Abstract
Antibiotic resistance is a serious threat to public health. Significant efforts are currently directed toward containment of the spread of resistance, finding new therapeutic options concerning resistant human and animal pathogens, and addressing the gaps in the fundamental understanding of mechanisms of resistance. Experimental data and kinetic modeling revealed a major factor in resistance, the synergy between active efflux and the low permeability barrier of the outer membrane, which dramatically reduces the intracellular accumulation of many antibiotics. The structural and mechanistic particularities of trans-envelope efflux pumps amplify the effectiveness of cell envelopes as permeability barriers. An important feature of this synergism is that efflux pumps and the outer membrane barriers are mechanistically independent and select antibiotics based on different physicochemical properties. The synergism amplifies even weak polyspecificity of multidrug efflux pumps and creates a major hurdle in the discovery and development of new therapeutics against Gram-negative pathogens.
Keywords: Permeability barrier, Hyperporination, Kinetic model, Active drug efflux, Substrate specificity
1. Trans-envelope efflux complexes
Trans-envelope (spanning the two membranes of Gram-negative bacteria) multidrug efflux pumps of Gram-negative bacteria have been in the focus of intense investigation as the major contributors to antibiotic resistance in Gram-negative bacteria, as well as potential targets for inhibition. In the past ten years, we have witnessed an unprecedented advance in structural and functional understanding of efflux pump components and their assemblies. On one hand, these pumps share trans-envelope properties and features. On the other hand, they impress by their structural and mechanistic diversity.
All trans-envelope multidrug efflux pumps share a three-component composition and comprise an active transporter located in the inner membrane (IM), a periplasmic membrane fusion protein (MFP) and an outer membrane channel/factor (OMF) [1–3]. Transporters in efflux pumps may belong to one of the three superfamilies of proteins: RND, ABC or MF, and vary dramatically in membrane topology, oligomeric state and biochemical mechanisms. ABC-type transporters utilize the energy of ATP hydrolysis, whereas RND and MF are secondary transporters driven by the proton-motive force. RND transporters are homo- or heterotrimers that function through a conformational rotation mechanism [4–9]. ABC transporters are functional dimers (intra- or inter-peptides) that enable a conformational switch mechanism [10–14]. Finally, MF transporters are functional monomers that transport substrates by an alternating access mechanism [15–18].
These structurally and mechanistically diverse transporters engage MFPs that share certain structural features: i) an elongated asymmetric shape of a protomer; ii) 3–4 domains that are linearly arranged, creating flexible structures; and iii) oligomerization [19–24]. The functional unit of MFPs is a dimer, in which each protomer engages a specific interface with a transporter and an OMF [25, 26]. Trimerization of MFP dimers leads to formation of inverted funnel-like structures that provide a path for various substrates to cross the periplasmic space [27, 28]. A significant body of evidence supports a notion that MFPs interact with substrates of efflux pumps and enable allosteric regulation of efflux. How this regulation is achieved in trans-envelope complexes of different stoichiometries remains unclear. However, it appears to depend on the structure and mechanism of the transporter [29–33]. This regulation of efflux is further integrated with the engagement and opening of OMFs.
In the presence of substrates, MFPs bridge efflux transporters with structurally conserved OMFs and assemble a tightly sealed trans-envelope protein complex that prevents escape of various substrates into the periplasm and transports them across the outer membrane and out of the cell [27, 28]. Despite significant sequence variability, OMFs are structurally conserved [34–37]. The periplasmic domains of these proteins extend into the periplasm to meet MFPs and to create a protein conduit for substrates to diffuse from the periplasmic binding sites in transporters and MFPs, all the way across the periplasm and the outer membrane. The engagement of OMFs into complexes is likely driven by affinities of MFPs to OMFs and their propensity to oligomerization [25, 27, 30, 38, 39]. Importantly, each protomer in the MFP dimer binds to a specific binding site on OMF, and the two binding sites are not functionally equivalent [26, 31]. Apparently, one MFP subunit grasps OMF, whereas another is responsible for opening of the channel [40, 41]. Opening and closing of the channel could also be part of the trans-envelope transport mechanism that does not require disengagement of the complex [30]. Interestingly, no experimental evidence exists for the diffusion of substrates through the channel. The strongest evidence for interactions between OMFs and substrates is the interaction between TolC and hemolysin, which assist this protein toxin in folding during secretion [42].
The above summarized structural, computational and functional advances identified similarities and differences in the structure and mechanism of trans-envelope efflux complexes. These studies further refined the model of trans-envelope drug transport and brought us closer to understanding how transport reactions separated into two different membranes are coupled together.
2. Synergy between efflux pumps and the permeability barrier of the outer membrane
Trans-envelope efflux pumps of Gram-negative bacteria function in the context of two membranes that differ dramatically in their structures and compositions [43]. The outer membrane is an asymmetric bilayer composed of lipopolysaccharides (LPS) in the outer leaflet and glycerophospholipids (PL) in the inner leaflet [44]. The major features of the LPS structure, such as the presence of lipid A, core and O-antigen chains, are conserved among various species, while specific chemical structures vary broadly. The different LPSs aggregate into species-specific LPS-PL bilayers with a variable number of LPS molecules, thicknesses, surface charge distributions and dynamics [45]. These features, in turn, translate into variations of the permeability properties of LPS-PL bilayers [46–48]. The asymmetric structure of the outer membrane bilayer creates a formidable barrier for permeation of most amphiphilic drugs. Therefore, the presence and the size of general or specific porins largely define the permeability properties of outer membranes in Gram-negative bacteria.
In contrast, the inner membrane composed of PLs is relatively permeable for most amphiphilic drug molecules. Despite this leakiness, the inner membrane provides a major contribution to bacterial defenses against drugs by housing a variety of multidrug transporters. The role of multidrug efflux pumps in binding and expulsion of drugs from the periplasm and across the outer membrane is critical for protection of the intracellular drug targets. Together, the slow influx of antibiotics across the outer membrane and the specificities and efficiencies of efflux pumps define the intracellular steady-state concentrations of antibiotics and the observed differences in antibiotic susceptibilities between different Gram-negative bacteria [49–51].
The exceptional efficiency of multidrug efflux pumps is the result of a complex interplay between the two opposing fluxes of drugs across the two membranes [50, 52, 53]. Recent mathematical modeling provided novel insights into the synergistic character of active drug efflux and the low permeability barrier of the outer membrane in Gram-negative bacteria, and revealed the need for a new kinetic formalism [50, 51]. The drug uptake patterns are non-linear, contain bifurcations and do not conform to Fick’s Law of diffusion or Michaelis-Menten kinetics, although both are integrated into the model [50]. The behavior of the system can be described in terms of two kinetic parameters, the efflux constant KE and the barrier constant B, which relate the rates of active and diffusional drug efflux at, respectively, low and high drug concentrations, and therefore, describe different aspects of drug uptake [50] (Fig. 1).
Fig. 1.
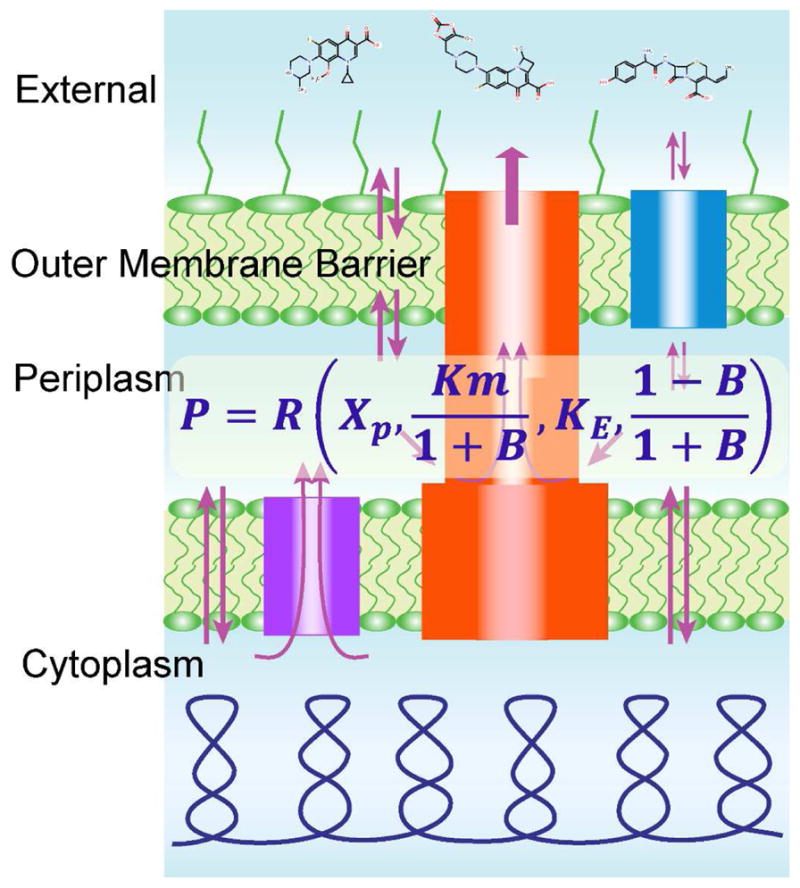
Schematic representation of drug fluxes across the two-membrane cell envelopes of Gram-negative bacteria and the mathematical model that describes this permeability barrier. Modified from [50].
The efflux constant reports by how much the intracellular concentration of the drug is reduced compared to thermodynamic equilibrium, under conditions during which the transporter operates below saturation. Even in E. coli, which is relatively susceptible to antibiotics, this number proved to be remarkably large and was estimated at 420 for the fluorescent drug Hoechst 33342. The barrier constant compares maximal attainable drug fluxes across the outer membrane and via the transporter, and thereby describes what happens to the system at high drug concentrations. The magnitude of B discriminates conditions of efficient and inefficient efflux. When B is smaller than 1, the transporter becomes saturated at high external concentrations of the drug; this cannot happen when B is greater than 1. At the latter condition, B > 1, efflux pumps operate below saturation even at high extracellular concentrations of substrates owing to their slow penetration across the outer membrane. Against such compounds, efflux pumps are very efficient, even though they could be poor substrates in biochemical terms.
Curiously, this kinetic analysis predicts that drug efflux does not provide equal protection against all compounds. Instead, its effect depends on the potency of the drug. In general, more toxic compounds are better protected against than compounds with high inhibition constants. Moreover, even small variations in the efficiency of the barrier can have dramatic effects on antibiotic susceptibility [50].
Further kinetic and experimental analyses of active efflux in the context of two-membrane barriers demonstrated that changes in drug uptake and antibacterial activities in cells compromised in both efflux and the outer membrane often exceed the sum of changes caused by each of the two factors alone [49, 50, 54]. These results suggested that active efflux and the outer membrane barrier act in synergy with each other. Indeed, should these two factors be independent, then the changes in rates of uptake and drug susceptibilities caused by the combination of the two effects should equal the product of the increases caused by each factor alone. Importantly, the synergistic interaction between active efflux and the outer membrane conveys new properties to the cell. In certain cases, when B is greater than one, the cell envelope effectively blocks access to the cell for a variety of external chemicals. This occurs even though, at a molecular level, the membrane itself might remain permeable to these compounds. Thus, the collective behavior of the individual components gives rise to a novel quality.
3. Experimental tools to analyze synergistic permeability barriers
To understand this complex interplay between active efflux and the outer membrane barrier, one must be able to analyze the contributions of both factors to the intracellular accumulation and activities of antibiotics. Two complementary approaches have emerged to analyze the contribution of the outer membrane. The first approach, the titrable outer membrane permeability assay system (TOMAS), enables analyses of drug permeation through specific porins [55]. For this purpose, Eschericia coli cells deficient in general porins are modified to express a porin of interest, for example OprD, an amino-acid-specific porin from P. aeruginosa. If a porin is essential for permeation of an antibiotic, one can expect that its expression in the outer membrane of E. coli will increase the influx and reduce the B constant for this antibiotic. Therefore, cell susceptibility to this antibiotic will be strongly affected by the amounts of the specific porin present in the outer membrane.
The second approach, controlled hyperporination (HYPE), manipulates the permeability barrier of the outer membrane by expressing a large, ~ 2.4 nm in diameter pore, which effectively and non-selectively reduces the B-factor and allows rapid influx of even very large antibiotics such as vancomycin into the cell [54]. The controlled hyperporination approach was applied to several Gram-negative species, including E. coli, P. aeruginosa, Acinetobacter baumannii and Burkholderia spp., and enabled the comparison of the outer membrane barriers that differ not only in the composition of porins, but also in the structure and composition of LPS [49, 54]. In all these species, hyperporination of the outer membrane leads to a significant increase in compound uptake across the outer membrane and, as a result, an increase in their periplasmic concentrations at the site of recognition and binding by efflux pumps. Importantly, hyperporination of cells does not inactivate efflux pumps. Furthermore, it enables kinetic analyses of drug efflux at saturating concentrations of substrates, and hence provides insight into the substrate specificity and efficiency of transporters.
The barrier and efflux constants can also be controlled by changes in the amounts or activities of efflux pumps. It is well established that inactivation of efflux pumps sensitizes bacterial cells to antibacterial activities of antibiotics and increases their intracellular concentrations [56–58]. Accordingly, overproduction of efflux pumps reduces antibiotic susceptibilities of bacterial cells. However, a simple task of varying the amounts of a transporter is not so simple after all. In E. coli and other enterobacteria, a single OMF, TolC, is required for activities of all trans-envelope pumps, and its inactivation largely eliminates efflux across the outer membrane [34]. However, TolC is involved in additional physiological functions, and comparison of TolC-deficient E. coli cells and cells lacking nine TolC-dependent transporters shows some notable differences in antibiotic activities and accumulation [54]. These differences primarily affect antibiotics that use the TolC channel to permeate the outer membrane. The situation is even more complicated in P. aeruginosa, where efflux pumps are co-expressed and assembled with specific OMFs [3]. Strains with multiple efflux pump knockouts ensure the lack of active efflux and, at the same time, decrease the influx of antibiotics across the outer membrane [49]. Combining efflux pump inactivation and hyperporination of the outer membrane with controlled expression of a single pump of interest allows evaluation of kinetic properties of the pump and its substrate specificity [59].
An additional, often overlooked, aspect of the synergy between the outer membrane barrier and efflux is the dilution effect due to bacterial growth and change in cell volume [51]. To some extent, this dilution behaves as an efflux. Antibiotics, for which diffusion across the outer membrane is slower than bacterial growth, are effectively “expelled” from the cells, even in the absence of active efflux pumps.
4. Substrate specificities of multidrug efflux pumps
The existing heuristics emphasize the size and polarity of compounds as important determinants of permeation across the outer membrane and the propensity to be recognized by efflux pumps [60–62]. In general, very polar and low MW compounds and zwitterionic and high MW compounds are thought to avoid efflux, whereas hydrophobicity of compounds positively correlates with the presence of active efflux [61–63]. However, most of these conclusions are based on the comparison of antibacterial activities in efflux-proficient and efflux-deficient cells with intact outer membranes. As a result, the outer membrane barrier defines which antibiotics can reach the periplasm and become accessible to efflux pumps and which antibiotics accumulate either above or below transporter saturation levels. The HYPE approach removes these limitations and enables characterization of efflux pump specificities.
Analyses of antibacterial activities of various compounds in hyperporinated cells demonstrated that substrate specificities and efficiencies of the RND-type efflux pumps vary dramatically among Gram-negative species [49]. Importantly, RND pumps universally affect activities of all tested antibiotics, even those considered to be outside of the efflux recognition space [62, 64]. For example, the activity of zeocin a glycopeptide antibiotic with the mass of 1428 Da, is strongly affected by active efflux in Gram-negative bacteria [49]. This result demonstrates that the size of a compound is not a predictive characteristic of an efflux substrate. In B. thailandensis, but not in E. coli or P. aeruginosa, active efflux plays an important role in protection against polycationic antibiotics such as aminoglycosides and polymyxin [49, 65, 66]. However, this protection, again, is enabled by synergistic interactions with the outer membrane. In this and other Burkholderia spp., LPS is constitutively modified with 4-amino-4-deoxy-L-arabinose moieties and, as a result, polycationic antibiotics fail to permeabilize the outer membrane [67].
In addition to efflux pumps responsible for intrinsic antibiotic resistance, chromosomes of most Gram-negative bacteria contains multiple operons encoding so called “minor” efflux pumps. The inactivation of these pumps does not lead to changes in bacterial antibiotic resistance [58, 68]. A recent comparison of the two RND-type efflux pumps MexEF-OprN and MexHI-OpmD from P. aeruginosa further highlights the importance of separation of contributions of the outer membrane barrier and active efflux in analyses of substrate specificities of “minor” efflux pumps [59]. In a laboratory PAO1 strain, these two pumps are expressed at very low levels and do not contribute to the intrinsic antibiotic resistance of P. aeruginosa. Although both pumps, when overexpressed, provide clinical levels of fluoroquinolone resistance, only MexEF-OprN is selected in clinical multidrug-resistant isolates [69, 70]. Furthermore, when the two overexpressed pumps are compared in their abilities to protect against 15 structurally different fluoroquinolones, the two pumps generate almost identical 64–128 fold changes in MICs [59]. However, this apparent lack of structural selectivity disappears in hyperporinated cells. MexEF-OprN remains effective against most fluoroquinolones, but a certain substrate selectivity emerges, whereas MexHI-OpmD fails with most fluoroquinolones. Further analyses identified phenazines, such as an endogenous pigment pyocyanin, as specific substrates of MexHI-OpmD [59]. A side-by-side comparison of the best substrates of the two pumps highlights their structural preferences toward substrates (Fig. 2). MexHI-OpmD is selective for rigid tricyclic phenazines, whereas MexEF-OprN prefers small flexible structures such as those of chloramphenicol and triclosan. This study also showed that expression of the majority of efflux pumps is low in the wild type strains, but not because their functions are not important under laboratory conditions. These pumps are engaged in specific physiological functions [2, 71], such as control of steady-state concentrations of pyocyanin by MexGHI-OpmD [59]. The low expression levels enforce substrate specificities of efflux pumps, so that only the substrates with high affinities are expelled from the cells.
Fig. 2.
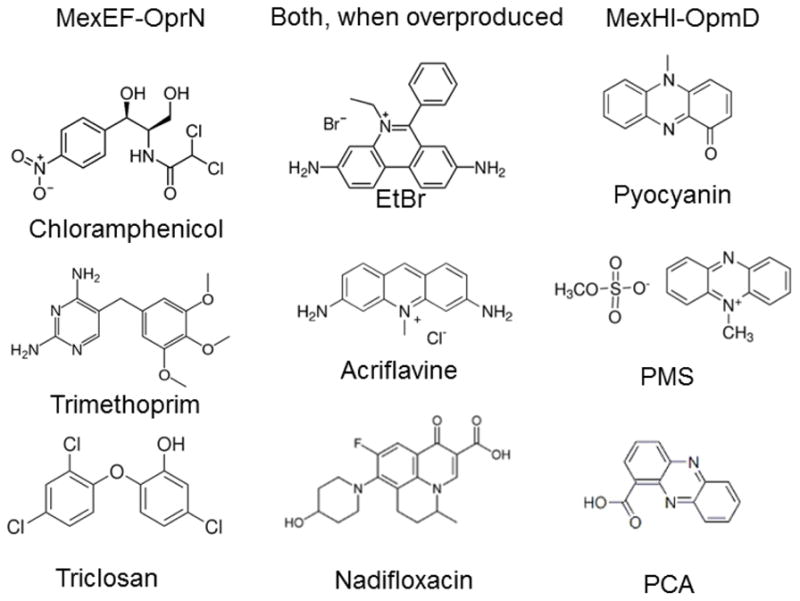
Substrate specificities of MexEF-OprN and MexHI-OpmD from P. aeruginosa. When overproduced, and in the context of the outer membrane permeability barrier, the two pumps have overlapping substrate specificities and provide the same levels of resistance to structurally diverse fluoroquinolones. Their actual substrate specificities are distinct, with MexHI-OpmD selective for phenazines [59]. EtBr, ethidium bromide; PMS, phenazine methosulfate; PCA, phenazine-1-carboxylic acid.
5. Conclusions
Experimental data and kinetic modeling agree that Gram-negative cell envelopes serve to dramatically reduce the intracellular concentration of many antibiotics [50, 51, 54]. The structural and mechanistic particularities of trans-envelope efflux pumps enable the synergistic character and effectiveness of cell envelopes. An important feature of this synergism is that active efflux pumps and the outer membrane barriers are mechanistically independent and select antibiotics based on different physicochemical properties [49]. The synergism amplifies the weak polyspecificity and multidrug characteristics of efflux pumps, creating a major hurdle in the discovery and development of new therapeutics against Gram-negative pathogens [43, 72, 73].
Acknowledgments
Studies in H.I.Z. and V.V.R. labs are sponsored by the Department of the Defense, Defense Threat Reduction Agency and by the NIH grant AI132836. The content of the information does not necessarily reflect the position or the policy of the federal government, and no official endorsement should be inferred.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nikaido H, Zgurskaya HI. Antibiotic efflux mechanisms. Curr Opin Infect Dis. 1999;12:529–36. doi: 10.1097/00001432-199912000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Piddock LJ. Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol. 2006;4:629–36. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 3.Poole K, Srikumar R. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr Top Med Chem. 2001;1:59–71. doi: 10.2174/1568026013395605. [DOI] [PubMed] [Google Scholar]
- 4.Murakami S. Multidrug efflux transporter, AcrB--the pumping mechanism. Curr Opin Struct Biol. 2008;18:459–65. doi: 10.1016/j.sbi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Seeger MA, Diederichs K, Eicher T, Brandstatter L, Schiefner A, Verrey F, et al. The AcrB efflux pump: conformational cycling and peristalsis lead to multidrug resistance. Curr Drug Targets. 2008;9:729–49. doi: 10.2174/138945008785747789. [DOI] [PubMed] [Google Scholar]
- 6.Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–8. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 7.Bolla JR, Su CC, Do SV, Radhakrishnan A, Kumar N, Long F, et al. Crystal structure of the Neisseria gonorrhoeae MtrD inner membrane multidrug efflux pump. PLoS One. 2014;9:e97903. doi: 10.1371/journal.pone.0097903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu EW, Aires JR, Nikaido H. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J Bacteriol. 2003;185:5657–64. doi: 10.1128/JB.185.19.5657-5664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramaswamy VK, Vargiu AV, Malloci G, Dreier J, Ruggerone P. Molecular Rationale behind the Differential Substrate Specificity of Bacterial RND Multi-Drug Transporters. Sci Rep. 2017;7:8075. doi: 10.1038/s41598-017-08747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin HT, Bavro VN, Barrera NP, Frankish HM, Velamakanni S, van Veen HW, et al. MacB ABC transporter is a dimer whose ATPase activity and macrolide-binding capacity are regulated by the membrane fusion protein MacA. J Biol Chem. 2009;284:1145–54. doi: 10.1074/jbc.M806964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modali SD, Zgurskaya HI. The periplasmic membrane proximal domain of MacA acts as a switch in stimulation of ATP hydrolysis by MacB transporter. Mol Microbiol. 2011;81:937–51. doi: 10.1111/j.1365-2958.2011.07744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tikhonova EB, Devroy VK, Lau SY, Zgurskaya HI. Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol Microbiol. 2007;63:895–910. doi: 10.1111/j.1365-2958.2006.05549.x. [DOI] [PubMed] [Google Scholar]
- 13.Balakrishnan L, Venter H, Shilling RA, van Veen HW. Reversible transport by the ATP-binding cassette multidrug export pump LmrA: ATP synthesis at the expense of downhill ethidium uptake. J Biol Chem. 2004;279:11273–80. doi: 10.1074/jbc.M308494200. [DOI] [PubMed] [Google Scholar]
- 14.van Veen HW, Margolles A, Muller M, Higgins CF, Konings WN. The homodimeric ATP-binding cassette transporter LmrA mediates multidrug transport by an alternating two-site (two-cylinder engine) mechanism. EMBO J. 2000;19:2503–14. doi: 10.1093/emboj/19.11.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adler J, Bibi E. Determinants of substrate recognition by the Escherichia coli multidrug transporter MdfA identified on both sides of the membrane. J Biol Chem. 2004;279:8957–65. doi: 10.1074/jbc.M313422200. [DOI] [PubMed] [Google Scholar]
- 16.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–80. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gbaguidi B, Mazurkiewicz P, Konings WN, Driessen AJ, Ruysschaert JM, Vigano C. Proton motive force mediates a reorientation of the cytosolic domains of the multidrug transporter LmrP. Cell Mol Life Sci. 2004;61:2646–57. doi: 10.1007/s00018-004-4298-2. [DOI] [PubMed] [Google Scholar]
- 18.Yardeni EH, Zomot E, Bibi E. The fascinating but mysterious mechanistic aspects of multidrug transport by MdfA from Escherichia coli. Res Microbiol. 2017 doi: 10.1016/j.resmic.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14:577–87. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akama H, Matsuura T, Kashiwagi S, Yoneyama H, Narita S, Tsukihara T, et al. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J Biol Chem. 2004;279:25939–42. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 21.Greene NP, Hinchliffe P, Crow A, Ababou A, Hughes C, Koronakis V. Structure of an atypical periplasmic adaptor from a multidrug efflux pump of the spirochete Borrelia burgdorferi. FEBS Lett. 2013;587:2984–8. doi: 10.1016/j.febslet.2013.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins MK, Bokma E, Koronakis E, Hughes C, Koronakis V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc Natl Acad Sci U S A. 2004;101:9994–9. doi: 10.1073/pnas.0400375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Sim SH, Nam KH, Jin XL, Kim HM, Hwang KY, et al. Crystal structure of the periplasmic region of MacB, a noncanonic ABC transporter. Biochemistry. 2009;48:5218–25. doi: 10.1021/bi900415t. [DOI] [PubMed] [Google Scholar]
- 24.Hinchliffe P, Greene NP, Paterson NG, Crow A, Hughes C, Koronakis V. Structure of the periplasmic adaptor protein from a major facilitator superfamily (MFS) multidrug efflux pump. FEBS Lett. 2014;588:3147–53. doi: 10.1016/j.febslet.2014.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tikhonova EB, Yamada Y, Zgurskaya HI. Sequential mechanism of assembly of multidrug efflux pump AcrAB-TolC. Chem Biol. 2011;18:454–63. doi: 10.1016/j.chembiol.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weeks JW, Nickels LM, Ntreh AT, Zgurskaya HI. Non-equivalent roles of two periplasmic subunits in the function and assembly of triclosan pump TriABC from Pseudomonas aeruginosa. Mol Microbiol. 2015;98:343–56. doi: 10.1111/mmi.13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, et al. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–5. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daury L, Orange F, Taveau JC, Verchere A, Monlezun L, Gounou C, et al. Tripartite assembly of RND multidrug efflux pumps. Nat Commun. 2016;7:10731. doi: 10.1038/ncomms10731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fitzpatrick AWP, Llabres S, Neuberger A, Blaza JN, Bai XC, Okada U, et al. Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump. Nat Microbiol. 2017;2:17070. doi: 10.1038/nmicrobiol.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Fan G, Hryc CF, Blaza JN, Serysheva, Schmid MF, et al. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. Elife. 2017:6. doi: 10.7554/eLife.24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zgurskaya HI, Weeks JW, Ntreh AT, Nickels LM, Wolloscheck D. Mechanism of coupling drug transport reactions located in two different membranes. Front Microbiol. 2015;6:100. doi: 10.3389/fmicb.2015.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janganan TK, Bavro VN, Zhang L, Borges-Walmsley MI, Walmsley AR. Tripartite efflux pumps: energy is required for dissociation, but not assembly or opening of the outer membrane channel of the pump. Mol Microbiol. 2013;88:590–602. doi: 10.1111/mmi.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Symmons MF, Marshall RL, Bavro VN. Architecture and roles of periplasmic adaptor proteins in tripartite efflux assemblies. Front Microbiol. 2015;6:513. doi: 10.3389/fmicb.2015.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zgurskaya HI, Krishnamoorthy G, Ntreh A, Lu S. Mechanism and Function of the Outer Membrane Channel TolC in Multidrug Resistance and Physiology of Enterobacteria. Front Microbiol. 2011;2:189. doi: 10.3389/fmicb.2011.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersen C, Koronakis E, Bokma E, Eswaran J, Humphreys D, Hughes C, et al. Transition to the open state of the TolC periplasmic tunnel entrance. Proc Natl Acad Sci U S A. 2002;99:11103–8. doi: 10.1073/pnas.162039399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–9. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 37.Akama H, Kanemaki M, Yoshimura M, Tsukihara T, Kashiwagi T, Yoneyama H, et al. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J Biol Chem. 2004;279:52816–9. doi: 10.1074/jbc.C400445200. [DOI] [PubMed] [Google Scholar]
- 38.Tikhonova EB, Dastidar V, Rybenkov VV, Zgurskaya HI. Kinetic control of TolC recruitment by multidrug efflux complexes. Proc Natl Acad Sci U S A. 2009;106:16416–21. doi: 10.1073/pnas.0906601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez CA, Travers T, Pos KM, HIZ, Gnanakaran S. Dynamics of Intact MexAB-OprM Efflux Pump: Focusing on the MexA-OprM Interface. Sci Rep. 2017;7:16251. doi: 10.1038/s41598-017-16497-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ntreh AT, Weeks JW, Nickels LM, Zgurskaya HI. Opening the Channel: the Two Functional Interfaces of Pseudomonas aeruginosa OpmH with the Triclosan Efflux Pump TriABC. J Bacteriol. 2016;198:3176–85. doi: 10.1128/JB.00535-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bavro VN, Pietras Z, Furnham N, Perez-Cano L, Fernandez-Recio J, Pei XY, et al. Assembly and channel opening in a bacterial drug efflux machine. Mol Cell. 2008;30:114–21. doi: 10.1016/j.molcel.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vakharia H, German GJ, Misra R. Isolation and characterization of Escherichia coli tolC mutants defective in secreting enzymatically active alpha-hemolysin. J Bacteriol. 2001;183:6908–16. doi: 10.1128/JB.183.23.6908-6916.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zgurskaya HI, Lopez CA, Gnanakaran S. Permeability Barrier of Gram-Negative Cell Envelopes and Approaches To Bypass It. ACS Infect Dis. 2015;1:512–22. doi: 10.1021/acsinfecdis.5b00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henderson JC, Zimmerman SM, Crofts AA, Boll JM, Kuhns LG, Herrera CM, et al. The Power of Asymmetry: Architecture and Assembly of the Gram-Negative Outer Membrane Lipid Bilayer. Annu Rev Microbiol. 2016;70:255–78. doi: 10.1146/annurev-micro-102215-095308. [DOI] [PubMed] [Google Scholar]
- 46.Nascimento A, Jr, Pontes FJ, Lins RD, Soares TA. Hydration, ionic valence and cross-linking propensities of cations determine the stability of lipopolysaccharide (LPS) membranes. Chem Commun (Camb) 2014;50:231–3. doi: 10.1039/c3cc46918b. [DOI] [PubMed] [Google Scholar]
- 47.Dias RP, da Hora GC, Ramstedt M, Soares TA. Outer Membrane Remodeling: The Structural Dynamics and Electrostatics of Rough Lipopolysaccharide Chemotypes. J Chem Theory Comput. 2014;10:2488–97. doi: 10.1021/ct500075h. [DOI] [PubMed] [Google Scholar]
- 48.Faunce CA, Reichelt H, Paradies HH, Quitschau P, Zimmermann K. The liquidlike ordering of lipid A-diphosphate colloidal crystals: the influence of Ca2+, Mg2+, Na+, and K+ on the ordering of colloidal suspensions of lipid A-diphosphate in aqueous solutions. J Chem Phys. 2005;122:214727. doi: 10.1063/1.1913477. [DOI] [PubMed] [Google Scholar]
- 49.Krishnamoorthy G, Leus IV, Weeks JW, Wolloscheck D, Rybenkov VV, Zgurskaya HI. Synergy between Active Efflux and Outer Membrane Diffusion Defines Rules of Antibiotic Permeation into Gram-Negative Bacteria. MBio. 2017:8. doi: 10.1128/mBio.01172-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westfall DA, Krishnamoorthy G, Wolloscheck D, Sarkar R, Zgurskaya HI, Rybenkov VV. Bifurcation kinetics of drug uptake by Gram-negative bacteria. PLoS One. 2017;12:e0184671. doi: 10.1371/journal.pone.0184671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nichols WW. Modeling the kinetics of the permeation of antibacterial agents into growing bacteria and its interplay with efflux. Antimicrob Agents Chemother. 2017 doi: 10.1128/AAC.02576-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim SP, Nikaido H. Kinetic parameters of efflux of penicillins by the multidrug efflux transporter AcrAB-TolC of Escherichia coli. Antimicrob Agents Chemother. 2010;54:1800–6. doi: 10.1128/AAC.01714-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagano K, Nikaido H. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci U S A. 2009;106:5854–8. doi: 10.1073/pnas.0901695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krishnamoorthy G, Wolloscheck D, Weeks JW, Croft C, Rybenkov VV, Zgurskaya HI. Breaking the Permeability Barrier of Escherichia coli by Controlled Hyperporination of the Outer Membrane. Antimicrob Agents Chemother. 2016;60:7372–81. doi: 10.1128/AAC.01882-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iyer R, Sylvester MA, Velez-Vega C, Tommasi R, Durand-Reville TF, Miller AA. Whole-Cell-Based Assay To Evaluate Structure Permeation Relationships for Carbapenem Passage through the Pseudomonas aeruginosa Porin OprD. ACS Infect Dis. 2017 doi: 10.1021/acsinfecdis.6b00197. [DOI] [PubMed] [Google Scholar]
- 56.Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, Hearst JE. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li XZ, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–53. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lomovskaya O, Lee A, Hoshino K, Ishida H, Mistry A, Warren MS, et al. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:1340–6. doi: 10.1128/aac.43.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolloscheck D, Krishnamoorthy G, Nguyen J, Zgurskaya HI. Kinetic control of quorum sensing in Pseudomonas aeruginosa by multidrug efflux pumps. ACS Infect Dis. 2017 doi: 10.1021/acsinfecdis.7b00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silver LL. A Gestalt approach to Gram-negative entry. Bioorg Med Chem. 2016;24:6379–89. doi: 10.1016/j.bmc.2016.06.044. [DOI] [PubMed] [Google Scholar]
- 61.Brown DG, May-Dracka TL, Gagnon MM, Tommasi R. Trends and exceptions of physical properties on antibacterial activity for Gram-positive and Gram-negative pathogens. J Med Chem. 2014;57:10144–61. doi: 10.1021/jm501552x. [DOI] [PubMed] [Google Scholar]
- 62.Manchester JI, Buurman ET, Bisacchi GS, McLaughlin RE. Molecular determinants of AcrB-mediated bacterial efflux implications for drug discovery. J Med Chem. 2012;55:2532–7. doi: 10.1021/jm201275d. [DOI] [PubMed] [Google Scholar]
- 63.O’Shea R, Moser HE. Physicochemical properties of antibacterial compounds: implications for drug discovery. J Med Chem. 2008;51:2871–8. doi: 10.1021/jm700967e. [DOI] [PubMed] [Google Scholar]
- 64.Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov. 2015;14:529–42. doi: 10.1038/nrd4572. [DOI] [PubMed] [Google Scholar]
- 65.Podnecky NL, Rhodes KA, Schweizer HP. Efflux pump-mediated drug resistance in Burkholderia. Front Microbiol. 2015;6:305. doi: 10.3389/fmicb.2015.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jassem AN, Forbes CM, Speert DP. Investigation of aminoglycoside resistance inducing conditions and a putative AmrAB-OprM efflux system in Burkholderia vietnamiensis. Ann Clin Microbiol Antimicrob. 2014;13:2. doi: 10.1186/1476-0711-13-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Madala NE, Leone MR, Molinaro A, Dubery IA. Deciphering the structural and biological properties of the lipid A moiety of lipopolysaccharides from Burkholderia cepacia strain ASP B 2D, in Arabidopsis thaliana. Glycobiology. 2010;21:184–94. doi: 10.1093/glycob/cwq146. [DOI] [PubMed] [Google Scholar]
- 68.Sulavik MC, Houseweart C, Cramer C, Jiwani N, Murgolo N, Greene J, et al. Antibiotic Susceptibility Profiles of Escherichia coli Strains Lacking Multidrug Efflux Pump Genes. Antimicrob Agents Chemother. 2001;45:1126–36. doi: 10.1128/AAC.45.4.1126-1136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Linares JF, Lopez JA, Camafeita E, Albar JP, Rojo F, Martinez JL. Overexpression of the multidrug efflux pumps MexCD-OprJ and MexEF-OprN is associated with a reduction of type III secretion in Pseudomonas aeruginosa. J Bacteriol. 2005;187:1384–91. doi: 10.1128/JB.187.4.1384-1391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morita Y, Tomida J, Kawamura Y. Efflux-mediated fluoroquinolone resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7: identification of a novel MexS variant involved in upregulation of the mexEF-oprN multidrug efflux operon. Front Microbiol. 2015;6:8. doi: 10.3389/fmicb.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poole K. Efflux pumps as antimicrobial resistance mechanisms. Ann Med. 2007;39:162–76. doi: 10.1080/07853890701195262. [DOI] [PubMed] [Google Scholar]
- 72.Silver LL. Challenges of antibacterial discovery. Clin Microbiol Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lewis K. Antibiotics: Recover the lost art of drug discovery. Nature. 2012;485:439–40. doi: 10.1038/485439a. [DOI] [PubMed] [Google Scholar]


