 | 0.017 | 0.031 | 0.634 |
| Unclassified Clostridiales | 0.040 | 0.034 | 0.011 | 0.692 |
| Porphyromonadaceae | 0.024 | 0.030 | 0.015 | 0.763 |
| [Mogibacteriaceae] | 0.023 | 0.029 | 0.008 | 0.593 |
| Bacillaceae | 0.009 | 0.031 | 0.020 | 0.405 |
| Desulfovibrionaceae | 0.022 | 0.022 | 0.008 | 0.955 |
| Fibrobacteraceae | 0.009 | 0.038 | 0.015 | 0.154 |
| Fusobacteriaceae | 0.013 | 0.025 | 0.009 | 0.342 |
| Dethiosulfovibrionaceae | 0.010 | 0.016 | 0.005 | 0.388 |
| Sphaerochaetaceae | 0.014 | 0.011 | 0.007 | 0.781 |
| Anaeroplasmataceae | 0.013 | 0.012 | 0.004 | 0.949 |
| Unclassified Alphaproteobacteria | 0.011 | 0.003 | 0.006 | 0.256 |
aValues are least squares means ± SEM.
bHRFI, high RFI; LRFI, low RFI.
cResidual feed intake was determined between days 42 and 91 pw (n = 8/RFI rank).
dRelative abundance presented amount to 98.4% and 97.9% of all reads at the cecal mucosa of LRFI and HRFI pigs, respectively.
eMain effect of RFI rank.
Residual Feed Intake Associated Variation in Cecal Mucosal Bacterial Community
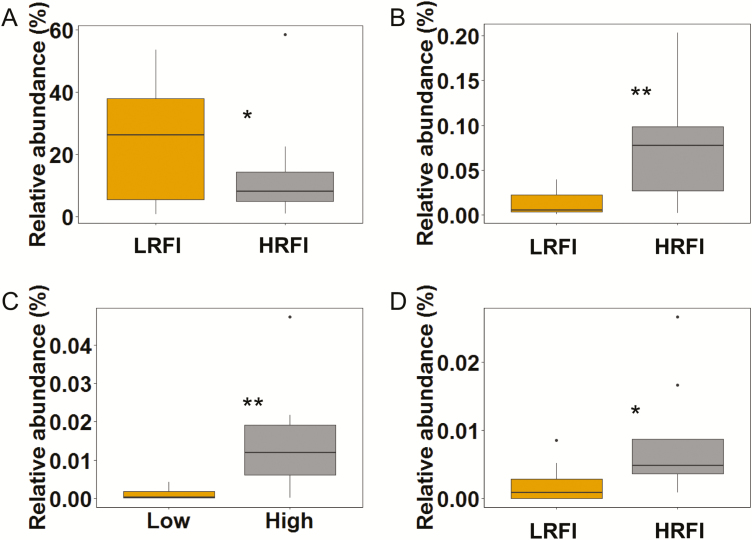
Total 16S rRNA gene copy numbers of the metabolically active mucosal bacteria were similar for LRFI and HRFI pigs (Table 3). According to β-diversity analysis using weighted and unweighted Unifrac analysis, the community structure of the metabolically active mucosal bacteria was also similar for LRFI and HRFI pigs (data not shown). This was also reflected by the α-diversity, showing similar index values for species richness (Chao1) and evenness (Shannon and Simpson) of the cecal mucosal community for LRFI and HRFI pigs. Differences in the metabolically active bacterial composition between RFI ranks were evident at family, genus, and OTU level. High-abundant Campylobacteraceae tended (P = 0.057) to be 0.3-more abundant in LRFI than in HRFI pigs (Table 5). The same trend was found for the genus Campylobacter and within this genus the most predominant OTU (97% sequence identity to C. hyointestinalis; Fig. 1A; Table 6). Other mucosal taxa that differed between RFI ranks were of low abundance (Tables 3 and 6; Fig. 1). Accordingly, the LRFI pigs had 3.4-fold less Enterobacteriaceae (Table 5) and within this family the genus Escherichia/Shigella than HRFI pigs (P < 0.05; Fig. 1B). Two further low abundant genera, Ruminobacter (P < 0.05; Fig. 1C) and Veillonella (P < 0.10; Fig. 1D) were 6.6- and 4.4-fold less abundant in LRFI compared to HRFI pigs, respectively. Comparison at OTU level for these 3 genera also showed that each of these differences in genera abundances between LRFI and HRFI pigs were mainly caused by 1 OTU (Table 6), which were identified as Escherichia coli/Shigella flexneri (100% sequence identity to reference strains), an uncultured Ruminobacter sp. (95% sequence identity to reference strain) and Veillonella dispar (100% sequence identity to reference strain).
Figure 1.
Relative abundance (% of total reads) of selected metabolically active bacterial genera at the cecal mucosa of finishing pigs ranked on RFI. (A) Campylobacter; (B) Escherichia/Shigella; (C) Ruminobacter; (D) Veillonella. **P < 0.05; *P < 0.10. LRFI, low RFI; HRFI, high RFI; n = 8/RFI rank.
Table 6.
Relative abundance (% of total reads) of selected OTUs at the cecal mucosa of finishing pigs ranked on RFIa–c
| Item | Closest reference strain (% identity similarity) | LRFI | HRFI | SEM | P-valued |
|---|---|---|---|---|---|
| OTU2 | Campylobacter hyointestinalis (97%) | 22.81 | 12.63 | 3.642 | 0.061 |
| OTU173 | Escherichia coli/Shigella flexneri (100%) | 0.017 | 0.068 | 0.0145 | 0.023 |
| OTU490 | Ruminobacter sp. (95%) | 0.002 | 0.016 | 0.0037 | 0.017 |
| OTU719 | Veillonella dispar (100%) | 0.001 | 0.006 | 0.0021 | 0.085 |
aValues are least squares means ± SEM.
bHRFI, high RFI; LRFI, low RFI.
cResidual feed intake was determined between days 42 and 91 pw (n = 8/RFI rank).
dMain effect of RFI rank.
Residual Feed Intake Associated Variation in Predicted Bacterial Metagenomic Functions
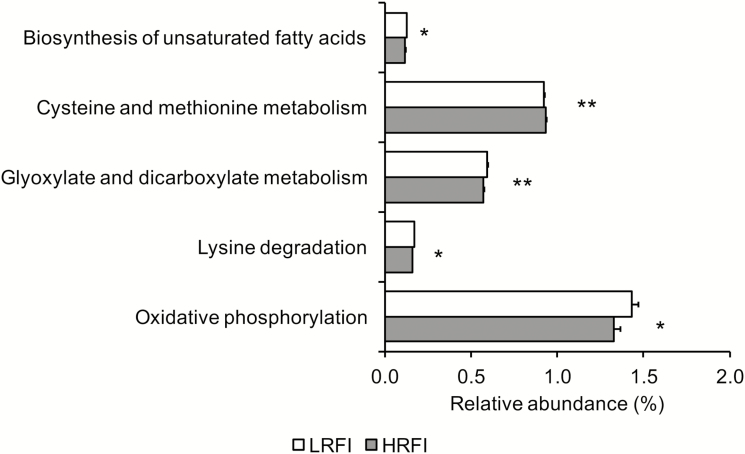
In total 5 predicted metagenomic function genes of the mucosal microbiome in the cecum were or tended to be differentially abundant in LRFI and HRFI pigs (Fig. 2). As such, LRFI pigs had a higher relative abundance of the KEGG pathway genes “biosynthesis of glyoxylate and dicarboxylate metabolism” (P < 0.05), “biosynthesis of unsaturated fatty acids,” “lysine degradation,” and “oxidative phosphorylation” compared to HRFI pigs (P < 0.10). In contrast, the predicted pathway “cysteine and methionine metabolism” was more abundant in HRFI compared to LRFI pigs (P < 0.05).
Figure 2.
Relative abundance of predicted metabolomic functions of the metabolically active bacterial microbiota at the cecal mucosa of finishing pigs ranked on RFI. **P < 0.05; *P < 0.10. LRFI, low RFI; HRFI, high RFI; n = 8/RFI rank.
Mucosal Gene Expression
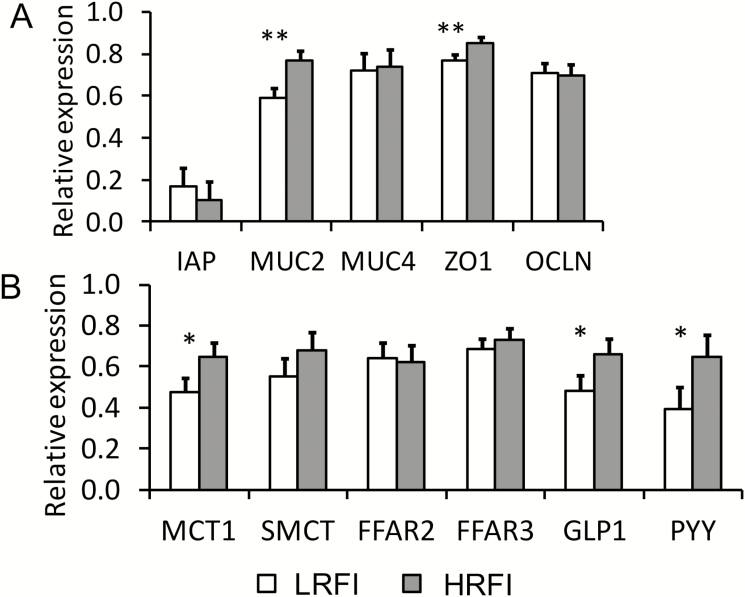
Many genes linked to the innate immune response, including genes for intestinal alkaline phosphatase, Toll-like receptors (TLR), cytokines, suppressors of cytokine signaling, and inflammation-related transcription factors, were similarly expressed in the cecal mucosa of LRFI and HRFI pigs (Table 7). In contrast, LRFI pigs had a 0.3-fold lower (P = 0.021) MUC2 expression compared to HRFI pigs (Fig. 3A). Similarly, of the 2 investigated tight-junction proteins ZO1 was 0.1-fold less expressed (P = 0.049) in LRFI compared to HRFI pigs. Moreover, genes related to SCFA transport and signaling MCT1, GLP1, and PYY tended (P < 0.10) to be 0.4-, 0.4-, and 0.7-fold less expressed in the cecal mucosa of LRFI pigs compared to HRFI pigs, respectively (Fig. 3B).
Table 7.
Relative expression of genes for glucose transporters, innate immune signaling and nuclear transcription factors in the cecal mucosa of finishing pigs ranked on RFIa–c
| Gene of interestd | LRFI | HRFI | SEM | P-valuese |
|---|---|---|---|---|
| Glucose transporters | ||||
| SGLT1 | 0.443 | 0.488 | 0.0707 | 0.633 |
| GLUT2 | 0.362 | 0.421 | 0.0997 | 0.657 |
| Innate immune signaling | ||||
| TLR2 | 0.858 | 0.835 | 0.0538 | 0.745 |
| TLR4 | 0.750 | 0.826 | 0.0514 | 0.282 |
| NFKB | 0.665 | 0.677 | 0.0394 | 0.817 |
| TNFA | 0.567 | 0.599 | 0.0674 | 0.720 |
| IL1B | 0.613 | 0.676 | 0.0779 | 0.542 |
| IL6 | 0.642 | 0.680 | 0.0587 | 0.624 |
| IL8 | 0.566 | 0.559 | 0.0721 | 0.944 |
| IL10 | 0.591 | 0.649 | 0.0675 | 0.522 |
| INFG | 0.544 | 0.682 | 0.0658 | 0.138 |
| TGFB1 | 0.484 | 0.501 | 0.0583 | 0.832 |
| SOCS1 | 0.649 | 0.628 | 0.0758 | 0.833 |
| SOCS2 | 0.691 | 0.659 | 0.0581 | 0.683 |
| SOCS3 | 0.754 | 0.764 | 0.0460 | 0.877 |
| SOCS4 | 0.616 | 0.701 | 0.0639 | 0.330 |
| SOCS5 | 0.782 | 0.832 | 0.0557 | 0.508 |
| SOCS6 | 0.653 | 0.757 | 0.0582 | 0.199 |
| Transcription factors | ||||
| PPARG | 0.450 | 0.575 | 0.0615 | 0.149 |
| SRBEF1 | 0.729 | 0.773 | 0.0673 | 0.628 |
| FASN | 0.700 | 0.772 | 0.0616 | 0.388 |
| AMPK | 0.623 | 0.706 | 0.0570 | 0.289 |
aValues are least squares means ± SEM.
b HRFI, high RFI; LRFI, low RFI.
cResidual feed intake was determined between days 42 and 91 pw (n = 8/RFI rank).
dGene names: AMPK, AMP-activated protein kinase; FASN, fatty acid synthase; GLUT2, solute carrier family 2 (facilitated glucose transporter), member 2; IL, interleukin; SRBEF1, sterol regulatory element binding transcription factor 1; PPARG, peroxisome proliferator-activated receptor-γ; SGLT1, solute carrier family 5 (sodium/glucose cotransporter), member 1; SOCS, suppressor of cytokine signaling; TGFB1, transforming growth factor-β 1; TNFA, tumor necrosis factor-α.
eMain effect of RFI rank.
Figure 3.
Relative expression of first luminal defense genes (A) and VFA transport and signaling genes (B) in the cecal mucosa of finishing pigs ranked on RFI. **P < 0.05; *P < 0.10. LRFI, low RFI; HRFI, high RFI; n = 8/RFI rank. Gene names: FFAR, free fatty acid receptor; GIP, gastric inhibitory polypeptide; GLP1, glucagon-like peptide-1; IAP, intestinal alkaline phosphatase; IL, interleukin; INFG, interferon-γ; MCT1, solute carrier family 16 (monocarboxylate transporter-1), member 1; MUC, mucin; OCLN, occludin; PYY, peptide YY/peptide tyrosine tyrosine; SMCT, solute carrier family 5 (sodium-monocarboxylate transporter), member 12; ZO1, zonula occludens-1.
Relevance Networks
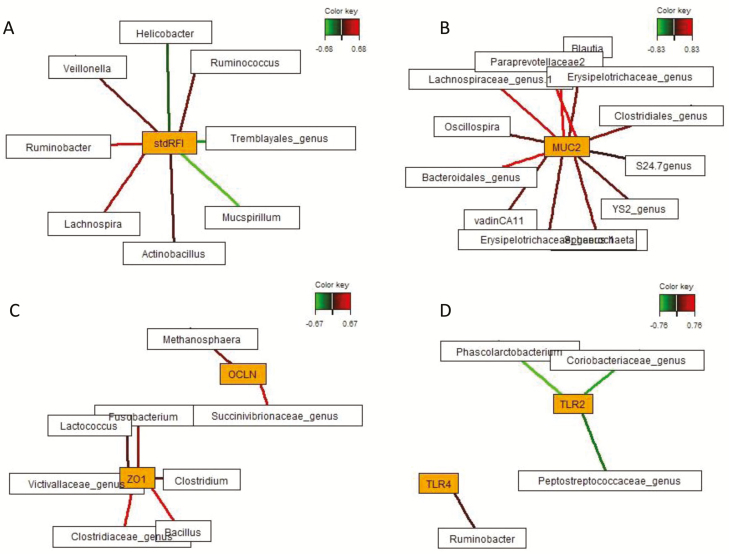
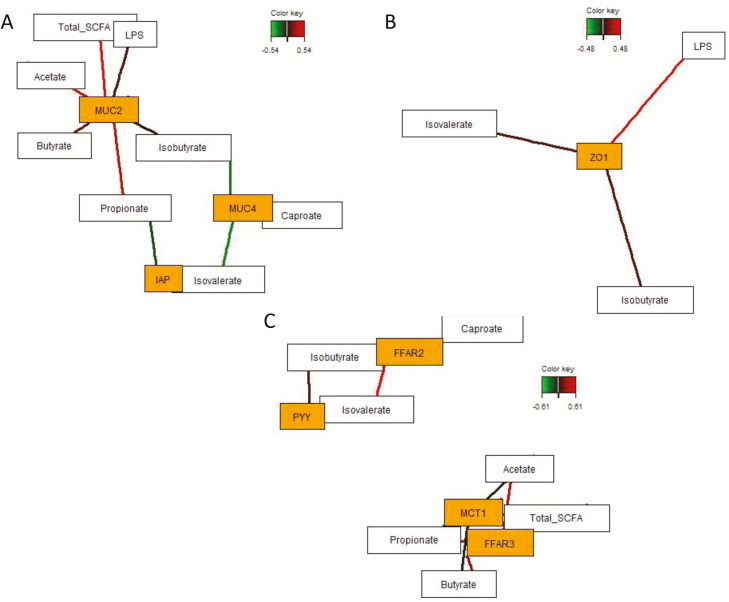
Sparse partial least squares regression and relevance networking analysis were performed based on the relative abundance of genera to discover influential bacterial taxa and microbial metabolites affecting mucosal gene expression response in the cecum relative to pig’s individual RFI. Only the strongest pairwise associations as the most influential are presented for each network (Figs. 4 and 5; Supplementary Table A3). Relevance networking analysis identified 3 and 5 bacterial genera which were negatively and positively associated with RFI, respectively. Correspondingly, increased mucosal abundance of Helicobacter, Mucspirillum, and an unclassified Tremblayales were discriminative for LRFI, whereas Ruminobacter, Veillonella, Ruminococcus, Lachnospira, and Actinobacillus were associated with HRFI (Fig. 4; Supplementary Table A3). Host receptor-mediated bacterial networking was indicated by the positive association between Ruminobacter and TLR4 expression as well as by the negative relationship between Phascolarctobacterium belonging to the family Veillonellaceae (Firmicutes), unclassified Peptostreptococcaceae (Firmicutes) and Coriobacteriaceae (Actinobacteria), and TLR2 expression levels. Moreover, different bacterial taxa were associated with expression levels of ZO1, OCLN, and MUC2, whereby all relationships were positive and comprised genera within the phyla Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria, Lentisphaera, and Euryarchaeota. Associations between mucosal gene expression levels and luminal SCFA concentrations were identified for FFAR3 expression which was positively associated with acetate and propionate, whereas caproate and branched-chain fatty acids stimulated FFAR2 and MUC4 expression (Fig. 5; Supplementary Table A3). Branched-chain fatty acids were further positively correlated with PYY expression levels, whereas higher concentrations of acetate, propionate, and butyrate were negatively associated with MCT1 expression. In addition, cecal luminal LPS concentrations were positively associated with enhanced TLR4 and ZO1 expression levels. Most SCFA and LPS stimulated MUC2 expression, whereas propionate was negatively associated with IAP expression.
Figure 4.
Determination of potential key mucosal genera for RFI and cecal mucosal gene expression in pigs ranked on RFI. Covariations between the relative abundances of mucosal bacterial genera (rel. abundances > 0.002) and RFI and relative gene expression in the cecum using sPLS regression. The network is displayed graphically as nodes (genera and expression of genes) and edges (biological relationship between nodes), with the edge color intensity indicating the level of the association: red = positive, and green = negative. Only the strongest pairwise associations were projected, with score threshold depending on the respective association. The score of the association (Pearson’s correlation) is provided in Supplementary Table A3. Relevance networks for (A) standardized RFI (stdRFI), (B) relative MUC2 expression, (C) relative expression of tight-junction proteins, and (D) relative expression of Toll-like receptors. Gene names: MUC2, mucin 2; OCLN, occludin; TLR, Toll-like-receptor; ZO1, zonula occludens-1.
Figure 5.
Determination of the influential effect of luminal VFA and lipopolysaccharide for cecal mucosal gene expression in pigs ranked on RFI. Covariations between VFA and lipopolysaccharides in cecal content and relative gene expression using sPLS regression. The network is displayed graphically as nodes (genera and expression of genes) and edges (biological relationship between nodes). The edge color intensity indicates the level of the association: red = positive, and green = negative. Only the strongest pairwise associations were projected, with score threshold depending on the respective association. The score of the association (Pearson’s correlation) is presented in Supplementary Table A3. Relevance networks for (A) relative expression of luminal defense genes, (B) relative ZO1 expression, (C) relative expression of VFA transporter and signaling genes. Gene names: FFAR, free-fatty acid receptor; IAP, intestinal alkaline phosphatase; MCT1, monocarboxylic acid transporter 1; MUC, mucin; PYY, peptide YY/peptide tyrosine tyrosine; ZO1, zonula occludens-1.
DISCUSSION
So far, few studies evaluated FE-related differences in the bacterial community of the gastrointestinal content in pigs using DNA-based approaches (McCormack et al., 2017; Tan et al., 2017; Yang et al., 2017), whereas a dearth of information is available for the contribution of the gut mucosal microbiota to variation in FE. Because RNA correlates better to microbial growth than DNA (Lettat et al., 2013), we used RNA to target the metabolically active bacteria only in the current study and not dead or dormant bacterial cells. Present results are among the first to show that certain shifts in the composition of the mucosal bacterial community were either indicative of or contributed to the variation in FE of the present pigs, at the end of the finishing period. Notably, the second most predominant genus Campylobacter, a pathobiont, appeared to be indicative for LRFI. This was accompanied by greater mucosal expression of genes for mucin production (MUC2) and barrier function (ZO1) as well as trends for greater expression of VFA transporter MCT1 and VFA signaling genes (GLP1 and PYY). In line with previous findings (Vigors et al., 2016), results further demonstrated that, in clinically healthy pigs, the cecum did not show a disproportionately high innate immune response to microbial activity in HRFI compared to LRFI pigs. Nonetheless, the established relevance networks supported a unique “microbiome signature” of the mucosal bacterial community for host RFI category and gene expression, emphasizing that certain mucosal bacteria and microbial metabolites in cecal content were more associated than others with the RFI category, cecal mucosal expression of VFA sensing, and innate immune response genes in our pigs.
Similar to Kelly et al. (2017), we observed the predominance of only a small number of distinct high-abundant genera and OTUs at the cecal mucosa. Environmental conditions and the bacterial substrate at the mucosa largely differ from those in luminal digesta and favor the growth of oxygen-tolerant populations and mucolytic species adept at living on host glycoproteins (Naughton et al., 2013; Albenberg et al., 2014). Correspondingly, the cecal mucosal community was predominated by microaerophilic Helicobacteraceae and Campylobacteraceae in our finishing pigs. Other highly abundant families at the cecal mucosa comprised Veillonellaceae, Lachnospiraceae, Prevotellaceae, Lactobacillaceae, and Ruminococcaceae which are often associated with the fermentation of complex carbohydrates (Kelly et al., 2017) and the cross-feeding of primary fermentation metabolites (Flint et al., 2015).
Similar to the bacterial community in cecal content (McCormack et al., 2017; Tan et al., 2017), the overall mucosal bacterial community structure and total bacterial numbers were equal between LRFI and HRFI pigs. Rather, RFI-associated variations were only detectable for few specific bacterial community members. Although Campylobacter species such as C. hyointestinalis belong to the normal commensal gut microbiota of pigs (Oporto and Hurtado, 2011; Mann et al., 2014), Campylobacter are enteric pathogens with the capability to cause proliferative enteritis in pigs (Kamei et al., 2015). Therefore, it was rather an unexpected finding that LRFI pigs harbored considerably more Campylobacter at the cecal mucosa; also because they fill a closely related niche with Helicobacter (Naughton et al., 2013; Mann et al., 2014). Both Helicobacter and Campylobacter are adept at survival in intestinal mucus by binding to the glycosylated epitopes in the glycolipid fraction of the outer mucus layer; however, they seem to have diverging binding affinities to the different mucins (Naughton et al., 2013). The outer gut mucus layer mostly consists of mucin 2, a large gel-forming glycoprotein that is modified with O-glycans, with levels and patterns of glycosylation varying with the localization (Quintana-Hayashi et al. 2015; Tailford et al., 2015). The lower mucosal MUC2 but equal MUC4 expression in LRFI pigs compared to HRFI pigs may confirm host-derived differences in mucin synthesis as a reason for the greater Campylobacter abundance in LRFI pigs. Two causal scenarios are plausible. The C. hyointestinalis-OTU may have directly decreased host mucin expression in LRFI pigs like other opportunistic pathogens (e.g., Helicobacter pylori and Campylobacter jejuni), which were shown to modulate host glycan expression following invasion (Cooke et al., 2009; Day et al., 2012). Alternatively, it may be conceivable that HRFI pigs showed a greater mucosal immune response to the residing cecal microbiota than LRFI pigs by expressing more MUC2 in order to counteract mucosal Campylobacter invasion (Quintana-Hayashi et al. 2015). This would have diverted energy for growth to the metabolically costly innate immune response (Rauw, 2012), thereby lowering pig’s FE. Changes in cecal mucosal MUC2 expression may further explain the lower abundance of mucin-adherent Escherichia/Shigella in LRFI compared to HRFI pigs. The 3-O-sulfo-galactosyl moiety on host mucin acts as one of the prime adherence targets for the gut commensal E. coli (Al-Saedi et al., 2017). In this regard, relevance networking indicated the dependency of other low abundant mucosal bacteria (Blautia, Oscillospira, and Bacteroides) on mucosal MUC2 expression. It is also conceivable that Escherichia/Shigella benefitted from the higher substrate flow to the cecum and release of monosaccharides, amino acids, and dipeptides by other bacteria in HRFI pigs. A similar reason may explain the higher mucosal abundance of Ruminobacter in HRFI pigs, which commonly utilize easily fermentable carbohydrates, such as starch and maltodextrins (Anderson, 1995). Likewise, enhanced cross-feeding of lactate and succinate (Flint et al., 2015) may have raised the mucosal abundance of Veillonella species, including V. dispar, in HRFI compared to LRFI pigs. Relevance networking supported a strong association between cecal mucosal enrichment with low abundant Veillonella and Ruminobacter and HRFI in the present study.
Due to their molecular patterns on their cell wall, the high mucosal abundance of ε-Proteobacteria (i.e., Helicobacter- and Campylobacter-OTUs) may have caused an elevated mucosal immune response which may have compromised the FE of the pigs in the present study. Intriguingly, the higher abundance of Campylobacter in LRFI pigs and the negative association between RFI and Helicobacter indicated an opposite relationship. For this reason, LRFI pigs may have developed a greater immune-tolerance toward the present Campylobacter than HRFI pigs (Frosali et al., 2015). The constant exposure of the gut mucosa to their TLR ligands may have induced a basal state of activation of downstream signaling pathways which probably limited inflammatory responses (Frosali et al., 2015). This may also explain why Ruminobacter was the most decisive genus associated with TLR4 expression in the respective relevance network. Strong mucosal immune stimulation via TLR-4 and nuclear factor NF-κB activation is commonly reported for E. coli-derived LPS (Wyns et al., 2015). However, this may have been of minor importance for the microbe–host interactions in our HRFI pigs due to the predominance of ε-Proteobacteria. Upregulation of NFKB expression by luminal triggers induces signaling cascades, ultimately leading to the enforcement of the intestinal barrier (Lawrence, 2009). Accordingly, the luminal LPS was positively associated with mucosal ZO1 expression in the present relevance network. Moreover, mucosal bacteria other than the RFI-associated genera appeared to be more associated with enhanced ZO1 expression in HRFI pigs, including genera such as Fusobacterium, Clostridium, Lactococcus, and Bacillus, some of them comprising pathobionts. Nonetheless, it is the complex interplay between taxa at the outer mucus layer that will cause alterations in host mucosal gene expression. Moreover, commensals in luminal digesta probably interfere in the cross talk between mucosal bacteria and the host (Frosali et al., 2015).
Although LRFI and HRFI pigs had similar cecal VFA concentrations, the higher feed intake in HRFI compared to LRFI pigs most likely increased the daily production of VFA as previously reported for pig lines divergent for RFI (Montagne et al., 2014). Therefore, the observed trends for higher mucosal expression of VFA uptake (MCT1) and signaling genes (GLP1 and PYY) in HRFI pigs could be the consequence of their higher feed intake compared to LRFI pigs. About 40% of intestinally produced VFA are actively taken up by the enterocyte via substrate-induced monocarboxylate transporters (Sepponen et al., 2007). Accordingly, enhanced cecal mucosal expression of MCT1 in HRFI pigs could reflect increased luminal VFA availability (Sepponen et al., 2007). Absorbed VFA contribute to energy homeostasis and de novo lipogenesis, thereby modulating body composition and potentially FE in the host (Li et al., 2017). The present relevance networks confirmed mucosal signaling of luminal VFA via activation of the G-protein receptors free fatty acid receptor (FFAR) 2 and FFAR3 (McKenzie et al., 2017). VFA-dependent FFAR signaling enhances host insulin sensitivity by promoting glucagon-like peptide-1 secretion (Tolhurst et al., 2012), whereas VFA-dependent secretion of the gut hormone peptide YY induces satiety, thereby reducing food intake (Samuel et al., 2008). Because 1 phenotypic characteristic of HRFI compared to LRFI pigs is their higher feed intake, we would have rather expected lower PYY expression in HRFI pigs. Clearly, the complex interactions between nervous, endocrine, and immune processes at the gut mucosa and systemic level need to be considered as a whole (Li et al., 2017) to explain pig’s feeding behavior and definitely not the expression of single genes. Moreover, mucosal VFA sensing via G-protein receptors downregulates the expression of pro-inflammatory genes (McKenzie et al., 2017). Therefore, it may be conceivable that a higher daily cecal VFA production might have balanced the mucosal innate immune response in HRFI compared to LRFI pigs.
Overall, differences in the mucosal bacterial metabolic pathways predicted to be either more or less abundant in feed efficient pigs were small and potentially of minor importance to host FE. Reactive oxygen species, such as superoxide and hydrogen peroxide, produced during oxidative phosphorylation have been proposed to play critical roles in microbial metabolism in response to environmental stressors (Brynildsen et al., 2013; Zhao and Drlica, 2014) and may therefore indicate differences in cecal mucosal microbe-to-microbe interactions between LRFI and HRFI pigs. By contrast, the trend for a greater relative abundance of biosynthesis of unsaturated fatty acid genes in LRFI pigs might support certain protective effects of the mucosal microbiota on epithelial functioning, such as barrier function (Miyamoto et al., 2016).
In conclusion, present results demonstrated FE-associated compositional differences within the cecal mucosal microbiota of finishing pigs. In particular, high-abundant Campylobacter tended to be indicative for LRFI. However, results do not allow the determination of whether mucosal bacterial changes were a contributing factor for variation in FE and mucosal gene expression or rather a consequence of FE-related changes in host physiology or behavior (e.g., feed intake). Restricting the feed intake of LRFI and HRFI pigs may be 1 optional strategy in future studies to investigate whether the feeding behavior provoked the observed changes in the mucosal bacterial community of LRFI and HRFI pigs. Relevance networks confirmed that certain mucosal bacteria and luminal microbial metabolites were more associated with RFI and the cecal mucosal gene expression. Using a cDNA-based method was probably more discriminative than using DNA for the characterization of FE-associated taxa at the cecal mucosa as it only measured active and not inactive and dead cells. However, differences in mucosal gene expression levels are only the initial steps in adaptive processes and do not necessarily reflect functional protein patterns.
SUPPLEMENTARY DATA
Supplementary data are available at Journal of Animal Science online.
ACKNOWLEDGMENTS
This project (ECO-FCE) has received funding from the European Union’s Seventh Framework Programme for research, technological development, and demonstration (project no. 311794). The authors would like to thank the staff of the research and teaching farm, C. Knecht, C. Unterweger, E. Sassu, and U. Ruzcicka of the University Clinic for Swine for assisting with pig management and sampling, as well as A. Sener, S. Sharma, A. Dockner, and M. Wild for excellent laboratory assistance (University of Veterinary Medicine). All authors disclose no conflict of interests.
LITERATURE CITED
- Albenberg L., T. V., Esipova C. P., Judge K., Bittinger J., Chen A., Laughlin S., Grunberg R. N., Baldassano J. D., Lewis H., Li et al. 2014. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147:1055–1063.e8. doi:10.1053/j.gastro.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Saedi F., Vaz D. P., Stones D. H., and Krachler A. M.. 2017. 3-sulfo-galactosyl dependent adhesion of Escherichia coli HS multivalent adhesion molecule is attenuated by sulfatase activity. J. Biol. Chem. 292:19792–19803. doi:10.1074/jbc.M117.817908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K. L. 1995. Biochemical analysis of starch degradation by Ruminobacter amylophilus 70. Appl. Environ. Microbiol. 61:1488–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen C. L., J. L. Jensen, and Ørntoft T. F.. 2004. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64:5245–5250. doi:10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- Brynildsen M. P., Winkler J. A., Spina C. S., MacDonald I. C., and Collins J. J.. 2013. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat. Biotechnol. 31:160–165. doi:10.1038/nbt.2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I.,. et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. doi:10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. R., Q., Wang E., Cardenas J., Fish B., Chai R. J., Farris A. S., Kulam-Syed-Mohideen D. M., McGarrell T., Marsh G. M., Garrity et al. 2009. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. doi:10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C. L., An H. J., Kim J., Canfield D. R., Torres J., Lebrilla C. B., and Solnick J. V.. 2009. Modification of gastric mucin oligosaccharide expression in rhesus macaques after infection with Helicobacter pylori. Gastroenterology 137:1061–1071, 1071.e1. doi:10.1053/j.gastro.2009.04.014 [DOI] [PubMed] [Google Scholar]
- Day C. J., Semchenko E. A., and Korolik V.. 2012. Glycoconjugates play a key role in Campylobacter jejuni infection: interactions between host and pathogen. Front. Cell. Infect. Microbiol. 2:9. doi:10.3389/fcimb.2012.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckardt K., Metzler-Zebeli B. U., and Zebeli Q.. 2016. Processing barley grain with lactic acid and tannic acid ameliorates rumen microbial fermentation and degradation of dietary fibre in vitro. J. Sci. Food Agric. 96:223–231. doi:10.1002/jsfa.7085 [DOI] [PubMed] [Google Scholar]
- de la Cruz-Leyva M. C., Zamudio-Maya M., Corona-Cruz A. I., González-de la Cruz J. U., and Rojas-Herrera R.. 2011. A method for isolating RNA from metabolically active bacterial flora associated with octopus. Lett. Appl. Microbiol. 53:8–13. doi:10.1111/j.1472-765X.2011.03057.x [DOI] [PubMed] [Google Scholar]
- Edgar R. C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi:10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C., and Knight R.. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi:10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint H. J., Duncan S. H., Scott K. P., and Louis P.. 2015. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 74:13–22. doi:10.1017/S0029665114001463 [DOI] [PubMed] [Google Scholar]
- Frosali S., Pagliari D., Gambassi G., Landolfi R., Pandolfi F., and Cianci R.. 2015. How the intricate interaction among Toll-like receptors, microbiota, and intestinal immunity can influence gastrointestinal pathology. J. Immunol. Res. 2015:489821. doi:10.1155/2015/489821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiko G. E. and Stappenbeck T. S.. 2014. Host-microbe interactions shaping the gastrointestinal environment. Trends Immunol. 35:538–548. doi:10.1016/j.it.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei K., N. Hatanaka M. Asakura S. Somroop W. Samosornsuk A. Hinenoya N. Misawa S. Nakagawa, and Yamasaki S.. 2015. Campylobacter hyointestinalis isolated from pigs produces multiple variants of biologically active cytolethal distending toxin. Infect. Immun. 83:4304–4313. doi:10.1128/IAI.00997-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J., K. Daly A. W. Moran S. Ryan D. Bravo, and Shirazi-Beechey S. P.. 2017. Composition and diversity of mucosa-associated microbiota along the entire length of the pig gastrointestinal tract; dietary influences. Environ. Microbiol. 19:1425–1438. doi:10.1111/1462-2920.13619 [DOI] [PubMed] [Google Scholar]
- Kononoff P. J. and Hanford K. J.. 2006. Technical note: estimating statistical power of mixed models used in dairy nutrition experiments. J. Dairy Sci. 89:3968–3971. doi:10.3168/jds.S0022-0302(06)72439-0 [DOI] [PubMed] [Google Scholar]
- Langille M. G., J., Zaneveld J. G., Caporaso D., McDonald D., Knights J. A., Reyes J. C., Clemente D. E., Burkepile R. L., Vega Thurber R., Knight et al. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31:814–821. doi:10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T. 2009. The nuclear factor NF-kappab pathway in inflammation. Cold Spring Harb. Perspect. Biol. 1:a001651. doi:10.1101/cshperspect.a001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê Cao K. A., I. González, and Déjean S.. 2009a. Integromics: an R package to unravel relationships between two omics datasets. Bioinformatics 25:2855–2856. doi:10.1093/bioinformatics/btp515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê Cao K. A., P. G. Martin C. Robert-Granié, and Besse P.. 2009b. Sparse canonical methods for biological data integration: application to a cross-platform study. BMC Bioinformatics 10:34. doi:10.1186/1471-2105-10-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leser T. D. and Mølbak L.. 2009. Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ. Microbiol. 11:2194–2206. doi:10.1111/j.1462-2920.2009.01941.x [DOI] [PubMed] [Google Scholar]
- Lettat A., F. Hassanat, and Benchaar C.. 2013. Corn silage in dairy cow diets to reduce ruminal methanogenesis: effects on the rumen metabolically active microbial communities. J. Dairy Sci. 96:5237–5248. doi:10.3168/jds.2012-6481 [DOI] [PubMed] [Google Scholar]
- Li X., Y. Shimizu, and Kimura I.. 2017. Gut microbial metabolite short-chain fatty acids and obesity. Biosci. Microbiota Food Health 36:135–140. doi:10.12938/bmfh.17-010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach N., M., Berri J., Estellé F., Levenez G., Lemonnier C., Denis J. J., Leplat C., Chevaleyre Y., Billon J., Doré et al. 2015. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 7:554–569. doi:10.1111/1758-2229.12285 [DOI] [PubMed] [Google Scholar]
- Mani V., Harris A. J., Keating A. F., Weber T. E., Dekkers J. C., and Gabler N. K.. 2013. Intestinal integrity, endotoxin transport and detoxification in pigs divergently selected for residual feed intake. J. Anim. Sci. 91:2141–2150. doi:10.2527/jas.2012-6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann E., Schmitz-Esser S., Zebeli Q., Wagner M., Ritzmann M., and Metzler-Zebeli B. U.. 2014. Mucosa-associated bacterial microbiome of the gastrointestinal tract of weaned pigs and dynamics linked to dietary calcium-phosphorus. PLoS One 9:e86950. doi:10.1371/journal.pone.0086950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer E. A., Tillisch K., and Gupta A.. 2015. Gut/brain axis and the microbiota. J. Clin. Invest. 125:926–938. doi:10.1172/JCI76304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack U. M., Curião T., Buzoianu S. G., Prieto M. L., Ryan T., Varley P., Crispie F., Magowan E, Metzler-Zebeli B. U., Berry D.,. et al. 2017. Exploring a possible link between the intestinal microbiota and feed efficiency in pigs. Appl. Environ. Microbiol. 83:e00380-17. doi:10.1128/AEM.00380-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie C., J. Tan L. Macia, and Mackay C. R.. 2017. The nutrition-gut microbiome-physiology axis and allergic diseases. Immunol. Rev. 278:277–295. doi:10.1111/imr.12556 [DOI] [PubMed] [Google Scholar]
- Metzler-Zebeli B. U., P. G. Lawlor E. Magowan U. M. McCormack T. Curião M. Hollmann R. Ertl J. R. Aschenbach, and Zebeli Q.. 2017. Finishing pigs that are divergent in feed efficiency show small differences in intestinal functionality and structure. PLoS One 12:e0174917. doi:10.1371/journal.pone.0174917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler-Zebeli B. U., Lawlor P. G., Magowan E., and Zebeli Q.. 2016. Effect of freezing conditions on fecal bacterial composition in pigs. Animals 6:18. doi:10.3390/ani6030018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler-Zebeli B. U., Mann E., Schmitz-Esser S., Wagner M., Ritzmann M., and Zebeli Q.. 2013. Changing dietary calcium-phosphorus level and cereal source selectively alters abundance of bacteria and metabolites in the upper gastrointestinal tracts of weaned pigs. Appl. Environ. Microbiol. 79:7264–7272. doi:10.1128/AEM.02691-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto J., S. Hasegawa M. Kasubuchi A. Ichimura A. Nakajima, and Kimura I.. 2016. Nutritional signaling via free fatty acid receptors. Int. J. Mol. Sci. 17:450. doi:10.3390/ijms17040450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne L., F. Loisel T. Le Naou F. Gondret H. Gilbert, and Le Gall M.. 2014. Difference in short-term responses to a high-fiber diet in pigs divergently selected for residual feed intake. J. Anim. Sci. 92:1512–1523. doi:10.2527/jas.2013-6623 [DOI] [PubMed] [Google Scholar]
- Muyzer G., E. C. de Waal, and Uitterlinden A. G.. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton J. A., K., Mariño B., Dolan C., Reid R., Gough M. E., Gallagher M., Kilcoyne J. Q., Gerlach L., Joshi P., Rudd et al. 2013. Divergent mechanisms of interaction of Helicobacter pylori and Campylobacter jejuni with mucus and mucins. Infect. Immun. 81:2838–2850. doi:10.1128/IAI.00415-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oporto B. and Hurtado A.. 2011. Emerging thermotolerant campylobacter species in healthy ruminants and swine. Foodborne Pathog. Dis. 8:807–813. doi:10.1089/fpd.2010.0803 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C., and Neuvians T. P.. 2004. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26:509–515. [DOI] [PubMed] [Google Scholar]
- Quintana-Hayashi M. P., M., Mahu N., De Pauw F., Boyen F., Pasmans A., Martel P., Premaratne H. R., Fernandez O., Teymournejad L., Vande Maele et al. 2015. The levels of Brachyspira hyodysenteriae binding to porcine colonic mucins differ between individuals, and binding is increased to mucins from infected pigs with de novo MUC5AC synthesis. Infect. Immun. 83:1610–1619. doi:10.1128/IAI.03073-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauw W. M. 2012. Immune response from a resource allocation perspective. Front. Genet. 3:267. doi:10.3389/fgene.2012.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel B. S., Shaito A., Motoike T., Rey F. E., Backhed F., Manchester J. K., Hammer R. E., Williams S. C., Crowley J., Yanagisawa M.,. et al. 2008. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. USA 105:16767–16772. doi:10.1073/pnas.0808567105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepponen K., M. Ruusunen J. A. Pakkanen, and Pösö A. R.. 2007. Expression of CD147 and monocarboxylate transporters MCT1, MCT2 and MCT4 in porcine small intestine and colon. Vet. J. 174:122–128. doi:10.1016/j.tvjl.2006.05.015 [DOI] [PubMed] [Google Scholar]
- Tailford L. E., Crost E. H., Kavanaugh D., and Juge N.. 2015. Mucin glycan foraging in the human gut microbiome. Front. Genet. 6:81. doi:10.3389/fgene.2015.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z., T. Yang Y. Wang K. Xing F. Zhang X. Zhao H. Ao S. Chen J. Liu, and Wang C.. 2017. Metagenomic analysis of cecal microbiome identified microbiota and functional capacities associated with feed efficiency in landrace finishing pigs. Front. Microbiol. 8:1546. doi:10.3389/fmicb.2017.01546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhurst G., H. Heffron Y. S. Lam H. E. Parker A. M. Habib E. Diakogiannaki J. Cameron J. Grosse F. Reimann, and Gribble F. M.. 2012. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 61:364–371. doi:10.2337/db11-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigors S., O’Doherty J. V., Kelly A. K., O’Shea C. J., and Sweeney T.. 2016. The effect of divergence in feed efficiency on the intestinal microbiota and the intestinal immune response in both unchallenged and lipopolysaccharide challenged ileal and colonic explants. PLoS One 11:e0148145. doi:10.1371/journal.pone.0148145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- Wyns H., E. Plessers P. De Backer E. Meyer, and Croubels S.. 2015. In vivo porcine lipopolysaccharide inflammation models to study immunomodulation of drugs. Vet. Immunol. Immunopathol. 166:58–69. doi:10.1016/j.vetimm.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Yang H., Huang X., Fang S., He M., Zhao Y., Wu Z., Yang M., Zhang Z., Chen C., and Huang L.. 2017. Unraveling the fecal microbiota and metagenomic functional capacity associated with feed efficiency in pigs. Front. Microbiol. 8:1555. doi:10.3389/fmicb.2017.01555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. and Drlica K.. 2014. Reactive oxygen species and the bacterial response to lethal stress. Curr. Opin. Microbiol. 21:1–6. doi:10.1016/j.mib.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.