Abstract
Dietary fiber content and composition affect microbial composition and activity in the gut, which in turn influence energetic contribution of fermentation products to the metabolic energy supply in pigs. This may affect feed efficiency (FE) in pigs. The present study investigated the relationship between the fecal microbial composition and FE in individual growing-finishing pigs. In addition, the effects of diet composition and sex on the fecal microbiome were studied. Fecal samples were collected of 154 grower-finisher pigs (3-way crossbreeds) the day before slaughter. Pigs were either fed a diet based on corn/soybean meal (CS) or a diet based on wheat/barley/by-products (WB). Fecal microbiome was characterized by 16S ribosomal DNA sequencing, clustered by operational taxonomic unit (OTU), and results were subjected to a discriminant approach combined with principal component analysis to discriminate diets, sexes, and FE extreme groups (10 high and 10 low FE pigs for each diet by sex-combination). Pigs on different diets and males vs. females had a very distinct fecal microbiome, needing only 2 OTU for diet (P = 0.020) and 18 OTU for sex (P = 0.040) to separate the groups. The 2 most important OTU for diet, and the most important OTU for sex, were taxonomically classified as the same bacterium. In pigs fed the CS diet, there was no significant association between FE and fecal microbiota composition based on OTU (P > 0.05), but in pigs fed the WB diet differences in FE were associated with 17 OTU in males (P = 0.018) and to 7 OTU in females (P = 0.010), with 3 OTU in common for both sexes. In conclusion, our results showed a diet and sex-dependent relationship between FE and the fecal microbial composition at slaughter weight in grower-finisher pigs.
Keywords: feed efficiency, fecal microbiome, diet, sex, pig
INTRODUCTION
In pork production, feed efficiency (FE) is very important, as feed is the main component of the cost prize. The gut microbiota can play an important role in FE, as pigs do not produce digestive enzymes that allow them to digest the fiber fraction in the diet. Instead, they depend on microbiota residing in the gastrointestinal tract, in particular in the hindgut, to break down the dietary fiber in fermentation processes. VFA are resulting by-products of the fermentation activity of the microbiota and they serve, after absorption from the gut, as energy sources in systemic metabolism (Ingerslev et al., 2014). In pigs, efficiency of energy utilization is lower when energy comes from fiber instead of starch (Noblet and Le Goff, 2001). Thus, for improving FE in pigs, low fiber, high starch diets have been favored (Zijlstra and Beltranena, 2013). However, dietary fiber has shown to reduce stereotypic behavior and aggression (Meunier-Salaün et al., 2001) and improve fecal consistency (Mateos et al., 2006; Wellock et al., 2008). Combined with the increasing competition of feed with human edible products for amongst others arable land (Van Kernebeek et al., 2016), this has caused the agricultural sector to move increasingly towards the formulation of diets with higher fiber contents. Therefore, the importance of intestinal microbiota and their fermentation activity in relation to FE in pigs is likely to increase.
The aim of this study was to investigate the association between FE and fecal microbial composition in commercial grower-finisher pigs. In addition, two factors affecting FE were investigated for their effect on the fecal microbiome: diet composition and sex.
MATERIALS AND METHODS
This study was carried out in strict accordance with the recommendations in the European Guidelines for accommodation and care of animals. The protocol was approved by the Animal Care and Use Committee of Schothorst Feed Research, The Netherlands (Protocol Number: AVD 246002015120/132). The dataset is available on request from the corresponding author.
Animals and Experimental Design
Pigs used in this study originated from a three-way cross (Synthetic boar × [Large White × Landrace]). Phenotypic data were available for 160 three-breed cross pigs, 81 males and 79 females, coming from 20 litters. All pigs were kept at the experimental facilities of Schothorst Feed Research B.V. (Lelystad, The Netherlands) under commercial conditions. Up until the start of the trial the animals were housed per litter and all animals were fed the same diet. The pigs were put on test at 8 to 9 wk of age (day 0), in two groups of 80, and experimental groups were set 13 wk apart. Distribution was as follows: ten pigs per pen and eight pens per compartment; one compartment was used per entrance date. Littermates were split randomly over the two diets and sexes were housed in separate pens, resulting in two pens per diet per sex per entrance date. All animals were used for the evaluation of the effects of diet composition and sex on fecal microbiota composition, except for six animals of which no fecal sample was obtained. The FE was defined as the ratio of body weight gain to cumulated feed intake from start of the test until the day of slaughter. For evaluation of the effect of fecal microbiota composition on FE, the 25% pigs with the highest and the 25% with the lowest individual FE per diet per sex (20 animals per combination) were used. Data of one animal were excluded, since it had a very low feed intake and body weight gain during the second half of the test. At the start of the experiment, the pigs had an average BW of 23.0 kg and were kept in the facilities until they reached a live weight at slaughter of approximately 120 kg. Pigs were allowed a minimal space of 1 m2 per pig, and the pens were equipped with 60% concrete floor and 40% slatted floor.
Feeding Strategy
Two different diets were studied, a diet based on corn/soybean meal (CS) as typically fed to commercial grower-finisher pigs in The America’s and a diet based on wheat/barley/by-products (WB) as typically fed in Europe (Table 1). For both diets, the pigs were fed ad libitum according to a three-phase feeding program. The first phase (Tstarter) was from days 0 to 25 on test and pigs were fed a starter diet. The second phase (Tgrower) was from days 26 to 67 on test and pigs were fed a grower diet. The third phase (Tfinisher) was from day 68 on test until the pigs reached slaughter weight and they were fed a finisher diet. The diets were custom-made diets based on commonly used commercial diets and were formulated on a fixed ratio of net energy to digestible lysine (NE:SID lysine). Each of the three phases had a different NE:SID lysine, being 0.89 J/g at Tstarter, 1.06 J/g at Tgrower, and 1.37 J/g at Tfinisher. The increase of NE:SID lysine in grower and finisher diets was mainly achieved by exchanging soybean meal with corn, and peas with wheat for the CS and WB diets, respectively. The experimental diets were produced in the feed plant of ABZ Animal Nutrition, Leusden, The Netherlands.
Table 1.
Ingredient and calculated nutrient composition of the diets, as-fed basis
| Item | Starter (days 0 to 25) | Grower (days 26 to 67) | Finisher (days 68 to end) | |||
|---|---|---|---|---|---|---|
| CS | WB | CS | WB | CS | WB | |
| Ingredient, g/kg | ||||||
| Corn | 647.1 | – | 698.4 | – | 755.1 | – |
| Corn gluten | 18.1 | – | 25.0 | 50.0 | 50.0 | 50.0 |
| Soybean meal | 240.5 | 100.0 | 180.5 | 21.5 | 98.3 | – |
| Soybean hull | – | – | – | 14.3 | – | 50.0 |
| Soybean oil | – | 25.0 | – | 0.3 | – | – |
| Barley | – | 200.0 | – | 100.0 | – | 150.0 |
| Wheat | – | 321.9 | – | 400.0 | – | 350.0 |
| Wheat middlings | – | – | – | 50.0 | – | 125.0 |
| Rapeseed meal | – | 63.0 | – | 80.0 | – | 100.0 |
| Sunflower meal | – | 80.0 | – | 80.0 | – | 21.9 |
| Palmkernel meal | – | – | – | 50.0 | – | 50.0 |
| Palm oil | 5.0 | 17.3 | 5.0 | 16.0 | 5.0 | 5.0 |
| Peas | – | 120.0 | – | 29.4 | – | – |
| Sugarcane molasses | 40.0 | 30.0 | 50.0 | 50.0 | 50.0 | 50.0 |
| Animal fat | – | – | – | 27.5 | – | 29.4 |
| Monocalcium phosphate | 6.7 | 5.3 | 2.0 | – | 0.7 | – |
| Salt | 2.7 | 2.1 | 2.4 | 1.8 | 1.8 | 2.1 |
| Calcium carbonate | 11.6 | 10.9 | 9.4 | 8.9 | 9.9 | 4.0 |
| Sodium bicarbonate | – | 1.1 | 1.0 | 1.0 | 3.4 | – |
| Phytase | 5.0 | 5.0 | 5.0 | 5.0 | 5.0 | 1.9 |
| L-lysine HCl | – | 3.8 | – | 4.3 | – | – |
| DL-methionine | – | 1.3 | – | 0.7 | – | – |
| L-threonine | – | 1.7 | – | 1.6 | – | – |
| Lysine + thrypophan | 7.7 | 4.3 | 8.2 | 3.6 | 9.1 | – |
| Lysine HC | 3.0 | – | 2.7 | – | 2.3 | 4.0 |
| Methionine HC | 2.8 | – | 2.5 | – | 1.5 | 0.3 |
| Threonine HC | 3.8 | – | 3.9 | – | 3.8 | 2.4 |
| Valine | – | 1.4 | – | – | – | – |
| Vitamin premixa | 1.0 | 1.0 | – | – | – | – |
| Vitamin-trace mineral premix 1b | 1.0 | 1.0 | – | – | – | – |
| Vitamin-trace mineral premix 2c | 4.0 | 4.0 | 4.0 | 4.0 | 4.0 | 4.0 |
| Nutrient composition, g/kgd | ||||||
| NE, MJ/kg | 9.9 | 9.9 | 10.1 | 9.7 | 10.3 | 9.3 |
| Moisture | 127 | 126 | 130 | 126 | 130 | 129 |
| Ash | 51 | 52 | 42 | 47 | 38 | 42 |
| Crude protein | 182 | 190 | 159 | 166 | 128 | 147 |
| Crude fat | 34 | 58 | 35 | 64 | 36 | 57 |
| Crude fiber | 24 | 45 | 24 | 60 | 25 | 71 |
| Starch | 437 | 360 | 471 | 335 | 512 | 334 |
| Sugar | 44 | 50 | 46 | 58 | 42 | 59 |
| NSP | 135 | 170 | 130 | 216 | 126 | 246 |
| Ca | 6.9 | 6.9 | 5.2 | 5.5 | 5.0 | 3.8 |
| P | 4.8 | 5.5 | 3.6 | 4.7 | 3.2 | 4.7 |
| SID Lys | 11.1 | 11.1 | 9.5 | 9.1 | 7.5 | 6.8 |
| SID Met + Cys | 6.6 | 6.6 | 5.9 | 5.6 | 4.6 | 4.6 |
| SID Thr | 7.1 | 7.1 | 6.3 | 6.0 | 5.2 | 4.7 |
| SID Trp | 2.1 | 2.1 | 1.8 | 1.7 | 1.4 | 1.3 |
aSupplied per kilogram of feed: 2500 IU of vitamin A, 500 IU of vitamin D3, and 5 IU of vitamin E (Mervit AD3E; PreMervo, Utrecht, The Netherlands).
bSupplied per kilogram of feed: 12 mg of Fe (ferrous sulfate), 10 mg of Mn (manganous oxide), 0.04 mg of Co cobalt oxide), 0.12 g of Ca, 0.0501 g of P, 0.04 mg of I (potassium iodide), 1000 IU of vitamin A, 100 IU of vitamin D3, 5 IU of vitamin E, 0.4 mg of vitamin B1, 0.8 mg of vitamin B2, 2 mg of pantothenic acid, 4 mg of niacine, 0.4 mg of vitamin B6, 0.2 mg of folate, 0.003 mg of vitamin B12, 10 mg of vitamin C, 0.01 mg of biotine, 0.2 mg of vitamin K3, and 40 mg of choline (Mervit Sporavit; PreMervo).
cSupplied per kilogram of premix: 0.4 g of Ca, 15 mg of Cu (copper sulfate)0, 80 mg of Fe (ferrous sulfate), 24 mg of Mn (manganous oxide), 62 mg of Zn (zinc oxide), 0.04 mg of Co (cobalt oxide), 0.4 mg of I (potassium iodide), 0.2 mg of Se (sodium selenite), 7500 IU of vitamin A, 1500 IU of vitamin D3, 25 IU of vitamin E, 4 mg of vitamin B2, 6 mg of pantothenate, 30 mg of niacin, 0.02 mg of vitamin B12, and 0.752 mg of vitamin K3 (Mervit START M220; PreMervo, Utrecht, the Netherlands).
dBased on chemical composition, digestibility, and energy values for pigs from the Centraal Veevoeder Bureau livestock feed table (CVB, 2011).
Measurements and Sampling
The experimental facilities of Schothorst Feed Research B.V. were equipped with IVOG feeding stations (INSENTEC, Marknesse, The Netherlands) that register individual feed intake of group-housed animals. All animals had ear tags with unique incremental numbering; therefore, individual feed intake records were available for all pigs for each day on test. Animals were weighted at days 0, 56, and at the end of the test. At the end of the feeding trial (1 d before slaughter), individual fecal samples were collected directly at defecation by hand, with gloves, mixed in the glove, and put in small tubes. The samples were immediately frozen in liquid nitrogen and stored at −80 °C. The ADFI was calculated as the cumulated individual feed intake records throughout the trial divided by the length of the trial. The ADG was calculated as the difference between BW measurements divided by the duration of the trial.
Fecal Microbiota Analysis
Fecal samples were used for ribosomal 16S DNA gene sequencing and analysis. Bead beating lyzed the microbial cells and the DNA was purified using the ZR-96 Soil Microbe DNA kit (Zymo Research, Irvine, CA) according to the manufacturer description (Frese et al., 2015). The V3-V4 region was amplified from purified genomic DNA with the primers F343 (CTTTCCCTACACGACGCTCTTC CGATCTTACGGRAGGCAGCAG) and R784 (GGAGTTCAGACGTGTGC TCTTCCGATCTTACCAGGG TATCTAATCCT) using 30 amplification cycles with an annealing temperature of 65 °C (an amplicon of 510 bp, although length varies depending on the organisms). Full length reads of the V3-V4 region were obtained using Illumina Miseq 250-bp paired end reads. Single multiplexing was performed using in-house 6 bp index, which were added to R784 during a second PCR with 12 cycles using forward primer (AATGATACGGCGA CCACCGAGATCTACA CTCTTTCCCTACACGAC) and reverse primer (CAAGCAGAAGACGGCA TACGAGAT-index-GTGACT GGAGTTCAGACGTGT). The resulting PCR products were purified and loaded onto the Illumina MiSeq cartridge according to the manufacturer instructions. The quality of the run was checked internally using PhiX control as recommended by manufacturer, and then each pair-end sequence was assigned to its sample with the help of the previously integrated index. Each pair-end sequence was assembled using Flash software (Magoč and Salzberg, 2011) using at least a 10-bp overlap between the forward and reverse sequences, allowing 10% of mismatch (Lluch et al., 2015). The absence of contamination was checked with a negative control during the PCR (water as template). The quality of the stitching procedure was controlled using four bacterial samples that are run routinely in the sequencing facility in parallel to the current samples.
Statistical Analysis
The resulting sequences of the 154 samples were clustered with Usearch (Edgar, 2010) using the Uparse pipeline (Edgar, 2013) to create operational taxonomic units (OTU). The OTU table of abundance was analyzed by discriminant analysis using principal components (DAPC) (Jombart et al., 2010), to test the association of OTU abundance with a number of factors. Number of dimensions to be included in further analyses was chosen based on stability of the results, determined by adding increasingly more dimensions. In case the stability test gave a range of dimensions, a threshold value of 99% of the original variance was used to decide the number of dimensions. The OTU were sorted based on their contribution to the separation of tested factors in the discriminant analysis, which echoes the weight of each OTU in separating the groups. Using this order, increasingly more OTU were added to separate the groups, until the separation reached significance at P < 0.05. The built-in a-score method of the DAPC was used to determine the statistical significance of the separation based on a permutation test. Briefly, 1,000 simulations with randomized group labels were performed to evaluate if the discriminant analysis could separate the samples in any of those random configurations. The a-score obtained with the true groups was compared with the distribution of the a-scores obtained with the 1,000 simulations to determine whether the separation was due to chance (Jombart et al., 2010). The method was repeated to test separation for diet, sex, and FE groups, and to test association of phylum, class, and genera abundance (based on OTU taxonomy) with these factors. To test whether the results extrapolated to the whole dataset, the OTU relevant for separating the FE groups were used for partial least squares regression (PLSr) (Mevik and Wehrens, 2007) on all animals within the groups and not only the FE extreme pigs. The number of components kept was based on the lowest root-mean-squares error of prediction after leave-one-out cross validation.
After rarefying the data (McMurdie and Holmes, 2013), Bray–Curtis distances between diet, sex, and FE extreme groups were calculated using a maximum of 200 iterations for diet and sex and 100 iterations for FE groups and tested with ADONIS for significance (Oksanen et al., 2017). Shannon Index, Simpson diversity index, and chao1 richness estimator were calculated using the vegan package (Oksanen et al., 2017). Significance of difference in the diversity estimates between the diets, sexes, and FE groups was determined using a generalized linear model (SAS 9.3; SAS Inst. Inc., Cary, NC) with diet, sex, and FE groups as fixed effect. Significance of difference in ADG, ADFI, and FE between the high and low FE groups was determined using a mixed model (SAS 9.3; SAS Inst. Inc., Cary, NC) with animal as experimental unit, FE groups and pen as fixed effect, and BW at start of the test as covariable. For the least squares means calculations, BW at start of the test was fixed at 22 kg.
Taxonomy
To investigate biological functionality of differences between groups, the taxonomy was determined for each OTU by the SILVA Incremental Aligner (SINA) software (Pruesse et al., 2012), which aligns the OTU with the rRNA gene databases provided by the SILVA ribosomal RNA project (Quast et al., 2013). Default SINA settings were used to assign the taxonomy of each OTU, with the minimum identity with query sequence set at 0.97 and number of neighbors per query sequence set at 10. Group level information within genera classification was deleted. In addition, OTU found by DAPC analysis were blasted against the NCBI 16S ribosomal RNA sequences (Bacteria and Archeae) database using BLASTn (McGinnis and Madden, 2004) to determine the bacteria with closest sequence similarity. Default megaBLAST settings were used.
RESULTS
Within the DAPC analysis, it is not possible to account for the pen effect directly. However, when doing a DAPC analysis for all the piglets across all the pens, the cohoused piglets did not group together (results not shown).
Differences Between Diets
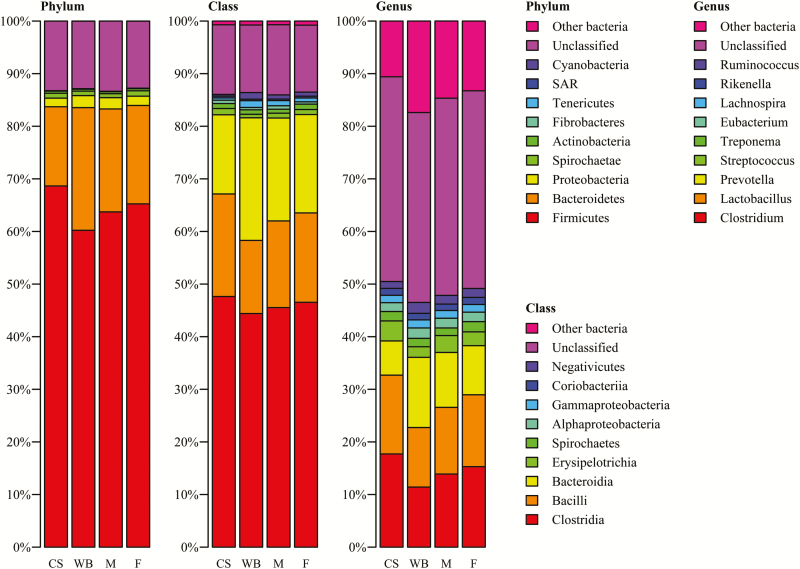
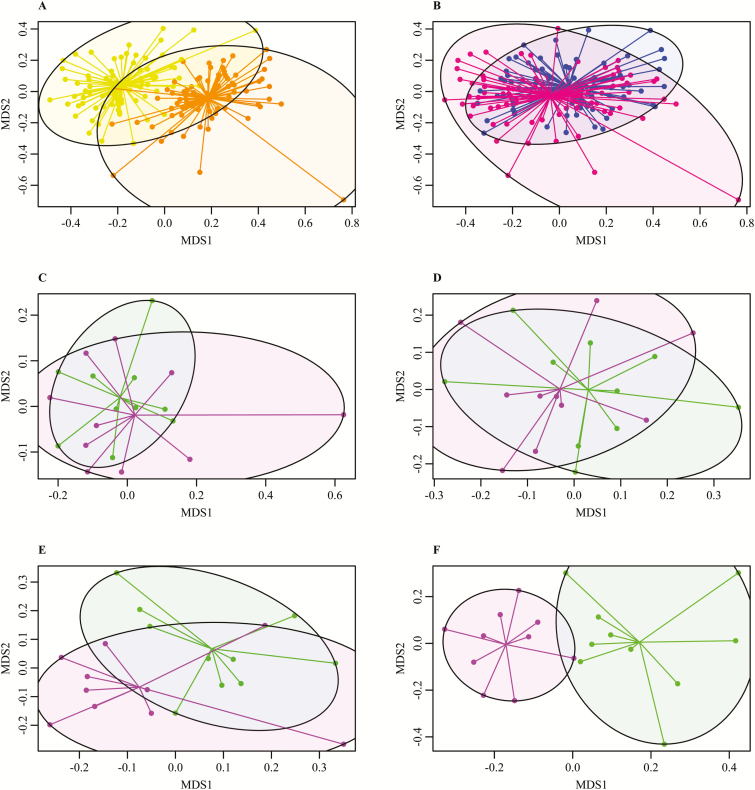
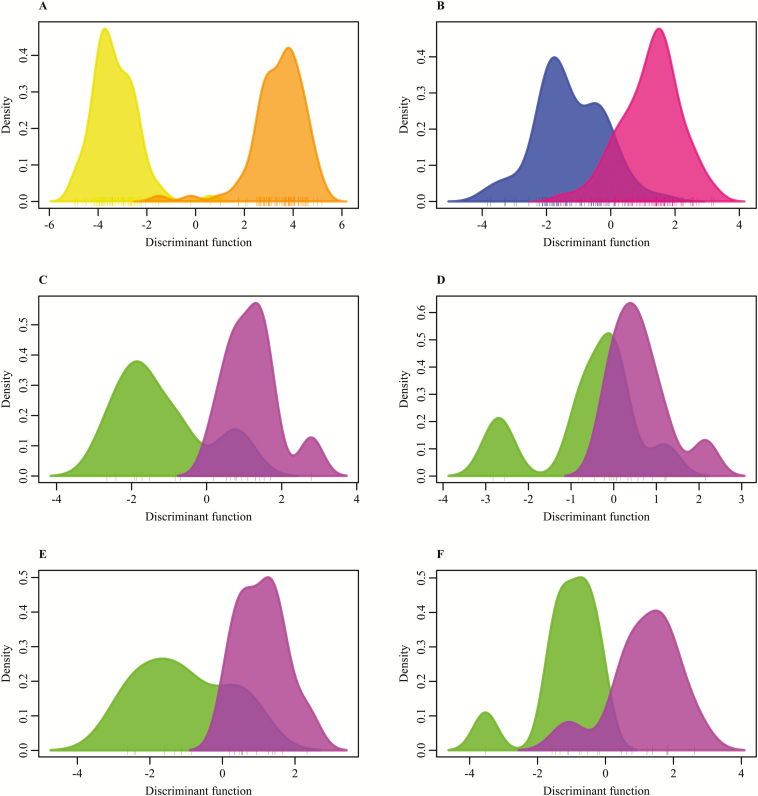
Between the two diets, differences in the relative abundance of the nine major phyla, classes, and genera for both diets were observed (Figure 1). This was reflected in the Bray–Curtis distances at phylum and OTU level (Figure 2), which were significantly different (P < 0.001), but not on class and genera level. The DAPC analysis gave a clear separation in fecal microbiota composition between the two diets based on phyla, classes, genera, and OTU (Figure 3) (P < 0.001). The separation was based on 3, 4, 10, and 55 dimensions for phyla, classes, genera, and OTU, respectively, which represented at least 99% of the original variance in microbiota composition. Keeping the two phyla (Bacteroidetes and Proteobacteria), three classes (Gammaproteobacteria, Spirochaetes, and Bacteroidia), two genera (Ruminococcus and Blautia), and two OTU (OTU 33 and OTU 16) with the highest contribution to the separation was sufficient to discriminate pigs on different diets. Blasting the sequence of the two most contributing OTU to NCBI gave a 95% identity with 99% query coverage with the bacterium Butyricicoccus pullicaecorum. The second most important OTU resulted in the same bacterium, with 96% identity and 99% query coverage. This difference between the diets, however, was not depicted in the measures for diversity. The CS diet had a higher Shannon index than the WB diet (P = 0.021), but the Simpson index and the chao1 index were similar for both diets.
Figure 1.
Relative abundance of 9 major bacterial phyla, classes, and genera in the feces male (M) and female (F) of pigs fed a corn/soybean meal diet (CS) or a wheat/barley/by-products diet (WB). Data are mean percentage of total identified sequences.
Figure 2.
Bray–Curtis distances for tested groups based on operational taxonomic units. (A) Diet. Yellow = corn/soybean meal diet, orange = wheat/barley/by-products diet. (B) Sex. Blue = male pigs, pink = female pigs. (C–F) Feed efficiency. Green = high feed efficiency, purple = low feed efficiency. (C) Male pigs fed a corn/soybean meal diet, (D) female pigs fed a corn/soybean meal diet, (E) male pigs fed a wheat/barley/by-products diet, and (F) female pigs fed a wheat/barley/by-products diet.
Figure 3.
Gaussian kernel density estimation of the discriminant function as a result of the discriminant analysis of principle components for tested groups based on operational taxonomic units. (A) Diet. Yellow = corn/soybean meal diet, orange = wheat/barley/by-products diet. (B) Sex. Blue = male pigs, pink = female pigs. (C–F) Feed efficiency. Green = high feed efficiency, purple = low feed efficiency. (C) Male pigs fed a corn/soybean meal diet, (D) female pigs fed a corn/soybean meal diet, (E) male pigs fed a wheat/barley/by-products diet, and (F) female pigs fed a wheat/barley/by-products diet.
Differences Between Sexes
In contrast to the diets, the overview of the relative abundance of the nine major phyla, classes, and genera (Figure 1) does not indicate obvious differences between the sexes. This is reflected by the results of the Bray–Curtis distances, which were only significant at OTU level (P = 0.037) (Figure 2). The DAPC analysis gave somewhat similar results, as it indicated no separation between the two sexes based on phyla (7 dimensions) and needed 22 out of 45 classes to reach a significant difference between the male and female pigs using 16 dimensions and 100.0% of the original variance. However, there was a highly significant distinction for sex based on genera (P = 0.003) and OTU (P = 0.001) (Figure 3), based on 38 and 60 dimensions (100.0% and 99.2% of the original variance), respectively. There were six genera and 18 OTU required to reach a significant separation between sexes. For nine out of those 18 OTU, it was possible to reliably assign the genus, for 8 it was possible to reliably assign the family, and for one OTU it was not possible to assign any taxonomy (Table 2). The main class differing between the sexes was Methanobacteria and the main genera differing was Bifidobacterium. The most important OTU for sex separation was the same as for diet, which was associated with B. pullicaecorum. There was no difference in any of the diversity indexes between the sexes.
Table 2.
Abundancy and taxonomy (genus level) of the OTU in order of statistical contribution to the separation between sexes
| OTU ID | Classification | Percentage of total sequences | |
|---|---|---|---|
| Boar | Gilt | ||
| OTU16 | Unclassified Ruminococcaceaea | 0.57 | 0.38 |
| OTU35 | Unclassified Ruminococcaceaea | 0.84 | 0.76 |
| OTU12472 | Clostridium | 0.77 | 0.77 |
| OTU373 | Subdoligranulum | 0.16 | 0.23 |
| OTU191 | Unclassifiedb | 0.05 | 0.14 |
| OTU174 | Unclassified Bacteroidalesa | 0.05 | 0.15 |
| OTU22 | Roseburia | 0.26 | 0.33 |
| OTU71 | Ruminococcus | 0.15 | 0.21 |
| OTU33 | Unclassified Ruminococcaceaea | 0.28 | 0.27 |
| OTU19 | Coprococcus | 0.34 | 0.35 |
| OTU136 | Prevotella | 0.06 | 0.13 |
| OTU29 | Unclassified Succinivibrionaceaea | 0.28 | 0.32 |
| OTU20 | Ruminococcus | 0.39 | 0.42 |
| OTU8 | Turicibacter | 0.88 | 0.91 |
| OTU38 | Unclassified Prevotellaceaea | 0.44 | 0.42 |
| OTU127 | Unclassified Prevotellaceaea | 0.20 | 0.13 |
| OTU1050 | Unclassified Prevotellaceaea | 0.42 | 0.32 |
| OTU44 | Ruminococcus | 0.32 | 0.19 |
aReliable depth of taxonomy is limited to family level (query sequence identical for at least 97%).
bNo taxonomic classification available (query sequence identical for at least 97%).
Differences Between FE Extremes
As there was a strong effect of diet and sex on the fecal microbial composition, the dataset was split into 4 groups to estimate the association between FE and microbiome within diet by sex combination. There was a 0.062 to 0.078 g/g difference between the FE groups in FE (Table 3) and there was no pen effect in any of the groups.
Table 3.
Least squares means of the high and low FE groups during the experimental period (overall mean BW at start = 22 kg, overall mean BW at end = 121 kg) per diet by sex combination
| Item | FE groups | P-value | |||
|---|---|---|---|---|---|
| Low | High | SEM | BW start | FE group | |
| CSMa | |||||
| ADG, g/d | 894 | 1028 | 24 | 0.255 | 0.001 |
| ADFI, kg/d | 2.28 | 2.19 | 0.07 | 0.123 | 0.357 |
| FE, g/g | 0.39 | 0.47 | 0.01 | 0.057 | <0.001 |
| CSFb | |||||
| ADG, g/d | 909 | 1045 | 25 | 0.004 | 0.001 |
| ADFI, kg/d | 2.41 | 2.38 | 0.06 | 0.001 | 0.724 |
| FE, g/g | 0.38 | 0.44 | 0.00 | 0.243 | <0.001 |
| WBMc | |||||
| ADG, g/d | 899 | 1016 | 23 | 0.008 | 0.001 |
| ADFI, kg/d | 2.27 | 2.18 | 0.06 | 0.003 | 0.274 |
| FE, g/g | 0.40 | 0.47 | 0.00 | 0.051 | <0.001 |
| WBFd | |||||
| ADG, g/d | 931 | 992 | 27 | 0.499 | 0.120 |
| ADFI, kg/d | 2.60 | 2.27 | 0.06 | 0.305 | 0.002 |
| FE, g/g | 0.36 | 0.44 | 0.00 | 0.471 | <0.001 |
aCSM = male pigs fed a corn/soybean meal diet.
bCSF = female pigs fed a corn/soybean meal diet.
cWBM = male pigs fed a wheat/barley/by-products diet.
dWBF = female pigs fed a wheat/barley/by-products diet
There was no difference in diversity index between the high and low FE animals in any of the diet by sex combinations. In addition, there were only significant Bray–Curtis distances at OTU level for the pigs fed the WB diet (Figure 2). Compared with the diet and sex analyses, the separation between the FE groups using the DAPC analysis was not as clear (Figure 3). At phylum level, only the male pigs fed a WB diet could be separated using five dimensions (100.0% of the original variance). Two phyla were necessary for significant separation, Actinobacteria and Proteobacteria, which were both highest in the high FE pigs. Also at class level, the male pigs fed a WB diet could be significantly separated, based on five dimensions explaining 99.7% of the original variance. Gammaproteobacteria was the first out of the nine contributing classes used for the separation. In addition, the male pigs fed a CS diet could be significantly separated (P = 0.008) and there were16 classes used for the separation. At genera level, the analysis only showed significant separation between high and low female pigs fed the CS diet (P = 0.009) and male pigs fed the WB diet (P = 0.038). Four dimensions were used, explaining 98.7% and 98.3% of the original variance, respectively, and keeping only two genera was sufficient for the separation in the female pigs. These genera were Prevotella and Streptococcus. There were 11 genera needed for the separation in the male pigs with the main genera being Roseburia.
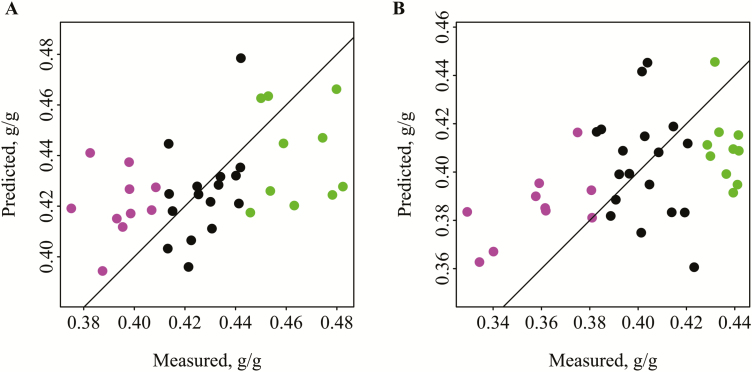
In the pigs fed the CS diet, there was no significant separation for either of the sexes when using OTU, based on eight dimensions for male animals and three for females, explaining 96.7% and 83.2% of the original variance, respectively. In the pigs fed the WB diet, when five dimensions were used (82.7% of the original variance), the low FE (P = 0.016), but not the high FE (P = 0.690), could be identified in the pool of males. In the females fed the WB diet, the high FE pigs were identified (P = 0.016), but not the low FE animals (P = 0.094), based on five dimensions (87.8% of the original variance). In total, 17 OTU were necessary to discriminate the low FE male pigs (P = 0.018) (Table 4) and seven OTU to distinguish the high FE female pigs (P = 0.010) fed a WB diet (Table 5). Putting these OTU in PLSr resulted in an R2 of 0.14 (two components) and 0.11 (three components) for male and female pigs fed the WB diet, respectively (Figure 4). Three of the OTU significant for discriminating high and low FE pigs were common for the male and female pigs. Strikingly, the effects of OTU 4 and 2 had different directions in male and female pigs, as higher abundance was associated with high FE in males and low FE in females.
Table 4.
Abundancy and taxonomy (genus level) of the OTU in order of statistical contribution to the separation between high and low FE boars fed a wheat/barley/by-product diet
| OTU ID | Percentage of total sequences | ||
|---|---|---|---|
| Classification | Low FE | High FE | |
| OTU4 | Lactobacillus | 1.75 | 4.36 |
| OTU24 | Roseburia | 0.23 | 1.36 |
| OTU2 | Unclassified Peptostreptococcaceaea | 4.59 | 5.10 |
| OTU12 | Unclassified Prevotellaceaea | 0.89 | 1.34 |
| OTU3 | Lactobacillus | 1.71 | 1.60 |
| OTU244 | Prevotella | 3.96 | 2.60 |
| OTU5 | Streptococcus | 1.80 | 2.41 |
| OTU8955 | Roseburia | 0.01 | 0.45 |
| OTU1050 | Unclassified Prevotellaceaea | 0.95 | 0.16 |
| OTU9 | Prevotella | 6.43 | 4.73 |
| OTU3132 | Roseburia | 0.03 | 0.43 |
| OTU1 | Clostridium | 8.96 | 7.28 |
| OTU22 | Roseburia | 0.29 | 0.67 |
| OTU12472 | Clostridium | 0.47 | 0.95 |
| OTU41 | Unclassified Prevotellaceaea | 1.82 | 1.34 |
| OTU180 | Ruminococcus | 0.07 | 0.29 |
| OTU13 | Roseburia | 3.27 | 2.50 |
aReliable depth of taxonomy is limited to family level (query sequence identical for at least 97%).
Table 5.
Abundancy and taxonomy (genus level) of the OTU in order of statistical contribution to the separation between high and low FE gilts fed a wheat/barley/by-product diet
| OTU ID | Percentage of total sequences | ||
|---|---|---|---|
| Classification | Low FE | High FE | |
| OTU2 | Unclassified Peptostreptococcaceaea | 5.35 | 5.27 |
| OTU10 | Prevotella | 0.65 | 2.00 |
| OTU55 | Ruminococcus | 0.24 | 0.82 |
| OTU13 | Roseburia | 2.80 | 1.76 |
| OTU4 | Lactobacillus | 4.25 | 3.14 |
| OTU49 | Prevotella | 0.97 | 0.32 |
| OTU6 | Lactobacillus | 4.21 | 2.45 |
aReliable depth of taxonomy is limited to family level (query sequence identical for at least 97%).
Figure 4.
Measured vs. predicted feed efficiency by partial least squares regression based on significant operational taxonomic units found by discriminant analysis of principal components. (A) Male pigs fed a wheat/barley/by-products diet. R2 = 0.14. (B) Female pigs fed a wheat/barley/by-products diet. R2 = 0.11. Green = pigs in high feed efficiency group, purple = pigs in low feed efficiency group.
DISCUSSION
The aim of this study was to investigate the association between FE and the composition of the fecal microbiome in commercial grower-finisher pigs. In the present experiment, the fecal microbiome was used as an indicator for the microbiome in the gastro-intestinal tract during the whole grower-finisher period. However, extrapolation of results of the microbial composition in the feces to other compartments of the gastro-intestinal tract might not be valid. Microbial composition in the ileum, cecum, and colon differs, with the ileal intestinal microbiome being most different from that in other compartments (Looft et al., 2014). Moreover, microbial composition in digesta in the lumen of the gut is different from the mucosa-associated microbiota (Looft et al., 2014). It is also questionable whether the fecal samples, taken at the end of the grower-finisher period, are representative for the whole grower finisher period, as the microbial composition in the feces might change with age (Kim et al., 2011). As the microbial composition at the start and at other time points of the experiment was not measured, the age at which differences in the microbiome for the tested effects appear is unknown. Nevertheless, the fecal microbiome seems most similar to both luminal and mucosal microbiome in the mid-colon (Looft et al., 2014) and is most similar for pigs aged 10 and 13 wk, and for pigs aged 16, 19, and 22 wk (Kim et al., 2011). Therefore, when interpreting our results in terms of relationships between microbial composition and performance of the pig, it should be considered that the fecal microbiome measured in the present study is likely most representative for the microbial composition in the colon, in particular during the last part of the growth trajectory considered.
Diets
From the literature, it is well known that diet composition affects the microbial composition in the gastrointestinal tract (Bauer et al., 2006) and the current study confirms these observations. Worldwide there are two mainstream diets fed to grower-finishers based on the availability of main ingredients: a diet based on corn and soybean meal as is common in North and South America, and a diet based on wheat, barley, and by-products from the agro-food sector as is common in Europe and parts of China. Both diets are used to grow pigs as fast and cost-efficient as possible, even though the ingredient composition is rather different. The diets studied differed mostly in dietary fiber content and composition. The main fiber components in wheat, barley, and corn are arabinoxylans, β-glucans, and cellulose, whereas in soybean meal, the fiber mainly contains pectic substances in the form of rhamnogalacturonan (Choct, 1997). This is reflected in the observed differences in microbiome between the two diets in the current study, as B. pullicaecorum, comprising the two OTU with highest abundance in the CS diet, is highly efficient in fermenting starch (Eeckhaut et al., 2008). This most likely relates to the high starch content in the CS diet. Additionally, the third most important OTU was found to be Blautia wexlera and had the highest abundancy in the WB diet. This bacterium mainly ferments arabinose, glucose, mannose and xylose (Liu et al., 2008), which relates to the high arabinoxylans content of the WB diet. So the most contributing OTU to discriminate pigs on the different diets resemble the source of dietary fiber.
Sexes
Our results are in accordance with a recent study of Xiao et al. (2016), which also showed a difference between male and female finisher pigs in fecal microbial composition. Both studies found differences in bacteria belonging to the Prevotella and Ruminococcus genus. Previously, most of the research in pigs investigated changes in intestinal microbiota related to digestive problems and diarrhea post-weaning in weaners (Konstantinov et al., 2006; Pajarillo et al., 2014). These studies in weaners did not find a sex effect on the microbiome (Mach et al., 2015). Sex steroids hormones might partially explain this, as levels of some sex-steroid hormones rapidly increase at onset of puberty (Camous et al., 1985; Zamaratskaia et al., 2004). In mice, gonadectomy of males and females resulted in a change in microbial composition of the feces, but testosterone treatment of the castrated males resulted in a microbiome similar to that of intact males (Org et al., 2016). Metabolism residues of sex-steroid hormones are excreted through bile into the lumen of the small intestine (Goymann, 2012), resulting in a different bile composition between sexes (Org et al., 2016). Mainly the Firmicutes, Proteobacteria, and Actinobacteria can metabolize and degrade steroid hormones (García-Gómez et al., 2013), which is reflected in the difference in OTU between the sexes in our study, where 11 out of the 18 OTU belonged to the Firmicutes phyla. Other pathways through which sex-steroid hormones might influence microbiota are the mucosal immune activation (Sankaran-Walters et al., 2013) and expression of steroid receptors (Menon et al., 2013). The observed limited effect of sex on microbial composition in the feces of weaners and the substantial effect at slaughter age is likely because sex-steroid hormones only start to play a large role in finisher pigs.
Feed Efficiencies
There are several ways via which the intestinal microbiota could influence FE of pigs, including competition between the host and the microbiota for nutrients in the small intestine and activation of the immune system through stimulation of the development of the mucus layer, epithelial cells, and lamina propria (Dibner and Richards, 2005). The latter could possibly induce changes in nutrient partitioning between utilization for immune system functioning and for deposition, e.g., in muscle protein, but this is likely to be primarily a juvenile phenomenon (Dibner and Richards, 2005). In addition, quantitative production of VFA by intestinal microbiota can relate to FE. Approximately 68% of the gross energy in fermentable carbohydrates can be transformed into VFA (Williams et al., 2001). The VFA composition depends amongst others on the composition of the substrates, microbial composition and activity, and absorption of the VFA across the large intestinal wall (Williams et al., 2001). Butyrate is the preferred energy source for colonocytes and 76% of the mucosal absorbed butyrate is metabolized in these cells (Herrmann et al., 2011). Once absorbed across the intestinal wall, the VFA are available as a precursor and energy substrate in organs and tissues in the body. Propionate is a precursor for glucose and is almost fully extracted by the liver (Ingerslev et al., 2014), whereas acetate and butyrate are used for acetyl-CoA production. Next to being direct energy substrates, VFA are also involved as regulators in fatty acid, glucose, and cholesterol metabolism (den Besten et al., 2013). Therefore, the microbiota might influence FE by the amount and composition of VFA produced.
There was a significant relationship between microbiome and FE in pigs fed the WB diet, but there was no significant relationship in pigs fed the CS diet on OTU level. The fiber level in the diets might explain this difference. When assuming the VFA production to contribute to the FE of the pigs, the difference in performance between the high and low FE pigs due to microbial composition differences is expected to be more pronounced at a higher content of fermentation substrate in the diet. As the finisher WB diet contained 2.8 times more crude fiber than the CS diet, there was more substrate available for fermentation in the WB diet. Consequently, in our study, the amount of substrate available might not have been sufficient to detect a relationship between microbiome and FE in the pigs fed the CS diet, whereas it was sufficient in the pigs fed the WB diet.
In male pigs fed the WB diet, the most contributing OTU to separate the FE groups was taxonomically classified as Lactobacillus, the high FE group having a higher abundance of this OTU. In contradiction to our results, Vigors et al. (2016) only showed a difference in Lactobacilli spp. in the cecum, and not in the colon, between divergent groups in residual feed intake in pigs. Nevertheless, the direction of the effect was similar in both studies, with an increase in Lactobacillus having a positive effect on FE. The species related to this OTU only produce D- and L-lactate (Roos et al., 2005; Slavica et al., 2015). In contrast, in the female pigs fed the WB diet, the same OTU was higher in the low FE group, but the difference was smaller between the FE groups in the female pigs. In accordance with the results of McCormack et al. (2017), the Clostridium abundancy in feces was important to distinguish between the high and low FE pigs. However, this was only the case in the male pigs fed the WB diet, and the two OTU classified as Clostridium had opposite effects. In addition, the other five genera important for distinguishing pigs divergent in residual feed intake discovered by McCormack et al. (2017) were not found in our study. An explanation may lie in the difference between the diets of the studies. Everything considered, the microbiota associated with FE in grower-finisher pigs might consist of several crucial species and other species only relevant in certain situations, e.g., when certain diets are fed.
Implications
Results of the present study suggest possibilities to improve FE of grower-finisher pigs by altering microbial composition in the distal part of the intestinal tract. Modification of diet composition might be an option to change microbiota composition, e.g., by changing fiber source or inclusion level, or by including specific additives such as probiotics, prebiotics, organic and inorganic acids, and essential oils (De Lange et al., 2010). In summary, FE might be improved by changing the nutrition of pigs partly through resulting changes in microbiota composition.
CONCLUSION
There is a sex-dependent relationship between the fecal microbial composition and FE in grower-finisher pigs fed a WB diet, having a higher concentration of dietary fiber than a CS diet. The exact interplay between the fecal microbial composition, composition and concentration of fiber, and production of VFA by intestinal microbiota remains to be determined. Furthermore, results on the relationship between microbiota composition in the digestive tract and FE remain to be confirmed in more and larger scale studies. Results of the present experiment suggest that there are possibilities to modify the intestinal microbial composition by means of nutrition (e.g., by use of specific additives such as pro- and prebiotics) in order to improve FE of grower-finisher pigs.
LITERATURE CITED
- Bauer E., B. A. Williams H. Smidt R. Mosenthin, and Verstegen M. W.. 2006. Influence of dietary components on development of the microbiota in single-stomached species. Nutr. Res. Rev. 19:63–78. doi:10.1079/NRR2006123. [DOI] [PubMed] [Google Scholar]
- den Besten G., K. van Eunen A. K. Groen K. Venema D. J. Reijngoud, and Bakker B. M.. 2013. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. j. Lipid Res. 54:2325–2340. doi:10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camous S., Prunier A., and Pelletier J.. 1985. Plasma prolactin, LH, FSH and estrogen excretion patterns in gilts during sexual development. J. Anim. Sci. 60: 1308–1317. doi:10.2527/jas1985.6051308x. [DOI] [PubMed] [Google Scholar]
- Choct M. 1997. Feed non-starch polysaccharides: chemical structures and nutritional significance. Feed milling international 191: 13–26. [Google Scholar]
- CVB 2011. Chemical compositions and nutritional values of feed ingredients. Centraal Veevoeder Bureau, Lelystad, The Netherlands. [Google Scholar]
- De Lange C., Pluske J., Gong J., and Nyachoti C.. 2010. Strategic use of feed ingredients and feed additives to stimulate gut health and development in young pigs. Livest. Sci. 134: 124–134. doi:10.1016/j.livsci.2010.06.117. [Google Scholar]
- Dibner J. J., and Richards J. D.. 2005. Antibiotic growth promoters in agriculture: history and mode of action. Poult. Sci. 84: 634–643. doi:10.1093/ps/84.4.634. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. 2010. Search and clustering orders of magnitude faster than blast. Bioinformatics. 26:2460–2461. doi:10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. 2013. uparse: highly accurate otu sequences from microbial amplicon reads. Nat. Methods. 10:996–998. doi:10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Eeckhaut V., F. Van Immerseel E. Teirlynck F. Pasmans V. Fievez C. Snauwaert F. Haesebrouck R. Ducatelle P. Louis, and Vandamme P.. 2008. Butyricicoccus pullicaecorum gen. Nov., sp. Nov., an anaerobic, butyrate-producing bacterium isolated from the caecal content of a broiler chicken. Int. j. Syst. Evol. Microbiol. 58(Pt 12):2799–2802. doi:10.1099/ijs.0.65730-0. [DOI] [PubMed] [Google Scholar]
- Frese S. A., K. Parker C. C. Calvert, and Mills D. A.. 2015. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome 3:28. doi:10.1186/s40168-015-0091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Gómez E., B. González-Pedrajo, and Camacho-Arroyo I.. 2013. Role of sex steroid hormones in bacterial-host interactions. Biomed Res. Int. 2013:928290. doi:10.1155/2013/928290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goymann W. 2012. On the use of non‐invasive hormone research in uncontrolled, natural environments: the problem with sex, diet, metabolic rate and the individual. Methods Ecol. Evol. 3: 757–765. doi:10.1111/j.2041-210X.2012.00203.x. [Google Scholar]
- Herrmann J., R. Hermes, and Breves G.. 2011. Transepithelial transport and intraepithelial metabolism of short-chain fatty acids (scfa) in the porcine proximal colon are influenced by scfa concentration and luminal ph. Comp. Biochem. Physiol. a. Mol. Integr. Physiol. 158:169–176. doi:10.1016/j.cbpa.2010.10.018. [DOI] [PubMed] [Google Scholar]
- Ingerslev A. K., P. K. Theil M. S. Hedemann H. N. Lærke, and Bach Knudsen K. E.. 2014. Resistant starch and arabinoxylan augment scfa absorption, but affect postprandial glucose and insulin responses differently. Br. j. Nutr. 111:1564–1576. doi:10.1017/S0007114513004066. [DOI] [PubMed] [Google Scholar]
- Jombart T., S. Devillard, and Balloux F.. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. bmc Genet. 11:94. doi:10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. B., K. Borewicz B. A. White R. S. Singer S. Sreevatsan Z. J. Tu, and Isaacson R. E.. 2011. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet. Microbiol. 153:124–133. doi:10.1016/j.vetmic.2011.05.021. [DOI] [PubMed] [Google Scholar]
- Konstantinov S. R., A. A. Awati B. A. Williams B. G. Miller P. Jones C. R. Stokes A. D. Akkermans H. Smidt, and de Vos W. M.. 2006. Post-natal development of the porcine microbiota composition and activities. Environ. Microbiol. 8:1191–1199. doi:10.1111/j.1462-2920.2006.01009.x. [DOI] [PubMed] [Google Scholar]
- Liu C., S. M. Finegold Y. Song, and Lawson P. A.. 2008. Reclassification of clostridium coccoides, ruminococcus hansenii, ruminococcus hydrogenotrophicus, ruminococcus luti, ruminococcus productus and ruminococcus schinkii as blautia coccoides gen. Nov., comb. Nov., blautia hansenii comb. Nov., blautia hydrogenotrophica comb. Nov., blautia luti comb. Nov., blautia producta comb. Nov., blautia schinkii comb. Nov. And description of blautia wexlerae sp. Nov., isolated from human faeces. Int. j. Syst. Evol. Microbiol. 58(Pt 8):1896–1902. doi:10.1099/ijs.0.65208-0. [DOI] [PubMed] [Google Scholar]
- Lluch J., F., Servant S., Païssé C., Valle S., Valière C., Kuchly G., Vilchez C., Donnadieu M., Courtney R., Burcelin et al. 2015. The characterization of novel tissue microbiota using an optimized 16s metagenomic sequencing pipeline. Plos One 10:e0142334. doi:10.1371/journal.pone.0142334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looft T., H. K. Allen B. L. Cantarel U. Y. Levine D. O. Bayles D. P. Alt B. Henrissat, and Stanton T. B.. 2014. Bacteria, phages and pigs: the effects of in-feed antibiotics on the microbiome at different gut locations. Isme J. 8:1566–1576. doi:10.1038/ismej.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach N., M., Berri J., Estellé F., Levenez G., Lemonnier C., Denis J. J., Leplat C., Chevaleyre Y., Billon J., Doré et al. 2015. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 7:554–569. doi:10.1111/1758-2229.12285. [DOI] [PubMed] [Google Scholar]
- Magoč T., and Salzberg S. L.. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27: 2957–2963. doi:10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos G. G., Martín F., Latorre M. A., Vicente B., and Lázaro R.. 2006. Inclusion of oat hulls in diets for young pigs based on cooked maize or cooked rice. Anim. Sci. 82: 57–63. doi:10.1079/ASC20053. [Google Scholar]
- McCormack U. M., Curiao T., Buzoianu S. G., Prieto M. L., Ryan T., Varley P., Crispie F., Magowan E., Metzler-Zebeli B. U., Berry D., O. O’Sullivan, P. D. Cotter, G. E. Gardiner, and P. G. Lawlor. 2017. Exploring a possible link between the intestinal microbiota and feed efficiency in pigs. Appl. Environ. Microbiol. 83:e00380–17. doi:10.1128/aem.00380-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis S. and Madden T. L.. 2004. blast: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32(Web Server issue):W20–W25. doi:10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P. J. and Holmes S.. 2013. Phyloseq: an r package for reproducible interactive analysis and graphics of microbiome census data. Plos One 8:e61217. doi:10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon R., S. E. Watson L. N. Thomas C. D. Allred A. Dabney M. A. Azcarate-Peril, and Sturino J. M.. 2013. Diet complexity and estrogen receptor β status affect the composition of the murine intestinal microbiota. Appl. Environ. Microbiol. 79:5763–5773. doi:10.1128/AEM.01182-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier-Salaün M., Edwards S., and Robert S.. 2001. Effect of dietary fibre on the behaviour and health of the restricted fed sow. Anim. Feed Sci. Technol. 90: 53–69. doi:10.1016/S0377-8401(01)00196-1. [Google Scholar]
- Mevik B.-H., and Wehrens R.. 2007. The pls package: principal component and partial least squares regression in R. J. Stat. Softw. 18:1–23. doi:10.18637/jss.v018.i02 [Google Scholar]
- Noblet J., and Le Goff G.. 2001. Effect of dietary fibre on the energy value of feeds for pigs. Anim. Feed Sci. Technol. 90: 35–52. doi:10.1016/S0377-8401(01)00195-X. [Google Scholar]
- Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., Minchin P. R., O’Hara R. B., Simpson G. L., Solymos P., Stevens M. H. H., E Szoecs, and H Wagner.2017. https://CRAN.R-project.org/package=vegan Vegan: Community Ecology Package. R Package Version 2.4–2. Retrieved from.
- Org E., M. Mehrabian B. W. Parks P. Shipkova X. Liu T. A. Drake, and Lusis A. J.. 2016. Sex differences and hormonal effects on gut microbiota composition in mice. Gut. Microbes. 7:313–322. doi:10.1080/19490976.2016.1203502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajarillo E. A. B., Chae J.-P., Balolong M. P., Bum Kim H., and Kang D.-K.. 2014. Assessment of fecal bacterial diversity among healthy piglets during the weaning transition. J. Gen. Appl. Microbiol. 60: 140–146. doi:10.2323/jgam.60.140. [DOI] [PubMed] [Google Scholar]
- Pruesse E., J. Peplies, and Glöckner F. O.. 2012. sina: accurate high-throughput multiple sequence alignment of ribosomal rna genes. Bioinformatics 28:1823–1829. doi:10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., E. Pruesse P. Yilmaz J. Gerken T. Schweer P. Yarza J. Peplies, and Glöckner F. O.. 2013. The silva ribosomal rna gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41(Database issue):D590–D596. doi:10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos S., L. Engstrand, and Jonsson H.. 2005. Lactobacillus gastricus sp. Nov., lactobacillus antri sp. Nov., lactobacillus kalixensis sp. Nov. And lactobacillus ultunensis sp. Nov., isolated from human stomach mucosa. Int. j. Syst. Evol. Microbiol. 55(Pt 1):77–82. doi:10.1099/ijs.0.63083-0. [DOI] [PubMed] [Google Scholar]
- Sankaran-Walters S., M., Macal I., Grishina L., Nagy L., Goulart K., Coolidge J., Li A., Fenton T., Williams M. K., Miller et al. 2013. Sex differences matter in the gut: effect on mucosal immune activation and inflammation. Biol. Sex Differ. 4:10. doi:10.1186/2042-6410-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavica A., A. Trontel N. Jelovac Ž. Kosovec B. Šantek, and Novak S.. 2015. Production of lactate and acetate by lactobacillus coryniformis subsp. Torquens dsm 20004(t) in comparison with lactobacillus amylovorus dsm 20531(t). j. Biotechnol. 202:50–59. doi:10.1016/j.jbiotec.2015.01.014. [DOI] [PubMed] [Google Scholar]
- Van Kernebeek H. R., Oosting S. J., Van Ittersum M. K., Bikker P., and De Boer I. J.. 2016. Saving land to feed a growing population: consequences for consumption of crop and livestock products. Int. J. Life Cycle Assess. 21: 677–687. doi:10.1007/s11367-015-0923-6. [Google Scholar]
- Vigors S., T. Sweeney C. J. O’Shea A. K. Kelly, and O’Doherty J. V.. 2016. Pigs that are divergent in feed efficiency, differ in intestinal enzyme and nutrient transporter gene expression, nutrient digestibility and microbial activity. Animal. 10:1848–1855. doi:10.1017/S1751731116000847. [DOI] [PubMed] [Google Scholar]
- Wellock I. J., P. D. Fortomaris J. G. Houdijk J. Wiseman, and Kyriazakis I.. 2008. The consequences of non-starch polysaccharide solubility and inclusion level on the health and performance of weaned pigs challenged with enterotoxigenic escherichia coli. Br. j. Nutr. 99:520–530. doi:10.1017/S0007114507819167. [DOI] [PubMed] [Google Scholar]
- Williams B. A., M. W. Verstegen, and Tamminga S.. 2001. Fermentation in the large intestine of single-stomached animals and its relationship to animal health. Nutr. Res. Rev. 14:207–228. doi:10.1079/NRR200127. [DOI] [PubMed] [Google Scholar]
- Xiao L., Estellé J., Kiilerich P., Ramayo-Caldas Y., Xia Z., Feng Q., Liang S., Pedersen A. Ø., Kjeldsen N. J., and Liu C.. 2016. A reference gene catalogue of the pig gut microbiome. Nat. Microbiol. 1: 16161. doi:10.1038/nmicrobiol.2016.161. [DOI] [PubMed] [Google Scholar]
- Zamaratskaia G., J. Babol A. Madej E. J. Squires, and Lundström K.. 2004. Age-related variation of plasma concentrations of skatole, androstenone, testosterone, oestradiol-17 beta, oestrone sulphate, dehydroepiandrosterone sulphate, triiodothyronine and igf-1 in six entire male pigs. Reprod. Domest. Anim. 39:168–172. doi:10.1111/j.1439-0531.2004.00496.x. [DOI] [PubMed] [Google Scholar]
- Zijlstra R., and Beltranena E.. 2013. Swine convert co-products from food and biofuel industries into animal protein for food. Animal Frontiers 3: 48–53. doi:10.2527/af.2013-0014. [Google Scholar]