Abstract
The human cerebral cortex is distinguished by its large size and abundant gyrification, or folding, yet the evolutionary mechanisms driving cortical size and structure are unknown. While genes essential for cortical developmental expansion have been identified from the genetics of human primary microcephaly (“small head”, associated with reduced brain size and intellectual disability)1, studies of these genes in mice, whose smooth cortex is one thousand times smaller than that of humans, have provided limited insight. Mutations of abnormal spindle-like microcephaly-associated (ASPM), the most common recessive microcephaly gene, reduce cortical volume by ≥50% in humans2–4, but have little effect in mice5–9, likely reflecting evolutionarily divergent functions of ASPM10,11. We used genome editing to create a germline knockout (KO) of Aspm in the ferret (Mustela putorius furo), a species with a larger, gyrified cortex and greater neural progenitor cell (NPC) diversity12–14 than mice, and closer Aspm protein sequence homology to human. Aspm KO ferrets exhibit severe microcephaly (25–40% decreases in brain weight), reflecting reduced cortical surface area without significant change in cortical thickness, as in human patients3,4, suggesting loss of “cortical units”. The mutant ferret fetal cortex displays a massive premature displacement of ventricular radial glial cells (VRG) to the outer subventricular zone (OSVZ), where many resemble outer radial glia (ORG), an NPC subtype essentially absent in mice and implicated in cerebral cortical expansion in primates12–16. These data suggest an evolutionary mechanism whereby Aspm regulates cortical expansion by controlling the affinity of VRG for the ventricular surface, thus modulating the ratio of VRG, the most undifferentiated cell type, to ORG, a more differentiated progenitor.
Main text
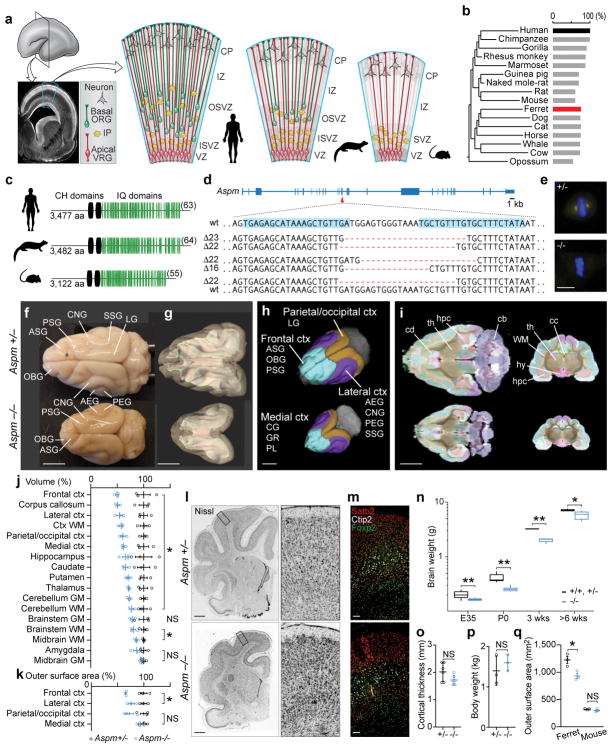
We injected 148 ferret zygotes with genome editing constructs targeting Aspm exon 15, mutations in which cause severe microcephaly in humans17, and recovered 11 kits born at full term, all carrying insertions or deletions in the targeted exon (Fig. 1a–d). We established three stable Aspm germline KO ferret lines, which showed comparable phenotypes. Loss of Aspm protein was confirmed in embryonic fibroblasts (Fig. 1e).
Figure 1. Aspm KO ferrets robustly model human microcephaly.
a, NPC diversity in humans, ferrets, and mice. b, c, ASPM protein is highly similar between humans and ferrets, including the number of IQ domains (c, in parentheses). d, Ferret Aspm gene showing targeted sequences (blue highlights) and founder frameshift deletions. e, Loss of Aspm in KO embryonic fibroblasts. f, Aspm+/− and −/− littermate brains. g–k, MRI segmentations of grey and white matter (g), gyri grouped into 4 regions (h), horizontal and coronal sections (i), and quantification of volume (j) and cortical surface area (k). For abbreviations, see Methods. (*, P < 0.05; n = 3/genotype). l–p, Aspm−/− ferrets show reduced brain weight (n, **, P < 0.005; *, P < 0.01; n = 3–17/genotype/age group) but cytoarchitecture (l), laminar organization (m), cortical thickness (o, n = 6/genotype) and body weight (p, n = 3/genotype) are preserved. q, Loss of Aspm decreases outer cortical surface area in ferrets, not in mice (n = 3/genotype) (*, P = 0.0217). Except the box plot (n), graphs show mean ± s.e.m. See Methods, Extended Data Table 1, and Source Data (graphs) for statistics and reproducibility. Scale bars: e, 10 μm; f–i, 5 mm; l, 1 mm; m, 100 μm.
Aspm KO ferrets displayed robust microcephaly (Fig. 1f–i), with up to 40% reduced brain weight (Fig. 1n) but no change in body weight (Fig. 1p), closely modeling the effects of human mutations2–4,17. Magnetic resonance imaging18 (MRI) showed that, as in humans4, loss of cortical volume and surface area followed an anterior-to-posterior gradient, with the frontal cortex most affected (Fig. 1f–k and Extended Data Table 1). However, the thickness of the KO cortex was preserved, similar to the cortex of human ASPM patients2–4, as was the cytoarchitecture and lamination of neurons (Fig. 1l, m, o and Extended Data Fig. 1). This phenotype is distinct from Aspm KO mice, which show ~10% reduced brain weight, variable body weight reduction, variable cortical thinning, and no discernable change in cortical surface area (Fig. 1q)5–9. Thus, the Aspm loss-of-function phenotype is more similar in ferrets than in mouse to that of human ASPM patients.
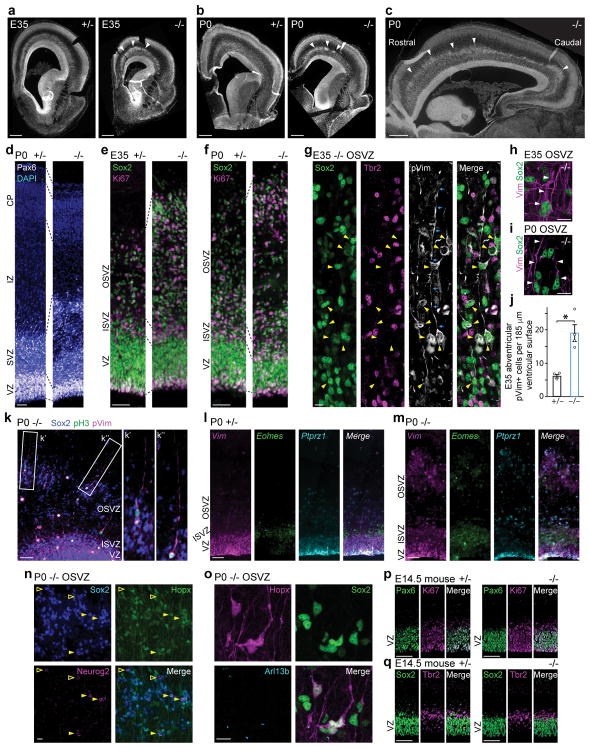
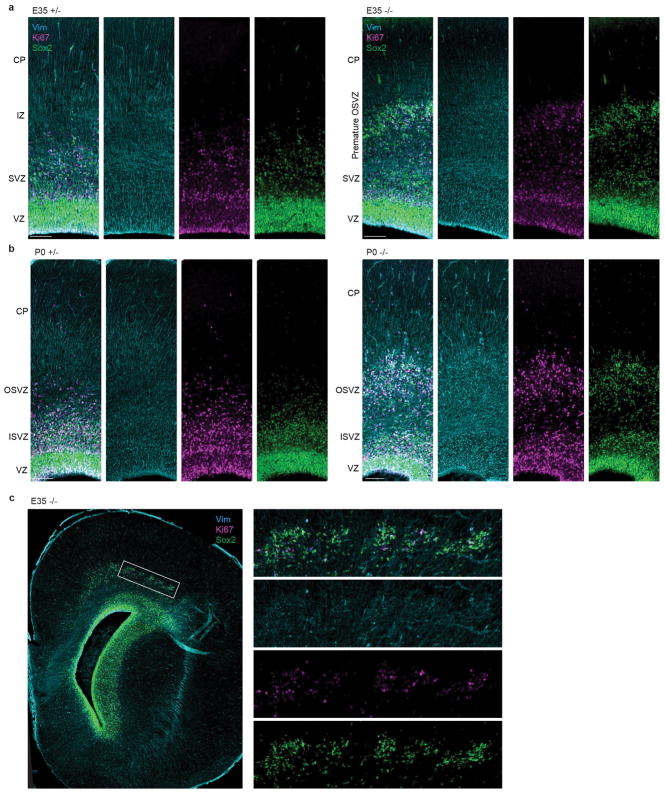
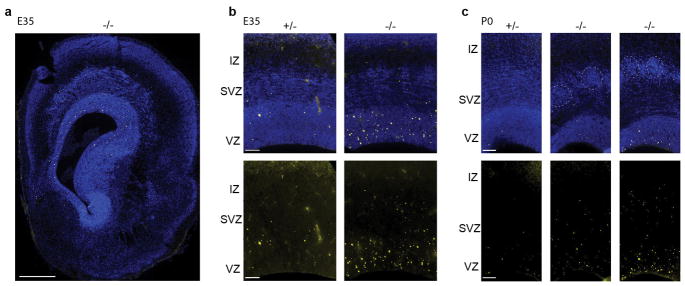
To elucidate the developmental mechanism of microcephaly, we examined Aspm KO ferrets during cortical neurogenesis (Fig. 2a–o and Extended Data Fig. 2–3), which begins around embryonic day 24 (E24) and continues for two weeks after birth, at E41. In the wild-type (WT) embryonic cortex, undifferentiated VRG divide symmetrically to expand the pool of VRG, or divide asymmetrically to produce two distinct, more differentiated progenitor subtypes, intermediate progenitors (IP) and ORG (Fig. 1a). ORG are multipotent, proliferative, unipolar progenitors abundant in the OSVZ that express molecular markers in common with VRG, including Sox2, Pax6, and vimentin (Vim); whereas IP are neuronally-fated, multipolar transit amplifying cells that predominate in the inner subventricular zone (ISVZ) and express Tbr2 (Eomes)12–14,16. All three NPC populations express the mitotic marker Ki67 and produce neurons that migrate radially into the cortical plate (CP)12–14,16,19. Thus, the Aspm+/− cortex at E35 and postnatal day 0 (P0) displayed a ventricular zone (VZ) densely packed with Pax6+ or Sox2+ VRG, and a less-dense zone of Ki67+ NPC expressing Sox2, Tbr2, or both in the SVZ (Fig. 2d–g and Extended Data Fig. 2–3). In contrast, the KO cortex contained overabundant Ki67+ NPC in the basal SVZ and intermediate zone (IZ) (Fig. 2e, f), reminiscent of the positioning of ORG that normally populate the OSVZ12–14,20. Discontinuous clusters of basal NPC were accompanied by thinning of the VZ, suggesting that precocious OSVZ progenitors are derived by premature withdrawal from the VZ (Fig. 2d–f and Extended Data Fig. 2–3). Displaced OSVZ progenitors were more abundant frontally and dorsally (Fig. 2a–c), matching the topography of cortical volume reduction in the adult (Fig. 1f–k).
Figure 2. Aspm KO ferrets show displaced NPC.
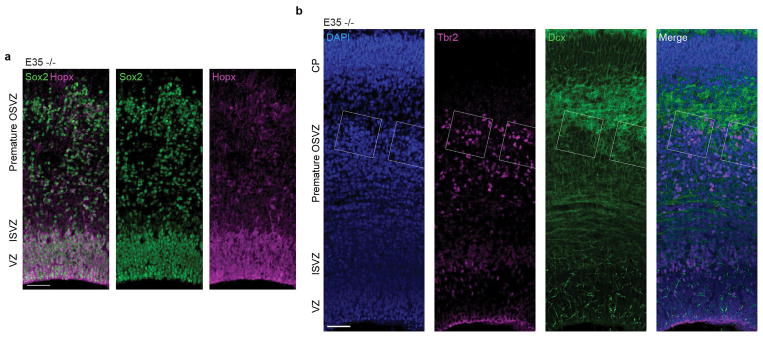
a–f, Nuclear staining of Aspm−/− ferrets shows a premature OSVZ-like zone (a–c, arrowheads) containing NPC that express Pax6, Sox2, and Ki67 (d–f). g–k, Displaced NPC include Sox2+/pVim+ ORG (g, arrowheads) with a basal process (g, arrows; h, i, k), and Tbr2+ IP. Abventricular pVim+ NPC are increased 3-fold in Aspm−/− ferrets (j) (*, P = 0.006 by one-tailed t-test; mean ± s.e.m; n = 3 +/− and n = 4 −/− animals). l–o, Displaced NPC express Vim, Eomes, or Ptprz1 (l, m), have Arl13b+ cilia (o), and are either Sox2+/Neurog2+/Hopx+ (filled arrowheads) or Sox2+/Neurog2+/Hopx- (open arrowheads) (n). p, q, Aspm−/− mice lack displaced NPC. See Methods for statistics and reproducibility. Scale bars: a–c, 500 μm; d–f, 50 μm; g–i, n, o, 10 μm; k–m, p, q, 100 μm.
Many displaced progenitors in the Aspm KO ferret OSVZ expressed VRG/ORG markers including Vim, phospho-vimentin (pVim), phospho-histone H3 (pH3), Sox2, and Pax6; as well as the ciliary marker Arl13b and the human ORG-enriched genes Ptprz1 and Hopx21, while others expressed the IP marker Tbr2 (Eomes) (Fig. 2g–o and Extended Data Fig. 2, 3, 6). Some displaced cells exhibited ORG-like unipolar morphology, with basal radial fibers immunoreactive for Vim, pVim, or Hopx (Fig. 2g–i, k, o). Quantification of pVim+ mitotic NPC revealed a three-fold increase in ORG-like progenitors in the KO at E35 (P = 0.006; 3 Aspm+/− and 4 Aspm−/− littermates) (Fig. 2j). The intermingled presence of Neurog2+/Hopx+ ORG, Tbr2+ IP, and Dcx+ newborn neurons together indicated preserved neurogenesis within clusters of displaced NPC (Fig. 2n and Extended Data Fig. 6). These data demonstrate that loss of Aspm in the ferret cortex causes VRG to prematurely detach from the VZ and relocate to the OSVZ where many dislocated cells exhibit ORG morphology, molecular profile, and neurogenic potential.
These dramatic changes in NPC populations in the mutant ferret contrast with 6 previously reported Aspm KO mouse lines5–9, which consistently show limited changes in NPC identity and organization. Aspm KO mice show a trend towards increased IP at the expense of VRG9, but lack ectopic basal Sox2+ or Pax6+ NPC (Fig. 2p, q and Extended Data Fig. 4). KO ferrets also showed increased apoptosis in telencephalic germinal zones not seen in Aspm KO mice5–7,9 (Extended Data Fig. 5), further highlighting that loss of Aspm elicits divergent brain phenotypes in ferrets and mice.
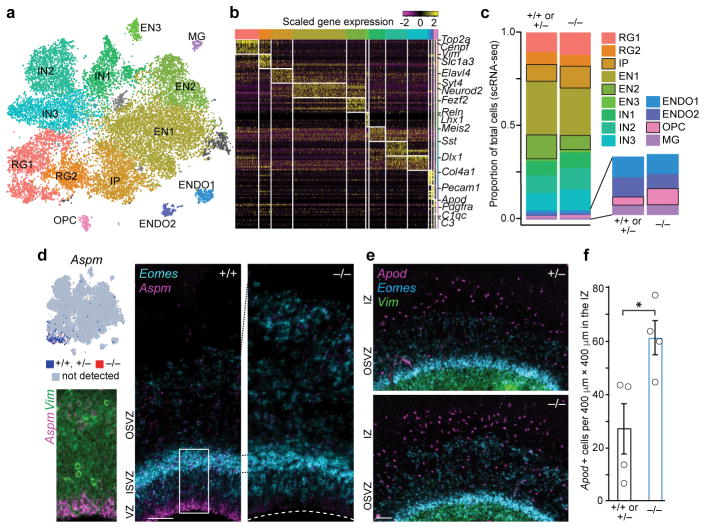
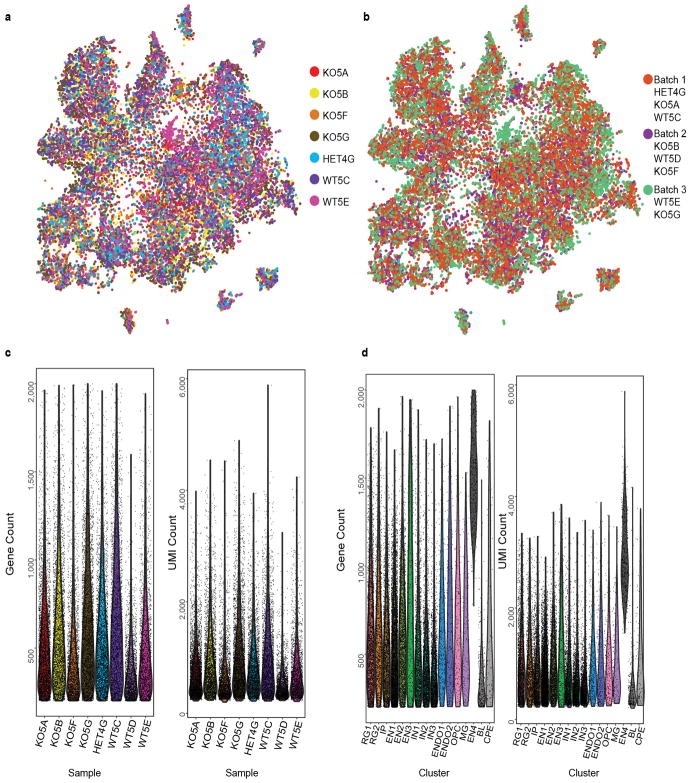
Single-cell RNA sequencing22 (scRNA-seq) of ~21,000 cells from the telencephalons of seven E35 embryos (3 Aspm+/+ or Aspm+/−, and 4 Aspm−/− animals) reinforced the conclusion that NPC proportions were altered in the Aspm mutant, though their transcriptional programs were largely preserved (Fig. 3, Extended Data Fig. 7, and Extended Data Table 2). We identified cell clusters corresponding to excitatory and inhibitory progenitor and neuronal subtypes, as well as non-neural cells (Fig. 3a, b) and found that the cell type composition of the E35 Aspm KO forebrain was significantly altered (Chi-squared = 267.27, df = 12, P = 2.2×10−16), yet cells still clustered by cell type, not by genotype or batch (Extended Data Fig. 7). Consistent with immunohistochemical observations, scRNA-seq suggested that VRG, WT ORG, and prematurely displaced KO ORG were transcriptionally indistinguishable, with the total proportion of radial glial cells (RG1+RG2) unchanged in the mutant (Extended Data Table 2). A 30% increase in the proportion of IP in the KO (P = 0.0002, FDR < 0.01; Fig. 3c and Extended Data Table 2) was consistent with increased IP detected by immunostaining (Fig. 2l, m and Extended Data Fig. 6) and was further validated by single-molecule fluorescent in situ hybridization (smFISH) (Fig. 3d, e). A doubling of the small proportion of cells expressing the oligodendrocyte precursor cell (OPC) markers Apod and Olig1 suggested limited but significant premature differentiation towards the glial lineage (Fig. 3c, e, f and Extended Data Table 2). These scRNA-seq data suggest that the gene expression programs of cortical neurogenesis are largely preserved, but the proportions of developmental cell types are changed, in ASPM-associated microcephaly.
Figure 3. Loss of Aspm changes cell type proportions but not transcriptional programs.
a, scRNA-seq identifies major cell types at E35. (For abbreviations and statistics, see Extended Data Table 2.) b, Cell type markers for each cluster. c, Proportions of each cell type, with the largest changes indicated by black outlines (bootstrap FDR < 0.01). d, Aspm is enriched at the VZ apical surface; Eomes+ IP are increased in the KO SVZ. e,f, Apod+ OPC are increased in Aspm−/− ferrets (*, P = 0.012 by one-tailed t-test; n = 4 animals/genotype). See Methods for statistics and reproducibility. The graph shows mean ± s.e.m. Asterisks in d, blood vessels. Scale bars: 100 μm.
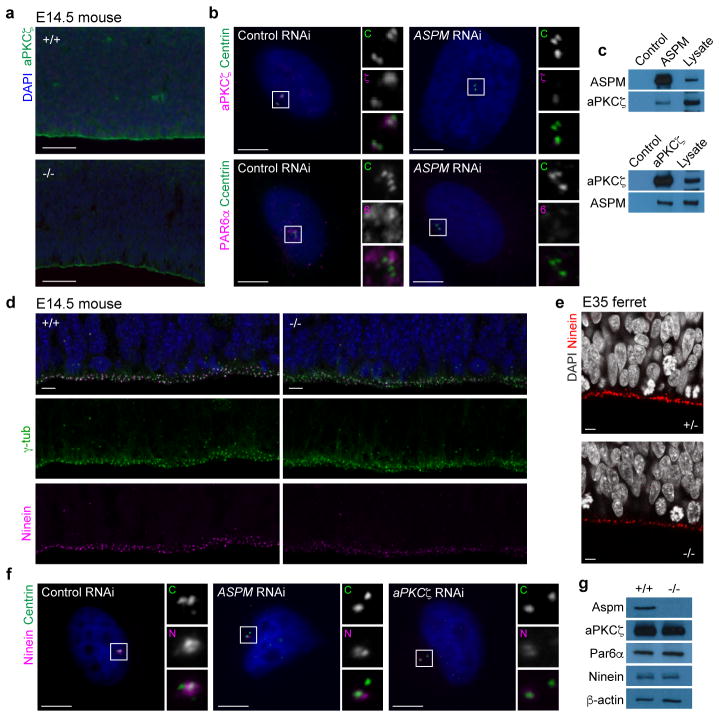
While examining potential molecular mechanisms for the detachment of VRG from the ventricular surface in the KO ferret, we identified a novel interaction between Aspm, which is localized to the centrosome, and the apical polarity complex (Fig. 4). Together with other centrosomal proteins, Aspm is essential for normal stem cell behaviors including centriole biogenesis and maternal centriole structure (Extended Data Fig. 8), and interactions between the mother centriole and the apical membrane have been implicated in the maintenance of NPC stem cell character9,23–25. In VRG, the centrosome is localized to the ventricular endfeet, which are linked by adherens junctions to form a polarized neuroepithelium expressing apical polarity complex proteins at the ventricular surface26. Aspm−/− mice showed abrogated staining of the core apical complex protein aPKCζ along the disrupted ventricular surface at E14.5 (Fig. 4a). Intriguingly, we found that depletion of ASPM by RNAi in H4 human neuroglioma cells resulted in loss of aPKCζ and another critical apical complex protein, PAR6α, from the centrosome (Fig. 4b). Furthermore, we found an interaction between ASPM and aPKCζ, reflected in mutual co-immunoprecipitation (Fig. 4c), that may mediate centrosomal recruitment of the apical polarity complex, providing new mechanistic insight into the link between centrosomal microcephaly-related proteins and apical progenitor identity.
Figure 4. Aspm controls localization of apical polarity complex proteins to the centrosome.
a, aPKCζ at the ventricular surface is decreased in Aspm−/− mice. b, Depletion of ASPM in H4 cells prevents recruitment of aPKCζ and PAR6α to the centrosomes. c, ASPM and aPKCζ co-immunoprecipitate from Hela cells. d, e, Loss of Aspm decreases ventricular surface staining for Ninein. f, Depletion of ASPM and aPKCζ prevents recruitment of Ninein to the centrosomes in H4 cells. g, Levels of aPKCζ, Par6α, and Ninein are unchanged in Aspm−/− mouse embryonic fibroblasts. See Methods for reproducibility. For gel source data, see Supplementary Data. Scale bars: a, 50 μm; b–f, 5 μm.
Finally, we found sharply reduced staining for Ninein, another microcephaly-associated centrosomal protein27, at both the E14.5 mouse and E35 ferret ventricular surface (Fig. 4d, e). Ninein localizes to the mother centriole and is critical for NPC maintenance25,28, and depletion of either ASPM or aPKCζ in H4 cells reduced centrosomal localization of Ninein (Fig. 4f). Importantly, Aspm−/− mouse embryonic fibroblasts expressed normal levels of aPKCζ, Par6, and Ninein proteins, suggesting that loss of aPKCζ and Ninein from the Aspm KO ventricular surface is due primarily to mislocalization, rather than downregulation. These data show that loss of Aspm disturbs the organization and function of the centrosome at multiple levels, and suggest disruption of the centrosome–apical polarity complex interface as a mechanism underlying the displacement of VRG from the VZ in the Aspm KO ferret.
Collectively, our data show that Aspm regulates the affinity of VRG for the ventricular surface. Displaced mutant progenitors show many features of ORG, indicating a central role for Aspm in regulating the normal timing of transition from VRG to ORG, and thus the ratio of VRG to ORG over the course of development. Premature basal displacement deprives VRG of proliferation-inducing factors received from the cerebrospinal fluid29, increases the proportions of less-proliferative ORG and IP, and results in a smaller cerebral cortex. The frontal predominance that characterizes both the loss of cortical surface area and VRG displacement further indicates that the premature transformation of VRG to ORG leads directly to reduced cortical “units” and surface area.
Our results support the idea that expansion of cortical surface area during human evolution may have arisen in part from changes to the proliferative time window of VRG. Changes in the amino acid sequence of Aspm and other microcephaly-associated centrosomal proteins10 may have impacted the timing of the VRG proliferative window by altering interactions between maternal centriole components and the apical polarity complex. Finally, we find that for human brain disorders that are poorly recapitulated in the mouse or in cell culture, the ferret is an efficient and faithful genetic model that demonstrates robust phenotypes and reveals novel mechanisms.
METHODS
We have complied with all relevant ethical regulations and performed the experiments approved by the Institutional Animal Care and Use Committees (IACUCs) at the University of Iowa, Boston Children’s Hospital, Yale School of Medicine, and Marshall BioResources.
ASPM protein homology and domain analysis
ASPM protein sequences of 16 mammals were extracted from NCBI Genbank. Global pairwise alignment was performed with EMBOSS Needleall (http://www.ebi.ac.uk/Tools/psa/emboss_needle/). The percent homology to human ASPM was calculated for each animal based on the alignment score utilizing the Needleman-Wunsch algorithm (gap opening penalty, 10; gap extension penalty, 0.5). The phylogenetic tree was generated using Timetree.org. Calponin homology (CH) and calmodulin binding (IQ) domains were counted using Simple Modular Architecture Research Tool (http://smart.embl.de/), NP_060606.3 (human), NP_033921.3 (mouse), and ENSMPUT00000010205.1 (ferret).
TALEN assembly and mRNA synthesis
We assembled three pairs of transcription activator-like effector nucleases (TALENs) that target exon 15 of ferret Aspm, which encodes the second CH domain, and cloned into a mammalian expression vector with the CMV and T7 promoters through a commercial service (PNA Bio). Gene targeting efficiency of each TALEN pair was tested in HEK 293T cells using a split GFP-based reporter30. The most efficient pair, targeting TGAGAGCATAAAGCTGTTGATGGAGTGGGTAAATGCTGTTTGTGCTTTCTATA (target-spacer-target) was chosen for genome editing in vivo. These plasmids are available through Addgene.org. For mRNA synthesis, endotoxin-free TALEN plasmids were prepared using NucleoBond Xtra Midi EF kit (Clontech), ethanol-precipated 3 times, linearized with ScaI digestion (New England BioLabs), and gel purified. mRNAs were synthesized using mMessage mMachine T7 ULTRA kit (ThermoFisher Scientific), cleared by MEGAclear transcription clean-up kit (ThermoFisher Scientific). Of note, we performed the optional ammonium acetate precipitation to improve the quality of the mRNAs. The TALEN mRNAs were diluted in sterile EmbryoMax injection buffer (Millipore) at 50 ng/μl, aliquoted, and kept frozen at −150 °C until used.
Embryo targeting of the ferret Aspm gene
Zygotes were collected from the mating of sable coat color ferrets (Marshall BioResources) as previously described31. TALEN mRNA (50 ng/μl) was injected into the cytoplasm of zygotes using a micromanipulator and injector (Eppendorf) and a phase contrast microscope. Initially 79 ferret zygotes were injected and cultured in vitro for 5 days for them to reach the blastocyst stage. Twelve zygotes developed to blastocysts, from each of which genomic DNA was extracted and whole genome multiple displacement amplification was performed. The targeted genomic region was amplified by PCR (primers 5′-TTTGTGTGTGTTTCAGGTGGA-3′ and 5′-TGCATTATACAACTGGTGACAGA-3′ with a 430-bp product size), gel purified and cloned using TOPO-TA cloning kit (ThermoFisher Scientific). Twelve plasmid clones were sequenced from individual bacterial colonies from each blastocyst. These studies demonstrated an 87% targeting efficiency (14 insertions/deletions in 16 alleles or 8 blastocysts that were amenable to analysis). Next, we injected 148 zygotes, incubated at 39°C for 24 h, and transferred 116 two-cell-stage embryos into the oviduct of pseudopregnant female sable ferrets as previously described31. Twenty-three ferrets were born and 11 survived. All 11 ferrets had insertions/deletions (100% efficiency). The F0 ferrets suckled and swallowed milk normally and grew without gross abnormalities.
Germline transmission
Six Aspm mutant ferrets were shipped to Marshall BioResources at 3 months of age and maintained according to the protocol approved by IACUC. Two compound heterozygous males, Δ23;Δ22 (c.3364_3386del;c.3363_3384del) and Δ16;Δ22 (c.3367_3388del;c.3364_3379del), and one heterozygous female, Δ22;wt (c.3363_3384del;wt), (Fig. 1d) were chosen as founders because they have similar frameshift, early truncating mutations. They were bred to each other or WT ferrets. Germline transmission was confirmed by T7 endonuclease I assay (New England BioLabs) and sequencing of both alleles. Eventually animals with a specific Δ22 mutation (c.3363_3384del) were maintained for breeding. Routine genotyping was carried out with PCR (primers 5′-ATCAATAAGAAAAAAGACAAAAGAAATAGTGG-3′ and 5′-CTTAAGTCAGTGAGCTTAAACAGAAAT-3′ with a 150-bp product size from the WT allele and 128-bp from the KO allele). Aspm KO males mated successfully, and KO kits were born at expected Mendelian ratios.
Semen analysis
Every mating was closely monitored at Marshall BioResources. Sperm samples were collected from mated females directly after mating. The concentration, motility, and morphology of the sperm were analyzed by an experienced technician. Each male received a sperm check evaluation at least once a month. Aspm+/+, +/− and −/− samples showed similar sperm counts.
Ferret colony management and tissue handling
The Aspm KO ferret colony was maintained at Marshall BioResources. For embryonic ages and <P8, timed pregnant jills were shipped to Boston Children’s Hospital and euthanized prior to embryo extraction, at which point brains were removed from the embryos and drop-fixed in 4% paraformaldehyde (PFA) at +4°C overnight. For ≥P8 ferrets, all animals were deeply anesthetized and weighed prior to transcardial perfusion with cold PBS followed by 4% PFA, then brains were extracted and placed in 4% PFA at +4°C overnight, then washed and stored in PBS prior to processing for immunohistochemistry. All brain weight measurements were made post-fixation, prior to sucrose infiltration.
MRI
Ferrets
Three Aspm+/− and 3 −/− ferrets (>8 months of age) were perfused using 4% PFA/PBS. The brains were dissected and post-fixed in PFA/PBS containing 4 mM gadolinium contrast Magnevist (Bayer) at +4°C for 2 weeks. The brains were scanned using a 9.4T at Yale Magnetic Resonance Center and the Martinos Center for Biomedical Imaging, Massachusetts General Hospital. At Yale, a custom-made 1H radiofrequency coil (40-mm diameter) was used for diffusion tensor imaging (DTI). DTI acquisition was obtained with Stejskal and Tanner spin-echo diffusion-weighted sequence with a diffusion gradient δ = 5 ms and a delay Δ = 15 ms between diffusion gradients32. Sixty-four slices of 500 μm thickness, field of view of 25.6 mm × 25.6 mm and 128 × 128 resolution were acquired with a repetition time (TR) of 4s, echo ime (TE) of 30 ms and 4 averages. Each of the 6 MR images was first corrected for B1 shading artifact using a slice inhomogeneity correction33 and an inverse covariance mapping of grey matter density (unpublished). Next, the Ferret Atlas18 was registered to each of the MRI images using a tensor b-spline normalized mutual information nonlinear intensity-based registration algorithm34,35 with a control point spacing of 1 mm. The result of the registration was used to warp the atlas regions to each individual MRI, and from this we calculated the volume of each of the warped regions as reported in Fig. 1 and Extended Data Table 1. ASG, OBG, and PSG were considered as the frontal cortex; AEG, CNG, PEG, and SSG were considered as the lateral cortex; CG, GR, and PL were considered as the medical cortex; and LG was considered as the parietal/occipital cortex. The name of each brain part in Fig. 1 is as previously described36: GM, grey matter; WM, white matter; AEG, anterior ectosylvian gyrus; ASG, anterior sigmoid gyrus; CG, cingulate gyrus; CNG, coronal gyrus; GR, gyrus rectus; LG, lateral gyrus; OBG, orbital gyrus; PEG, posterior ectosylvian gyrus; PL, piriform lobe; PSG, posterior sigmoid gyrus; SSG, suprasylvian gyrus; cb, cerebellum; cc, corpus callosum; cd, caudate; hpc, hippocampus; hy, hypothalamus; th, thalamus. For DTI tensor measurement, a total of 15 different non-collinear diffusion weighted directions (b = 1000 s/mm2) and 1 without diffusion weighting were obtained. The six elements of the diffusion tensor were calculated from the signal intensity of the diffusion-weighed images. Tensor eigenvalues and their corresponding eigenvectors were computed, along with fractional anisotropy, at each voxel. The images were color-coded by the principal direction (eigenvector) of diffusion using BioImage Suite37 (http://www.bioimagesuite.org/). At Massachusetts General Hospital, we acquired anatomically accurate brain volume images with minimal distortion using FLASH (fast low angle shot) MRI sequence with TR = 100 ms, TE = 30 ms, and 150 μm isotropic resolution. Cortical GM and WM were manually segmented using the FreeView (http://surfer.nmr.mgh.harvard.edu), and their volumes were measured. Mice. Three Aspm+/+ and 3 −/− mouse brains were dissected from P30 perfused animals and post-fixed as described above. Brains were submerged into perfluorocarbon oil (Fomblin, Fisher Scientific) at +4°C for 3 days, and imaged in this oil using a Bruker BioSpec 70/30 7T MRI scanner (a sub-millimeter MRI with a 30 cm bore and 450 mT/m gradient) in the Small Animal Imaging Facility at Boston Children’s Hospital. MRI scans were isotropic 63 μm voxels across the entire brain. Cortical surface area was visualized and measured using the FreeView, Osyrix, and ImageJ 3D projection.
Fluorescent immunohistochemistry
Fixed ferret brains were infiltrated with a series of 10%, then 20%, and finally 30% w/v sucrose solutions in PBS until sunk, then embedded in optimal cutting temperature (OCT) compound and frozen in isopentane cooled to −40°C, then stored long-term at −80°C. Brains were sectioned at 10 to 20 μm thickness on a Leica Cryostat, mounted immediately onto warm charged SuperFrost Plus slides (Fisher Scientific), then dried at 37°C for 10 to 30 min before storage at −80°C. After applying a hydrophobic barrier around the tissue (ImmEdge Pen, Vector Labs), slides were washed in cold 0.1M PBS followed by antigen retrieval in Retrievagen A pH 6.0 (BD Biosciences) at 80–90°C in a hybridization oven for 45 min. Sections were then cooled to RT in Retrievagen, washed in cold 0.1M PBS, and blocked for 1 h at RT (5% normal donkey serum, 1% w/v BSA, 0.2% w/v each glycine/lysine, in PBS). Slides were incubated with primary antibodies for two nights on a rotary shaker at +4°C in blocking buffer plus 0.3% Triton X-100. Sections were then washed in PBS and incubated for 2 h at RT in blocking buffer containing secondary antibodies at 1:500 (Jackson Immunoresearch). Finally, slides were washed in PBS, counterstained with DAPI at 1 μg/ml in PBS for 15 min, washed again, and coverslipped with Fluoromount-G (Southern Biotech). Images were obtained with a Zeiss LSM700 confocal microscope and Leica MZ16 F fluorescence stereomicroscope. The following antibodies were used at 1:200 – 1:2000: Pax6 (Abcam ab5790), Foxp2 (Abcam ab16046), Ctip2 (Abcam ab18465), Satb2 (Bethyl A301-864A), Satb2 (Abcam ab51502), Sox2 (SCBT sc-17320), Tbr2 (Millipore AB15894), Ki67 (BD 550609), phospho-Vimentin (MBL D076-3), Vimentin (Abcam ab8978), phospho-histone H3 (Millipore 07-145), Hopx (SCBT sc-30216), Arl13b (Abcam ab136648), Arl13b (ProteinTech 17711-1-AP), Neurog2 (R&D MAB3314), Dcx (SCBT sc-8066).
Apoptosis assay
We examined apoptosis on cryosections using the ApopTag Red In Situ Apoptosis Detection Kit (Millipore) according to the manufacturer’s protocol.
Single-cell RNA-sequencing
Cell capture and sequencing were performed by the DropSeq method22 (http://mccarrolllab.com/dropseq/). Forebrain tissue was isolated from 2 Aspm+/+, 1 Aspm+/−, and 4 Aspm−/− E35 ferret embryos and cryopreserved38, then shipped to Cornell and processed there for single-cell capture, library preparation, and sequencing. Read alignment and digital gene expression matrix generation. Ferret reference gene annotations were expanded using bulk RNA-sequencing data from the cortex of two P2 ferrets. Bulk data were first mapped to the Ensembl ferret reference genome and transcriptome using TopHat2, and a transcriptome assembled with cufflinks; this assembled transcriptome and the Ensembl reference transcriptome version 1.0.85 were merged using cuffmerge39. The Drop-Seq Core Computational Protocol version 1.0.1 was followed22. Fastq reads were converted to BAM using the “FastqToSam” command in Picard (http://broadinstitute.github.io/picard/). Read pairs where more than one base in the barcode had a quality below 10 were discarded. Adapter sequences were trimmed from the 5′ end of the read, along with polyA tails. Star-2.5.2a (Ref. 40) was used to map reads to the custom transcriptome reference. The digital gene expression matrix was extracted using the “DigitalExpression” program of the Drop-seq protocol22, keeping only cells with at least 200 reads per cell for clustering analysis. Single cell clustering. Seurat software was used for dimensionality reduction, clustering, and obtaining cluster markers41. Cells from Aspm+/+ or Aspm+/− and Aspm−/− were merged in a single matrix. An initial run showed that one of the Aspm+/− samples contained low Unique Molecular Identifier (UMI) and gene counts compared to the other samples, and clustered differently, so this sample was removed from the downstream analysis (Extended Data Fig. 7). Genes were included if they were expressed in ≥3 cells and cells were included if they expressed >200 genes and <2000 genes. This resulted in 22,211 cells and 21,962 genes from 8,037 Aspm+/+ or +/− cells and 14,174 Aspm−/− cells. The data were log-normalized per cell, scaling each cell to 10,000 molecules as performed previously22. The “MeanVarPlot” Seurat function was used to identify the most variable genes, obtaining 3,555 variable genes. Negative binomial regression was performed on the variable genes, using the number of UMIs per cell as a confounder variable before clustering. The “PCAFast” function on Seurat was used to implement Principal Component Analysis using the IRLBA package. Twenty-five principal components were selected for clustering and as input for t-distributed stochastic neighbor embedding (tSNE) in Seurat. These were selected by plotting the standard deviation of the principal components and setting a cutoff at the “elbow” of the graph using the “PCElbowPlot” function in Seurat. Clustering was performed using the Seurat function “FindClusters”, which implements a shared nearest neighbor modularity optimization based algorithm using k.param = 30 for defining the k for the k-nearest neighbor algorithm and a resolution of 0.5 (Ref. 41,42). The Barnes-hut implementation of tSNE was used for visualizing the clusters using the “RunTSNE” and “TSNEPlot” Seurat functions. We observed a co-localization in the tSNE plot of cells clustered together by the graph-based clustering algorithm (Fig. 3a). Cluster markers were obtained with the “FindAllMarkers” Seurat function using a likelihood-ratio test, testing only genes expressed in at least 0.25 of cells in either all cells or the cells in a specific cluster, and testing only genes with at least 0.25-fold difference on a log-scale between cells in a cluster and all cells. P-values were adjusted for multiple comparison testing using the p.adjust function in R for Benjamini-Hochberg False Discovery Rate (FDR), selecting an FDR threshold of 0.01. Known markers were used to determine the corresponding cell type to each cluster. The heatmap in Fig. 3b displays expression data for the top ten cluster markers for each cluster, in a random sample of 10% of the cells of each cluster. Plotting cells onto the tSNE plot based on their batch (3 batches with both Aspm+/+, or +/−, and −/− each) suggested that batches did not strongly influence clustering (Extended Data Fig. 7). Plotting cells from each sample onto the tSNE plot suggested that two non-neuronal clusters of blood and choroid plexus epithelial cells were primarily from a single sample (a likely dissection artifact) and these two clusters were removed from further analysis (grey clusters in Fig. 3a) (Extended Data Fig. 7c, d). Plotting the number of genes and UMIs in each cluster revealed that one of the excitatory neuronal clusters had almost three times as many genes and UMIs per cell (Extended Data Fig. 7c, d). This, along with the fact that this cluster expressed a combination of markers from the other two excitatory neuronal clusters suggested that this cluster may contain doublets or other technical or batch artifacts; therefore we also removed this cluster from further analysis. Statistical analysis of cell type composition by genotype. A Chi-square test was performed using the “chisq.test” function in R to test the association of cluster composition with genotype. We also quantified the fraction of cells corresponding to each cluster under the assumption that Drop-Seq captures and sequences cells in an unbiased manner, and that the frequencies of cells are representative of their frequency in tissue22. The fraction of cells corresponding to each cluster was obtained by counting the number of cells assigned to each cluster for Aspm+/+ or +/− and Aspm−/− samples and dividing over the total number of cells that passed the filters described above and excluding the three clusters that were removed, for a total of 7,645 Aspm+/+ and +/− cells and 13,725 Aspm−/− cells. Empirical p-values were obtained by permuting genotype 10,000 times and obtaining the fraction of cells corresponding to each cluster for each permutation. These fractions were sorted, and the p-value was obtained by counting the number of times a fraction was more extreme or equal to the observed fraction in the non-permuted data divided by 10,000 and multiplied by 2 for a two-tailed test. P-values were adjusted for multiple comparison testing using the p.adjust function in R for Benjamini-Hochberg FDR, selecting an FDR threshold of 0.01.
Code availability
The code is available upon request.
smFISH
Using RNAscope fluorescent detection assays and probes (ACDbio), we performed smFISH according to the manufacturer’s protocols. Cryosections on SuperFrost Plus slides (Fisher Scientific) were dried at −20°C, rather than RT or 37°C, for 15 min after mounting, and were best used within a week of sectioning. Target retrieval was performed at 80°C in a hybridization oven for 30–40 min prior to proceeding with the RNAscope multiplex fluorescent detection protocol.
Cell culture and siRNA transfection
H4 and Hela cells authenticated by short tandem repeat profiling were obtained from ATCC, cultured in Advanced DMEM (ThermoFisher Scientific) supplemented with 3% FBS (Altantis) and Glutamax-I (ThermoFisher Scientific), and used within 5 passages with routine mycoplasma screening. Ferret embryonic fibroblasts (FEFs) and mouse embryonic fibroblasts (MEFs) were derived from post fertilization day 35 and 14.5 embryos, respectively. FEFs and MEFs were cultured in AmnioMAX (ThermoFisher Scientific). Cells were transfected with validated siRNA to ASPM or PRKCZ encoding aPKCζ (ThermoFisher Scientific) using Oligofectamine and OptiMEM (ThermoFisher Scientific) according to manufacturer’s instructions and analyzed 48 h later.
Immunoprecipitation and immunoblotting
Immunoprecipitation experiments were performed as previously described9. In brief, Hela cells were harvested in Dulbecco’s PBS (DPBS, ThermoFisher Scientific) and lysed in lysis buffer (50 mM Tris-HCl pH7.4, 266 mM NaCl, 2.27 mM KCl, 1.25 mM KH2PO4, 6.8 mM Na2HPO4-7H2O and 1% NP-40) supplemented with EDTA-free protease inhibitors cocktail set III (Calbiochem). For each immunoprecipitation, 1 mg of lysate was incubated with 2 μg of antibody for 2 h and then incubated with magnetic protein G-sepharose beads (GE Healthcare Life Sciences) for another 1 h at 4°C. Complexes were washed and then boiled in 2× Laemmli reducing buffer with β-mercaptoethanol (Bio-Rad). Samples were separated on 4–15% TGX gels (Bio-Rad), transferred onto BA85 supported nitrocellulose (GE Healthcare Life Sciences) at 100V for 30–45 min using a plate electrode Trans-Blot cell with cooling coil (Bio-Rad) and then subjected to immunoblot analysis using ECL Lightening Plus (Perkin-Elmers) or Western Pico (ThermoFisher Scientific). All the immunoprecipitation and immunoblotting were replicated three times.
Fluorescent immunocytochemistry
Cells were fixed in ice-cold methanol for 3 min, permeabilized in blocking buffer (2.5% BSA or FBS, 0.1% TritonX-100, 0.03% NaN3 in DPBS). Primary and secondary antibodies were diluted in blocking buffer and incubated for 2 h at RT. Coverslips were mounted using Gelvatol, or Prolong Diamond (ThermoFisher Scientific) and imaged with the inverted confocal microscope Zeiss LSM700. Images were processed with ImageJ/FIJI. For 3D-structured illumination microscopy (SIM) (Fig. 1e), WT and KO FEFs were plated on 1.5-mm coverslips and immunostained as above. Coverslips were mounted with Vectashield (Vectorlabs). 3D-SIM imaging was performed on a Zeiss Elyra PS.1 microscope equipped with a 100x/1.40 oil objective lens. Exciting light was directed through a movable optical grating to generate a fine-striped interference pattern on the same plane. Z-stacks of 15 optical sections with a step size of 0.1 μm were acquired to generate images in maximum intensity projection. The epitope of the ASPM (216-1) antibody43, NDNYGLNQDLESES, is located prior to the TALEN target site. The following antibodies were used at 1:100 – 1:2000: Centrin (Millipore 20H5), Par6α (SCBT sc-14405), Par6α (Abcam ab180159), β-actin (Proteintech 20536-1-AP), ASPM (SCBT sc-98903), ASPM (gift from J. Bond, 216-1), Ninein (Biolegend Poly6028), aPKCζ (SCBT sc-216).
Statistics and reproducibility
All the experiments in Fig. 1e, 1l–m, 2a–i, 2l–q, 3d, 4a–g were repeated 3 times independently with similar results. No statistical method was employed to predetermine sample size. At least 3 animals or samples were generally analyzed per genotype/age. Two-tailed t-tests were performed for most data using Prism 7, unless otherwise stated. Ferret kits were born at Mendelian ratio but individual kit’s genotype was random, which inherently randomized our experiments. To perform blinded experiments, genotype of each animal was revealed only after the analysis was completed. In ferrets, sex was undifferentiated up to P21, after which only male ferrets were analyzed. In mice, sex was undifferentiated up to P0, after which only male mice were analyzed. (Fig. 1j, k) n = 3 male ferrets/genotype of >8 months of age. Individual P values are in Source Data (graphs). (Fig. 1n) Box plot elements mean maximum, third quartile, median, first quartile, and minimum. E35: n = 17 +/+ or +/− and n = 10 −/− from three litters (P = 0.0023); P0: n = 9 +/+ or +/− and n = 6 −/− from one litter (P = 0.0003); P21/22 (“3 weeks”): n = 8 +/+ or +/− and n = 3 −/− from two litters (P = 0.0010); P41 and older animals (“>6 weeks”): n = 7 +/+ or +/− and n = 7 −/− (P = 0.0094). As brain weight is not significantly different after P41, both adult and P41 animals were combined for presentation as “>6 wks” group. Ferrets display considerable variability in body weight and brain weight at birth, related to variance in the exact time of birth post-conception and to litter size, which can vary from 3 to 15 kits. Thus, one small litter of three P0 kits, including one WT and two Aspm+/−, which had body weights ~2× the other P0 litters collected, were excluded from brain weight analysis. Because the y-axis is log scale, overlaying each data point as dot plots for n < 10 does not indicate the distribution of the data efficiently. Instead, brain weight of individual animals can be found in Source Data (graphs). (Fig. 1o) Using the whole brain images of coronal sections stained with Nissl or DAPI from n = 6 animals/genotype, we manually measured mean cortical thickness of PSG. No significant difference was found (P = 0.0843). (Fig. 1p) The same animals used for MRI (3 Aspm+/− and 3 −/− as described above) were used for body weight analysis. No significant difference was found (P = 0.4481). (Fig. 2j) Immunofluorescence images were coded and counted blind to genotype by four individuals, and the four independent counts were then averaged for each brain section. The inter-rater correlation was r≥0.89. Four to six brain sections were imaged and counted per animal, with n = 3 Aspm+/+ or +/− and n = 4 Aspm−/− littermate E35 animals analyzed. (Fig. 3f) Apod+ cells from smFISH were segmented and counted using ImageJ, in an area of dorsal cortex 400 μm × 400 μm centered on the IZ, in multiple sections per animal, with n = 4 Aspm+/+ or +/−, and n = 4 −/−. Per brain average counts were then compared by one-tailed t-test.
Data availability
scRNA-seq data have been deposited in Gene Expression Omnibus (GEO) under accession number GSE110010. All other data are included within the paper.
Extended Data
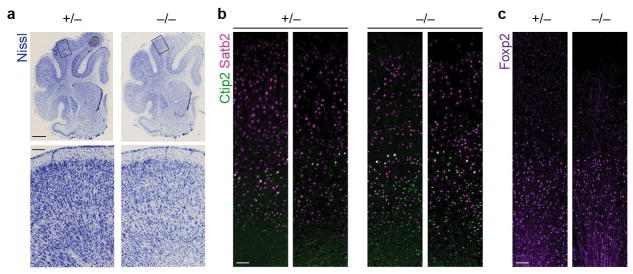
Extended Data Figure 1. Cytoarchitecture and neuronal subtype lamination in the mature Aspm KO ferret cortex.
a, Nissl stains of P41 littermate coronal sections, as shown in Fig. 1l, with additional Aspm+/− and Aspm−/− littermates shown. b, c, P41 littermates immunostained for cortical layer-specific projection neurons including Satb2 (layer II–IV), Ctip2 (layer V), and FoxP2 (layer VI). The experiments were repeated 3 times independently with similar results. Scale bars: 2 mm (a, top), 200 μm (a, bottom), 100 μm (b–c).
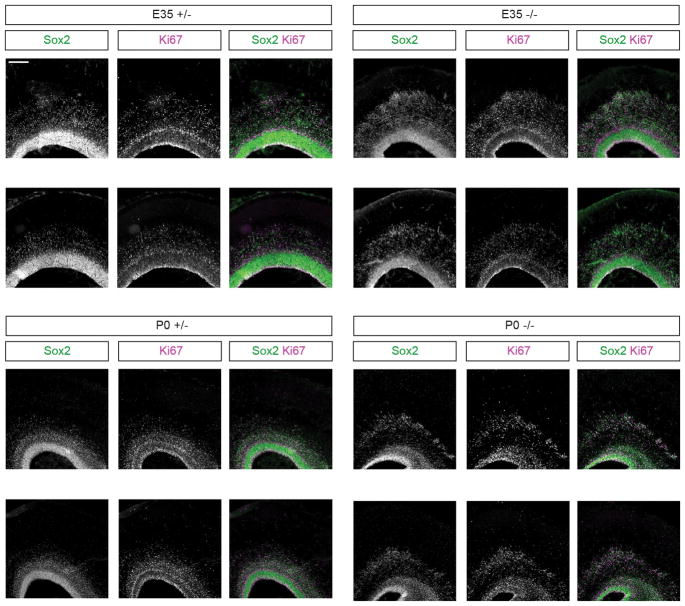
Extended Data Figure 2. Sox2/Ki67 immunostaining in additional E35 and P0 littermates dorsal cortex.
Extending findings of Fig. 2e, f, each set of panels is from a separate littermate, showing the high penetrance of the neural progenitor cell basal displacement phenotype. The experiments were repeated 3 times independently with similar results. Scale bar: 200 μm.
Extended Data Figure 3. Displaced progenitors in Aspm KO ferrets have basal fibers.
Extending findings of Fig. 2h, i, immunostaining of Sox2, Ki67, and Vim shows that displaced neural progenitors have basal radial fibers. The experiments were repeated 3 times independently with similar results. Scale bar: 100 μm.
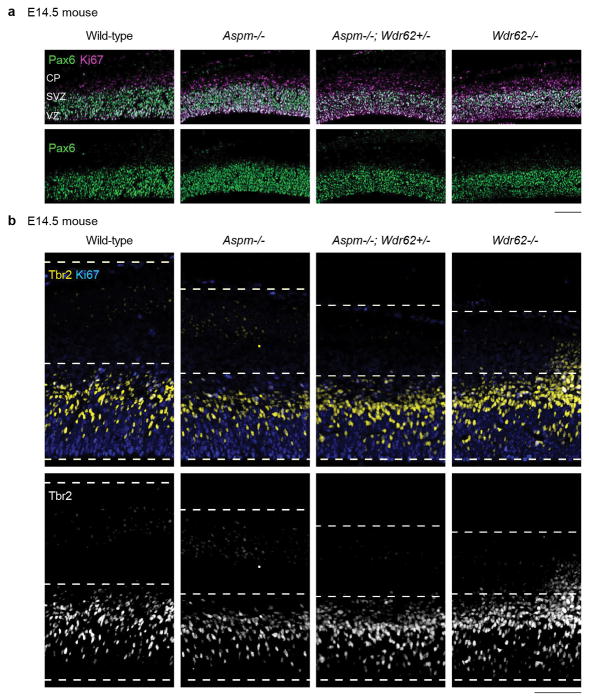
Extended Data Figure 4. Aspm KO mice do not demonstrate displaced progenitors in the IZ.
Unlike Aspm−/− ferrets, Aspm−/− mice do not demonstrate displaced NPC in the IZ. However, they show a variable increase in IP (Pax6-/Ki67+ cells in a and Tbr2+ cells in b), which is enhanced by heterozygous, compound mutation in Wdr62, a microcephaly gene causing more severe microcephaly9. The experiments were repeated 3 times independently with similar results. Scale bar: 100 μm.
Extended Data Figure 5. Modest increase in apoptosis throughout the germinal zones of the Aspm KO telencephalon.
Apoptotic cells (yellow) are indicated by enzymatic fluorescent detection of double-stranded DNA damage with DAPI nuclear counterstaining (blue). The experiments were repeated 3 times independently with similar results. Scale bars: 500 μm (a, whole section) and 100 μm (b, c, cortical wall columns).
Extended Data Figure 6. Additional immunohistochemical analyses of displaced progenitors in the Aspm KO cortex.
a, E35 KO cortex stained for VRG/ORG markers Sox2 and Hopx reveals extensive co-labeling in both the VZ and SVZ, including in displaced OSVZ progenitors. b, In the E35 KO OSVZ, clusters of supernumerary displaced neural progenitor cells include numerous Tbr2+ IP and are surrounded by Dcx+ newborn neurons, indicating preserved neurogenesis within the precocious OSVZ niche of the Aspm KO cortex. The experiments were repeated 3 times independently with similar results. Scale bar: 50 μm.
Extended Data Figure 7. Single-cell RNA-seq batch, sample, and cluster analyses.
a, tSNE plot from Fig. 3a with cells colored by biological replicate (i.e., animal). Most clusters include cells from all samples, except for a cluster expressing blood genes and a cluster expressing choroid plexus epithelial cells that are mostly from animal WT5E. These two cell clusters were not included in the downstream analysis. b, tSNE plot from Fig. 3a with cells colored by the batch they were processed in. Clusters are composed of cells from all batches. c, Per cell gene count and unique molecular identifier (UMI) count per sample. Each violin plot is one biological replicate and each dot is one cell. Sample WT5D was not included in the analysis due to the lower gene and UMI count compared to other samples as well as the inconsistent clustering compared to other WT samples (data not shown). d, Per cell gene count and UMI count for identified clusters. Each violin plot is one cell cluster and each dot is one cell. The three clusters in grey (EN4, BL, CPE) were not included in the downstream analysis. See Methods for details. This scRNA-seq experiment was performed once with n = 22,211 cells (8,037 cells from 2 Aspm+/+ and 1 +/− ferrets, and 14,174 cells from 4 Aspm−/− ferrets). RG1, cycling radial glial progenitors; RG2, interphase radial glial progenitors; IP, intermediate progenitors; EN1, upper-layer excitatory neurons; EN2, deep-layer excitatory neurons; EN3, Cajal-Retzius cells; IN1, immature inhibitory neurons; IN2, SST+ inhibitory neurons; IN3, ventral/inhibitory progenitors; ENDO1, endothelial cells 1; ENDO2, endothelial cells 2; OPC, oligodendrocyte precursors; MG, microglia; EN4, mixed excitatory neuron identity; BL, blood cells; CPE, choroid plexus epithelial cells.
Extended Data Figure 8. Loss of Aspm disrupts centriole duplication in ferret embryonic fibroblasts.
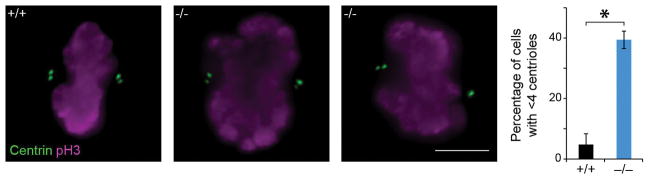
Mitotic Aspm KO FEF, identified by staining for pH3 and co-stained for the centriolar marker Centrin, display a significant loss of centrioles. The percentage of cells with an abnormal number (less than 4) of centrioles is increased 8-fold in Aspm−/− FEF compared to Aspm+/+ FEF (n = 100 cells/genotype/independent experiments; P = 0.003). The experiments were repeated 3 times independently with similar results. Statistical analysis was performed by two-tailed t-test and the graph shows mean ± s.e.m.
Extended Data Table 1. Region-specific changes in volume and surface area by loss of Aspm in ferrets.
a, Multiple brain regions are significantly decreased in volume with most reduction in the frontal cortex of adult Aspm−/− ferrets (n = 3 per genotype). Subcortical regions are relatively preserved. b, Outer cortical surface is most reduced in the frontal cortex followed by the lateral cortex. The parietal/occipital cortex is also decreased but the difference is not as significant. The medial cortex shows no discernible decrease. c, d, DTI shows that the orientation of white matter tracts or connectivity is fundamentally unchanged in Aspm KO ferrets except in the frontal cortex, which show a modest decrease in fractional anisotropy (d). The directional map (c) shows white matter orientation with red, green, and blue indicating the medial-lateral, superior-inferior, and anterior-posterior components respectively. Statistical analysis was performed by two-tailed t-test. Mean ± s.e.m.
| a | |||
|---|---|---|---|
| Volume (mm3)
| |||
| Region | Aspm+/− | Aspm−/− | P-value |
| Frontal ctx | 475.0 ± 58.4 | 235.0 ± 11.0 | 0.0156 |
| Corpus callosum | 79.3 ± 6.7 | 40.3 ± 2.3 | 0.0048 |
| Lateral ctx | 666.9 ± 48.4 | 364.8 ± 33.9 | 0.0069 |
| Ctx WM | 701.4 ± 46.1 | 402.9 ± 19.0 | 0.0039 |
| Parietal/occipital ctx | 421.6 ± 17.5 | 256.6 ± 17.5 | 0.0026 |
| Medial ctx | 639.4 ± 58.5 | 397.3 ± 14.3 | 0.0159 |
| Hippocampus | 319.7 ± 49.8 | 208.4 ± 18.5 | 0.0136 |
| Caudate | 113.9± 13.3 | 74.8 ± 4.5 | 0.0478 |
| Putamen | 14.3 ± 0.9 | 10.1 ± 0.4 | 0.0147 |
| Thalamus | 239.7 ± 18.5 | 172.4 ± 3.4 | 0.0238 |
| Cerebellum GM | 895.0 ± 32.0 | 650.3 ± 20.7 | 0.0030 |
| Cerebellum WM | 182.5 ± 12.1 | 141.5± 6.2 | 0.0382 |
| Brainstem GM | 263.8 ± 23.1 | 207.0 ± 8.4 | 0.0836 (NS) |
| Brainstem WM | 149.8 ± 6.5 | 121.3 ± 5.7 | 0.0298 |
| Midbrain WM | 43.2 ± 0.6 | 37.3 ± 0.8 | 0.0018 |
| Amygdala | 23.2 ± 1.1 | 20.2 ± 1.4 | 0.1868 (NS) |
| Midbrain GM | 106.0 ± 3.2 | 101.8 ± 2.6 | 0.3146 (NS) |
| b | |||
|---|---|---|---|
| Outer surface area
(mm2) | |||
| Region | Aspm+/− | Aspm−/− | P-value |
| Cerebral ctx (total) | 1,225.4 ± 64.1 | 933.0 ± 47.5 | 0.0217 |
| Frontal ctx | 309.6 ± 18.5 | 203.5 ± 2.8 | 0.0048 |
| Lateral ctx | 391.9 ± 18.5 | 301.4 ± 20.5 | 0.0305 |
| Parietal/occipital ctx | 298.4 ± 20.1 | 217.1 ± 22.7 | 0.0551 (NS) |
| Medial ctx | 225.5 ± 9.7 | 211.0 ± 9.5 | 0.3476 (NS) |
| c |
|---|
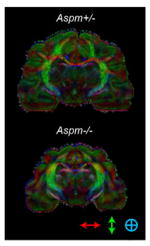
|
| d | |||
|---|---|---|---|
| Fractional anisotropy
| |||
| Region | Aspm+/− | Aspm−/− | P-value |
| Frontal ctx | 0.272 ± 0.002 | 0.240 ± 0.007 | 0.0130 |
| Medial ctx | 0.255 ± 0.004 | 0.237 ± 0.009 | 0.1377 (NS) |
| Lateral ctx | 0.253 ± 0.011 | 0.252 ± 0.008 | 0.9198 (NS) |
| Parietal/occipital ctx | 0.267 ± 0.014 | 0.246 ± 0.014 | 0.3421 (NS) |
| Ctx WM | 0.431 ± 0.031 | 0.379 ± 0.018 | 0.2285 (NS) |
| Corpus callosum | 0.487 ± 0.034 | 0.434 ± 0.048 | 0.4170 (NS) |
| Cingulum | 0.415 ± 0.063 | 0.342 ± 0.053 | 0.4267 (NS) |
| Fornix | 0.458 ± 0.041 | 0.435 ± 0.022 | 0.6542 (NS) |
| Anterior commisure | 0.437 ± 0.069 | 0.336 ± 0.052 | 0.3046 (NS) |
| Optic tract | 0.523 ± 0.059 | 0.435 ± 0.045 | 0.3013 (NS) |
| Corticospinal tract | 0.550 ± 0.059 | 0.470 ± 0.041 | 0.3330 (NS) |
| Brainstem WM | 0.501 ± 0.042 | 0.423 ± 0.026 | 0.1925 (NS) |
| Cerebellum WM | 0.454 ± 0.031 | 0.368 ± 0.033 | 0.1321 (NS) |
| Midbrain WM | 0.398 ± 0.058 | 0.338 ± 0.043 | 0.4496 (NS) |
| Cerebellum GM | 0.301 ± 0.010 | 0.258 ± 0.007 | 0.0211 |
| Brainstem GM | 0.353 ± 0.011 | 0.285 ± 0.015 | 0.0230 |
| Inferior colliculus | 0.289 ± 0.001 | 0.276 ± 0.009 | 0.2078 (NS) |
| Superior colliculus | 0.270 ± 0.008 | 0.262 ± 0.008 | 0.4870 (NS) |
| Periaqueductal gray | 0.273 ± 0.010 | 0.268 ± 0.003 | 0.6517 (NS) |
| Midbrain GM | 0.291 ± 0.017 | 0.270 ± 0.015 | 0.3974 (NS) |
| Thalamus | 0.286 ± 0.015 | 0.269 ± 0.003 | 0.3413 (NS) |
| Hippocampus | 0.270 ± 0.011 | 0.254 ± 0.002 | 0.2276 (NS) |
| Hypothalamus | 0.257 ± 0.021 | 0.225 ± 0.011 | 0.2533 (NS) |
| Globus pallidus | 0.291 ± 0.020 | 0.288 ± 0.021 | 0.9115 (NS) |
| Putamen | 0.285 ± 0.007 | 0.292 ± 0.023 | 0.7631 (NS) |
| Caudate | 0.216 ± 0.005 | 0.221 ± 0.013 | 0.7161 (NS) |
| Septum | 0.267 ± 0.022 | 0.245 ± 0.010 | 0.4183 (NS) |
| Amygdala | 0.206 ± 0.013 | 0.213 ± 0.019 | 0.7790 (NS) |
Extended Data Table 2.
Cluster IDs of E35 ferret cerebral cortical cells analyzed by single-cell RNA sequencing. The three clusters highlighted in blue represent the largest proportional changes with empirical FDR < 0.01, and are similarly indicated in Figure 3. Statistical analysis was performed by two-tailed Chi-square test.
| Color | Cluster ID |
Top 10 Marker Transcripts | Inferred Cell Type | Cell Count
in Aspm+/− |
Cell Count
in Aspm−/− |
Proportion
of Aspm+/− |
Proportion
of Aspm−/− |
Fold- change in proportion |
Empirical P- value |
|---|---|---|---|---|---|---|---|---|---|
| RG1 | TOP2A; HMGB2; CENPF; CENPE; TPX2; XLOC_000183; 2810417H13Rik; SMC4; XLOC_036181; KIF11 | Cycling radial glial progenitors | 803 | 1,661 | 10.5% | 12.1% | +15% | 0.0002 (FDR<0.01) | |
| RG2 | VIM; HES1; SLC1A3; GDPD2; SFRP1; PON2; PTN; SMPDL3A; NES; PAX6 | Interphase radial glial progenitors | 495 | 800 | 6.5% | 5.8% | −10% | 0.0696 | |
| IP | NRN1; PTPDC1; TENM4; ELAVL4; ROBO2; FBXW7; IGFBP2; NELL2; PRKX; TTC28 | Intermediate progenitors | 686 | 1,603 | 9.0% | 11.7% | +30% | 0.0002 (FDR<0.01) | |
| EN1 | SYT4; CSRP2; NEUROD6; NEUROD2; UNC5D; NTM; NSG2; LIMCH1; SORBS2; ISLR2 | Upper-layer excitatory neurons | 2,203 | 3,490 | 28.8% | 25.4% | −12% | 0.0002 (FDR<0.01) | |
| EN2 | NEFM; FEZF2; NEFL; GRIA2; GUCY1B3; DYNC1I1; ARPP21; NEUROD6; B3GALT2; KCTD12 | Deep-layer excitatory neurons | 981 | 1,053 | 12.8% | 7.7% | −40% | 0.0002 (FDR<0.01) | |
| EN3 | RELN; LHX1; NDNF; TP73; NHLH2; VSNL1; ENSMPUG00000012124; PLCL1; ENSMPUG00000009767; SEMA6A | Cajal-Retzius cells | 103 | 227 | 1.3% | 1.7% | +23% | 0.0704 | |
| IN1 | PBX3; MEIS2; XLOC_008478; ENSMPUG00000024751; INA; MAP1B; ATP1B1; XLOC_021005; RUNX1T1; JAKMIP2 | Immature inhibitory neurons | 591 | 1,167 | 7.7% | 8.5% | +10% | 0.0444 | |
| IN2 | SST; XLOC_014564; PDZRN4; NXPH1; NXPH2; SYT1; XLOC_026835; GRIA1; ARX; PDE4DIP | SST interneurons | 734 | 1,568 | 9.6% | 11.4% | +19% | 0.0002 (FDR<0.01) | |
| IN3 | XLOC_026835; DLX1; XLOC_026893; CCDC88A; ZNF704; PFN2; EPHA5; XLOC_007250; INA; NR2F1 | Ventral/inhibitory progenitors | 710 | 1,521 | 9.3% | 11.1% | +19% | 0.0002 (FDR<0.01) | |
| ENDO1 | APOA1; COL4A1; CALD1; SPARC; FN1; IGFBP7; XLOC_010971; ENSMPUG00000012145; LAMA4; MGP | Endothelial cells | 122 | 210 | 1.6% | 1.5% | −4% | 0.7418 | |
| ENDO2 | SPARCL1; LYZ; PECAM1; IFNAR1; SPARC; IFI27; XLOC_010971; EMB; S100A6; IGFBP7 | Endothelial cells | 113 | 150 | 1.5% | 1.1% | −26% | 0.0166 (FDR<0.05) | |
| OPC | APOD; XLOC_017682; SCRG1; DBI; SPARCL1; PDGFRA; SERPINE2; ENSMPUG00000011077; PTPRZ1; LHFPL3 | Oligodendrocyte precursors | 48 | 170 | 0.6% | 1.2% | +97% | 0.0002 (FDR<0.01) | |
| MG | ENSMPUG00000001122; C1QC; C3; RGS10; XLOC_039347; CCL8; XLOC_039690; ZFP36; CCL4; SPP1 | Microglia | 56 | 105 | 0.7% | 0.8% | +4% | 0.7234 |
Supplementary Material
Uncropped scans with size marker indications for Fig. 4c, g.
Acknowledgments
We dedicate this paper to the late R. W. Guillery, who first introduced ferrets as a model for developmental neuroscience. We thank J. K. Joung for advice on genome editing; J. Bond for ASPM antibody; L. Vasung, P. Herman, J. Neil and C. D. Kroenke for advice on ferret brain MRI; A. Lee, the G. M. Church lab (S. Biwas), the S. McCarroll lab (S. Burger), the P. Kharchenko lab (J. Fan), and the R. Satija lab (A. Butler) for advice on scRNA-seq; S. Wasiuk, E. Feiner, A.S. Kamumbu, and M. Lee for technical assistance; Marshall BioResources for animal husbandry; and E. Pollack and the veterinary staff at Boston Children’s Hospital and Yale School of Medicine for surgical support. Animal silhouettes in Fig. 1 were designed by Freepik from Flaticon.com. X.P. is a consultant for Electrical Geodesics Inc. This work was supported by P30NS052519 (F.H. and Yale’s QNMR Core Center), 2R01MH067528 (F.H.), 1R24MH114805 (X.P.), R21HD083956 (K.I.), R01EB017337 (P.E.G.), R24HL123482 (J.F.E.), 5R01NS032457 (C.A.W.), 5R21NS091865 (B.-I.B.), and the Allen Discovery Center program through The Paul G. Allen Frontiers Group. C.A.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions B.-I.B., C.A.W., and J.F.E conceived the project. B.-I.B. generated genome editing reagents. X.S., Z.Y., and B.L. injected ferret zygotes under J.F.E.’s supervision. M.B.J., K.M.G., P.P.W., and D.M.G. performed immunohistochemistry. R.S.S., M.C., K.I., J.B.M., P.E.G., D.C., X.P., L.S., and F.H. performed MRI analyses. M.B.J., R.B.-M., Y.M.W., and H.K. performed scRNA-seq. M.B.J. and R.B.-M. performed smFISH. A.K. and S.C.R. characterized molecular defects with K.P. and B.-I.B. With input from all authors, M.B.J., B.-I.B., and C.A.W interpreted the data and wrote the paper.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Faheem M, et al. Molecular genetics of human primary microcephaly: an overview. BMC medical genomics. 2015;8(Suppl 1):S4. doi: 10.1186/1755-8794-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bond J, et al. ASPM is a major determinant of cerebral cortical size. Nature genetics. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 3.Passemard S, et al. Abnormal spindle-like microcephaly-associated (ASPM) mutations strongly disrupt neocortical structure but spare the hippocampus and long-term memory. Cortex; a journal devoted to the study of the nervous system and behavior. 2016;74:158–176. doi: 10.1016/j.cortex.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Desir J, Cassart M, David P, Van Bogaert P, Abramowicz M. Primary microcephaly with ASPM mutation shows simplified cortical gyration with antero-posterior gradient pre- and post-natally. American journal of medical genetics. Part A. 2008;146A:1439–1443. doi: 10.1002/ajmg.a.32312. [DOI] [PubMed] [Google Scholar]
- 5.Pulvers JN, et al. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16595–16600. doi: 10.1073/pnas.1010494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujimori A, et al. Disruption of Aspm causes microcephaly with abnormal neuronal differentiation. Brain & development. 2014;36:661–669. doi: 10.1016/j.braindev.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Capecchi MR, Pozner A. ASPM regulates symmetric stem cell division by tuning Cyclin E ubiquitination. Nature communications. 2015;6:8763. doi: 10.1038/ncomms9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams SE, et al. Aspm sustains postnatal cerebellar neurogenesis and medulloblastoma growth in mice. Development. 2015;142:3921–3932. doi: 10.1242/dev.124271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jayaraman D, et al. Microcephaly Proteins Wdr62 and Aspm Define a Mother Centriole Complex Regulating Centriole Biogenesis, Apical Complex, and Cell Fate. Neuron. 2016;92:813–828. doi: 10.1016/j.neuron.2016.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montgomery SH, Mundy NI. Microcephaly genes evolved adaptively throughout the evolution of eutherian mammals. BMC evolutionary biology. 2014;14:120. doi: 10.1186/1471-2148-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bae BI, Jayaraman D, Walsh CA. Genetic changes shaping the human brain. Dev Cell. 2015;32:423–434. doi: 10.1016/j.devcel.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fietz SA, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nature neuroscience. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- 13.Reillo I, de Juan Romero C, Garcia-Cabezas MA, Borrell V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex. 2011;21:1674–1694. doi: 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- 14.Johnson MB, et al. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nature neuroscience. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smart IH, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex. 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 17.Bond J, et al. Protein-truncating mutations in ASPM cause variable reduction in brain size. American journal of human genetics. 2003;73:1170–1177. doi: 10.1086/379085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchinson EB, et al. Population based MRI and DTI templates of the adult ferret brain and tools for voxelwise analysis. NeuroImage. 2017;152:575–589. doi: 10.1016/j.neuroimage.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Cerdeno V, et al. Comparative analysis of the subventricular zone in rat, ferret and macaque: evidence for an outer subventricular zone in rodents. PloS one. 2012;7:e30178. doi: 10.1371/journal.pone.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Martinez MA, et al. A restricted period for formation of outer subventricular zone defined by Cdh1 and Trnp1 levels. Nature communications. 2016;7:11812. doi: 10.1038/ncomms11812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollen AA, et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kodani A, et al. Centriolar satellites assemble centrosomal microcephaly proteins to recruit CDK2 and promote centriole duplication. eLife. 2015;4 doi: 10.7554/eLife.07519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paridaen JT, Wilsch-Brauninger M, Huttner WB. Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell. 2013;155:333–344. doi: 10.1016/j.cell.2013.08.060. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh S, Solecki DJ. Polarity transitions during neurogenesis and germinal zone exit in the developing central nervous system. Frontiers in cellular neuroscience. 2015;9:62. doi: 10.3389/fncel.2015.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dauber A, et al. Novel microcephalic primordial dwarfism disorder associated with variants in the centrosomal protein ninein. The Journal of clinical endocrinology and metabolism. 2012;97:E2140–2151. doi: 10.1210/jc.2012-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, et al. Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell. 2016;166:1147–1162e1115. doi: 10.1016/j.cell.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehtinen MK, et al. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mashiko D, et al. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Scientific reports. 2013;3:3355. doi: 10.1038/srep03355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Sun X, Chen J, Leno GH, Engelhardt JF. Factors affecting the efficiency of embryo transfer in the domestic ferret (Mustela putorius furo) Theriogenology. 2006;66:183–190. doi: 10.1016/j.theriogenology.2005.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chahboune H, et al. Neurodevelopment of C57B/L6 mouse brain assessed by in vivo diffusion tensor imaging. NMR in biomedicine. 2007;20:375–382. doi: 10.1002/nbm.1130. [DOI] [PubMed] [Google Scholar]
- 33.Petersen KF, et al. The role of skeletal muscle insulin resistance in the pathogenesis of the metabolic syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12587–12594. doi: 10.1073/pnas.0705408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rueckert D, et al. Nonrigid registration using free-form deformations: application to breast MR images. IEEE transactions on medical imaging. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 35.Papademetris X, Jackowski AP, Schultz RT, Staib LH, Duncan JS. Integrated Intensity and Point-Feature Nonrigid Registration. Medical image computing and computer-assisted intervention : MICCAI … International Conference on Medical Image Computing and Computer-Assisted Intervention. 2001;3216:763–770. doi: 10.1901/jaba.2001.3216-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawada K, Watanabe M. Development of cerebral sulci and gyri in ferrets (Mustela putorius) Congenital anomalies. 2012;52:168–175. doi: 10.1111/j.1741-4520.2012.00372.x. [DOI] [PubMed] [Google Scholar]
- 37.Joshi A, et al. Unified framework for development, deployment and robust testing of neuroimaging algorithms. Neuroinformatics. 2011;9:69–84. doi: 10.1007/s12021-010-9092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman AS, Parvinjah S, Hanna MA, Helguera PR, Busciglio J. Cryopreservation of cortical tissue blocks for the generation of highly enriched neuronal cultures. Journal of visualized experiments : JoVE. 2010 doi: 10.3791/2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature biotechnology. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villani AC, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017:356. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higgins J, et al. Human ASPM participates in spindle organisation, spindle orientation and cytokinesis. BMC cell biology. 2010;11:85. doi: 10.1186/1471-2121-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Uncropped scans with size marker indications for Fig. 4c, g.
Data Availability Statement
scRNA-seq data have been deposited in Gene Expression Omnibus (GEO) under accession number GSE110010. All other data are included within the paper.