Abstract
Introduction:
The Agatston score is a well-established metric of cardiovascular disease related to clinical outcomes. It is computed from CT scans by a) measuring the volume and intensity of the atherosclerotic plaques and b) aggregating such information in an index.
Objective:
To generate a convolutional neural network that inputs a non-contrast chest CT scan and outputs the Agatston score associated with it directly, without a prior segmentation of Coronary Artery Calcifications (CAC).
Materials and methods:
We use a database of 5973 non-contrast non-ECG gated chest CT scans where the Agatston score has been manually computed. The heart of each scan is cropped automatically using an object detector. The database is split in 4973 cases for training and 1000 for testing. We train a 3D deep convolutional neural network to regress the Agatston score directly from the extracted hearts.
Results:
The proposed method yields a Pearson correlation coefficient of r = 0.93; p ≤ 0.0001 against manual reference standard in the 1000 test cases. It further stratifies correctly 72.6% of the cases with respect to standard risk groups. This compares to more complex state-of-the-art methods based on prior segmentations of the CACs, which achieve r = 0.94 in ECG-gated pulmonary CT.
Conclusions:
A convolutional neural network can regress the Agatston score from the image of the heart directly, without a prior segmentation of the CACs. This is a new and simpler paradigm in the Agatston score computation that yields similar results to the state-of-the-art literature.
Keywords: Agatston score, pulmonary CT, computed aided detection, deep learning
1. INTRODUCTION
One third (31%) of the 62 million CT scans performed in the United States are pulmonary chest CT studies. Patients suffering from chronic pulmonary diseases are at risk of cardiovascular events, such as coronary artery disease (CAD) 1. The Agatston score is a well stablished metric used to measure the extent of CAD disease in ECG-gated CT studies2. Recently, a clinically meaningful correlation has been shown between the Agatston score computed in cardiac ECG-gated CT and in non ECG-gated chest CTs3, enabling the analysis of cardiovascular disease on chest CT studies4.
The Agatston score is computed by finding coronary artery calcifications (CAC) in the image, calculating their volume and multiplying it by a factor related to the maximum intensity of the CAC, adding the perlesion scores to compute a global biomarker. Several studies have automated the Agatston score from cardiac CT images. They all follow the same general framework: first, a region of interest (ROI) around the heart is located, either using anatomy-based approaches5, atlas-based location strategies6 or 2.5D object detection strategies.7,8 Second, CACs are found in the ROI and categorized as relevant or not using their relative position in the image9, their texture and size features10 or a combination of both11,12. The latest works of Ref. 13, 14 uses a deep-learning approach towards CAC detection. These techniques use a database of segmented CACs to learn a lesion-based or a voxel-based classifier, where a per-lesion label of whether the candidate CAC should be included or not is present. In contrast, the work of Ref. 8 generates the inclusion and exclusion rules of the CACs by optimizing the global correlation coefficient directly.
We propose to further develop this latter strategy by regressing directly the CAC score from an ROI that encompasses the whole heart. We use a large database of non-contrast non-ECG gated chest CT scans for whom reference standard Agatston score is available to train and evaluate the proposed system.
2. MATERIALS AND METHODS
2.1. Evaluation Database
The evaluation database, the COPDGene study, is a multicenter observational study designed to understand the evolution and genetic signature of COPD in smokers15. 10, 000 subjects have been enrolled in the study and undergone pulmonary non-ECG gated CT scanning with a scanner of at least 16 detectors. The Agatston score was computed in 6983 of such images, forming the database in which we train and evaluate the proposed method.
2.2. Image preprocessing
We select a region of interest (ROI) centered around the heart in each CT scan using the method of Ref. 7, 8. Each heart ROI is normalized to a canonical size of 64 × 64 × 64 voxels to enable their comparison using the convolutional neural network. The reference Agatston score is normalized accordingly to enable training, resulting in the training set. The images are clamped to the range [−500, 2000] to highlight the lesions and discard lung structures.
2.3. Deep Neural Network Agatston Score Computation
We use a deep convolutional neural network of Fig. 1. It consists of three 3D convolutional layers with rectified linear activation followed by 3D max-pooling operations. We linearize the output and forward it to a fully connected and the output layers. We use dropout layers before the linearization operation and the output layers to prevent overfitting.
Figure 1.
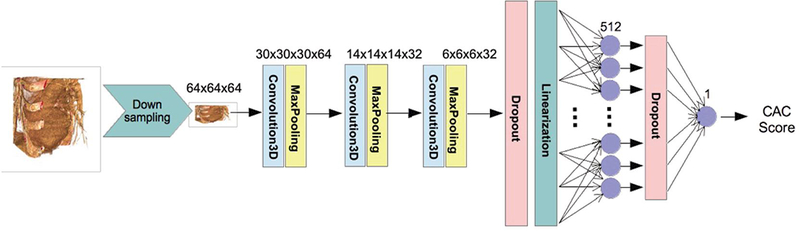
Regression network structure used.
We eliminated the images in which the automated ROI placement failed, resulting in a database of 5973 images with their corresponding Agatston score. The database is split between a training set (n = 4973) and a test set (n = 1000). The optimization of the network parameters is performed by minimizing the root mean squared error between the computed score and the reference standard using an ADAM16 optimizer with an exponential decay rate. We augment the training set by displacing the ROI a given set of pixels in each axial direction. Training is performed for 250 epochs, taking 2 days on a Maxwell TitanX GPU.
2.3.1. Implementation.
The software is implemented in Python using the SimpleITK17 (Kitware Inc., Clifton Park, NY, USA), Keras18 and Tensorflow19 libararies. Statistical analysis was performed with JMP (JMP, Version 11. SAS Institute Inc., Cary, NC, USA).
3. RESULTS
The object detector of Section 2.2 is run on the 6983 cases of the database, detecting correctly the four planes in 5973 scans. We use 4973 cases of such scans to train the deep neural network and leave 1000 aside for testing. Examples of such detections are shown in Fig. 2.
Figure 2.
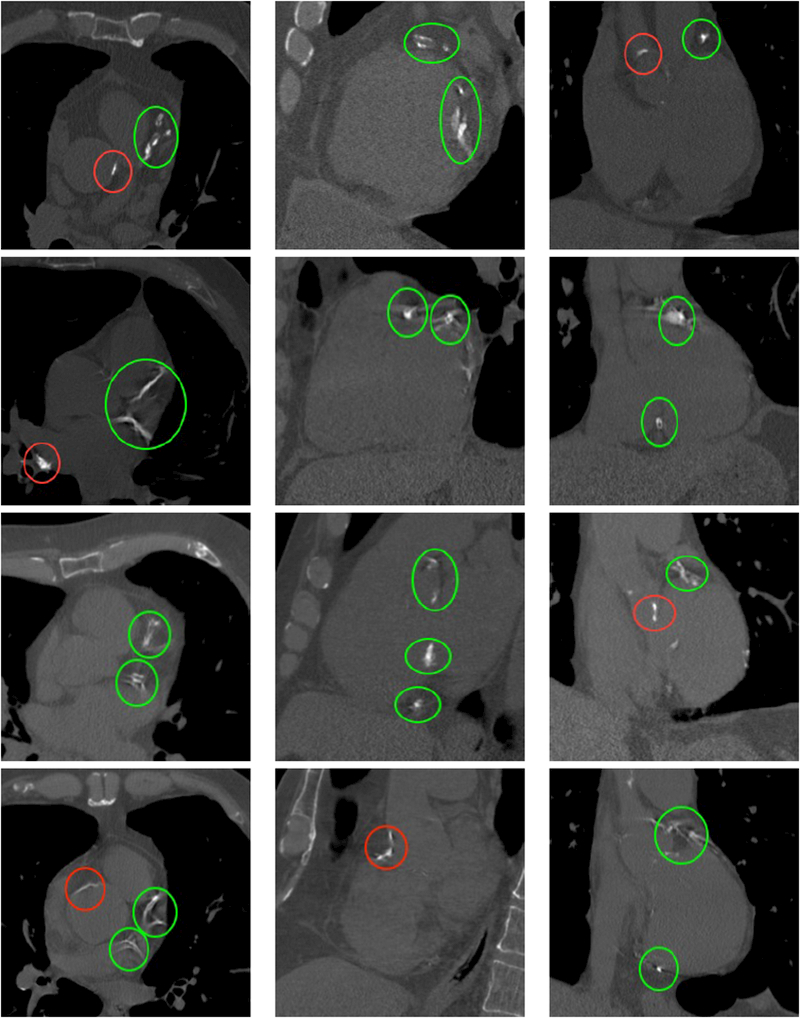
Cut planes of the images of four different subjects with calcifications. First column: axial, second column: sagittal, third column: coronal. The subjects have Agatston scores of 3578, 3151, 4147 and 4217 respectively. Calcifications appear as bright structures within the coronary arteries and are highlighted with green ellipses. Please note the presence of extra calcifications in the aorta and heart valves, highlighted with red ellipses, and bone structures, such as the sternum or the vertebrae.
The Pearson correlation coeffcient between the reference standard and the computed scores on the test set is 0.932; p < 0.0001 and depicted in Fig. 3. The Bland-Altman plot is shown in Fig. 3. The mean difference is 7.67 units, the standard deviation of the difference is 140.13, and the limits of agreement are [−266.98, 282.34].
Figure 3.
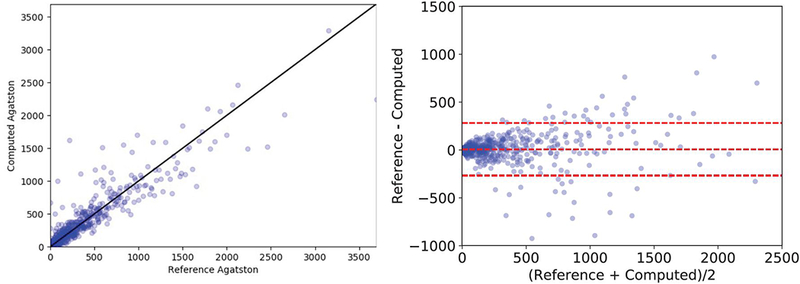
Evaluation of the automated Agatston score computation method. Left: correlation plot between the reference standard and the automatically computed score. Right: Bland-Altman plot.
We further perform the evaluation methodology of Ref. 20, which consists in estimate the correct placing of subjects according to their risk group. Five risk groups are defined depending on the value of the Agatston score. Group I is defined as subjects with a score between [0, 10), Group II ranges between [10, 100), Group III is defined in the range [100, 400), Group IV in [400, 1000) and finally Group V in cases with a score >1000 units. The proposed method correctly classifies the risk group in 75.6% of the cases and classifies the subjects in the same group or with a diffrence of one in 99.3%. The confusion matrix is shown in Table 1.
Table 1.
Confusion matrix.
| Automated | ||||||
| Manual | I | II | III | IV | V | |
| I | 279 | 114 | 2 | 1 | 0 | |
| II | 25 | 180 | 26 | 1 | 1 | |
| III | 0 | 17 | 179 | 16 | 2 | |
| IV | 0 | 0 | 16 | 83 | 12 | |
| V | 0 | 0 | 0 | 11 | 35 | |
4. DISCUSSION
We have presented a deep learning regression framework to estimate the Agatston score directly from images of the heart. The proposed method is trained using 4973 scans and tested in 1000 cases, archiving a high Pearson correlation coeffcient concerning the reference standard (ρ = 0.932; p < 0.0001) and shows no systematic bias in Bland-Altmann analysis. The results reflect a good patient risk stratification, with 75.6% of cases being assigned to the right group. The proposed method has been trained and tested in non-contrast non-ECG gated pulmonary scans, which are known to have substantial noise and motion artifacts in comparison to ECG-gated CT scans21. Prior art in such higher quality images has obtained better correlation coeffcients. For instance, Ref. 12 obtained (ρ = 0.94) in cardiac ECG-gated scans. In cardiac calcium scoring CT protocols the work of Ref. 11 and Ref. 14 achieved ρ = 0.95. It is unknown how the proposed method would perform under those image characteristics.
The proposed network structure is simple in comparison to state-of-the-art detection networks, which use more convolutional and fully connected layers. Further research will focus on network architecture and transfer learning to improve the results.
The main advantage of the proposed method with respect to prior art is that it does not require a database of annotated calcifications, but require only the input CT scan and the measured Agatston score. Such information is readily available in the healthcare databases of the hospitals. The proposed network could leverage such datasets directly, without the need for experts labeling.
ACKNOWLEDGMENTS
This work has been funded by NIH NHLBI grants R01-HL116931 and R21HL140422. The Titan Xp used for this research was donated by the NVIDIA Corporation.
REFERENCES
- [1].Matsuoka S, Yamashiro T, Diaz A, Estépar RSJ, Ross JC, Silverman EK, Kobayashi Y, Dransfield MT, Bartholmai BJ, Hatabu H, et al. , “The relationship between small pulmonary vascular alteration and aortic atherosclerosis in chronic obstructive pulmonary disease: quantitative ct analysis,” Academic radiology 18(1), 40–46 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Agatston AS, Janowitz WR, Hildner FJ, Zusmer NR, Viamonte M, and Detrano R, “Quantification of coronary artery calcium using ultrafast computed tomography,” Journal of the American College of Cardiology 15(4), 827–832 (1990). [DOI] [PubMed] [Google Scholar]
- [3].Budoff MJ, Nasir K, Kinney GL, Hokanson JE, Barr RG, Steiner R, Nath H, Lopez-Garcia C, Black-Shinn J, and Casaburi R, “Coronary artery and thoracic calcium on noncontrast thoracic ct scans: comparison of ungated and gated examinations in patients from the copd gene cohort,” Journal of cardiovascular computed tomography 5(2), 113–118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rahaghi FN, Vegas-Sanchez-Ferrero G, Minhas JK, Come CE, De La Bruere I, Wells JM, González G, Bhatt SP, Fenster BE, Diaz AA, et al. , “Ventricular geometry from non-contrast nonecg-gated ct scans: An imaging marker of cardiopulmonary disease in smokers,” Academic Radiology 24(5), 594–602 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Reeves AP, Biancardi AM, Yankelevitz DF, Cham MD, and Henschke CI, “Heart region segmentation from low-dose ct scans: an anatomy based approach.,” in [Medical Imaging: Image Processing], 83142A (2012). [Google Scholar]
- [6].Isgum I, Staring M, Rutten A, Prokop M, Viergever MA, and Van Ginneken B, “Multi-atlas-based segmentation with local decision fusionapplication to cardiac and aortic segmentation in ct scans,” IEEE transactions on medical imaging 28(7), 1000–1010 (2009). [DOI] [PubMed] [Google Scholar]
- [7].Rodriguez-Lopez S, Jimenez-Carretero D, Estepar RSJ, Moreno EF, Kumamaru KK, Rybicki FJ, Ledesma-Carbayo MJ, and Gonzalez G, “Automatic ventricle detection in computed tomography pulmonary angiography,” in [Biomedical Imaging (ISBI) 2015 IEEE 12th International Symposium on], 1143–1146, IEEE (2015).
- [8].González G, Washko GR, and Estépar RSJ, “Automated agatston score computation in a large dataset of non ecg-gated chest computed tomography,” in [Biomedical Imaging (ISBI) 2016 IEEE 13th International Symposium on], 53–57, IEEE (2016). [DOI] [PMC free article] [PubMed]
- [9].Xie Y, Cham MD, Henschke C, Yankelevitz D, and Reeves AP, “Automated coronary artery calcification detection on low-dose chest ct images,” in [Proc. SPIE], 9035, 90350F (2014). [Google Scholar]
- [10].Isgum I, Prokop M, Niemeijer M, Viergever MA, and van Ginneken B, “Automatic coronary calcium scoring in low-dose chest computed tomography,” IEEE transactions on medical imaging 31(12), 2322–2334 (2012). [DOI] [PubMed] [Google Scholar]
- [11].Shahzad R, van Walsum T, Schaap M, Rossi A, Klein S, Weustink AC, de Feyter PJ, van Vliet LJ, and Niessen WJ, “Vessel specific coronary artery calcium scoring: an automatic system,” Academic radiology 20(1), 1–9 (2013). [DOI] [PubMed] [Google Scholar]
- [12].Wolterink JM, Leiner T, Takx RA, Viergever MA, and Išgum I, “An automatic machine learning system for coronary calcium scoring in clinical non-contrast enhanced, ecg-triggered cardiac ct,” in [Medical Imaging 2014: Computer-Aided Diagnosis], 90350E, International Society for Optics and Photonics (2014). [DOI] [PubMed] [Google Scholar]
- [13].Wolterink JM, Leiner T, Viergever MA, and Išgum I, “Automatic coronary calcium scoring in cardiac ct angiography using convolutional neural networks,” in [International Conference on Medical Image Computing and Computer-Assisted Intervention], 589–596, Springer (2015). [Google Scholar]
- [14].Wolterink JM, Leiner T, de Vos BD, van Hamersvelt RW, Viergever MA, and Išgum I, “Automatic coronary artery calcium scoring in cardiac ct angiography using paired convolutional neural networks,” Medical image analysis 34, 123–136 (2016). [DOI] [PubMed] [Google Scholar]
- [15].Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, and Crapo JD, “Genetic epidemiology of copd (copdgene) study design,” COPD: Journal of Chronic Obstructive Pulmonary Disease 7(1), 32–43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kingma D and Ba J, “Adam: A method for stochastic optimization,” arXiv preprint arXiv:1412.6980 (2014). [Google Scholar]
- [17].Lowekamp BC, Chen DT, Ibáñez L, and Blezek D, “The design of simpleitk,” Frontiers in neuroinformatics 7(December), 45 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chollet F, “Keras.” https://github.com/fchollet/keras (2015).
- [19].Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, Ghemawat S, Goodfellow I, Harp A, Irving G, Isard M, Jia Y, Jozefowicz R, Kaiser L, Kudlur M, Levenberg J, Mané D, Monga R, Moore S, Murray D, Olah C, Schuster M, Shlens J, Steiner B, Sutskever I, Talwar K, Tucker P, Vanhoucke V, Vasudevan V, Viégas F, Vinyals O, Warden P, Wattenberg M, Wicke M, Yu Y, and Zheng X, “TensorFlow: Large-scale machine learning on heterogeneous systems,” (2016). [Google Scholar]
- [20].Erbel R, Möhlenkamp S, Kerkho G, Budde T, and Schmermund A, “Non-invasive screening for coronary artery disease: calcium scoring,” Heart 93(12), 1620–1629 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schernthaner RE, Stadler A, Beitzke D, Homolka P, Weber M, Lammer J, Czerny M, and Loewe C, “Dose modulated retrospective ecg-gated versus non-gated 64-row ct angiography of the aorta at the same radiation dose: comparison of motion artifacts, diagnostic confidence and signal-to-noise-ratios,” European journal of radiology 81(4), e585–e590 (2012). [DOI] [PubMed] [Google Scholar]


