Abstract
Epigenetic effects of environmental chemicals are under intense investigation to fill existing knowledge gaps between environmental/occupational exposures and adverse health outcomes. Chromatin accessibility is one prominent mechanism of epigenetic control of transcription, and understanding of the chemical effects on both could inform the causal role of epigenetic alterations in disease mechanisms. In this study, we hypothesized that baseline variability in chromatin organization and transcription profiles among various tissues and mouse strains influence the outcome of exposure to the DNA damaging chemical 1,3-butadiene. To test this hypothesis, we evaluated DNA damage along with comprehensive quantification of RNA transcripts (RNA-seq), identification of accessible chromatin (ATAC-seq), and characterization of regions with histone modifications associated with active transcription (ChIP-seq for acetylation at histone 3 lysine 27, H3K27ac). We collected these data in the lung, liver and kidney of mice from two genetically divergent strains, C57BL/6J and CAST/EiJ, that were exposed to clean air or to 1,3-butadiene (~600 ppm) for 2 weeks. We found that tissue effects dominate differences in both gene expression and chromatin states, followed by strain effects. At baseline, xenobiotic metabolism was consistently more active in CAST/EiJ, while immune system pathways were more active in C57BL/6J across tissues. Surprisingly, even though all three tissues in both strains harbored butadiene-induced DNA damage, little transcriptional effect of butadiene was observed in liver and kidney. Toxicologically-relevant effects of butadiene in the lung were on the pathways of xenobiotic metabolism and inflammation. We also found that variability in chromatin accessibility across individuals (i.e., strains) only partially explains the variability in transcription. This study showed that variation in the basal states of epigenome and transcriptome may be useful indicators for individuals or tissues susceptible to genotoxic environmental chemicals.
Introduction
Chemical exposure-associated epigenetic events are gaining attention in toxicology as providing important mechanistic information for human health assessments (Chappell et al. 2016; Cote et al. 2016). The epigenetic footprints from exposure may also provide key insights into potential health outcomes. Most studies of epigenetics in toxicology have focused on chemical carcinogenesis (Pogribny and Rusyn 2013), including research into the linkages to other mechanisms such as genotoxicity. However, epigenetic signatures of carcinogens have yet to make an impact on regulatory decision-making in public health; additional data such as chromatin accessibility and transcriptomics could inform the causal role for the epigenetic alterations and connect exposures and outcomes.
1,3-Butadiene is a model genotoxic chemical that has been used to investigate the linkages between exposure, DNA damage and epigenetic alterations (Chappell et al. 2014; Chappell et al. 2017; Koturbash et al. 2011a; Koturbash et al. 2011b). Butadiene is a major industrial chemical used in the production of synthetic rubbers and polymers; it is also a ubiquitous environmental contaminant that is present in cigarette smoke and automobile exhaust (IARC 2008). United States and international regulatory and health agencies classify butadiene as a human carcinogen (IARC 2008; National Toxicology Program 2011). Epidemiological data support an association between butadiene exposure and cancers of the hematopoietic system, such as leukemia and lymphoma. Mice exposed to butadiene concentrations ranging from 6.25 to 1250 ppm developed tumors in multiple organs, including the lung and liver (IARC 2012).
The direct mechanism for butadiene carcinogenesis is well established: reactive epoxide metabolites directly interact with DNA to form DNA adducts that lead to mutations (Goggin et al. 2009; Swenberg et al. 2011). In addition to genotoxicity, butadiene also effects the epigenome, disrupting global DNA methylation and chromatin structure (Koturbash et al. 2011b). It was hypothesized that butadiene-induced tissue-specific epigenetic effects may explain mechanistically the lack of tumorigenesis in specific tissues (Chappell et al. 2014). Inter-strain differences in butadiene-induced genotoxic and epigenetic alterations were also identified by using a panel of mouse inbred strains (Koturbash et al. 2011a). Specifically, CAST/EiJ and C57BL/6J strains exhibited divergent responses to butadiene in terms of DNA damage, histone modifications, and chromatin states.
The inter-strain differences in transcriptional and gene-specific chromatin changes were further investigated in both CAST/EiJ and C57BL/6J mice in the lung (Chappell et al. 2017). Strain-specific differences in the baseline molecular states were identified and have been associated to the variability in butadiene effects among strains. Butadiene exposure also elicited a distinct chromatin and transcriptional response in each strain. Specifically, detoxification and DNA damage pathways were more highly expressed at the basal level in CAST/EiJ whereas these pathways were upregulated in C57BL/6J mice after butadiene exposure. This study indicated that butadiene exposure in C57BL/6J leads to reprograming of the transcriptome and chromatin, an effect that resembles baseline state of CAST/EiJ. Varying degrees of accessibility and thus transcriptional activity before exposure could be responsible for inter-individual differences in responses to toxicants.
In order to evaluate population epigenomic variability, informative assays and endpoints need to be incorporated into both toxicology and epigenetic studies. RNA-seq in conjunction with ATAC-seq represents a cost-effective method to sufficiently investigate the link between genetic variants and epigenetic landscape in the inter-individual responses to toxicant exposure (Lewis et al. 2017). This combination of techniques provides the opportunity to identify and resolve regions of active or repressed enhancers and promoters. In the present study, we assessed the chromatin landscape and transcriptional differences in response to butadiene in target (lung and liver) and non-target (kidney) tissues of carcinogenesis in mice. Our results indicate that the majority of the molecular differences occur in the lung as compared to the liver and kidney.
Materials and Methods
Animals and Exposures
The in vivo portion of the study and tissue collection was detailed in (Chappell et al. 2017). Butadiene is carcinogenic in mice at doses as low as 6.25 ppm, it has been tested up to 8,000 ppm (Melnick and Huff 1993). In brief, male C57BL/6J and CAST/EiJ mice (Jackson Laboratory, 9–13 weeks old) were exposed to clean air or to 1,3-butadiene (average concentration of 593±61ppm) for 6h a day, Monday-Friday, over a 2-week period. This exposure regimen is meant to model human occupational exposures to 1,3-butadiene and this experimental design was used by other laboratories (Goggin et al. 2011; Goggin et al. 2009; Meng et al. 2007) to characterize the genotoxicity of 1,3-butadiene and is similar to that used in a chronic cancer bioassay (National Toxicology Program 2011). Also, at this dose and duration of exposure we demonstrated robust and strain-specific differences in genotoxic, epigenetic and transcriptional responses (Chappell et al. 2014; Chappell et al. 2017; Koturbash et al. 2011a; Koturbash et al. 2011b). Immediately following the final exposure, mice were euthanized and lungs, liver and kidney tissues were collected. The animals were treated humanely and with regard for alleviation of suffering, and the procedures were approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill.
Determination of N7-guanine adduct formation
Genomic DNA was isolated from flash-frozen tissues using a Qiagen DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Measurement of N-7-(2,3,4-trihydroxybut-1-yl)-guanine (THB-Gua) was performed as described in (Goggin et al. 2009) with minor modifications.
mRNA sequencing and data processing
Total RNA was isolated from frozen tissues using a Qiagen miRNeasy Kit (Valencia, CA) according to the manufacturer’s protocol. Thermo Scientific Nanodrop 2000 (Waltham, MA) and an Agilent 2100 Bioanalyzer (Santa Clara, CA) were used to evaluate RNA purity and integrity. A minimum RNA integrity value of 7.0 was required for RNA samples to be used for library preparation and sequencing. Total RNA sequencing (RNA-seq) libraries were prepared using the Illumina TruSeq Total RNA Sample Prep Kit (Illumina, Inc., San Diego, USA) with ribosomal depletion. Paired-end (50bp) sequencing was performed using the Illumina HiSeq 2500 instrument; Supplemental Table 1). Reads were filtered (score of 20 or greater in at least 90 percent of nucleotides), adapters removed, and aligned to appropriate reference genomes (C57BL/6: NCBI Build 37; CAST/EiJ: Build 37 pseudogenome (http://csbio.unc.edu/CCstatus/index.py?run=Pseudo) using STAR (Wu and Nacu 2010). Transcript quantification was performed using RSEM (Li and Dewey 2011). Further details on data processing pipeline are described in (Chappell et al. 2017).
Assay for Transposase Accessible Chromatin (ATAC) sequencing and data processing
Frozen tissues were pulverized in liquid nitrogen using the BioPulverizer (Biospec), thawed in glycerol-containing nuclear isolation buffer, and then filtered through Miracloth (Calbiochem) to remove tissue debris. Nuclei were washed and directly used for treatment with Tn5 transposase. Single-end (50bp) sequencing was performed on the Illumina HiSeq 2500 instrument (Supplemental Table 1). Reads were quality-filtered (requiring a quality score of 20 or greater in at least 90% of nucleotides), adapters removed with cutadapt (Martin 2011), and aligned with GSNAP (Wu and Nacu 2010; Zhang et al. 2012) to the appropriate reference genome. Post-alignment filtering was performed as described in (Chappell et al. 2017).
H3k27ac Chromatin ImmunoPrecipitation (ChIP) sequencing and data processing
ChIP was performed as previously described (Lee et al. 2006) with the following modifications: For each ChIP reaction, approximately 25mg of pulverized tissue (stored at −80C) was used. Tissue samples were re-suspended in 10ml PBS at room temperature (RT). Crosslinking was performed by adding methanol-free formaldehyde (ThermoScientific) to a final concentration of 1%. Following cell lysis, nuclei pellets were collected by at 1350g for 5 min at 4C. Pellets were carefully re-suspended in 1ml covaris sonication buffer (10 mM Tris-HCl (pH 7.6), 1 mM EDTA, 0.25% SDS) and transferred to covaris tubes. Chromatin was sheared for an average 200–300 bp fragments using a Covaris S2 sonicator (Duty Factor: 5.0, Peak Power: 140, Cycles/Burst: 200). Cellular debris was removed by centrifugation at 15000×g for 5 min at 4C. Before ChIP, Triton-X and NaCl were added to the chromatin suspension for a final concentration of 1% Triton-X and 150mM NaCl. Each precipitation was performed with 1μg of an antibody against acetylated histone 3 lysine 27 (anti-H3K27ac, Abcam, ab4729), following the steps outlined in Ihn et al. DNA was isolated following the IP, and libraries were prepared using the Illumina TruSeq ChIP protocol. Single-end (50bp) sequencing was performed on the Illumina HiSeq 2500 instrument (Supplemental Table 1). ChIP-seq data was processed similarly to ATAC-seq data (see above), except that cutadapt was not used.
mRNA, chromatin accessibility, and histone acetylation differential analyses
Differentially expressed mRNAs were identified using the R package edgeR v. 3.16.5 (Robinson et al. 2010) (Supplemental Table 2). For accessible regions, the union set of the top 50,000 peaks, as determined by F-seq (Boyle et al. 2008), from all samples within a tissue group was identified. The number of mapped reads within 300bp overlapping windows (peaks smaller than 300 bp were expanded to 300 bp) within peaks was computed for each sample. Differential chromatin accessibility was detected using the R package csaw v. 1.2.1 (Lun and Smyth 2016), which uses methods from the edgeR package (Robinson et al. 2010). Normalization factors were computed to correct for compositional bias between samples with the windowCounts function, with width=10000, and the normOffsets function with default parameters. The function estimateDisp was used to estimate the dispersion and a generalized linear model was fit using glmQLFit with robust=TRUE. Following each test, significant neighboring windows (FDR<10%, overlap < 250 bp) were merged (Supplemental Table 3). This method was also used to determine differentially acetylated regions (Supplemental Table 4). A summary of differentially expressed genes, accessible chromatin, and acetylated regions is provided in Supplemental Table 5.
Chromatin state analysis and chord diagram visualizations
For each condition group (e.g., C57BL/6J lung control samples, CAST/EiJ liver butadiene-exposed, etc.), the top 50,000 and top 100,000 peaks were identified based on the average peak score across biological replicates, as determined by F-seq (Boyle et al. 2008). Peaks within 250 bp were merged, and coverage was calculated in 1000 bp windows centered on these peaks, and also in 1000 bp windows tiled across the genome to obtain a background coverage distribution. If a peak was identified in the top 50,000 peaks within a condition group, overlapped a differentially accessible region (edgeR FDR<10%), and it also had appreciable H3K27ac signal (95th percentile of background), then it was classified as “active”. In a comparison of two condition groups, if that peak remained within the top 100,000 peaks in the opposite group, and retained H3K27ac signal, it remained classified as active. However, if it was not within the top 100,000 peaks, it was classified as “closed”. Peaks within the top 50,000 without appreciable H3k27ac signal were classified as “poised”. Due to the small number of poised peaks identified (Supplemental Table 6), they were not considered in further analyses. Peaks that otherwise met the criteria for inclusion, but did not overlap a differentially accessible region (edgeR FDR<10%), were classified as a “no change”. Regions were associated with genes using GREAT v. 3.0.0 (McLean et al. 2010), and the direction of expression change for that gene was obtained from our analysis differential expression using edgeR. Visualization of chromatin state transitions was performed using circlize (Gu et al. 2014).
Pathway enrichment analysis
Significant enrichment of biological pathways associated with chromatin state transitions were determined using the runGSAhyper function of the R package Piano v.1.14.5 (Varemo et al. 2013) and the Reactome Pathway Database (http://baderlab.org/GeneSets) (Merico et al. 2010). Pathways with <15 genes or >500 genes were not included. Genomic regulatory regions were associated with genes using GREAT v. 3.0.0 (McLean et al. 2010).
Results
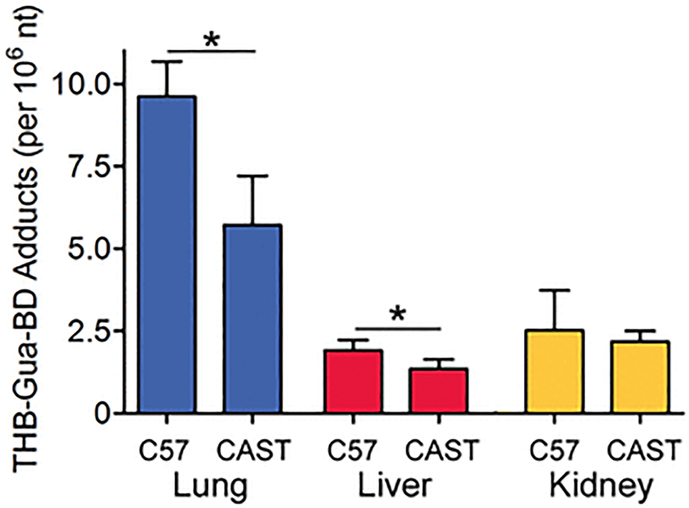
This study focused on characterizing tissue- and strain-specific effects of the genotoxic carcinogen 1,3-butadiene on chromatin and transcription. To explore the relationships between inherent variation in biological responses to molecular and/or cellular interactions with the chemical, we used an in vivo mouse model and two inbred strains known to have divergent responses to butadiene-induced DNA damage and epigenetic effects (Chappell et al. 2014; Chappell et al. 2017; Koturbash et al. 2011a). Exposure to butadiene results in the formation of a variety of DNA adducts (Swenberg et al. 2011), including THB-Gua adducts that are used as common biomarkers for butadiene exposure. Tissue variability in the formation of THB-Gua adducts was previously demonstrated in C57BL/6J mice with the highest levels in the lung, followed by kidney, and finally liver (Chappell et al. 2014). In this study, we also found that the lung was the organ with the highest THB-Gua adduct burden (Figure 1), with liver and kidney adduct levels 3–4 fold lower than those in the lung, in both strains. As reported previously for the liver (Koturbash et al. 2011a) and lung (Chappell et al. 2017), we also found strain-specific significant differences in THB-Gua adducts between C57BL/6J and CAST/EiJ mice. These data served as confirmation of the varying response to butadiene across tissues and strains in this exposure model, warranting further exploration of the variability in chromatin and transcription responses.
Figure 1.
Amounts of THB-Gua-BD adducts in tissues from mice exposed to ~600 ppm of 1,3-butadiene. Data are presented as mean±SD (n=3–6). Asterisks (*) denote significant (P<0.05) differences in the amount of the adducts within the same tissue between strains.
Global gene expression, chromatin accessibility, and histone acetylation patterns reveal striking differences between mouse tissues and strains at baseline
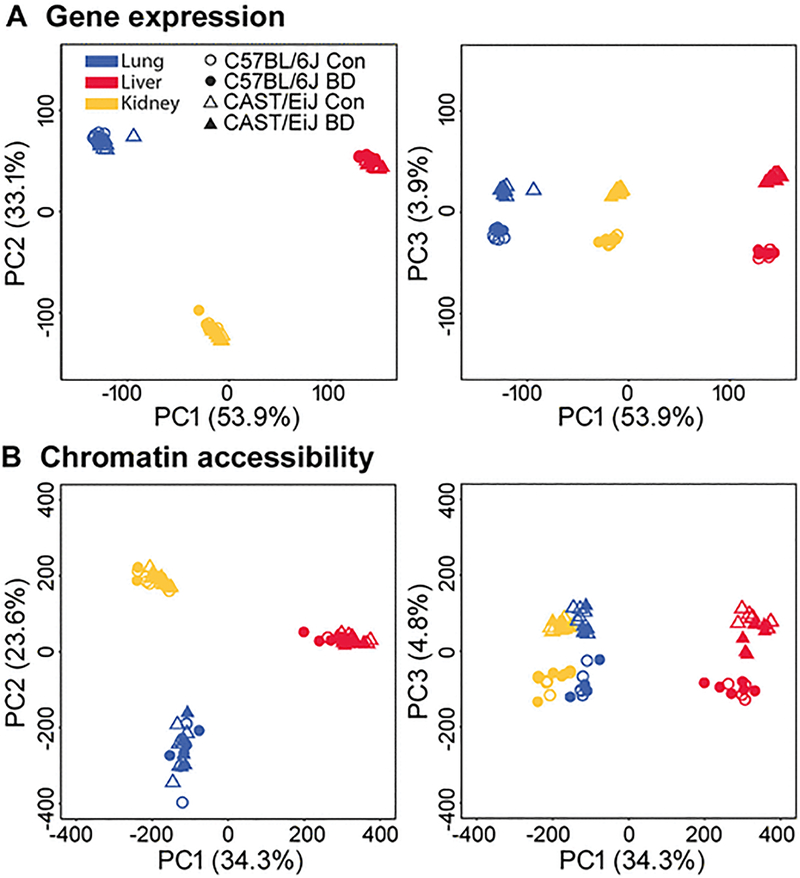
We compared transcriptional activity and chromatin accessibility between strains and tissues with or without 1,3-butadiene exposure using RNA-seq, the assay for transposase-accessible chromatin (ATAC-seq), and ChIP-seq for H3K27ac. We first performed a principal component analysis (PCA) to identify the major sources of variance in our RNA-seq, ATAC-seq, and ChIP-seq datasets. The first two principal components (PC1 and PC2) clearly separated samples according to tissue in both RNA-seq (Figure 2A) and ATAC-seq (Figure 2B) datasets, while PC3 further distinguished CAST/EiJ from C57BL/6J samples. Importantly, exposure to butadiene did not significantly contribute to variation in gene expression or chromatin accessibility. We also observed a similar trend for H3K27ac, in that tissue type was the main source of the variance, while strain was not a major contributor (Supplemental Figure 1). These results identify tissue and strain as the main sources of variation in global gene expression and chromatin accessibility, while tissue was the main source of variance in H3K27ac profiles among the samples we examined. In contrast, butadiene exposure did not significantly contribute to divergence in the transcriptome or epigenome as a whole. We also performed an analysis of the transcriptomic and epigenomic differences between tissues, strains, and exposure groups, in which we identified genes with significantly different expression (Supplemental Table 2), as well as genomic regions with significantly different chromatin accessibility and/or H3K27 acetylation (Supplemental Table 3–4). All three tissues exhibited striking differences in the transcriptome and epigenome between strains that were not subjected to chemical insult (i.e., basal differences); however, lung was the only tissue to exhibit numerous gene expression and chromatin accessibility differences in butadiene-exposed mice relative their clean air-exposed counterparts. In all three tissues, the effect of butadiene exposure on transcription and chromatin accessibility was more pronounced in C57BL/6J than in CAST/EiJ (Supplemental Table 5). Interestingly, we identified few, if any, differentially acetylated regions upon butadiene exposure in either strain (Supplemental Table 5).
Figure 2. Principal component (PC) analysis of normalized gene expression (A) and chromatin accessibility data (B).
For both datasets, PC1 and PC2 clearly separate lung (blue), liver (red) and kidney (yellow) samples, whereas PC3 separates CAST/EiJ (triangle) from C57BL/6J (circle) samples. In contrast to tissue and strain, treatment status did not significantly contribute to the overall variation observed in either gene expression or chromatin accessibility data.
The relationship between the epigenomic landscape and transcription is dynamic between strains at baseline
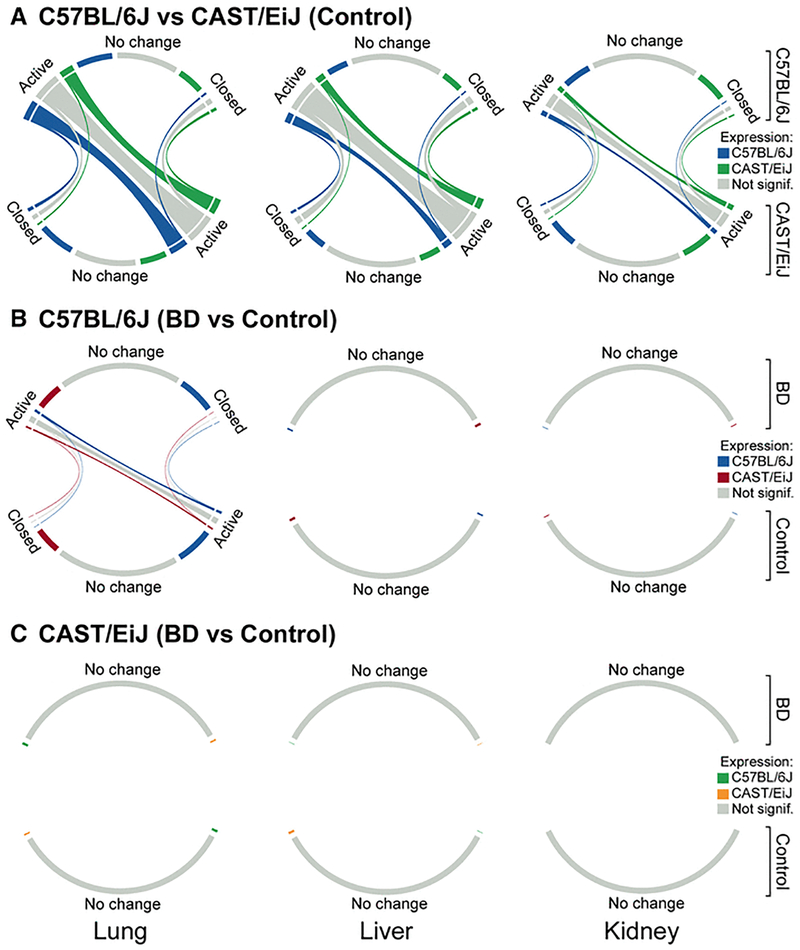
To further evaluate the relationship between the epigenomic landscape and gene expression, we classified chromatin state transitions between strains or exposure groups as “active-to-active” or “active-to-closed” (see Methods, Supplemental Table 6) and visualized these transitions as chord diagrams (Figure 3) (Gu et al. 2014). Regions of chromatin that did not change significantly between strains (FDR<10%) were classified as “no change”. Consistent with PCA, the majority of chromatin state transitions occurred between strains in unexposed mice. In contrast, strikingly few transitions occurred in response to butadiene exposure, except in the C57BL/6J lung. Further, for all comparisons, chromatin state transitions were primarily “active-to-active”, suggesting that the majority of differences in chromatin state, both between strains and in response to butadiene exposure, are not between “off” and “on” states but rather between degrees of activity.
Figure 3. Chord diagrams of chromatin state transitions between CAST/EiJ and C57BL/6J at baseline (A) and between control and exposed C57BL/6J (B) and CAST/EiJ (C) mice.
For each chord diagram, the size of the outer segments represents the number of individual chromatin regions classified as either “active”, “closed”, or “no change”. These are further sub-divided into 3 groups based on the direction of expression change for genes associated with those chromatin regions. The size of the ribbons between segments represents the number of chromatin regions undergoing a transition from one state to another. Segments and ribbons are colored according to the direction of expression change. For the number of regions within a particular state, see Supplemental Table 5.
It is evident from the chord diagrams that the relationship between chromatin state and expression is dynamic, and our results highlight several general trends regarding this relationship. We show that chromatin state transitions do not always result in differential expression (Figure 3, grey chord ribbons), and that differential expression is not always related to changes in the underlying chromatin state (Figure 3, outer chord segments labeled as “no change”). We also observed that, in general, more accessible chromatin was associated with higher expression (i.e., for the majority of “active-to-closed” chromatin state transitions, the strain or treatment group with active chromatin had higher expression of associated genes than the group with closed chromatin).
To explore if particular chromatin state transitions were associated with higher-level functional classes of genes, we performed pathway enrichment analysis (see Methods, Supplemental Table 6–7). Our results show that the tricarboxylic acid (TCA) cycle (all three tissues), as well as Phase I (lung and liver) and Phase II (all three tissues) metabolic pathways were enriched in gene sets associated with active chromatin regions in both strains but more highly expressed in CAST/EiJ compared to C57BL/6J at baseline. Phase I and Phase II metabolic pathways are critical for the bioactivation and detoxification of butadiene, respectively, and this result is consistent with our previous study of transcriptomic and epigenomic responses to butadiene in the lung (Chappell et al. 2017). In contrast, genes that were more highly expressed in C57BL/6J compared to CAST/EiJ that were associated with active chromatin regions in both strains were enriched for pathways related to immune signaling, including interferon signaling, in all three tissues.
Further, our results show that exposure to butadiene did not result in significant chromatin state transitions except in the lung of C57BL/6J mice, consistent with our previous study (Chappell, et al. 2017). Here, we found that in the lung, immune-related signaling pathways were enriched in gene sets associated with active chromatin regions in both control and exposed groups but more highly expressed in the control group than in the exposure group. Similar pathways were also enriched in gene sets associated with active chromatin regions in the control group, which showed relative decreased accessibility in butadiene-exposed mice.
Expression and chromatin changes in key pathways vary across tissues
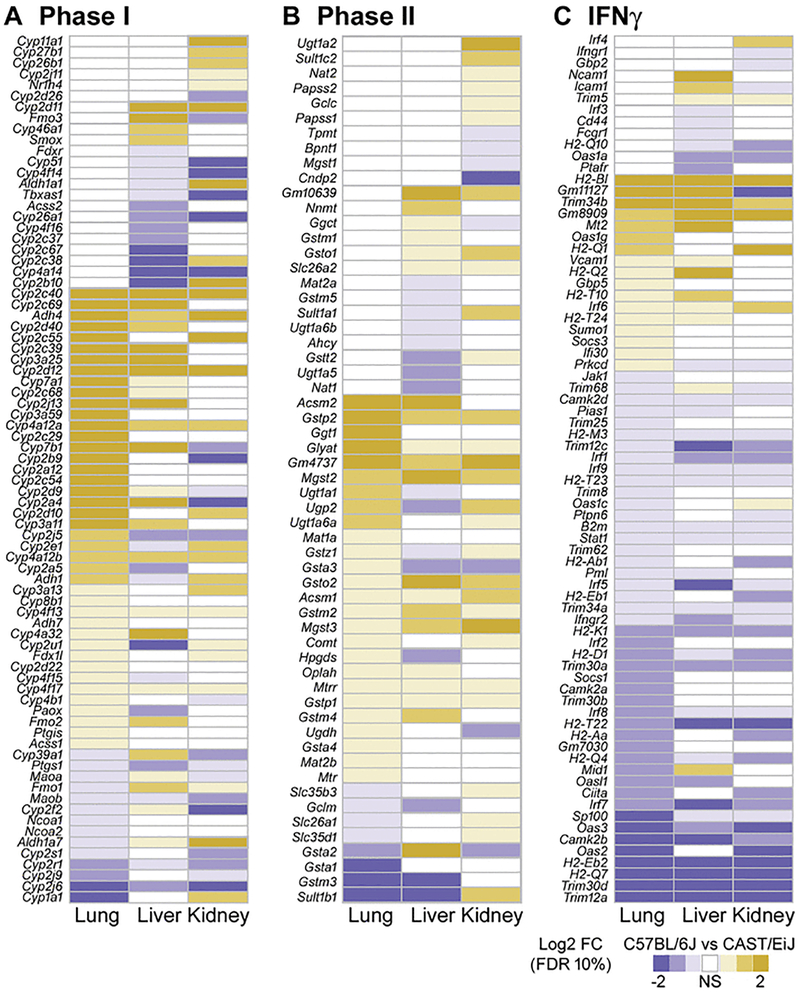
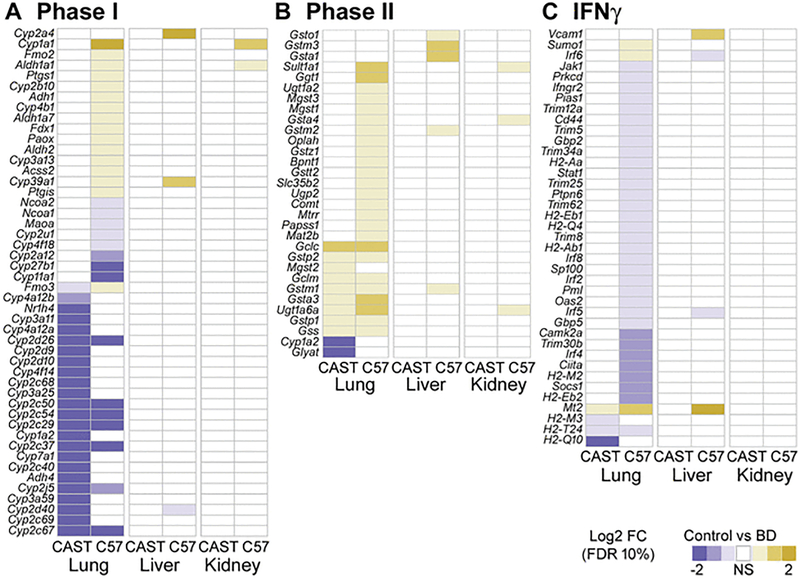
To better understand the pathways consistently enriched in expression and chromatin profile differences between strains at baseline and upon exposure in each tissue, we examined in detail the specific genes whose expression was significantly altered. In particular, we focused on Phase I and Phase II metabolism, which were generally more active in CAST/EiJ and showed increased activity upon butadiene exposure, as well as the interferon pathway, which was generally more active in C57BL/6J and decreased in activity upon exposure.
Figure 4 shows each gene that was significantly differentially expressed between strains in at least one tissue in unexposed mice, for each of the three pathways. The specific differentially expressed genes vary widely across tissues, although there is some overlap. Most genes in Phase I and Phase II metabolism show higher expression in CAST/EiJ at baseline, though very few are consistently higher in all tissues, such as Cyp3a25, Cyp2d12, and Adh4. Some genes are expressed higher in C57BL/6J, but again primarily in only one or two tissues, and to varying degrees. In addition, several genes were significantly differentially expressed in all three tissues, but the direction of this expression varies by tissue. For example, Cyp2a4 is expressed higher in CAST/EiJ in both lung and liver tissue, but is higher in C57BL/6J in kidney tissue. In contrast, most genes in the interferon pathway are more highly expressed in C57BL/6J in all tissues compared to that of CAST/EiJ, but again there is variability in tissues and strains as to the direction and extent of the differences.
Figure 4. Heat maps of gene expression (log2 fold-change) for genes involved with Phase I metabolism (A), Phase II metabolism (B) and IFNγ signaling (C) across all tissues and strains at baseline.
Within each heat map, genes are ordered by the first column. Only genes that were significantly differentially expressed in a given contrast (FDR <10%) are depicted in the relevant color scale. Non-significant expression differences are represented in white.
When comparing mice exposed to butadiene to unexposed mice (Figure 5), it is evident that most changes are restricted to lung tissue, and that there are few differentially expressed genes in liver and kidney, as described above. Across strains, we see some genes that either increase or decrease expression consistently upon exposure, such as Cyp2d26 and Cyp2c50 which are decreased in lung (Phase I), and Gclc which is increased in liver (Phase II). The majority of genes that are affected by butadiene exposure, though, are altered in only one strain and in only one tissue.
Figure 5. Heat maps of gene expression (log2 fold-change) for genes involved with Phase I metabolism (A), Phase II metabolism (B) and IFNγ signaling (C) across all tissues and strains at in response to butadiene exposure.
Within each heat map, genes are ordered by the first column. Only genes that were significantly differentially expressed in a given contrast (FDR<10%) are depicted in the relevant color scale. Non-significant expression differences are represented in white.
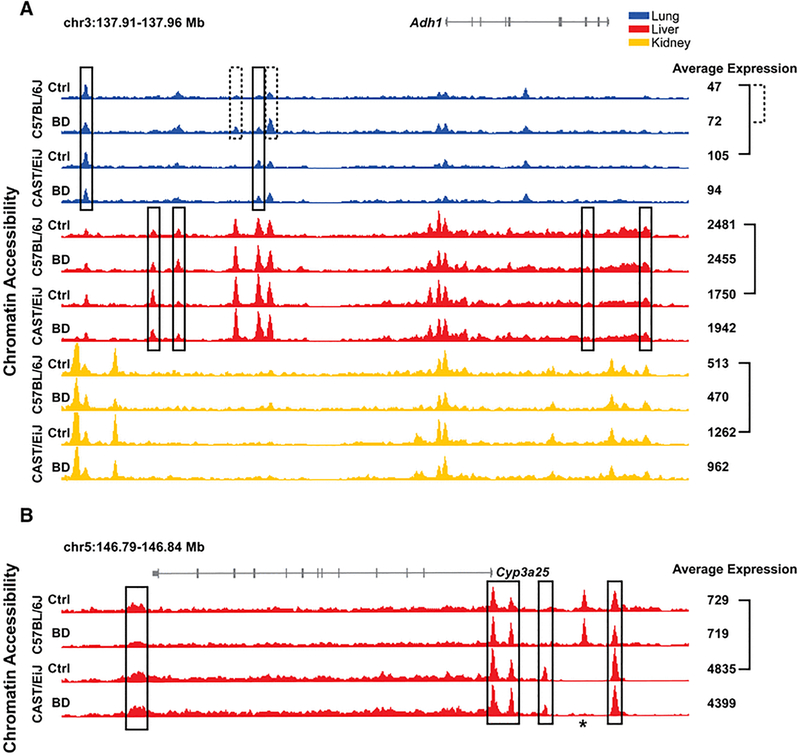
This complexity in expression activity across strains and tissues and in response to butadiene exposure can be explained in large part by variation in the underlying chromatin architecture that show tissue and condition-specific regulatory elements. As an example, Figure 6A shows the accessible chromatin signals and provide gene expression levels for one of the genes with a known role in butadiene metabolism, alcohol dehydrogenase 1 (Adh1) gene (Kemper and Elfarra 1996). As indicated in Figure 5 and shown more explicitly in Figure 6A, this gene is more highly expressed in CAST/EiJ at baseline in lung and kidney, is higher in C57BL/6J in liver at baseline, and is significantly increased in only lung in C57BL/6J upon exposure to butadiene. Across tissues, we see that in the proximal promoter regions immediately adjacent to the transcription start site (TSS), the strength of the signal corresponds with the general expression levels with the highest signal and expression in liver, and the lowest in lung. In addition, we see multiple distal accessible chromatin regions that are demonstrably different across tissues. Together, these help explain why the greatest variability seen in the PCA plots (Figure 2) is between tissues as even genes expressed in all three tissues can be highly distinct in their molecular patterns.
Figure 6. Adh1 (A) and Cyp3a25 (B) loci.
Each browser track represents the averaged, normalized signal across biological replicates for chromatin accessibility data. Averaged, normalized expression values are also shown to the right of the browser tracks (rounded to the nearest integer). Solid black boxes mark regions of differentially accessible chromatin between strains at baseline within a tissue, and dashed black boxes mark regions of differentially accessible chromatin between exposure groups within a strain (FDR<10%). Solid and dashed brackets mark significant differential expression between strains at baseline within a tissue and between exposure groups within a strain, respectively (FDR<10%). Accessible regions that met quality filtering criteria in one strain, but not the other, are marked by an asterisk and not evaluated for differential accessibility. Significant differences in gene expression are indicated by brackets.
Between strains and between exposure conditions in each tissue, differences in gene expression and chromatin profiles are subtler compared to across tissues. In fact, the promoter regions display relatively similar chromatin accessibility within each tissue group, indicating that strain and exposure-specific differences are primarily due to changes in distal regulatory elements. In addition, the specific regions that show change in accessibility at baseline across strains (Figure 6A, black boxes) are different across tissues. It is interesting to note that these changes are relatively subtle and involve regions that in general are accessible in both strains, as suggested by Figure 3. The same is true in the case of exposure in C57BL/6J lung, where two distal elements are subtly but significantly increased upon exposure, likely contributing to the increase in expression (Figure 6A, dashed boxes).
Even for genes that are tissue-specific, strain-specific differences in chromatin state can be more extensive and contribute to larger differences in expression. Among the three tissues, cytochrome P450 3A25 (Cyp3a25) is only appreciably expressed in liver, but expression is over six times higher in CAST/EiJ compared to C57BL/6J (Figure 6B). Indeed, CYP-mediated oxidation of butadiene to epoxybutene and other metabolites is well known (Himmelstein et al. 1997), albeit the role of individual CYPs is poorly characterized beyond the role of CYP2E1. Here, we find that accessibility in regions near the Cyp3a25 gene are significantly higher in CAST/EiJ, including both promoter and several distal regions (Figure 6B, black boxes), but again in most cases these regions are also accessible in C57BL/6J. One region upstream of the TSS that is present in CAST/EiJ appears to be largely absent in C57BL/6J. Interestingly, we also see one accessible region in C57BL/6J that may be completely absent in CAST/EiJ, as there were almost no read sequences that could be aligned and the signal is well below the background signal seen in even inaccessible chromatin (Figure 6B, asterisk).
Discussion
Seminal strides in understanding genome biology have been made by the human Encyclopedia of DNA Elements (ENCODE) project (Encode Project Consortium et al. 2012) and the Mouse ENCODE Consortium (Yue et al. 2014) which annotated functional elements encoded in the human and mouse genomes. These projects mapped gene expression, open chromatin regions, transcription factor binding, chromatin modifications and replication domains in diverse cell and tissue types in humans and in the C57BL/6 mouse. It was found that the chromatin state landscape is developmentally stable and evolutionarily conserved (Cheng et al. 2014), and is cell lineage/tissue-specific in both humans and mice; however, there is a large degree of divergence of sequences involved in transcriptional regulation, chromatin state and higher order chromatin organization that results in major transcriptional differences between species (Lin et al. 2014). In parallel, the Roadmap Epigenomics Project has examined over one hundred human tissues and cell types and produced reference epigenomes that provide information on key functional elements controlling gene expression (Roadmap Epigenomics Consortium et al. 2015). Together, these data open new opportunities to uncover the associations between tissue-specific chromatin states and genetic polymorphisms (Fagny et al. 2017), as well as to more deeply explore the molecular causes of human disease (Romanoski et al. 2015).
Still, little is known about how regulatory elements that were identified by the studies of natural or disease-associated variation are affected by exposure to environmental chemicals or drugs. Additional limitation of the current state of knowledge is that apart from the population variability being explored by GTEx consortium, large-scale epigenetic studies do not capture diversity across the population. Experimental studies that model chemical toxicity and encompass exploration of complex multi-dimensional datasets of inter-individual variation, gene expression, chromatin states and tissue-specific effects are few (Harrill and McAllister 2017). In studies of linkages between gene expression and genetic polymorphisms in a toxicological context, almost all of which were conducted in populations of mouse strains (Rusyn 2010), strain-specific effects on transcription predominate (Bradford et al. 2011; Harrill et al. 2009; Mosedale et al. 2017).
Extending the complexity of the experimental designs of Gene × Environment investigations to the Epigenome may uncover not only relationships between chemical exposures and gene expression, but also determine the regulatory mechanisms altered by exposures; however, the complexity and technical challenges make such studies difficult (Lewis et al. 2017). Our recent study examined effects of 1,3-butadiene on DNA damage, messenger RNA and microRNA expression, and genome-wide chromatin accessibility in the lung in the same C57BL/6J and CAST/EiJ mouse strains (Chappell et al. 2017). We found that the variability in and the extent of damaging effects of 1,3-butadiene may be due not only to genetic polymorphisms, but also may depend on the basal transcriptional and epigenetic states, also influenced by genetic background. Such differences may confer inherent variation in the ability to defend against and/or effectively respond to injury on the cellular and molecular level.
The study reported herein aimed to extend the findings of (Chappell et al. 2017) to other tissues that are a target (i.e., liver) or not a target (i.e., kidney) of butadiene-induced carcinogenesis in the mouse. Human relevance of this work is supported by the focus on a fundamental mechanism of toxicity and carcinogenesis, DNA damage by a known genotoxic agent, the fact that butadiene is a known human and rodent carcinogen, and our previous work demonstrating butadiene’s effects on chromatin, histone modifications and other epigenetic states in a strain- and tissue-dependent manner. We focused on three readouts that allow for explorations of the relationships between epigenetic and transcriptional activity in the context of inter-individual variability in responses to toxicants: RNA-seq to comprehensively quantify multiple types of RNA transcripts, ATAC-seq to identify “accessible chromatin”, and ChIP-seq for H3K27ac to characterize regions that are active for transcription. We posit that this combination of assays represents a cost-effective and comprehensive approach that enables assessment of chromatin accessibility coupled with various RNA levels to comprehensively characterize epigenetic and transcriptional activity in the context of inter-individual variability in responses to a toxicant (Lewis et al. 2017).
We found that tissue effects dominate both gene expression and chromatin states, followed by strain effects. These findings are concordant with recent reports from human tissue-specific gene expression studies in humans (GTEx Consortium 2015), as well as the findings of ENCODE (Thurman et al. 2012; Yue et al. 2014) and Epigenetic Roadmap (Roadmap Epigenomics Consortium et al. 2015). Surprisingly, the effect of butadiene treatment was non-discriminating in either of the tissues examined, except for lung. This result shows that exposure to a genotoxic chemical carcinogen had an overall minor effect, relative to tissue and strain differences, on the transcriptome and chromatin organization. This further suggests that the individual’s baseline transcriptional and epigenetic states play a key role in priming the responses to exposure, not the effect of the exposure itself. This leads to a model where individual response to chemical exposure is driven by the status of these molecular characteristics within relevant cells leading to variations in susceptibility to damaging effects of the chemical.
Tissue-specific exploration of transcription, chromatin and H3K27ac status also showed that the major differences are among strains, and not in response to exposure. A comparison between strains in untreated animals shows that (i) changes in chromatin do not always result in changes in expression, (ii) changes in expression are not always related to changes in underlying chromatin, and (iii) while open chromatin is associated with higher transcription of those genomic regions, such an association may not be generalizable to all loci. The lack of concordance between expression and chromatin is expected. For instance, increased levels of transcription factors that bind to existing accessible regions can modulate expression of target genes without necessitating a change in chromatin. Our data reinforces this property by indicating that variability in chromatin accessibility across individuals (i.e., strains) only partially explains the variability in transcription. One caution to this, however, is the identification of gene targets for open chromatin regions, as the procedure used in this study is one possible approximation for such a relationship.
It is also evident that H3K27ac data does not significantly add to our understanding of differential regulatory regions identified by ATAC-seq because almost all of these accessible chromatin regions were also marked by H3K27ac. While this study did not conduct an in-depth exploration of all key histone modifications, this finding is noteworthy in the context of an acute need to focus the scope of complex studies on epigenetics, gene expression, genetic variability and toxicity mechanisms. Most informative studies in toxicology include exploration of dose- and time-relationships between exposure and adverse effects, and may also include multiple tissues, strains, both sexes, etc. Exploration of all possible histone modifications in such studies may be prohibitive in terms of the cost and the amount of the biological sample required. Therefore, our study is instructive insofar as it advocates that RNA-seq and ATAC-seq may be sufficient for genome-wide, but locus-specific, characterization of the relationships between transcription and chromatin states, especially in studies of complex study designs and limited tissue availability.
We also note that while we find significant differences in gene expression and chromatin architecture in each tissue between the two strains, it is important to note that the vast majority of these differences do not represent on/off or presence/absence scenarios, which are preferred conditions to serve as biomarkers of effect. Rather, most differences are in the magnitude of change. In ATAC-seq data, we find that variable regions across strains in each tissue are considered significantly accessible within each strain itself, but comparatively more accessible in one of the strains. As these genome-wide assays reflect the average molecular state across a population of cells in a tissue, these differences in strength of signals likely reflect altered proportions of individual cells in the tissue that are accessible at a given locus. While in aggregate across all loci these features may be used to predict the effect/toxicity, this suggests individual genes and loci may not be good biomarkers given the absence of binary states.
Furthermore, we found that short-term (two weeks) inhalational exposure to butadiene, a potent human and rodent carcinogen and a DNA damaging agent, produced little transcriptional effect in all organs examined. With the exception of the lung, the tissue that is first in contact with inhaled butadiene, liver and kidney were virtually devoid of a transcriptional response to treatment. This result is notable as all three tissues harbor butadiene-induced DNA damage (Chappell et al. 2014), while not all three tissues developed tumors after longer term exposure (National Toxicology Program 2011). Few studies are available on the transcriptomic effects of 1,3-butadiene. Studies of inhalational exposure to butadiene soot and β-chloroprene (2-chloro-1,3-butadiene) examined lung transcriptional effects in rodents. Three weeks of inhalation exposure to butadiene soot particles resulted in up-regulation of biotransformation, oxidative response, and inflammatory genes in the lungs of BALB/c mice; however, there were only about 100 significantly differentially expressed genes (Noel et al. 2016). Exposure to β-chloroprene yields multi-organ tumors in mouse and rat cancer bioassays (Pagan 2007). Transcriptomic analysis of the lungs in mice and rats following exposure for up to 3 weeks also identified significant effects on pathways related to glutathione biosynthesis and metabolism; albeit the transcriptional responses were somewhat muted with up to several hundred transcripts changed significantly (Thomas et al. 2013). These findings are consistent with our observation of the effects on butadiene in the lung being confined to the pathways of xenobiotic metabolism and inflammation, but also show that carcinogenic chemicals that act through DNA damage may exhibit a very limited transcriptomic footprint. Therefore, studies of a genotoxic potential of chemicals in non-mammalian systems are likely more informative of the potential for the chronic health hazard and carcinogenic potential, then short-term exposure transcriptomic studies (Cote et al. 2016).
Overall, we conclude that this study demonstrates high tissue and strain specificity of the response to a genotoxic carcinogen at the transcriptional and chromatin levels even though DNA damage response is much less variable. It is difficult, however, to draw a link between the relative lack of responsiveness of the transcriptome and chromatin in kidney to the fact that the kidney is relatively insensitive to butadiene-induced carcinogenesis because little response was also observed in the liver, a target tissue. Future studies are needed to more extensively analyze the association between the chemical-induced adducts and/or mutations and chromatin states. These may include studies of longer exposures as well as an investigation of the location of DNA damage and mutations with respect to the basal chromatin state in different cell types and tissues. These questions are difficult to address in a study of two inbred strains, to draw confident conclusions about basal architecture and gene expression changes on exposure a larger study across many strains may provide the power to make such connections. Not only such studies will need to be performed on larger populations, but they also should incorporate dose- and time-response dimensions to prove that epigenetic endpoints may be used in decision-making, as surrogate biomarkers of toxicity mechanisms or susceptibility, and in dose-response aspects of risk assessment.
Supplementary Material
Acknowledgements
This work was supported, in part, by grants from National Institutes of Health (NIH R01ES023195 and P30ES025128). The views expressed in this article are those of the authors and do not necessarily reflect the views of NIH.
Footnotes
Conflict of Interest
G. Chappell is currently employed by ToxStrategies, Inc., a scientific consulting firm. G. Chappell received no funding from ToxStrategies or any of its clients for this project, nor was ToxStrategies or any of its clients involved in the development or execution of the research presented herein. The other authors declare that they have no actual or potential competing financial interests.
Data Access
All RNA-seq, ATAC-seq, and ChIP-seq data sets generated during this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and an accession ID is forthcoming.
References
- Boyle AP, Guinney J, Crawford GE, Furey TS (2008) F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics 24, 2537–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford BU, Lock EF, Kosyk O, Kim S, Uehara T, Harbourt D, DeSimone M, Threadgill DW, Tryndyak V, Pogribny IP, Bleyle L, Koop DR, Rusyn I (2011) Interstrain differences in the liver effects of trichloroethylene in a multistrain panel of inbred mice. Toxicol Sci 120, 206–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell G, Kobets T, O’Brien B, Tretyakova N, Sangaraju D, Kosyk O, Sexton KG, Bodnar W, Pogribny IP, Rusyn I (2014) Epigenetic events determine tissue-specific toxicity of inhalational exposure to the genotoxic chemical 1,3-butadiene in male C57BL/6J mice. Toxicol Sci 142, 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell G, Pogribny IP, Guyton KZ, Rusyn I (2016) Epigenetic alterations induced by genotoxic occupational and environmental human chemical carcinogens: A systematic literature review. Mutat Res Rev Mutat Res 768, 27–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell GA, Israel JW, Simon JM, Pott S, Safi A, Eklund K, Sexton KG, Bodnar W, Lieb JD, Crawford GE, Rusyn I, Furey TS (2017) Variation in DNA-Damage Responses to an Inhalational Carcinogen (1,3-Butadiene) in Relation to Strain-Specific Differences in Chromatin Accessibility and Gene Transcription Profiles in C57BL/6J and CAST/EiJ Mice. Environ Health Perspect 125, 107006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Ma Z, Kim BH, Wu W, Cayting P, Boyle AP, Sundaram V, Xing X, Dogan N, Li J, Euskirchen G, Lin S, Lin Y, Visel A, Kawli T, Yang X, Patacsil D, Keller CA, Giardine B, mouse EC, Kundaje A, Wang T, Pennacchio LA, Weng Z, Hardison RC, Snyder MP (2014) Principles of regulatory information conservation between mouse and human. Nature 515, 371–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote I, Andersen ME, Ankley GT, Barone S, Birnbaum LS, Boekelheide K, Bois FY, Burgoon LD, Chiu WA, Crawford-Brown D, Crofton KM, DeVito M, Devlin RB, Edwards SW, Guyton KZ, Hattis D, Judson RS, Knight D, Krewski D, Lambert J, Maull EA, Mendrick D, Paoli GM, Patel CJ, Perkins EJ, Poje G, Portier CJ, Rusyn I, Schulte PA, Simeonov A, Smith MT, Thayer KA, Thomas RS, Thomas R, Tice RR, Vandenberg JJ, Villeneuve DL, Wesselkamper S, Whelan M, Whittaker C, White R, Xia M, Yauk C, Zeise L, Zhao J, DeWoskin RS (2016) The Next Generation of Risk Assessment Multi-Year Study-Highlights of Findings, Applications to Risk Assessment, and Future Directions. Environ Health Perspect 124, 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Project Consortium,Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Dunham I, Ernst J, Furey TS, Gerstein M, Giardine B, Greven M, Hardison RC, Harris RS, Herrero J, Hoffman MM, Iyer S, Kelllis M, Khatun J, Kheradpour P, Kundaje A, Lassman T, Li Q, Lin X, Marinov GK, Merkel A, Mortazavi A, Parker SC, Reddy TE, Rozowsky J, Schlesinger F, Thurman RE, Wang J, Ward LD, Whitfield TW, Wilder SP, Wu W, Xi HS, Yip KY, Zhuang J, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, Pazin MJ, Lowdon RF, Dillon LA, Adams LB, Kelly CJ, Zhang J, Wexler JR, Green ED, Good PJ, Feingold EA, Bernstein BE, Birney E, Crawford GE, Dekker J, Elinitski L, Farnham PJ, Gerstein M, Giddings MC, Gingeras TR, Green ED, Guigo R, Hardison RC, Hubbard TJ, Kellis M, Kent WJ, Lieb JD, Margulies EH, Myers RM, Snyder M, Starnatoyannopoulos JA, Tennebaum SA, Weng Z, White KP, Wold B, Khatun J, Yu Y, Wrobel J, Risk BA, Gunawardena HP, Kuiper HC, Maier CW, Xie L, Chen X, Giddings MC, Bernstein BE, Epstein CB, Shoresh N, Ernst J, Kheradpour P, Mikkelsen TS, Gillespie S, Goren A, Ram O, Zhang X, Wang L, Issner R, Coyne MJ, Durham T, Ku M, Truong T, Ward LD, Altshuler RC, Eaton ML, Kellis M, Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Batut P, Bell I, Bell K, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena HP, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Li G, Luo OJ, Park E, Preall JB, Presaud K, Ribeca P, Risk BA, Robyr D, Ruan X, Sammeth M, Sandu KS, Schaeffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Hayashizaki Y, Harrow J, Gerstein M, Hubbard TJ, Reymond A, Antonarakis SE, Hannon GJ, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR, Rosenbloom KR, Sloan CA, Learned K, Malladi VS, Wong MC, Barber GP, Cline MS, Dreszer TR, Heitner SG, Karolchik D, Kent WJ, Kirkup VM, Meyer LR, Long JC, Maddren M, Raney BJ, Furey TS, Song L, Grasfeder LL, Giresi PG, Lee BK, Battenhouse A, Sheffield NC, Simon JM, Showers KA, Safi A, London D, Bhinge AA, Shestak C, Schaner MR, Kim SK, Zhang ZZ, Mieczkowski PA, Mieczkowska JO, Liu Z, McDaniell RM, Ni Y, Rashid NU, Kim MJ, Adar S, Zhang Z, Wang T, Winter D, Keefe D, Birney E, Iyer VR, Lieb JD, Crawford GE, Li G, Sandhu KS, Zheng M, Wang P, Luo OJ, Shahab A, Fullwood MJ, Ruan X, Ruan Y, Myers RM, Pauli F, Williams BA, Gertz J, Marinov GK, Reddy TE, Vielmetter J, Partridge EC, Trout D, Varley KE, Gasper C, Bansal A, Pepke S, Jain P, Amrhein H, Bowling KM, Anaya M, Cross MK, King B, Muratet MA, Antoshechkin I, Newberry KM, McCue K, Nesmith AS, Fisher-Aylor KI, Pusey B, DeSalvo G, Parker SL, Balasubramanian S, Davis NS, Meadows SK, Eggleston T, Gunter C, Newberry JS, Levy SE, Absher DM, Mortazavi A, Wong WH, Wold B, Blow MJ, Visel A, Pennachio LA, Elnitski L, Margulies EH, Parker SC, Petrykowska HM, Abyzov A, Aken B, Barrell D, Barson G, Berry A, Bignell A, Boychenko V, Bussotti G, Chrast J, Davidson C, Derrien T, Despacio-Reyes G, Diekhans M, Ezkurdia I, Frankish A, Gilbert J, Gonzalez JM, Griffiths E, Harte R, Hendrix DA, Howald C, Hunt T, Jungreis I, Kay M, Khurana E, Kokocinski F, Leng J, Lin MF, Loveland J, Lu Z, Manthravadi D, Mariotti M, Mudge J, Mukherjee G, Notredame C, Pei B, Rodriguez JM, Saunders G, Sboner A, Searle S, Sisu C, Snow C, Steward C, Tanzer A, Tapanan E, Tress ML, van Baren MJ, Walters N, Washieti S, Wilming L, Zadissa A, Zhengdong Z, Brent M, Haussler D, Kellis M, Valencia A, Gerstein M, Raymond A, Guigo R, Harrow J, Hubbard TJ, Landt SG, Frietze S, Abyzov A, Addleman N, Alexander RP, Auerbach RK, Balasubramanian S, Bettinger K, Bhardwaj N, Boyle AP, Cao AR, Cayting P, Charos A, Cheng Y, Cheng C, Eastman C, Euskirchen G, Fleming JD, Grubert F, Habegger L, Hariharan M, Harmanci A, Iyenger S, Jin VX, Karczewski KJ, Kasowski M, Lacroute P, Lam H, Larnarre-Vincent N, Leng J, Lian J, Lindahl-Allen M, Min R, Miotto B, Monahan H, Moqtaderi Z, Mu XJ, O’Geen H, Ouyang Z, Patacsil D, Pei B, Raha D, Ramirez L, Reed B, Rozowsky J, Sboner A, Shi M, Sisu C, Slifer T, Witt H, Wu L, Xu X, Yan KK, Yang X, Yip KY, Zhang Z, Struhl K, Weissman SM, Gerstein M, Farnham PJ, Snyder M, Tenebaum SA, Penalva LO, Doyle F, Karmakar S, Landt SG, Bhanvadia RR, Choudhury A, Domanus M, Ma L, Moran J, Patacsil D, Slifer T, Victorsen A, Yang X, Snyder M, White KP, Auer T, Centarin L, Eichenlaub M, Gruhl F, Heerman S, Hoeckendorf B, Inoue D, Kellner T, Kirchmaier S, Mueller C, Reinhardt R, Schertel L, Schneider S, Sinn R, Wittbrodt B, Wittbrodt J, Weng Z, Whitfield TW, Wang J, Collins PJ, Aldred SF, Trinklein ND, Partridge EC, Myers RM, Dekker J, Jain G, Lajoie BR, Sanyal A, Balasundaram G, Bates DL, Byron R, Canfield TK, Diegel MJ, Dunn D, Ebersol AK, Ebersol AK, Frum T, Garg K, Gist E, Hansen RS, Boatman L, Haugen E, Humbert R, Jain G, Johnson AK, Johnson EM, Kutyavin TM, Lajoie BR, Lee K, Lotakis D, Maurano MT, Neph SJ, Neri FV, Nguyen ED, Qu H, Reynolds AP, Roach V, Rynes E, Sabo P, Sanchez ME, Sandstrom RS, Sanyal A, Shafer AO, Stergachis AB, Thomas S, Thurman RE, Vernot B, Vierstra J, Vong S, Wang H, Weaver MA, Yan Y, Zhang M, Akey JA, Bender M, Dorschner MO, Groudine M, MacCoss MJ, Navas P, Stamatoyannopoulos G, Kaul R, Dekker J, Stamatoyannopoulos JA, Dunham I, Beal K, Brazma A, Flicek P, Herrero J, Johnson N, Keefe D, Lukk M, Luscombe NM, Sobral D, Vaquerizas JM, Wilder SP, Batzoglou S, Sidow A, Hussami N, Kyriazopoulou-Panagiotopoulou S, Libbrecht MW, Schaub MA, Kundaje A, Hardison RC, Miller W, Giardine B, Harris RS, Wu W, Bickel PJ, Banfai B, Boley NP, Brown JB, Huang H, Li Q, Li JJ, Noble WS, Bilmes JA, Buske OJ, Hoffman MM, Sahu AO, Kharchenko PV, Park PJ, Baker D, Taylor J, Weng Z, Iyer S, Dong X, Greven M, Lin X, Wang J, Xi HS, Zhuang J, Gerstein M, Alexander RP, Balasubramanian S, Cheng C, Harmanci A, Lochovsky L, Min R, Mu XJ, Rozowsky J, Yan KK, Yip KY, Birney E (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagny M, Paulson JN, Kuijjer ML, Sonawane AR, Chen CY, Lopes-Ramos CM, Glass K, Quackenbush J, Platig J (2017) Exploring regulation in tissues with eQTL networks. Proc Natl Acad Sci U S A 114, E7841–E7850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggin M, Sangaraju D, Walker VE, Wickliffe J, Swenberg JA, Tretyakova N (2011) Persistence and repair of bifunctional DNA adducts in tissues of laboratory animals exposed to 1,3-butadiene by inhalation. Chem Res Toxicol 24, 809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggin M, Swenberg JA, Walker VE, Tretyakova N (2009) Molecular dosimetry of 1,2,3,4-diepoxybutane-induced DNA-DNA cross-links in B6C3F1 mice and F344 rats exposed to 1,3-butadiene by inhalation. Cancer Res 69, 2479–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, Brors B (2014) circlize Implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 [DOI] [PubMed] [Google Scholar]
- Harrill AH, McAllister KA (2017) New Rodent Population Models May Inform Human Health Risk Assessment and Identification of Genetic Susceptibility to Environmental Exposures. Environ Health Perspect 125, 086002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill AH, Ross PK, Gatti DM, Threadgill DW, Rusyn I (2009) Population-based discovery of toxicogenomics biomarkers for hepatotoxicity using a laboratory strain diversity panel. Toxicol Sci 110, 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelstein MW, Acquavella JF, Recio L, Medinsky MA, Bond JA (1997) Toxicology and epidemiology of 1,3-butadiene. Critical Reviews in Toxicology 27, 1–108 [DOI] [PubMed] [Google Scholar]
- IARC (2008) IARC monographs on the evaluation of carcinogenic risks to humans. Volume 97. 1,3-butadiene, ethylene oxide and vinyl halides (vinyl fluoride, vinyl chloride and vinyl bromide). IARC Monogr Eval Carcinog Risks Hum 97, 3–471 [PMC free article] [PubMed] [Google Scholar]
- IARC (2012) Chemical agents and related occupations—a review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum 100F, 1–567 [PMC free article] [PubMed] [Google Scholar]
- Kemper RA, Elfarra AA (1996) Oxidation of 3-butene-1,2-diol by alcohol dehydrogenase. Chem Res Toxicol 9, 1127–1134 [DOI] [PubMed] [Google Scholar]
- Koturbash I, Scherhag A, Sorrentino J, Sexton K, Bodnar W, Swenberg JA, Beland FA, Pardo-Manuel Devillena F, Rusyn I, Pogribny IP (2011a) Epigenetic mechanisms of mouse interstrain variability in genotoxicity of the environmental toxicant 1,3-butadiene. Toxicol Sci 122, 448–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Scherhag A, Sorrentino J, Sexton K, Bodnar W, Tryndyak V, Latendresse JR, Swenberg JA, Beland FA, Pogribny IP, Rusyn I (2011b) Epigenetic alterations in liver of C57BL/6J mice after short-term inhalational exposure to 1,3-butadiene. Environ Health Perspect 119, 635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA (2006) Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1, 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L, Crawford GE, Furey TS, Rusyn I (2017) Genetic and epigenetic determinants of inter-individual variability in responses to toxicants. Curr Opin Toxicol 6, 50–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lin Y, Nery JR, Urich MA, Breschi A, Davis CA, Dobin A, Zaleski C, Beer MA, Chapman WC, Gingeras TR, Ecker JR, Snyder MP (2014) Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci U S A 111, 17224–17229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun AT, Smyth GK (2016) csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res 44, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011) Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet.journal 17, 10–12 [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28, 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick RL, Huff JE (1993) 1,3-Butadiene induces cancer in experimental animals at all concentrations from 6.25 to 8000 parts per million. IARC Sci Publ, 309–322 [PubMed] [Google Scholar]
- Meng Q, Walker DM, McDonald JD, Henderson RF, Carter MM, Cook DL Jr., McCash CL, Torres SM, Bauer MJ, Seilkop SK, Upton PB, Georgieva NI, Boysen G, Swenberg JA, Walker VE (2007) Age-, gender-, and species-dependent mutagenicity in T cells of mice and rats exposed by inhalation to 1,3-butadiene. Chem Biol Interact 166, 121–131 [DOI] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD (2010) Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5, e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale M, Kim Y, Brock WJ, Roth SE, Wiltshire T, Eaddy JS, Keele GR, Corty RW, Xie Y, Valdar W, Watkins PB (2017) Candidate Risk Factors and Mechanisms for Tolvaptan-Induced Liver Injury Are Identified Using a Collaborative Cross Approach. Toxicol Sci 156, 438–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Toxicology Program (2011) 1,3-Butadiene. Rep Carcinog 12, 75–77 [PubMed] [Google Scholar]
- Noel A, Xiao R, Perveen Z, Zaman HM, Rouse RL, Paulsen DB, Penn AL (2016) Incomplete lung recovery following sub-acute inhalation of combustion-derived ultrafine particles in mice. Part Fibre Toxicol 13, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan I (2007) Chloroprene: overview of studies under consideration for the development of an IRIS assessment. Chem Biol Interact 166, 341–351 [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Rusyn I (2013) Environmental toxicants, epigenetics, and cancer. Adv Exp Med Biol 754, 215–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M (2015) Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanoski CE, Glass CK, Stunnenberg HG, Wilson L, Almouzni G (2015) Epigenomics: Roadmap for regulation. Nature 518, 314–316 [DOI] [PubMed] [Google Scholar]
- Rusyn I (2010) Toxicogenetics: population-based testing of drug and chemical safety in mouse models. Mutat Res 11, 1127–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenberg JA, Bordeerat NK, Boysen G, Carro S, Georgieva NI, Nakamura J, Troutman JM, Upton PB, Albertini RJ, Vacek PM, Walker VE, Sram RJ, Goggin M, Tretyakova N (2011) 1,3-Butadiene: Biomarkers and application to risk assessment. Chem Biol Interact. 24, 809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RS, Himmelstein MW, Clewell HJ 3rd, Yang Y, Healy E, Black MB, Andersen ME (2013) Cross-species transcriptomic analysis of mouse and rat lung exposed to chloroprene. Toxicol Sci 131, 629–640 [DOI] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA (2012) The accessible chromatin landscape of the human genome. Nature 489, 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varemo L, Nielsen J, Nookaew I (2013) Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res 41, 4378–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, Shen Y, Pervouchine DD, Djebali S, Thurman RE, Kaul R, Rynes E, Kirilusha A, Marinov GK, Williams BA, Trout D, Amrhein H, Fisher-Aylor K, Antoshechkin I, DeSalvo G, See LH, Fastuca M, Drenkow J, Zaleski C, Dobin A, Prieto P, Lagarde J, Bussotti G, Tanzer A, Denas O, Li K, Bender MA, Zhang M, Byron R, Groudine MT, McCleary D, Pham L, Ye Z, Kuan S, Edsall L, Wu YC, Rasmussen MD, Bansal MS, Kellis M, Keller CA, Morrissey CS, Mishra T, Jain D, Dogan N, Harris RS, Cayting P, Kawli T, Boyle AP, Euskirchen G, Kundaje A, Lin S, Lin Y, Jansen C, Malladi VS, Cline MS, Erickson DT, Kirkup VM, Learned K, Sloan CA, Rosenbloom KR, Lacerda de Sousa B, Beal K, Pignatelli M, Flicek P, Lian J, Kahveci T, Lee D, Kent WJ, Ramalho Santos M, Herrero J, Notredame C, Johnson A, Vong S, Lee K, Bates D, Neri F, Diegel M, Canfield T, Sabo PJ, Wilken MS, Reh TA, Giste E, Shafer A, Kutyavin T, Haugen E, Dunn D, Reynolds AP, Neph S, Humbert R, Hansen RS, De Bruijn M, Selleri L, Rudensky A, Josefowicz S, Samstein R, Eichler EE, Orkin SH, Levasseur D, Papayannopoulou T, Chang KH, Skoultchi A, Gosh S, Disteche C, Treuting P, Wang Y, Weiss MJ, Blobel GA, Cao X, Zhong S, Wang T, Good PJ, Lowdon RF, Adams LB, Zhou XQ, Pazin MJ, Feingold EA, Wold B, Taylor J, Mortazavi A, Weissman SM, Stamatoyannopoulos JA, Snyder MP, Guigo R, Gingeras TR, Gilbert DM, Hardison RC, Beer MA, Ren B, Mouse EC (2014) A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wu Y, Schnable JC, Zeng Z, Freeling M, Crawford GE, Jiang J (2012) High-resolution mapping of open chromatin in the rice genome. Genome Res 22, 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.