Abstract
Despite the prevalence of sexual reproduction across eukaryotes, there is a remarkable diversity of sex determination mechanisms. The underlying causes of this diversity remain unclear, and it is unknown if there are convergent trends in the directionality of turnover in sex determination mechanisms. We used the recently assembled Tree of Sex database to assess patterns in the evolution of sex determination systems in the remarkably diverse vertebrate clades of teleost fish, squamate reptiles, and amphibians. Contrary to theoretical predictions, we find no evidence that the evolution of separate sexes is irreversible, as transitions from separate sexes to hermaphroditism occur at higher rates than the reverse in fish. We also find that transitions from environmental sex determination to genetic sex determination occur at higher rates than the reverse in both squamates and fish, suggesting that genetic sex determination is more stable. However, our data are not consistent with the hypothesis that heteromorphic sex chromosomes are an “evolutionary trap”. Rather, we find similar transition rates between homomorphic and heteromorphic sex chromosomes in both fish and amphibians, and to environmental sex determination from heteromorphic versus homomorphic sex chromosome systems in fish. Finally, we find that transitions between male and female heterogamety occur at similar rates in amphibians and squamates, while transitions to male heterogamety occur at higher rates in fish. Together, these results provide the most comprehensive view to date of the evolution of vertebrate sex determination in a phylogenetic context, providing new insight into longstanding questions about the evolution of sexual reproduction.
Keywords: sex determination, sex chromosome, phylogenetic comparative methods, fish, amphibians, squamate reptiles
Introduction
The vast majority of eukaryotes reproduce sexually, and male and female reproductive phenotypes are broadly conserved across a vast array of taxa. Despite this conservation, there is an enormous diversity in the mechanisms used to determine sex across eukaryotes (Bull, 1983; Bachtrog et al., 2014; Beukeboom & Perrin, 2014). This diversity of sex-determination mechanisms is encapsulated in vertebrates, a clade in which nearly all known mechanisms of sex determination are present (Bachtrog et al., 2014; The Tree of Sex Consortium, 2014). This extensive diversity begs the question of both how and why transitions among sex determination mechanisms occur. Although there is an extensive body of theoretical work predicting when and why we might expect transitions in sex determination systems, we have lacked sufficient empirical data to critically test these hypotheses (Bull, 1983; Bachtrog et al., 2014; Beukeboom & Perrin, 2014). Furthermore, little is known about the mechanisms that underlie transitions among sex determination mechanisms. As a first step, identifying whether particular transitions occur at a higher rate than others can provide into whether there are evolutionary or mechanistic constraints on transitions among sex determination systems.
One of the most fundamental transitions in sex determination mechanisms is between the presence of both sexes within the same individual (hermaphroditism) and the presence of two sexes in different individuals (called gonochorism in animals and dioecy in plants). Hermaphroditism is only found in 5% of animal species (Jarne & Auld, 2006; Eppley & Jesson, 2008) but is quite common in flowering plants, with some form of hermaphroditism observed in 94% of species (Renner & Ricklefs, 1995; Renner, 2014). Although the transition to separate sexes was once considered to be irreversible (Bull & Charnov, 1985), recent work has suggested that transitions from dioecy to hermaphroditism might commonly occur in plants (Barrett, 2013; Käfer et al., 2014, 2017; Renner, 2014). Indeed, a recent study in flowering plants revealed no consistent trends in the rates of transition between hermaphroditism and dioecy (Goldberg et al., 2017). However, the rates of transition between hermaphroditism and gonochorism have not yet been investigated in any group of animals. The prevalence of both hermaphroditism and gonochorism in fish makes it an excellent clade to investigate these transition rates.
Even across gonochoristic vertebrate species, there is still a large diversity of sex determination mechanisms (The Tree of Sex Consortium, 2014). Many fish and non-avian reptiles have environmental sex determination (ESD), in which environmental cues, such as temperature during development, are used to determine sex. Many other species of fish, reptiles and amphibians, as well as all known birds and mammals, have genetic sex determination (GSD). The evolution of ESD is thought to be favored when environmental variation has a differential effect on the fitness of males and females (Charnov & Bull, 1977; Warner & Shine, 2008). By contrast, the evolution of GSD is favored in unpredictable environments or in environments with low variability (Bull, 1983). In addition, because single locus GSD results in roughly even sex ratios, it has been assumed to be more stable than ESD, which can result in highly skewed sex ratios (Conover & Heins, 1987; Pen et al., 2010), particularly during periods of environmental instability (Jensen et al., 2018). Sex determining systems with unbalanced sex ratios are prone to invasions by mechanisms that restore balanced sex ratios (Fisher, 1930). Therefore, the balanced sex ratio in GSD might be more resilient to invasion by new sex determining mechanisms, while ESD might be prone to invasion and replacement by systems that restore equal sex ratios. As a result, we might expect a bias toward GSD systems. A recent analysis found mixed support for this hypothesis, with no differences in transition rates between GSD and ESD in turtles, but a higher rate of transition from ESD to GSD in squamate reptiles (Pokorna & Kratochvíl, 2009; Gamble et al., 2015; Sabath, Itescu et al., 2016). However, the rates of transitions between GSD and ESD have not been tested in other suitable groups, such as fish.
Although GSD can be polygenic (Moore & Roberts, 2013), it is more often under single locus control, resulting in systems where either the male or female is heterozygous at the sex-determination locus. In single-locus GSD systems, the genetic difference between males and females can be as small as a single nucleotide variant (Kamiya et al., 2012), however this difference has often progressed to heteromorphic sex chromosomes (Charlesworth et al., 2005; Bachtrog et al., 2013; Wright et al., 2016). In these cases, male heterozygosity progresses to distinct X and Y sex chromosomes, and female heterozygosity to Z and W sex chromosomes. Heteromorphic sex chromosomes are most notably found in the therian mammals (Bellott et al., 2014; Cortez et al., 2014), birds (Zhou et al., 2014; Bellott et al., 2017), and snakes (Matsubara et al., 2006; Vicoso et al., 2013). These clades exhibit a surprising degree of conservation (but see Gamble et al., 2017), given the rapid sex chromosome turnover exhibited by other vertebrate clades. This has led to the suggestion that highly heteromorphic sex chromosomes, once established, are an evolutionary trap, and that transitions from heteromorphic sex chromosomes will be rare due to the accumulation of recessive mutations and loss of essential genes on either the Y or W (Bull, 1983; Bull & Charnov, 1977; Pokorná & Kratochvíl, 2009). This is somewhat at odds with theoretical suggestions that sex chromosomes might cycle quickly due to a combination of sexually antagonistic selection (van Doorn & Kirkpatrick, 2007, 2010) and the accumulation of deleterious mutations on the non-recombining Y or W chromosome (Blaser et al., 2013, 2014). The latter will be more acute as sex chromosome divergence progresses, leading to the counterintuitive idea that older, more degenerate systems would be more prone to transitions. Limited empirical support for the evolutionary trap hypothesis was previously found in squamates (Pokorná & Kratochvíl, 2009; Gamble et al., 2015), but this hypothesis requires further investigation.
In species with GSD, there is further variation in whether the male or the female is the heterogametic sex (The Tree of Sex Consortium, 2014). Here, we will refer to species with male heterogamety as XY and female heterogamety as ZW, regardless of whether there are heteromorphic sex chromosomes present in that species. Transitions between XY and ZW systems have been proposed to result from a variety of evolutionary forces, including drift, selection on pleiotropic effects of sex-determination genes, selection on sex ratio, and sexually antagonistic selection (Bull & Charnov, 1977; Werren & Beukeboom, 1998; Jaenike, 2001; van Doorn & Kirkpatrick, 2010; Bachtrog et al., 2011; Veller et al., 2017). Theory suggests that if transitions between XY and ZW systems are driven by sexually antagonistic selection or even by drift, then the new sex-determination system should be epistatically dominant to the ancestral system (van Doorn & Kirkpatrick, 2010; Veller et al., 2017). Because new W chromosomes have been shown to be dominant to the ancestral Y chromosome in species of fish (Kallman, 1984; Ser et al., 2010) and amphibians (Ogata et al., 2008), it was suggested that there could be a bias in transitions from XY to ZW systems in these groups (van Doorn & Kirkpatrick, 2010). However, empirical tests of this hypothesis have been limited and the few studies that have been conducted have produced mixed results. While a previous study in amphibians found that transitions from ZW to XY systems are more common than the reverse (Hillis & Green, 1990), another found no support for a difference in transition rates between XY and ZW systems across amphibians (Evans et al., 2012). Additional work is clearly needed to determine whether there are any biases in the direction of transitions between male and female heterogamety.
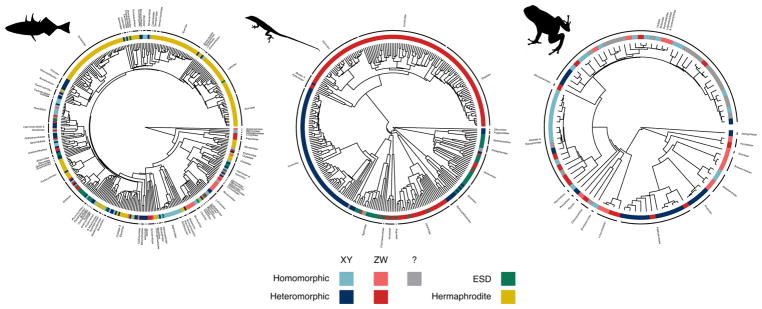
Here, we take advantage of the recently assembled Tree of Sex database (The Tree of Sex Consortium, 2014) and the diversity of sex-determination systems found in three major vertebrate groups (fish, amphibians, squamate reptiles; Figure 1) to examine transitions among sex determination systems. The complete dataset includes information on 705 species of fish, 173 amphibians and 487 squamate reptiles, and as such constitutes the broadest and most comprehensive analysis of vertebrate sex determination evolution conducted to date in a phylogenetic context. We used these data to compare the rates of transition between: (1) gonochorism and hermaphroditism in fish; (2) ESD and GSD in fish and squamates; (3) homomorphic and heteromorphic sex chromosomes in fish and amphibians; (4) homomorphic vs. heteromorphic sex chromosomes and ESD in fish; and (5) XY and ZW sex determination systems in fish, squamates and amphibians (Table 1).
Figure 1. The distribution of sex determination across three vertebrate clades.
Species are coded as being either XY heteromorphic (dark blue), XY homomorphic (light blue), ZW heteromorphic (dark red), ZW homomorphic (light red), unknown homomorphic (?, gray), having environmental sex determination (ESD, green), or being hermaphrodites (yellow). Species that had some degree of ESD were classified as such, regardless of their chromosomes. Note that in the actual analyses we estimated parameters across 10 different datasets, with slightly different taxonomic coverage; for the purposes of this figure, we selected one of these at random.
Table 1.
Summary of analysis
| Clade | State 1: # species in dataset (avg # tree-matched species) | State 2: # species in dataset (avg # tree-matched species) | State 3: # species in dataset (avg # tree-matched species) | Prob. of higher transition rate |
|---|---|---|---|---|
| 1. Transitions between gonochorism and hermaphroditism | ||||
| Fish | Gonochorism: 371 (178.8) | Hermaphroditism: 309 (165) | N/A | 0.949 gonochorism to hermaphroditism |
|
| ||||
| 2. Transitions between ESD and GSD | ||||
| Fish | ESD: 61 (22) | GSD: 310 (156.8) | N/A | 0.984 ESD to GSD |
| Squamates | ESD: 49 (22) | GSD: 389 (279) | N/A | 1.0 ESD to GSD |
|
| ||||
| 3. Transitions between homomorphic and heteromorphic sex chromosomes | ||||
| Fish | Homomorphic: 145 (82.2) | Heteromorphic: 126 (64.1) | N/A | 0.699 heteromorphic to homomorphic |
| Amphibians | Homomorphic: 94 (60.9) | Heteromorphic: 45 (31.1) | N/A | 0.697 heteromorphic to homomorphic |
|
| ||||
| 4. Transitions between homomorphic vs. heteromorphic sex chromosomes and ESD | ||||
| Fish | Homomorphic: 137 (76.7) | Heteromorphic: 125 (64.8) | ESD: 52 (16.4) | 0.437 heteromorphic to ESD |
|
| ||||
| 5. Transitions between XY and ZW sex determination systems | ||||
| Fish | XY: 204 (110.5) | ZW: 92 (47) | N/A | 0.999 ZW to XY |
| Squamates | XY: 116 (88) | ZW: 231 (160) | N/A | 0.779 ZW to XY |
| Amphibians | XY: 67 (46.2) | ZW: 32 (28) | N/A | 0.560 ZW to XY |
Methods
Data
We matched data from the Tree of Sex database (The Tree of Sex Consortium, 2014) to recently published large-scale phylogenies of ray-finned fishes (Rabosky et al., 2013, with 11 erroneously placed species removed from the original; M. Alfaro, pers. comm), squamate reptiles (Pyron et al., 2013, Pyron & Burbrink, 2014), and amphibians (Pyron & Wiens, 2011; Eastman et al., 2013). All trees were ultrametric with branch lengths in units of millions of years. The Tree of Sex dataset we used is the same as that in the original publication with the addition of recently discovered XY systems in two species (Boa imperator, Python bivittatus) of snakes (Gamble et al., 2017). However, other recent studies in squamates are not included in this dataset (Koubová et al. 2014; Pokorná, Rens et al., 2014; Pokorná, Rovatsos et al., 2014; Gamble et al. 2015; Rovatsos et al., 2014, 2015, 2016). In many cases, we had congeneric matches between species in the tree and species in the dataset. In these cases, we used a recently developed algorithm (Pennell et al., 2016), implemented in the R package ‘phyndr’ (https://github.com/traitecoevo/phyndr), to swap species in the phylogeny which were not included in our dataset with ‘phylogenetically equivalent’ species that were (see Pennell et al., 2016 for full details on the algorithm). Unlike data imputation approaches (see Rabosky, 2015), the algorithm is conservative such that it is guaranteed not to introduce any biases into analyses of character evolution, so long as the taxonomy is phylogenetically informative. Since there may be many combinations of phylogenetically equivalent swaps, we ran each analysis across ten different tree and trait combinations to ensure that our analyses were robust to sampling artifacts. Furthermore, for species in which there were multiple records in the dataset, we randomly selected one of these to include in each of the ten analyses. The alternative datasets gave essentially identical results.
Overview of analyses
We addressed five different questions in our analyses. For each analysis, we coded the characters as discrete states and fit a Markov model of trait evolution (Pagel, 1994) using a Markov chain Monte Carlo (MCMC) procedure implemented in the R package ‘diversitree’ (FitzJohn, 2012). For all rates, we set a broad exponential prior (mean of 0.1). We ran all chains for 50,000 generations and removed the first 10,000 samples as burn-in. As stated above, each analysis was run across ten related trait/tree datasets in order to mitigate sampling error, and the results from all individual analyses were summarized together. To examine the support (or lack thereof) for differences in transition rates, we computed differences in rates across the posterior and examined the extent to which the distribution of differences overlapped with zero. In the absence of a reliable procedure for estimating Bayes Factors for MK models in R, we think this is the most accurate way to present our results. All analyses were conducted in R v3.3.1. Code to reproduce all analyses and results is available at https://github.com/mwpennell/vert_trans.
Transitions between gonochorism and hermaphroditism
The prevalence of hermaphroditism in fish (Devlin & Nagahama, 2002; Mank et al., 2006) makes it possible to test for differences in rates of transition between gonochorism and hermaphroditism in this clade. In order to evaluate the relative transition rates between gonochorism and hermaphroditism, we fit a two state Markov model, collapsing all species with separate sexes into a single category of gonochorism. In our fish dataset, there were 371 records of gonochorism (178.8 of which matched to the tree on average across the ten runs) and 309 records of hermaphroditism (165 matched to the tree on average).
Transitions between ESD and GSD
For this analysis, we focused on fish and squamates, as both ESD and GSD have evolved repeatedly in both of these clades. In order to investigate transitions between GSD and ESD, we fit a two-state Markov model similar to the gonochorism/hermaphroditism analysis above. Species were coded as having one or the other form of sex determination. For any species that had some degree of both genetic and environmental sex determination, we coded these species as having environmental sex determination. We also repeated the analysis with these ambiguously coded species excluded, and this did not qualitatively affect our results (Supplemental Figure 1). For fish, there were 310 GSD records in the database (156.8 matched to the tree on average) and 61 ESD records (22 matched to the tree on average). For squamates, there were 389 GSD records (279 matched to the tree on average) and 49 ESD records (22 matched to the tree on average).
Transitions between homomorphic and heteromorphic sex chromosomes
For this analysis, we considered both fish and amphibians. We did not analyze the squamate data due to the rarity of conclusive evidence for homomorphic sex chromosomes in this group (Gamble et al., 2015; Gamble, 2016), precluding the estimation of meaningful character correlations (Maddison & FitzJohn, 2015). Restricting the analysis to gonochoristic species, we coded species for two variables: 1) whether they had cytogenetically visible (i.e. heteromorphic) sex chromosomes or not; and 2) whether they were male or female heterogametic. We removed the few species from the datasets (fish: 15 records; amphibians: 1 record) where the Y (or W) sex chromosome has been completely lost (i.e., XO and ZO systems) as we expect the evolutionary dynamics to be different from those of true XY or ZW systems (Bull, 1983; Maddison & Leduc-Robert, 2013; Blackmon & Demuth, 2014). Three fish species with polygenic sex determination were also excluded. Across fish, this coding scheme resulted in 83 XY homomorphic, 97 XY heteromorphic, 51 ZW homomorphic, 29 ZW heteromorphic, and 11 unknown homomorphic records (average tree-matched counts: 53.2, 47, 23.4, 17.1, and 5.6). For amphibians, these numbers are 37 XY homomorphic, 29 XY heteromorphic, 18 ZW homomorphic, 16 ZW heteromorphic, and 39 unknown homomorphic records (average tree-matched counts: 25.9, 18.3, 14, 12.8, and 21).
Transitions between homomorphic vs. heteromorphic sex chromosomes and ESD
We were only able to perform this analysis on fish, as we were not able to meaningfully estimate parameters in the other clades. For this analysis, we coded gonochoristic species as being ESD, homomorphic GSD, or heteromorphic GSD. For species with GSD, we did not distinguish between whether they were male or female heterogametic, in contrast to the previous analysis. Species that had both GSD and some level of ESD were not included. This resulted in 137 species with homomorphic GSD, 125 species with heteromorphic GSD, and 52 species with ESD. After matching to the tree, we ended up with an average of 76.7 homomorphic species, 64.8 heteromorphic species, and 16.4 ESD species per analysis. We then fit a simple 3-state Markov model and estimated the six (i.e., forward and reverse) transition rates between the three states. For simplicity, we only report the comparison of the transition rate from homomorphic GSD to ESD versus the transition rate from heteromorphic GSD to ESD.
Transitions between XY and ZW sex determination systems
For this analysis, we used fish, squamate and amphibian datasets. We again considered only gonochoristic species with single-locus GSD. We coded all species as being either male (XY) or female heterogametic (ZW) and assumed for simplicity that the probability of an invasion by a novel sex chromosome did not depend on whether a species had homomorphic or heteromorphic sex chromosomes. This assumption is consistent with our finding that the transition rates between homomorphic and heteromorphic sex chromosomes are similar in both XY and ZW systems (see below). There were 204 XY and 92 ZW systems (average tree-matched counts: 110.5 XY, 47 ZW) in the fish dataset, 116 XY and 231 ZW systems (average tree-matched counts: 88 XY, 160 ZW) in the squamate dataset, and 67 XY and 32 ZW systems (average tree-matched counts: 46.2 XY, 28 ZW) in the amphibian dataset. We fit a simple, two state Markov model and estimated net transition rates between XY and ZW systems.
Results
Transition rates from gonochorism to hermaphroditism are higher than the reverse
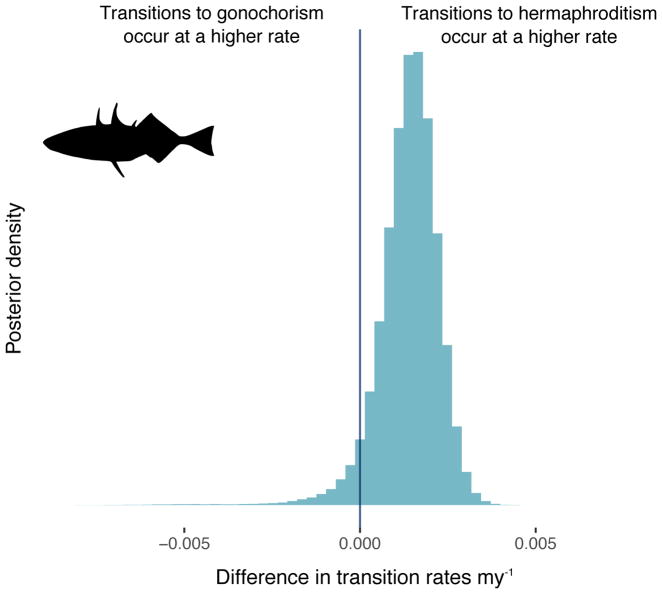
In our fish dataset, there were roughly equal numbers of gonochoristic (n = 371) and hermaphroditic (n = 309) species (across the ten datasets, there were on average approximately 179 gonochoristic and 165 hermaphroditic species matched to the tree). However, 94.9% of the posterior distribution supports that transitions to hermaphroditism occur at a higher rate than transitions to gonochorism (Figure 2; Table 1). The median rate of transition from gonochorism to hermaphroditism is 2.4 times higher than the reverse.
Figure 2. Transitions from gonochorism to hermaphroditism occur at a higher rate than the reverse in fish.
Posterior distribution of the difference in transition rates between gonochorism and hermaphroditism in fish. Across the 10 datasets, 94.9% of the posterior distribution supports the conclusion that transitions from gonochorism to hermaphroditism occur at a higher rate than the reverse.
Transition rates from ESD to GSD are higher than the reverse
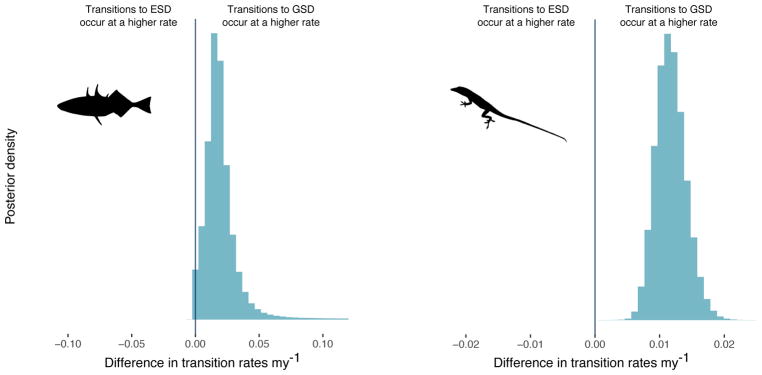
There is strong support for the conclusion that transitions from ESD to GSD occur at a higher rate than the opposite transition in both fish and squamates (Figure 3; Table 1; fish: 98.4% of the posterior distribution; squamates: 100% of the posterior distribution). The transition rate from ESD to GSD is six times higher than the reverse in fish, and 17.3 times higher than the reverse in squamates.
Figure 3. Transitions from environmental sex determination to genetic sex determination occur at a higher rate than the reverse in fish and squamates.
Posterior distribution of the difference in transition rates between genetic sex determination (GSD) and environmental sex determination (ESD) for fish (left panel) and squamates (right panel). Across the 10 datasets, 98.4% of the posterior distribution in fish and 100% of the posterior distribution in squamates supports the conclusion that transitions from ESD to GSD occur at a higher rate than the reverse. Indeed, across the posterior distribution, the rate of transition from ESD to GSD is, on average, around 6 (fish) and 17.3 (squamates) times higher than the reverse.
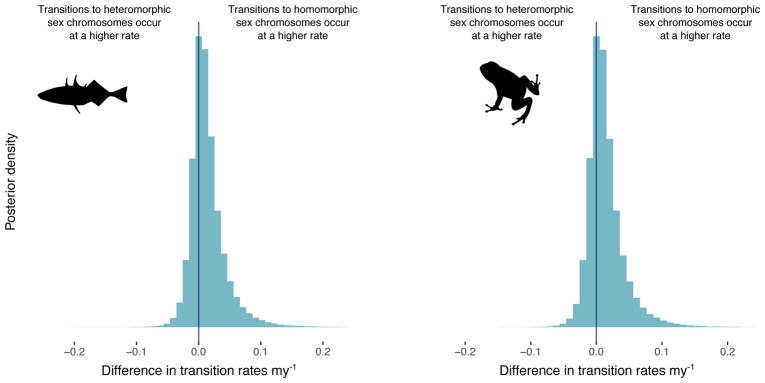
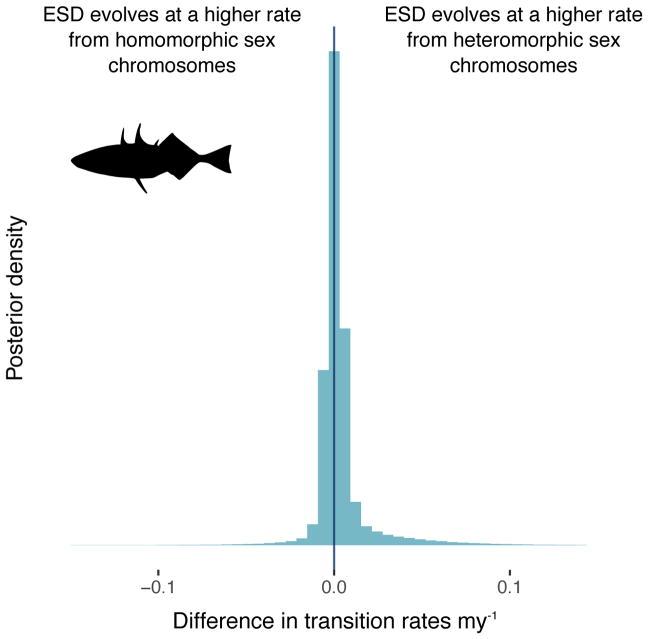
Similar rates of transition between homomorphic and heteromorphic sex chromosomes
We did not detect a significant difference in the rates of transitions between homomorphic and heteromorphic sex chromosomes; in both the fish and amphibian datasets approximately 70% of the posterior distribution supports the transition from heteromorphic to homomorphic as having occurred at higher rates and 30% suggests the reverse (Figure 4; Table 1). In fish and amphibians, both XY and ZW systems show similar rates of transitions from homomorphic to heteromorphic sex chromosomes (data not shown).
Figure 4. No differences in transition rates between homomorphic and heteromorphic sex chromosomes in either fish or amphibians.
Posterior distribution of the difference in transition rates between homomorphic and heteromorphic sex chromosomes in fish (left panel) and amphibians (right panel). Across the 10 datasets, 69.9% of the posterior distribution in fish and 69.7% of the posterior distribution in amphibians support a higher rate of transitions from heteromorphic to homomorphic sex chromosomes than the reverse, but these results are not significant.
Similar rates of transition to ESD from homomorphic vs. heteromorphic sex chromosomes
The transition rates from either homomorphic or heteromorphic sex chromosomes to ESD are significantly greater than zero (Supplemental Figures 2, 3). However, there is no significant difference between the transition rate from homomorphic sex chromosomes to ESD versus the transition rate from heteromorphic sex chromosomes to ESD (Figure 5; Table 1). These data suggest that the presence of heteromorphic sex chromosomes does not preclude transitions to ESD, at least in fish.
Figure 5. No difference in transition rate from homomorphic sex chromosomes to ESD versus heteromorphic sex chromosomes to ESD in fish.
Posterior distribution of the difference in transition rates between homomorphic sex chromosomes and ESD, and heteromorphic sex chromosomes and ESD. Across the 10 datasets and the entire analyses, 43.7% of the posterior distribution supports a higher rate of transitions from heteromorphic sex chromosomes to ESD, but this is not significant. The posterior distributions of the transition rates between heteromorphic sex chromosomes and ESD and between homomorphic sex chromosomes and ESD are significantly greater than zero (Supplemental Figures 2 and 3).
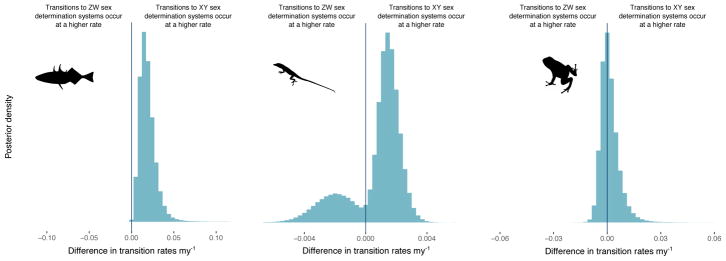
Rates of transition between XY and ZW systems differ among clades
In the fish dataset, 99.9% of the posterior distribution supports the conclusion that there is a higher rate of transition from ZW to XY systems than the reverse (Figure 6; Table 1). In squamates, 77.9% of the posterior distribution supports a higher rate of transition from ZW to XY systems. However, the bimodal distribution of posterior probabilities likely reflects that there are two different configuration of rates that produce the same distribution at the tips and suggests that our model may not adequately describe this data (Figure 6; Table 1). In amphibians, there is no significant difference in the rate of transition between ZW and XY systems (Figure 6; Table 1).
Figure 6. Transitions from ZW to XY sex determination systems occur at a higher rate than the reverse in fish, but not in squamates or amphibians.
Posterior distribution of the difference in transition rates between XY and ZW systems for fish (left panel), squamates (middle panel), and amphibians (right panel). Across the 10 datasets, 99.9% of the posterior distribution supports the conclusion that there is a higher rate of transitions from ZW to XY sex determination systems in fish. However, there is not significant support for this conclusion in other clades: only 77.9% of the posterior distribution in squamates and 56.0% of the posterior distribution in amphibians suggest a higher rate of transitions from ZW to XY sex determination systems than the reverse.
Discussion
Using the Tree of Sex database (The Tree of Sex Consortium, 2014), we compared the rates of transitions among different mechanisms of sex determination in three vertebrate clades with the most extensive variation in these fundamental traits. Here, we discuss how these results have provided new insight into the theoretical predictions about transitions in sex determination mechanisms, the caveats of our analyses, and the implications of the evolution of sex determination mechanisms, particularly sex chromosomes, for speciation.
The evolution of separate sexes is not always irreversible
In contrast to the hypothesis that the evolution of separate sexes is irreversible (Bull & Charnov, 1985), we find that transitions from gonochorism to hermaphroditism occur at higher rates than the reverse in fish. Thus, we conclude that the evolution of separate sexes is not always an evolutionary “one-way street”, consistent with recent studies in flowering plants (Barrett, 2013; Käfer et al., 2014, 2017; Renner, 2014; Goldberg et al., 2017). Nonetheless, our result is perhaps counter-intuitive, as it would suggest that there should be more hermaphroditic lineages in fish. However, there are actually fewer hermaphroditic species than gonochoristic species in our database (Figure 1; Table 1). Although it is possible that speciation rates are higher for gonochoristic lineages or extinction rates are higher for hermaphroditic lineages, we are unable to formally test these possibilities with the current dataset.
Hermaphroditism is particularly common in reef-dwelling fish (Ghislelin, 1969; Smith, 1975), and it is worth noting that the bright colors of many reef-fishes, which make them popular in the aquarium trade, may also have led to a relative over-sampling of these lineages in our dataset (see Caveats of our analyses). Nonetheless, there also may be extrinsic or intrinsic factors that select against gonochorism in reef environments. Given the relative rarity of dioecy in plants (Renner & Ricklefs, 1995; Renner, 2014), these factors might be shared between plants and reef fish. Alternatively, if gonochorism is the ancestral state in fish (still to be formally tested), then the genetic and developmental mechanisms that underlie the evolution of hermaphroditism from a gonochoristic ancestor might preclude a reversion to separate sexes. Such a constraint does not appear to be present in flowering plants, as the evolution of separate sexes from a hermaphroditic ancestor has occurred many times, and there are no differences in the rates of transition between hermaphroditism and dioecy (Barrett, 2013; Käfer et al., 2014, 2017; Renner, 2014; Goldberg et al., 2017). Future work is needed to identify the evolutionary and genetic mechanisms underlying transitions between hermaphroditism and separate sexes in plants and fish, as well as the potential ecological or life history conditions that might predispose these lineages to hermaphroditism.
ESD is less stable than GSD
Consistent with the hypothesis that species with ESD might have unequal sex ratios and therefore be prone to invasions by GSD to restore balanced sex ratios (Fisher, 1930), we find that transitions from ESD to GSD occur at higher rates than the reverse in both fish and squamates. Our results are also consistent with previous studies in squamates, which have also found that transitions from ESD to GSD are more common than the reverse (Pokorna & Kratochvíl, 2009; Gamble et al., 2015; Sabath, Itescu et al., 2016). However, there is no difference in transition rates between ESD and GSD in turtles, possibly due to the longer lifespan of turtles (Sabath, Itescu et al., 2016). Such longer-lived species are less affected by seasonal variation in the environment that could lead to biased sex ratios and extinction in short-lived species (Bull & Bulmer, 1989; Valenzuela & Lance, 2004). Indeed, turtles with ESD have longer average lifespans than turtles with GSD, and other lineages of reptiles including crocodylians and tuatara, which only exhibit ESD, are also long-lived (Sabath, Itescu et al., 2016). However, broader climatic shifts, such as human-induced climate change, have been shown to lead to major skews in sex-bias (Jensen et al., 2018), suggesting that ESD may be prone to invasion by GSD even in long-lived species. These results highlight that there are many sources of selection on sex determination mechanisms and that additional comparative studies across many systems are needed to shed further light on mechanisms driving these transitions.
Heteromorphic sex chromosomes are not always an evolutionary trap
The observation that some groups, including birds and mammals, have evolutionarily stable heteromorphic sex chromosomes has led to the suggestion that GSD, particularly heteromorphic sex chromosomes, acts as an evolutionary trap that prevents transitions to other mechanisms of sex determination due to the degeneration of either the Y or W (Bull, 1983; Bull & Charnov, 1977; Pokorná & Kratochvíl, 2009). Although previous analyses in squamates are consistent with this hypothesis, these analyses did not distinguish between GSD with homomorphic sex chromosomes and GSD with heteromorphic sex chromosomes (Pokorná & Kratochvíl, 2009; Gamble et al., 2015). This is in part due to the difficulty of reliably identifying systems with homomorphic sex chromosomes in squamates (Gamble et al. 2015; Gamble, 2016). Thus, we performed two complementary analyses to test the evolutionary trap hypothesis. In the first analysis, we found no differences in transition rate between homomorphic and heteromorphic sex chromosomes in either fish or amphibians. In the second analysis, we found that rates of transitions to ESD from homomorphic sex chromosomes were not significantly different than rates of transitions to ESD from heteromorphic sex chromosomes in fish. Our data are consistent with a recent study in Drosophila demonstrating that ancient sex chromosomes have reverted to autosomes (Vicoso & Bachtrog, 2013). Taken together, these data suggest that heteromorphic sex chromosomes might not always be an evolutionary trap that precludes transitions to other systems.
However, these results should be considered as preliminary. It is important to emphasize that we performed these analyses on a dataset in which the classification of heteromorphic sex chromosomes is based mostly on the presence of cytogenetically distinct sex chromosomes (The Tree of Sex Consortium, 2014). For the vast majority of species, this is the only data available. However, cytogenetic methods have low resolution and will greatly underestimate both the number of homomorphic and heteromorphic systems and therefore do not necessarily reveal the extent of degeneration found on a sex chromosome (Ross & Peichel, 2008; Gamble et al., 2015; Gamble, 2016). Testing whether there are differences in transition rates between sex chromosomes with high versus low levels of degeneration, as posited by the evolutionary trap hypothesis, will need to await detailed molecular analyses of sex chromosomes across many systems. Excellent efforts towards this goal have recently been made in some groups of squamates (e.g. Rovatsos et al., 2014, 2016; Gamble et al., 2015), but much more data are needed, which will be facilitated by new methods that rely on next-generation sequencing approaches to detect sex chromosomes (Vicoso & Bachtrog, 2013; Gamble, 2016; Muyle et al., 2016). Such analyses will also enable tests of whether high loads of deleterious mutations might actually promote turnover of degenerate sex chromosomes (Blaser et al., 2013, 2014).
Transition rates between XY and ZW systems differ among clades
We found clade-specific patterns in transition rates between female heterogametic (ZW) and male heterogametic (XY) systems. In fish, transitions from ZW to XY systems occur at higher rates than the reverse. By contrast, we found no differences in transition rates between XY and ZW systems in squamates or amphibians. Our results in amphibians are consistent with those of Evans et al. (2012), although an earlier study in amphibians found evidence for a bias from ZW to XY systems (Hillis & Green, 1990). However, both our study and Evans et al. (2012) used a larger dataset, updated phylogeny, and different methodologies.
Importantly, none of these results support the prediction that transitions from XY to ZW systems should occur at higher rates in these groups (van Doorn & Kirkpatrick, 2010). This prediction was based on both theoretical findings that transitions occur most readily when the new sex chromosome is dominant to the ancestral sex chromosome (van Doorn & Kirkpatrick, 2010; Veller et al., 2017), and empirical findings that new W chromosomes are dominant to ancestral Y chromosomes in multi-factorial sex determination systems found in fish and amphibians (Kallman, 1984; Ogata et al., 2008; Ser et al., 2010). To our knowledge, there are no systems in which a new Y chromosome is dominant to an ancestral W chromosome, but this could be because there are very few systems in which both types of sex chromosomes segregate. Furthermore, both W-linked and Y-linked sex determination loci are often dominant (Bachtrog et al., 2014), suggesting that there should not necessarily be a bias in the dominance relationships between W and Y-chromosomes. One alternative hypothesis to explain the prevalence of ZW to XY transitions in fish is that stronger sexual selection in males might promote transitions to XY systems if the sex-determination locus is linked to loci that are beneficial in males (Rice, 1986; Bachtrog et al., 2011). However, it is not known whether sexual selection is generally stronger in fish than in amphibians or squamates. A second alternative hypothesis is that transitions to XY systems might be favored because dominant masculinizing mutations on a Y chromosome can protect against female sex-ratio biases caused by cytoplasmic sex ratio distorters (Beukeboom & Perrin, 2014). The presence of numerous transitions between XY and ZW systems in vertebrates provide an excellent opportunity to further explore these hypotheses.
Caveats of our analyses
Because we were making inferences about the evolutionary dynamics of sex determination systems across large phylogenetic scales, there are a number of caveats to our analyses. First, it was necessary to assume homogeneous transitions rates within each clade that we studied (i.e., rates were assumed to be the same within fish but could differ between fish and squamates). Although transition rates between traits are often highly variable (e.g., Beaulieu et al., 2013) and several methods exist to estimate the phylogenetic position at which they change (e.g., Drummond & Suchard, 2010; Beaulieu et al., 2013; King & Lee, 2015), such methods have not been extended to the multi-state case we consider in some of our analyses, particular to the linear model formulation (FitzJohn, 2012). Therefore, for the sake of consistency and coherence, we maintained this rate-homogeneity assumption across all the analyses. As a result, ours is likely a conservative approach, as this assumption is will tend to obscure true differences rather than induce spurious ones.
Second, we coded all characters as having discrete states, even though this is artificial in some cases. In particular, in our analysis of transitions between homomorphic and heteromorphic sex chromosomes, we assumed that all heteromorphic chromosomes were alike, even though there in in fact a continuum of sex chromosome divergence, ranging from systems in which the two sex chromosomes are nearly identical in size and gene content to those in which the sex-limited Y or W chromosome is severely diminished. However this simplification is necessary in the absence of more refined data on the relative size of the sex chromosomes or a phylogenetic model that can adequately describe the process of chromosomal differentiation. Discrete models of evolution such as this one may make our analysis prone to phylogenetic pseudoreplication, wherein apparently strong evolutionary associations between characters resulted from only a few evolutionary events (Read & Nee, 1995; Maddison & FitzJohn, 2015; Uyeda et al., 2017). There is no clear solution to this problem, nor is there any reliable diagnostic test for phylogenetic pseudoreplication. However, there is a general increase in the robustness of estimates as the number of independent evolutionary transitions increases in a dataset. As apparent from Figure 1, each of the transitions we considered has occurred multiple times in different parts of the phylogeny such that we can be reasonably confident the associations we have found are unlikely to be spurious.
Third, our dataset is sparse. For example, in our largest dataset (gonochorism vs. hermaphroditism) we have 680 records for bony fish from a clade of more than 27,000 species. The squamate and amphibian datasets similarly comprise less than 5% of extant species. However, if our dataset represents a random sample (at least with regard to their sexual system) of the existing diversity, our estimates of transition rates will be unbiased (Pagel, 1994). However, if some taxonomic groups or states are disproportionately represented in our dataset, this could lead to erroneous estimates. For example, it is quite plausible that species with homomorphic sex chromosomes are underrepresented in our database; as discussed above, these are more challenging to recognize than heteromorphic sex chromosomes with mostly cytogenetic data. Likewise, some taxonomic groups are far more likely to be included in the database for a variety of reasons. While some methods have been developed to assess and mitigate the effect of such sampling biases on evolutionary inference from discrete characters (FitzJohn et al., 2014), the sampling in our database is still too low to apply these methods here. As such, we encourage readers to keep this important caveat in mind.
Fourth, our analyses make the assumption that the system of sex determination does not influence macroevolutionary rates of speciation or extinction. But as Maddison (2006) eloquently pointed out, failing to consider an association between a character and diversification rates when such an association actually exists can lead to estimates of transition rates being biased. A number of methods, most notably the *SSE (State-dependent speciation and extinction) family of models (e.g., Maddison et al., 2007; FitzJohn, 2012) have been developed to simultaneously model the evolution of traits and the diversification process, thus mitigating the potential for bias. Indeed, there are theoretical predictions that differences in sexual systems may lead to differences in the rates at which new species form or go extinct (see below). However, we chose to ignore speciation and extinction for two reasons. First, as we state above, the number of species included in our dataset is small relative to the number of species in these clades; as such, we would have essentially no power to detect any differences in diversification rates (FitzJohn et al., 2009). Second, our data spans large taxonomic groups, such that are likely to be many differences in speciation rates unrelated to the traits of interest; recent work has shown that BiSSE (and related methods) are susceptible to being misled by such background variation in rates (Rabosky & Goldberg, 2015). No general solution to this problem has yet been developed. When more data become available and/or novel approaches to studying state-dependent diversification are developed, it would be very interesting to revisit this work to understand the role of speciation and extinction in shaping the phylogenetic distribution of sexual systems.
Does the evolution of sex determination mechanisms influence speciation?
The evolution of sex determination, particularly sex chromosomes, also has important implications for speciation. In particular, two empirical patterns, Haldane’s Rule and the large-X effect, provide evidence that heteromorphic sex chromosomes play an important role in the evolution of post-zygotic hybrid sterility and inviability in animals (Haldane, 1922; Coyne and Orr, 1989; Presgraves, 2008). These patterns have led to the hypothesis that the presence of heteromorphic sex chromosomes might facilitate speciation, particularly the evolution of post-zygotic incompatibilities (Rieseberg, 2001; Philips & Edmands, 2012). Limited support for this hypothesis has been found. For example, higher levels of intrinsic post-zygotic isolation are found in species with sex chromosomes than in those without sex chromosomes, and in species with more heteromorphic or larger sex chromosomes (Turelli & Begun, 1997; Lima, 2014). Philips & Edmands (2012) tested this hypothesis using net diversification intervals as a proxy for speciation rate in reptiles; they found that speciation occurs more rapidly in squamates, in which heteromorphic sex chromosomes are common, than in turtles and crocodylians, in which heteromorphic sex chromosomes are rare or absent. However, these results were equivocal as net diversification intervals in birds, which have heteromorphic sex chromosomes, are similar to those in turtles and crocodylians (Philips & Edmands, 2012). Indeed, other analyses have found no association between diversification rates and the presence of ESD or GSD in turtles, squamates or birds (Organ & Janes, 2008; Sabath, Itescu et al. 2016). Thus, there is not strong evidence that the presence of heteromorphic sex chromosomes influences speciation rates in vertebrates.
It is also possible that speciation rates might instead reflect the rates of turnover in sex chromosomes (Demuth, 2014). Indeed, in stickleback fish, a fusion between an existing Y chromosome and an autosome created a neo-sex chromosome system that harbors loci involved in both behavioral isolation and hybrid male sterility between species, suggesting that sex chromosome turnover has facilitated speciation in this case (Kitano et al., 2009). However, testing the role of sex chromosome turnover or transitions in sex determination mechanisms in speciation using methods such as BiSSE requires more complete datasets than we have assembled here for vertebrates (FitzJohn et al., 2009). A few studies have been conducted in plants; however, no consistent association was found between sexual system and diversification rates (Leslie et al., 2013; McDaniel et al., 2013; Villareal & Renner, 2013; Sabath, Goldberg et al., 2016). Thus, the relative importance of sex chromosome turnover or transitions in sex determination mechanisms to the process of speciation is mostly unclear. Additional studies to characterize the diversity of sex determination mechanisms that we have highlighted here in vertebrates, as well as the diversity present in plants and invertebrates (The Tree of Sex Consortium, 2014), will provide a rich resource for future research to address this question.
Supplementary Material
Acknowledgments
We thank all members of the Tree of Sex Consortium, particularly Jun Kitano, Nicolas Perrin and Nicole Valenzuela for collection of the vertebrate dataset and Sally Otto for discussion and comments on the manuscript. The Tree of Sex Consortium was funded by the National Evolutionary Synthesis Center (NESCent) through a US National Science Foundation grant (EF-0905606). M.W.P. was funded by a NSERC Postdoctoral Fellowship, an Izaak Killam Memorial Postdoctoral Fellowship, and an NSERC Discovery Grant. J.E.M. gratefully acknowledges support from the European Research Council (grant agreements 260233 and 680951) and a Royal Society Wolfson Merit Award. C.L.P. was funded by a US National Institutes of Health grant (R01GM116853).
Footnotes
Author Contributions M.W.P., J.E.M. and C.L.P. designed the study; M.W.P. conducted the analysis; M.W.P., J.E.M. and C.L.P. wrote the paper.
Data Accessibility All trait and phylogenetic data used in this analysis has been previously published. Scripts to clean data, run analyses, and produce figures are available on GitHub (https://github.com/mwpennell/vert_trans).
References
- Bachtrog D. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nature Reviews Genetics. 2013;14:113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Kirkpatrick M, Mank JE, McDaniel SF, Pires JC, Rice WR, Valenzuela N. Are all sex chromosomes created equal? Trends in Genetics. 2011;27:350–357. doi: 10.1016/j.tig.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman TL, Hahn MW, Kitano J, Mayrose I, Ming R, Perrin N, Ross L, Valenzuela N, Vamosi JC. Sex determination: why so many ways of doing it? PLoS Biology. 2014;12:e1001899. doi: 10.1371/journal.pbio.1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, O’Meara BC, Donoghue MJ. Identifying hidden rate changes in the evolution of a binary morphological character: the evolution of plant habitat in Campanulid angiosperms. Systematic Biology. 2013;62:725–737. doi: 10.1093/sysbio/syt034. [DOI] [PubMed] [Google Scholar]
- Barrett SCH. The evolution of plant reproductive systems: how often are transitions irreversible? Proceedings of the Royal Society B: Biological Sciences. 2013;280:20130913. doi: 10.1098/rspb.2013.0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, Koutseva N, Zaghul S, Graves T, Rock S, Kremitzki C, Fulton RS, Dugan S, Ding Y, Morton D, Khan Z, Lewis L, Buhay C, Wang Q, Watt J, Holder M, Lee S, Nazareth L, Rozen S, Muzny DM, Warren WC, Gibbs RA, Wilson RK, Page DC. Mammalian Y chromosomes retain widely-expressed dosage-sensitive regulators. Nature. 2014;508:494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Skaletsky H, Cho TJ, Brown L, Locke D, Chen N, Galkina S, Pyntikova T, Koutseva N, Graves T, Kremitzki C, Warren WC, Clark AG, Gaginskaya E, Wilson RK, Page DC. Avian W and mammalian Y chromosomes convergently retained dosage-sensitive regulators. Nature Genetics. 2017;49:387–394. doi: 10.1038/ng.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukeboom L, Perrin N. The Evolution of Sex Determination. Oxford, UK: Oxford University Press; 2014. [Google Scholar]
- Blackmon H, Demuth JP. Estimating tempo and mode of Y chromosome turnover: explaining Y chromosome loss with the fragile Y hypothesis. Genetics. 2014;197:561–572. doi: 10.1534/genetics.114.164269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser O, Grossen C, Neuenschwander S, Perrin N. Sex chromosome turnovers induced by deleterious mutation load. Evolution. 2013;67:635–645. doi: 10.1111/j.1558-5646.2012.01810.x. [DOI] [PubMed] [Google Scholar]
- Blaser O, Neuenschwander S, Perrin N. Sex chromosome turnovers: The hot-potato model. American Naturalist. 2014;183:140–146. doi: 10.1086/674026. [DOI] [PubMed] [Google Scholar]
- Bull JJ. Evolution of Sex Determining Mechanisms. Menlo Park, CA: Benjamin Cummings; 1983. [Google Scholar]
- Bull JJ, Bulmer MG. Longevity enhances selection of environmental sex determination. Heredity. 1989;63:315–320. doi: 10.1038/hdy.1989.104. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Charnov EL. Changes in the heterogametic mechanism of sex determination. Heredity. 1977;39:1–14. doi: 10.1038/hdy.1977.38. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Charnov EL. On irreversible evolution. Evolution. 1985;39:1149–1155. doi: 10.1111/j.1558-5646.1985.tb00455.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Charnov EL, Bull JJ. When is sex environmentally determined? Nature. 1977;266:828–830. doi: 10.1038/266828a0. [DOI] [PubMed] [Google Scholar]
- Conover DO, Heins SW. Adaptive variation in environmental and genetic sex determination in a fish. Nature. 1987;326:496–498. doi: 10.1038/326496a0. [DOI] [PubMed] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grützner F, Kaessmann H. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014;508:488–493. doi: 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Two rules of speciation. In: Otte D, Endler J, editors. Speciation and Its Consequences. Sunderland, MA: Sinauer Associates; 1989. pp. 180–207. [Google Scholar]
- Demuth J. Do sex chromosomes affect speciation rate? Bio Essays. 2014;36:632. doi: 10.1002/bies.201400049. [DOI] [PubMed] [Google Scholar]
- Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- Drummond AJ, Suchard MA. Bayesian random local clocks, or one rate to rule them all. BMC Biology. 2010;8:114. doi: 10.1186/1741-7007-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman JM, Harmon LJ, Tank DC. Congruification: support for time scaling large phylogenetic trees. Methods in Ecology and Evolution. 2013;4:688–691. [Google Scholar]
- Eppley SM, Jesson LK. Moving to mate: the evolution of separate and combined sexes in multicellular organisms. Journal of Evolutionary Biology. 2008;21:727–736. doi: 10.1111/j.1420-9101.2008.01524.x. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Pyron RA, Wiens JJ. Polyploidization and sex chromosome evolution in amphibians. In: Soltis PS, Soltis DE, editors. Polyploidy and Genome Evolution. Berlin: Springer Verlag; 2012. pp. 385–410. [Google Scholar]
- Fisher RA. The Genetical Theory of Natural Selection. Oxford, UK: Oxford University Press; 1930. [Google Scholar]
- FitzJohn RG, Maddison WP, Otto SP. Estimating trait-dependent speciation and extinction from incompletely resolved phylogenies. Systematic Biology. 2009;58:595–611. doi: 10.1093/sysbio/syp067. [DOI] [PubMed] [Google Scholar]
- FitzJohn RG. Diversitree: comparative phylogenetic analyses of diversification in R. Methods in Ecology and Evolution. 2012;3:1084–1092. [Google Scholar]
- FitzJohn RG, Pennell MW, Zanne AE, Stevens PF, Tank DC, Cornwell WK. How much of the world is woody? Journal of Ecology. 2014;102:1266–1272. [Google Scholar]
- Gamble T, Coryell J, Ezaz T, Lynch J, Scantlebury DP, Zarkower D. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Molecular Biology and Evolution. 2015;32:1296–1309. doi: 10.1093/molbev/msv023. [DOI] [PubMed] [Google Scholar]
- Gamble T. Using RAD-seq to recognize sex-specific markers and sex chromosome systems. Molecular Ecology. 2016;25:2114–2116. doi: 10.1111/mec.13648. [DOI] [PubMed] [Google Scholar]
- Gamble T, Castoe TA, Nielson SV, Banks JL, Card DC, Schield DR, Schuett GW, Booth W. The discovery of XY sex chromosomes in a Boa and Python. Current Biology. 2017;27:1–6. doi: 10.1016/j.cub.2017.06.010. [DOI] [PubMed] [Google Scholar]
- Ghiselin MT. The evolution of hermaphroditism among animals. Quarterly Review of Biology. 1969;44:189–208. doi: 10.1086/406066. [DOI] [PubMed] [Google Scholar]
- Goldberg EE, Otto SP, Vamosi JC, Mayrose I, Sabath N, Ming R, Ashman TL. Macroevolutionary synthesis of flowering plant sexual systems. Evolution. 2017;71:898–912. doi: 10.1111/evo.13181. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. Sex ratio and unisexual sterility in animal hybrids. Journal of Genetics. 1922;12:101–109. [Google Scholar]
- Hillis DM, Green DM. Evolutionary changes of heterogametic sex in the phylogenetic history of amphibians. Journal of Evolutionary Biology. 1990;3:49–64. [Google Scholar]
- Jaenike J. Sex chromosome meiotic drive. Annual Review of Ecology and Systematics. 2001;32:25–49. [Google Scholar]
- Jarne P, Auld JR. Animals mix it up too: the distribution of self fertilization among hermaphroditic animals. Evolution. 2006;60:1816–1824. doi: 10.1554/06-246.1. [DOI] [PubMed] [Google Scholar]
- Jensen MP, Allen CD, Eguchi T, Bell IP, LaCasella EL, Hilton WA, Hof CAM, Dutton PH. Environmental warming and feminization of one of the largest sea turtle populations in the world. Current Biology. 2018;28:154–159. doi: 10.1016/j.cub.2017.11.057. [DOI] [PubMed] [Google Scholar]
- Kallman KD. A new look at sex determination in poeciliid fishes. In: Turner BJ, editor. Evolutionary Genetics of Fishes. New York, NY: Plenum Press; 1984. pp. 95–171. [Google Scholar]
- Kamiya T, Kai W, Tasumi S, Oka A, Matsunaga T, Mizuno N, Fujita M, Suetake H, Suzuki S, Hosoya S, Tohari S, Brenner S, Miyadai T, Venkatesh B, Suzuki Y, Kikuchi K. A trans-specific missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu) PLOS Genetics. 2012;8:e1002798. doi: 10.1371/journal.pgen.1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käfer J, de Boer HJ, Mousset S, Kool A, Dufay M, Marais GA. Dioecy is associated with higher diversification rates in flowering plants. Journal of Evolutionary Biology. 2014;27:1478–1490. doi: 10.1111/jeb.12385. [DOI] [PubMed] [Google Scholar]
- Käfer J, Marais GA, Pannell JR. On the rarity of dioecy in flowering plants. Molecular Ecology. 2017;26:1225–1241. doi: 10.1111/mec.14020. [DOI] [PubMed] [Google Scholar]
- King B, Lee MSY. Ancestral state reconstruction, rate heterogeneity, and the evolution of reptile viviparity. Systematic Biology. 2015;64:532–544. doi: 10.1093/sysbio/syv005. [DOI] [PubMed] [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, Absher DM, Grimwood J, Schmutz J, Myers RM, Kingsley DM, Peichel CL. A role for a neo-sex chromosome in stickleback speciation. Nature. 2009;461:1079–1083. doi: 10.1038/nature08441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koubová M, Johnson Pokorná M, Rovatsos M, Farkačová K, Altmanová M, Kratochvíl L. Sex determination in Madagascar geckos of the genus Paroedura (Squamata: Gekkonidae); are differentiated sex chromosomes indeed so evolutionary stable? Chromosome Research. 2014;22:441–452. doi: 10.1007/s10577-014-9430-z. [DOI] [PubMed] [Google Scholar]
- Leslie AB, Beaulieu JM, Crane PR, Donoghue MJ. Explaining the distribution of breeding and dispersal systems in conifers. Proceedings of the Royal Society B: Biological Sciences. 2013;280:20131812. doi: 10.1098/rspb.2013.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima TG. Higher levels of sex chromosome heteromorphy are associated with markedly stronger reproductive isolation. Nature Communications. 2014;5:4743. doi: 10.1038/ncomms5743. [DOI] [PubMed] [Google Scholar]
- Maddison WP. Confounding asymmetries in evolutionary diversification and character change. Evolution. 2006;60:1743–1746. [PubMed] [Google Scholar]
- Maddison WP, Midford PE, Otto SP. Estimating a binary character’s effect on speciation and extinction. Systematic Biology. 2007;56:701–710. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Leduc-Robert G. Multiple origins of sex chromosome fusions correlated with chiasma localization in Habronattus jumping spiders (Araneae: Salticidae) Evolution. 2013;67:2258–2272. doi: 10.1111/evo.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, FitzJohn RG. The unsolved challenge to phylogenetic correlation tests for categorical characters. Systematic Biology. 2015;64:127–136. doi: 10.1093/sysbio/syu070. [DOI] [PubMed] [Google Scholar]
- Mank JE, Promislow DEL, Avise JC. Evolution of alternative sex-determining mechanisms in teleost fishes. Biological Journal of the Linnean Society. 2006;87:83–93. [Google Scholar]
- Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, Agata K, Matsuda Y. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and stepwise differentiation of snake sex chromosomes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18190–18195. doi: 10.1073/pnas.0605274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel SF, Atwood J, Burleigh JG. Recurrent evolution of dioecy in bryophytes. Evolution. 2013;67:567–572. doi: 10.1111/j.1558-5646.2012.01808.x. [DOI] [PubMed] [Google Scholar]
- Moore EC, Roberts RB. Polygenic sex determination. Current Biology. 2013;23:R510–R512. doi: 10.1016/j.cub.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Muyle A, Käfer J, Zemp N, Mousset S, Picard F, Marais GAB. SEX-DETector: a probabilistic approach to study sex chromosomes in non-model organisms. Genome Biology and Evolution. 2016;8:2530–2543. doi: 10.1093/gbe/evw172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hasegawa Y, Ohtani H, Mineyama M, Miura I. The ZZ/ZW sex-determining mechanism originated twice and independently during evolution of the frog, Rana rugosa. Heredity. 2008;100:92–99. doi: 10.1038/sj.hdy.6801068. [DOI] [PubMed] [Google Scholar]
- Organ CL, Janes DE. Evolution of sex chromosomes in Sauropsida. Integrative and Comparative Biology. 2008;48:512–519. doi: 10.1093/icb/icn041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proceedings of the Royal Society B: Biological Sciences. 1994;255:37–45. [Google Scholar]
- Pen I, Uller T, Feldmeyer B, Harts A, While GM, Wapstra E. Climate-driven population divergence in sex-determining systems. Nature. 2010;468:436–438. doi: 10.1038/nature09512. [DOI] [PubMed] [Google Scholar]
- Pennell MW, FitzJohn RG, Cornwell WK. A simple approach for maximizing the overlap of phylogenetic and comparative data. Methods in Ecology and Evolution. 2016;7:751–758. doi: 10.1111/2041-210X.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips BC, Edmands S. Does the speciation clock tick more slowly in the absence of heteromorphic sex chromosomes? BioEssays. 2012;34:166–169. doi: 10.1002/bies.201100164. [DOI] [PubMed] [Google Scholar]
- Pokorná M, Kratochvíl L. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zoological Journal of the Linnean Society. 2009;156:168–183. [Google Scholar]
- Pokorná M, Rens W, Rovatsos M, Kratochvíl L. A ZZ/ZW sex chromosome system in the thick-tailed gecko (Underwoodisaurus milii; Squamata: Gekkota: Carphodactylidae), a member of the ancient gecko lineage. Cytogenetic and Genome Research. 2014;142:190–196. doi: 10.1159/000358847. [DOI] [PubMed] [Google Scholar]
- Pokorná M, Rovatsos M, Kratochvíl L. Sex chromosomes and karyotype of the (nearly) mythical creature, the Gila Monster, Heloderma suspectum (Squamata: Helodermatidae) PLoS One. 2014;9:e104716. doi: 10.1371/journal.pone.0104716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends in Genetics. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyron RA, Wiens JJ. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Molecular Phylogenetics and Evolution. 2011;61:543–583. doi: 10.1016/j.ympev.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Pyron RA, Burbrink FT, Wiens JJ. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evolutionary Biology. 2013;13:93. doi: 10.1186/1471-2148-13-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyron RA, Burbrink FT. Early origin of viviparity and multiple reversions to oviparity in squamate reptiles. Ecology Letters. 2014;17:13–21. doi: 10.1111/ele.12168. [DOI] [PubMed] [Google Scholar]
- Rabosky DL. No substitute for real data: A cautionary note on the use of phylogenies from birth-death polytomy resolvers for downstream comparative analyses. Evolution. 2015;69:3207–3216. doi: 10.1111/evo.12817. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, Goldberg EE. Model inadequacy and mistaken inferences of trait-dependent speciation. Systematic Biology. 2015;64:340–355. doi: 10.1093/sysbio/syu131. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, Santini F, Eastman J, Smith SA, Sidlauskas B, Chang J, Alfaro ME. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nature Communications. 2013;4:1958. doi: 10.1038/ncomms2958. [DOI] [PubMed] [Google Scholar]
- Read AF, Nee S. Inference from binary comparative data. Journal of Theoretical Biology. 1995;173:99–108. [Google Scholar]
- Renner SS. The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. American Journal of Botany. 2014;101:1588–1596. doi: 10.3732/ajb.1400196. [DOI] [PubMed] [Google Scholar]
- Renner SS, Ricklefs RE. Dioecy and its correlates in the flowering plants. American Journal of Botany. 1995;82:596–606. [Google Scholar]
- Rice WR. On the instability of polygenic sex determination: the effect of sex-specific selection. Evolution. 1986;40:633–639. doi: 10.1111/j.1558-5646.1986.tb00514.x. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends in Ecology and Evolution. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Ross JA, Peichel CL. Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback fish. Genetics. 2008;179:2173–2182. doi: 10.1534/genetics.108.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Altmanová M, Pokorná MJ, Kratochvíl L. Cretaceous park of sex determination: sex chromosomes are conserved across iguanas. Biology Letters. 2014;10:20131093. doi: 10.1098/rsbl.2013.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Pokorná MJ, Altmanová M, Kratochvíl L. Female heterogamety in Madagascar chameleons (Squamata: Chamaeleonidae: Furcifer): differentiation of sex and neo-nex chromosomes. Scientific Reports. 2015;5:13196. doi: 10.1038/srep13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Vukić J, Altmanová M, Pokorná MJ, Moravec J, Kratochvíl L. Conservation of sex chromosomes in lacertid lizards. Molecular Ecology. 2016;25:3120–3126. doi: 10.1111/mec.13635. [DOI] [PubMed] [Google Scholar]
- Sabath N, Goldberg EE, Glick L, Einhorn M, Ashman TL, Ming R, Otto SP, Vamosi JC, Mayrose I. Dioecy does not consistently accelerate or slow lineage diversification across multiple genera of angiosperms. New Phytologist. 2016;209:1223–1235. doi: 10.1111/nph.13696. [DOI] [PubMed] [Google Scholar]
- Sabath N, Itescu Y, Feldman A, Meiri S, Mayrose I, Valenzuela N. Sex determination, longevity, and the birth and death of reptilian species. Ecology and Evolution. 2016;6:5207–5220. doi: 10.1002/ece3.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ser JR, Roberts RB, Kocher TD. Multiple interacting loci control sex determination in Lake Malawi cichlid fish. Evolution. 2010;64:486–501. doi: 10.1111/j.1558-5646.2009.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL. The evolution of hermaphroditism in fishes. In: Reinboth R, editor. Intersexuality in the Animal Kingdom. Berlin: Springer; 1975. pp. 295–310. [Google Scholar]
- The Tree of Sex Consortium. Ashman T-L, Bachtrog D, Blackmon H, Goldberg EE, Hahn MW, Kirkpatrick M, Kitano J, Mank JE, Mayrose I, Ming R, Otto SP, Peichel CL, Pennell MW, Perrin N, Ross L, Valenzuela N, Vamosi JC. Tree of Sex: a database of sexual systems. Nature Scientific Data. 2014;1:140015. doi: 10.1038/sdata.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Begun DJ. Haldane’s rule and sex chromosome size in Drosophila. Genetics. 1997;147:1799–1815. doi: 10.1093/genetics/147.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda JC, Zenil-Ferguson R, Pennell MW. Rethinking phylogenetic comparative methods. 2017 doi: 10.1101/222729. [DOI] [PubMed]
- Valenzuela N, Lance VA. Temperature Dependent Sex Determination in Vertebrates. Washington, DC: Smithsonian Books; 2004. [Google Scholar]
- van Doorn GS, Kirkpatrick M. Turnover of sex chromosomes induced by sexual conflict. Nature. 2007;449:909–912. doi: 10.1038/nature06178. [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics. 2010;186:629–645. doi: 10.1534/genetics.110.118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veller C, Muraldihar P, Constable GWA, Nowak MA. Drift-induced selection between male and female heterogamety. Genetics. 2017;207:711–727. doi: 10.1534/genetics.117.300151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature. 2013;499:332–335. doi: 10.1038/nature12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biology. 2013;11:e1001643. doi: 10.1371/journal.pbio.1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villareal JC, Renner SS. Correlates of monoicy and dioecy in hornworts, the apparent sister group to vascular plants. BMC Evolutionary Biology. 2013;13:239. doi: 10.1186/1471-2148-13-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner DA, Shine R. The adaptive significance of temperature-dependent sex-determination in a reptile. Nature. 2008;451:566–568. doi: 10.1038/nature06519. [DOI] [PubMed] [Google Scholar]
- Werren JH, Beukeboom LW. Sex determination, sex ratios, and genetic conflict. Annual Review of Ecology and Systematics. 1998;29:233–261. [Google Scholar]
- Wright AE, Dean R, Zimmer F, Mank JE. How to make a sex chromosome. Nature Communications. 2016;7:12087. doi: 10.1038/ncomms12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhang J, Bachtrog D, An N, Huang Q, Jarvis ED, Gilbert MTP, Zhang G. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science. 2014;346:1246338. doi: 10.1126/science.1246338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.