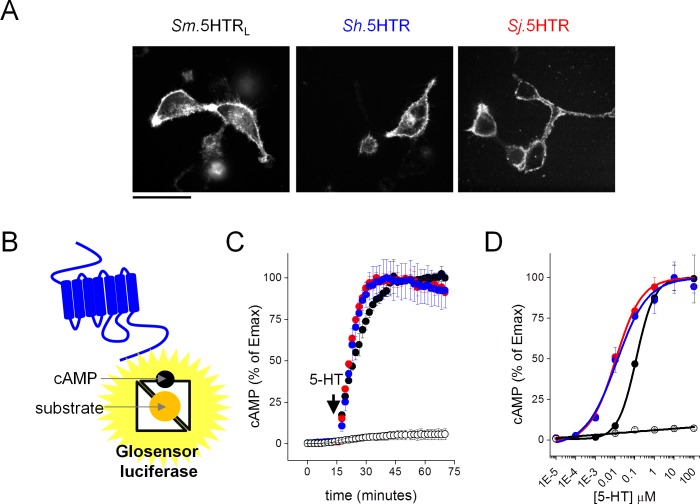
Figure 1. Functional expression of Schistosoma serotonin receptors.
(A) Confocal images showing cell surface expression in mammalian HEK293 cells of 5-HT receptors cloned from S. mansoni (Sm.5HTRL), S. haematobium (Sh.5HTR) and S. japonicum (Sj.5HTR) localized by COOH-terminally tagged eGFP. Scalebar, 50 µm. (B) Schematic of luminescent cAMP sensor bioassay. cAMP generated by schistosome 5-HTRs (blue) binds the engineered GloSensor luciferase, switching the sensor to a more active conformation resulting in enhanced luminescence output. (C) Kinetics of signal following the addition of 5-HT (1 µM) to HEK293 cells co-transfected with luminescent cAMP sensor and individual schistosome 5-HT receptors. Open circles, cells not transfected with 5-HT receptor, colored circles represent measurements in cells transfected with Sm.5HTRL (black), (Sh.5HTR (blue) and Sj.5HTR (red). (D) Serotonin dose-response curves for each of the three receptors (colored circles) and HEK293 cells expressing the cAMP sensor alone (open circles). Data reflect mean ± standard error of at least 3 biological replicates.
Figure 1—figure supplement 1. Schistosome 5-HT receptor protein alignment.
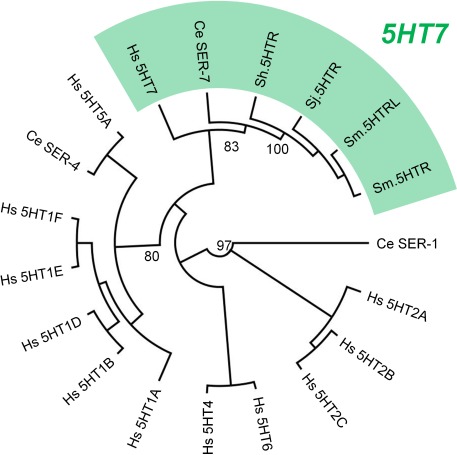
Figure 1—figure supplement 2. Cladogram of schistosome 5-HT receptors.