Abstract
The accumulation of reactive oxygen species (ROS) commonly occurs during normal aging and during some acute/chronic progressive disorders. In order to avoid oxidative damage, scavenging of these radicals is important. Previously, we identified zinc finger protein 179 (Znf179) as a neuroprotector that increases antioxidant enzymes against superoxide radicals. However, the molecular mechanisms involved in the activation and regulation of Znf179 remain unresolved. Here, by performing sequence alignment, bioinformatics analysis, immunoprecipitation using two specific acetyl-lysine antibodies, and treatment with the histone deacetylase (HDAC) inhibitor SAHA, we determined the lysine-specific acetylation of Znf179. Furthermore, we investigated Znf179 interaction with HDACs and revealed that peroxide insult induced a dissociation of Znf179-HDAC1/HDAC6, causing an increase in Znf179 acetylation. Importantly, HDAC inhibition by SAHA further prompted Znf179 hyperacetylation, which promoted Znf179 to form a transcriptional complex with Sp1 and increased antioxidant gene expression against oxidative attack. In summary, the results obtained in this study showed that Znf179 was regulated by HDACs and that Znf179 acetylation was a critical mechanism in the induction of antioxidant defense systems. Additionally, HDAC inhibitors may have therapeutic potential for induction of Znf179 acetylation, strengthening the Znf179 protective functions against neurodegenerative processes.
Keywords: Znf179, SAHA, HDACs, Oxidative stress, Antioxidants
Highlights
-
•
Zinc finger protein 179 (Znf179) is regulated by histone deacetylase (HDAC).
-
•
Acetylation of Znf179 is lysine specific.
-
•
Znf179 acetylation induces antioxidant defense systems.
-
•
HDAC inhibitors may have therapeutic potential as inducers of Znf179 acetylation.
1. Introduction
Reactive oxygen species (ROS), including singlet oxygen (1O2), superoxide anion (·O2-), hydroxyl radical (•OH), hydrogen peroxide (H2O2), and others are produced during normal oxidative processes related to cell metabolism [1]. Normally, ROS levels are tightly controlled by endogenous antioxidant enzymes but impaired mitochondrial function and environmental stresses, including inflammation, hypoxia/ischemia, and excitatory neurotransmitter release, cause a reduction in the antioxidant capacity and an increase in ROS production [2], [3], [4]. Left unchecked, the accumulated ROS will damage cellular organelles and induce apoptosis leading to tissue atrophy and dysfunction [5]. ROS has been proposed to play crucial roles in the pathogenesis of neurodegenerative disorders. Therefore, preventing ROS accumulation or enzymatically eliminating them is an important subject in the treatment of chronic and subacute degenerative diseases in the brain and other organ systems of the body.
Znf179, also known as RING finger protein 112 (Rnf112), brain finger protein (BFP), and neurolastin, is abundant in the nervous system and plays a critical role in the regulation of embryonic brain development [6]. Recently, we determined the neuroprotective effects of Znf179 and found that Znf179 interaction with sigma 1 receptor (Sig-1R), an inter-organelle protein chaperone involved in the stress response, is capable of reducing ROS-induced cellular damage [7]. Furthermore, to examine Znf179 expression and its protein sub-cellular localization, we demonstrated that nuclear import of Znf179 results in its ability to bind and transactivate its own promoter through a Sp1-dependent mechanism, causing the upregulation of endogenous neuroprotective factors against oxidative insults [8]. Although we have shown an elevation in the levels of Znf179 in response to oxidative stress, the regulatory mechanism of Znf179 interacting with target proteins to increase gene transcription remains unclear.
In eukaryotes, acetylation is one of the major post-translational modifications (PTMs) and it may induce new functions of targeted proteins [9], including protein-protein interactions, subcellular localization, and transcriptional potential [10], [11]. The acetyl group from acetyl coenzyme A is transferred to the ε-amino group of lysine residues on a polypeptide chain by histone acetyltransferases (HATs), but several lysine deacetylases such as histone deacetylases (HDACs) counteract HATs [10]. Previous studies have indicated that protein deacetylation and transcriptional initiation induced by HDACs are associated with a wide variety of brain disorders. Treatment with HDAC inhibitors like suberoylanilide hydroxamic acid (SAHA), which is also known as Vorinostat and is permeable to the blood-brain barrier, causes a net increase in protein acetylation and facilitates gene transcription, resulting in extended survival and improved pathological phenotypes in several animal models of neurodegeneration [12], [13].
In the current study, we investigated Znf179 acetylation and determined that oxidative stress slightly increased the amount of acetylated Znf179 and that SAHA co-treatment further enhanced the acetylation levels of Znf179. Additionally, we revealed that SAHA promoted Znf179 interaction with Sp1, leading to positive autoregulation of its own transcription and elevated expression of antioxidant enzymes that protected cells in a Znf179-dependent manner. These findings indicate potential therapeutic value in that HDAC inhibitors may induce an increase in Znf179 acetylation and strengthen the Znf179 protective functions against neurodegenerative processes.
2. Materials and methods
2.1. Materials
Common reagents, bovine serum albumin (BSA), streptavidin-agarose beads, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and anti-HDAC1 antibody were purchased from Sigma-Aldrich (St. Louis, MO, USA). Cell culture supplies, fetal bovine serum (FBS), Lipofectamine 2000, and anti-SUMO1 antibody were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Polyvinylidene difluoride (PVDF) membrane was purchased from Bio-Rad Laboratories (Hercules, CA, USA). Ultra ECL-HRP Substrate and the EasyPrep plasmid extraction kit were purchased from Biotools Co., Ltd. (Taipei, Taiwan). The Dual-Luciferase Reporter (DLR) Assay System was purchased from Promega (Madison, WI, USA). Anti-Sp1, anti-actin, anti-HDAC2, and anti-HDAC3 antibodies were purchased from Millipore (Billerica, MA, USA). Anti-acetyl-lysine, anti-SOD1, anti-SOD2, anti-catalase, anti-Prx3, and anti-GPx1 antibodies were purchased from Genetex Inc. (Irvine, CA, USA). Anti-p53, anti-phospho-p53 (Ser15), anti-acetyl-lysine, and anti-HDAC6 antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-Nrf2 and horseradish peroxidase (HRP)-conjugated anti-mouse and HPR-conjugated anti-rabbit antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The anti-Znf179 antibody was described previously [6].
2.2. Cell culture
As described in previous studies [7], [8], mouse neuroblastoma Neuro-2a (N2a) cells (ATCC; Manassas, VA, USA) were grown in complete culture Eagle's Minimum Essential Medium (EMEM) containing 10% FBS and were induced to differentiate into a neuron-like phenotype by serum deprivation in 0.2% BSA EMEM for 48 h. The differentiated N2a cells were used for all experiments. Cell monolayers at 80% confluency were used for drug treatment including H2O2 and SAHA. Cell monolayers at 50% confluency were used for transfection with plasmids including pGL2-Basic-znf179 [8], pEGFP, pEGFP-Znf179 [14], pLKO-shLuc, and pLKO-shZnf179 [6] using Lipofectamine 2000, which provided lower toxicity and higher than 80% of transfection efficiency in N2a cells (https://www.thermofisher.com), according to the manufacturer's protocol (Thermo Fisher Scientific). One µl of Lipofectamine was incubated with 1 μg of different plasmids in 0.2 ml of serum-free medium for 30 min at room temperature. Subsequently, the cells in a 3.5-cm dish were transfected by the DNA-Lipofectamine complexes in 2 ml of differentiation medium (0.2% BSA EMEM), and then incubated at 37 °C in 5% CO2 for 6 h. The cells were then incubated for an additional 36 h using 2 ml of fresh medium. The transfection efficiency was mostly greater than 80%. Znf179 silencing efficiency was determined by immunoblotting.
2.3. Immunoblot analysis
Total cell lysates were separated by electrophoresis on sodium dodecyl sulfate (SDS)-polyacrylamide gels, and transferred onto PVDF membranes. The membranes, after transfer, were blocked with 5% skim milk in TBST (10 mM Tris, pH 8.0; 150 mM NaCl; and 0.5% Tween 20) for 1 h and incubated with primary antibodies for 2 h at room temperature. After incubation with the primary antibodies, the membranes were washed with TBST buffer and incubated with HRP-conjugated secondary antibodies for 1 h. The membranes were again washed and the peroxidase was developed using the Ultra ECL-HRP Substrate system.
2.4. Immunoprecipitation (IP)
Differentiated neuron-like N2a cells (1 × 107) were lysed with cell lysis buffer (50 mM Tris, pH 7.6; 150 mM NaCl; 0.5% Triton-X100; 0.1% Nonidet P-40) containing 0.5 µM SAHA and protease inhibitors. Following centrifugation, the supernatants (600 µg) were subjected to IP using the antibody indicated with the immunoblotting being performed as described previously [7], [15].
2.5. Luciferase reporter assay
The mouse znf179 promoter fragment (− 533 to − 5) was cloned into the pGL2-Basic vector [8] to produce a promoter–luciferase construct. Subsequently, cells were transfected with the reporter construct and treated with reagents as indicated. After one day of incubation, the luciferase activity from the cell lysates was measured using the Dual-Luciferase Reporter Assay System (Promega) as per the manufacturer's instructions.
2.6. DNA affinity precipitation assay (DAPA)
The 5′-biotinylated double-stranded DNA probes (2 µg), Znf179-Sp1 probe (5′-GCTCTCCCCCTCCCCTCCCCCTCCCTGTCCTT-3′) and Znf179-Sp1 mutant probe (5′-GCTCTCaCaaTCaaCTCaCaaTCaCTGTCCTT-3′) were incubated with the cell extracts (300 µg) in 500 µl of binding buffer (20 mM Tris-HCl, pH 8.0; 60 mM KCl; 2 mM MgCl2; 5% glycerol; 0.05% Nonidet P-40; and protease inhibitors) for 2 h at 4 °C. Subsequently, the reaction mixture was combined with 30 µl of streptavidin-agarose beads and further incubated for 1 h at 4 °C to pull down the DNA-protein complexes.
2.7. MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay
Cells were washed with phosphate-buffered saline (PBS) and incubated with MTT solution (0.5 mg/ml in culture medium) at 37 °C for 1 h. After incubation, the MTT medium was removed and 300 µl of 100% dimethyl sulfoxide (DMSO) was added to dissolve the MTT formazan crystals to visualize the metabolic activity of the cells. The amount of resultant crystals was determined by measuring the absorbance at 570 nm using an iMark Microplate Absorbance Reader (Bio-Rad).
2.8. Statistical analysis
All experiments were performed more than three independent times with duplicate samples in each test. Statistical analyses of the data from the immunoblotting, IP, reporter, DAPA, and MTT assays were performed using the unpaired, two-tailed Student's t-test. The level for statistical significance was set at less than 0.05.
3. Results
3.1. Znf179 is acetylated
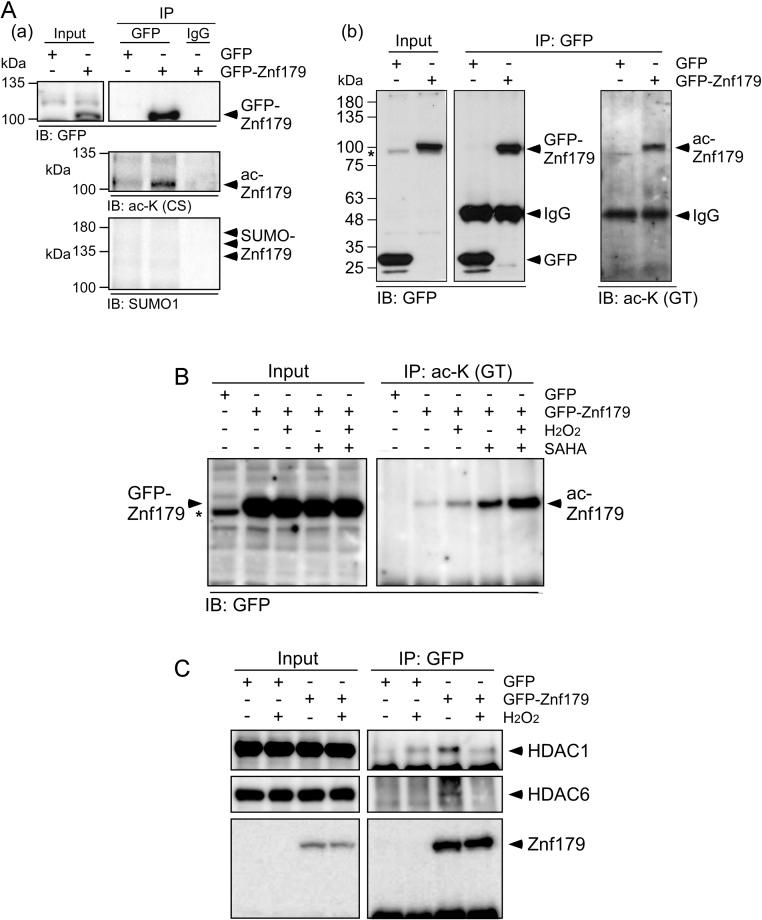
Protein sequences of the human and mouse Znf179 were aligned using the Basic Local Alignment Search Tool (BLAST) provided by the National Center for Biotechnology Information (NCBI) and the results revealed that both proteins were quite similar with 88% sequence identity (Fig. 1A). Meanwhile, 26 of the 28 lysine sites in the human Znf179 were shared by the mouse protein, suggesting these lysine residues were conserved in the Znf179 protein across humans and mice. Lysine acetylation is one of the most abundant PTMs in eukaryotic cells and plays important roles in regulating protein function, including protein-protein interaction and subcellular localization. To investigate whether Znf179 was acetylated, we performed bioinformatic prediction analysis on the mouse Znf179 protein sequence using the GPS-PAIL 2.0 software (http://pail.biocuckoo.org/online.php), which was developed for predicting substrates and 7 lysine-acetyl-transferases (KATs) sites including CREBBP, EP300, HAT1, KAT2A, KAT2B, KAT5, and KAT8 [16]. Five lysine residues were identified as candidate sites for acetylation targeted by KATs in the mouse Znf179 protein sequence (Table 1 and Fig. 1B). We subsequently examined Znf179 protein acetylation using two pan acetylated-lysine (ac-K) antibodies (Fig. 2A). The immunoblots revealed a robust band that specifically corresponded to Znf179, indicating that Znf179 was acetylated in the differentiated N2a cells. Since a lysine residue is also targeted by small ubiquitin-like modifier (SUMO) proteins, Znf179 sumoylation was further investigated using an anti-SUMO1 antibody. However, no SUMO-modified GFP-Znf179 bands of the expected 120, 140, or 160 kDa sizes were detected (Fig. 2A, panel a).
Fig. 1.
Sequence alignment of the mouse vs. human Znf179 protein motifs. (A) Comparison of mouse Znf179 protein sequence to its human counterpart. Identical amino acids, presented using their standard single-letter abbreviations, are in black background and similar amino acids (+) are in gray background. The conserved lysine residues are marked in yellow background. (B) Schematic visualization of the predicted lysine acetylation sites in the Znf179 protein sequence.
Table 1.
The acetylation of lysine residues of mouse Znf179 was predicted by the GPS-PAIL 2.0 software (http://pail.biocuckoo.org/online.php). Position: The position of the site which is predicted to be acetylated. HAT: The histone acetyltransferase which is predicted to acetylate Znf179. Score: The value calculated by group-based prediction and scoring (GPS) algorithm to evaluate the potential of acetylation.
| ID | Position | Peptide | HAT | Score |
|---|---|---|---|---|
| Mus-Znf179 | 17 | AFCHRLGKRESKRSF | EP300 | 0.452 |
| Mus-Znf179 | 17 | AFCHRLGKRESKRSF | KAT5 | 1.281 |
| Mus-Znf179 | 340 | ILQKLSGKYPKVQEL | KAT2A | 2.333 |
| Mus-Znf179 | 401 | STAPQHAKSRCQGYW | KAT5 | 0.781 |
| Mus-Znf179 | 465 | LRKVEAAKKEFEEYV | KAT2B | 1.67 |
| Mus-Znf179 | 613 | ATVGCMEKEEDERVQ | CREBBP | 1.577 |
Fig. 2.
Suberoylanilide hydroxamic acid (SAHA) increases Znf179 acetylation. (A) Differentiated neuron-like N2a cells were transiently transfected with pEGFP or pEGFP-Znf179 to overexpress GFP or GFP-Znf179, respectively. The cell lysates were then used in immunoprecipitation (IP) assays using anti-GFP antibodies and rabbit IgG, and analyzed via immunoblotting with anti-GFP, anti-acetyl-lysine (ac-K, CS: Cell Signaling Technology in panel a; GT: GeneTex in panel b), or anti-SUMO1 antibodies. (B and C) GFP-overexpressing cells and GFP-Znf179-overexpressing cells were treated with 3 µM SAHA (B) or 50 µM hydrogen peroxide (H2O2; B and C) alone or in combination (B) for 30 min. After treatment, the cells were used in IP assays using antibodies as indicated and analyzed by immunoblotting. The asterisk indicates a nonspecific band at 100 kDa recognized by the anti-GFP antibody in the blot of total input.
To examine whether oxidants altered Znf179 acetylation levels, the differentiated neuron-like N2a cells were treated with 50 µM H2O2 for 1 h with the results showing that Znf179 acetylation was induced by H2O2 treatment (Fig. 2B). A previous study revealed that oxidative stress reduces HDAC activity [17]. Thus, the interaction between Znf179 and class I HDACs, such as HDAC1, HDAC2, HDAC3, or a class II HDAC, HDAC6, was further investigated following oxidative stress. By performing co-IP assays we detected an obvious association between Znf179 and HDAC1 and HDAC6, but the association was significantly suppressed following H2O2 treatment (Fig. 2C). Moreover, treatment of cells with the HDAC inhibitor SAHA caused increased acetylation of Znf179, which was much more highly induced by the combined treatment of H2O2 and SAHA than that of their individual treatments separately (Fig. 2B). These results suggested that Znf179 acetylation was regulated by HDACs and that oxidative stress could attenuate the Znf179-HDACs interaction to elevate the levels of Znf179 acetylation.
3.2. SAHA causes the recruitment of Znf179 to the Sp1 bound znf179 promoter
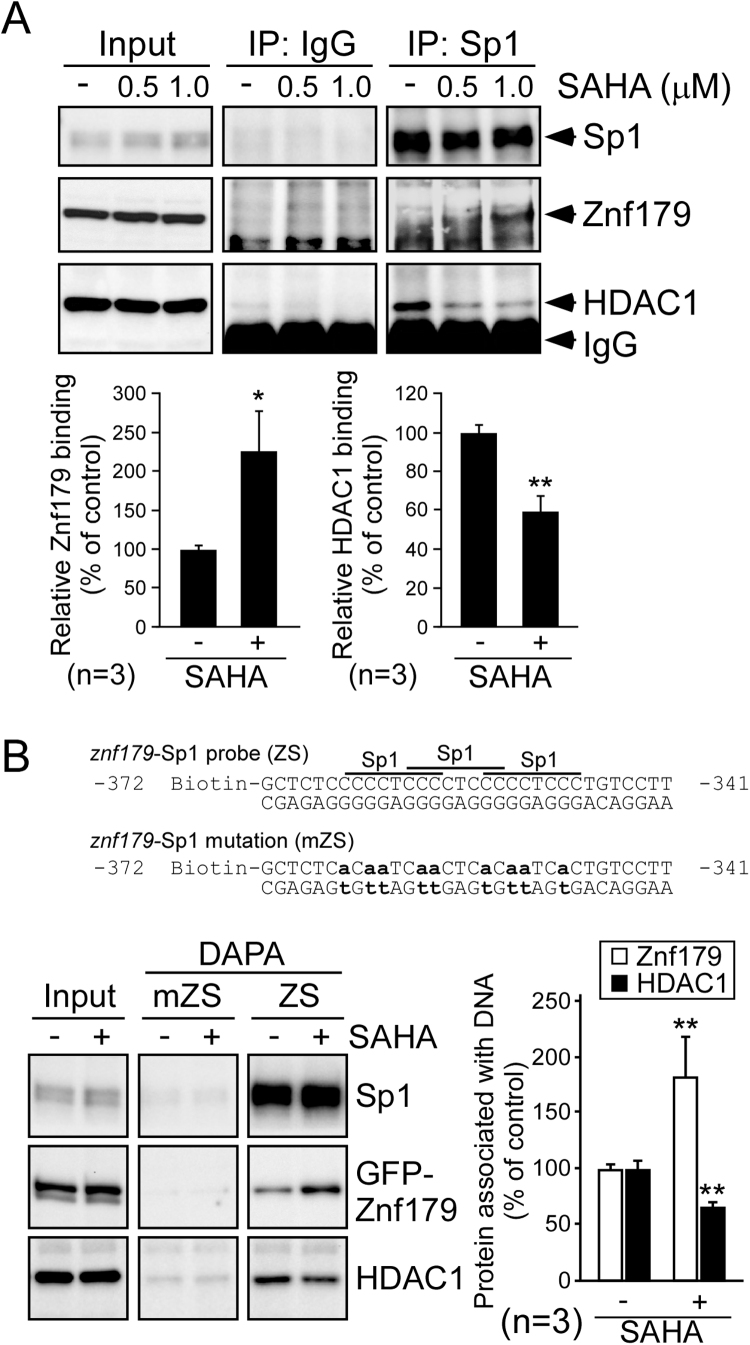
Lysine acetylation regulates protein functions, such as protein interactions and transcriptional activity [10]. Since both HDAC1 and Znf179 are cofactors that bind with Sp1 [8], [18], the interaction of Znf179, HDAC1, and Sp1 after SAHA treatment was further evaluated by co-IP. As shown in Fig. 3A, SAHA increased the level of Znf179-Sp1 association, but dissociated HDAC1 from the protein complexes. Since our previous study revealed that Znf179 is able to autoregulate its own transcription via binding to Sp1 [8], these cofactors on Sp1-binding sites of the znf179 promoter were investigated by performing DAPA assays. An Sp1-binding oligonucleotide that localized from − 372 to − 341 bp within the znf179 promoter was used to demonstrated that SAHA enhanced Znf179 binding to Sp1 elements but that it had no effect on Sp1 binding (Fig. 3B). Additionally, the presence of SAHA decreased the recruitment of HDAC1 to the DNA-protein complex. Taken together, these results provided the first demonstration of SAHA-mediated Znf179-Sp1 complex formation at Sp1-binding elements, which may activate promoter transcription.
Fig. 3.
SAHA treatment induces Sp1-Znf179 complex formation on the Znf179 promoter. (A) N2a cells were treated with different concentrations of SAHA (0, 0.5, and 1 μM) for 30 min. After treatment, the cells were used in IP assays and analyzed via immunoblotting using anti-Sp1, anti-Znf179, or anti-HDAC1 antibodies. The protein interaction of Sp1, Znf179, and HDAC1 were normalized to their total protein and quantified (t-test: * p < 0.05, ** p < 0.01). (B) GFP-Znf179-overexpressing cells were treated without or with 0.5 µM SAHA for 30 min. DNA affinity precipitation assay (DAPA) analysis was then performed using the cell extracts with a biotinylated DNA probe containing wild-type (ZS) or mutated (mZS) Sp1 binding sequence. The precipitated samples from DAPA were analyzed by immunoblotting. Bars represent means ± SEM from three independent experiments (t-test: ** p < 0.01).
3.3. SAHA increases Znf179 protein levels and protected cells against oxidative injury
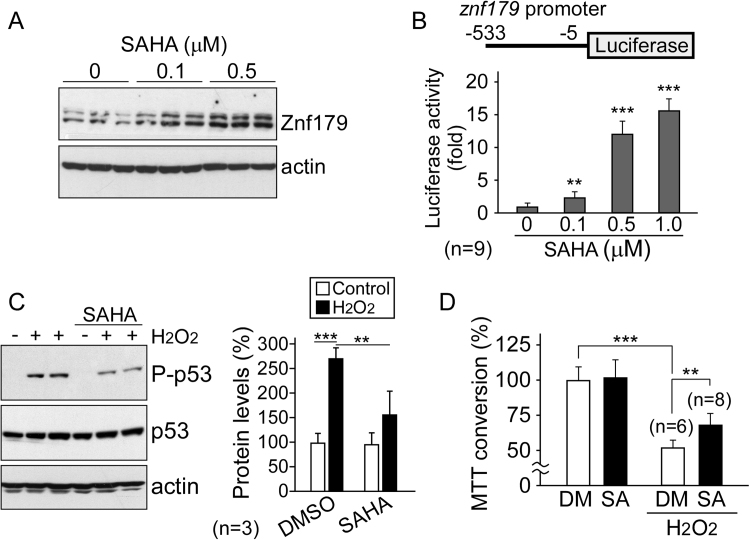
The expression of Znf179 following SAHA treatment was examined in differentiated N2a cells. Immunoblotting results illustrated that Znf179 levels significantly increased in response to SAHA exposure in a dose-dependent manner (Fig. 4A). To further investigate the Znf179 promoter activity, a luciferase reporter (pGL2-Basic-znf179) system was generated and the results demonstrated that SAHA induced Znf179 upregulation by increasing its promoter activity (Fig. 4B). Since Znf179 is a neuro-protector against oxidative stress [7], [8], we then examined the effect of SAHA in H2O2-treated cells. As shown in Fig. 4C, immunoblotting results verified that SAHA decreased the phosphorylation of p53 at Ser15, a general marker for the induction of apoptosis [19]. In addition, cell viability was also measured using MTT assays, which demonstrated that SAHA protected cells against H2O2-induced cytotoxicity (Fig. 4D). Taken together, these data suggested that Znf179 upregulation may play a role in the mitigation of cell death by SAHA under oxidative stress.
Fig. 4.
SAHA upregulates Znf179 transcription and protects cells against oxidative stress. (A) N2a cells were treated with SAHA for 1 day, and the levels of Znf179 and actin proteins were analyzed by immunoblotting. (B) pGL2-Basic-Znf179-transfected N2a cells were treated with different concentrations of SAHA as indicated for 1 day and the luciferase activity was measured. Bars represent luciferase activity (mean ± SEM) obtained in nine independent experiments (t-test: ** p < 0.01, *** p < 0.001). (C and D) N2a cells were treated with 100 µM H2O2 or co-treated with 0.5 µM SAHA for 1 day. After treatment, the phosphorylated levels of p53 (Ser15) was analyzed using immunoblotting and the results normalized to levels of total p53 (C). Cell viability was assessed using MTT assays (D). Bars represent means ± SEM from three to eight independent experiments as indicated (t-test: ** p < 0.01, *** p < 0.001).
3.4. SAHA stimulates Znf179-dependent expression of antioxidant defense enzymes
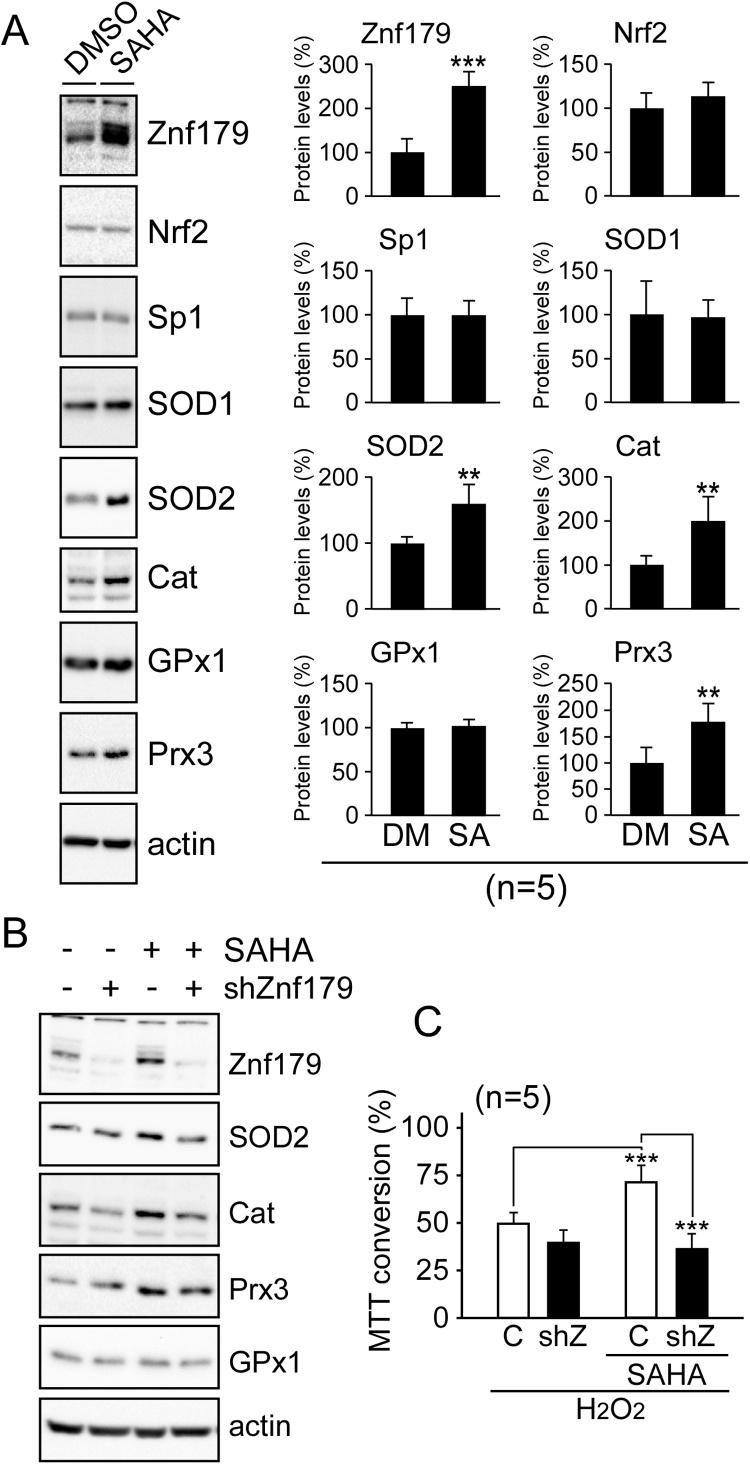
Our previous study indicated that Znf179 expression is correlated with protein levels of cellular antioxidant enzymes [7]. To investigate whether SAHA stimulated expression of cellular antioxidants through Znf179, we screened for the expression of multiple ROS-detoxifying enzymes, including superoxide dismutase 1 (SOD1), SOD2, catalase, glutathione peroxidase (GPx), peroxiredoxin (Prx), and oxidant response transcription factor Nrf2. We found three enzymes, SOD2, catalase, and Prx3 were significantly upregulated following treatment with SAHA (Fig. 5A). Additionally, we used a short hairpin RNA (shRNA) to knockdown Znf179 in order to further determine the role of Znf179 in the expression of these antioxidant proteins. The results revealed that reducing levels of Znf179 attenuated the SAHA-induced expression of all three enzymes (Fig. 5B). Moreover, knockdown of Znf179 also abolished the protective effects of SAHA (Fig. 5C). Taken together, the data suggested that Znf179-mediated its own transcription, as well as the expression of downstream antioxidants, which are novel targets of SAHA for ensuring cell survival following oxidative stress. For this protection, acetylated Znf179 interacting with Sp1 in the promoter region was a critical mechanism resulting in transcriptional upregulation following SAHA treatment (Fig. 6).
Fig. 5.
SAHA treatment increases expression of antioxidant enzymes via a Znf179-dependent mechanism. (A) N2a cells were treated with 0.5 µM SAHA for 1 day. Following treatment, protein levels of antioxidant enzymes and their upstream regulator were analyzed by immunoblotting. Bar graphs represent means ± SEM from five independent experiments (t-test: ** p < 0.01). Cat, catalase. (B) Cells were transfected with a Znf179-specific shRNA (shZnf179) or a control shRNA targeting luciferase (-) as indicated. After knockdown of Znf179, cells were treated with 0.5 µM SAHA for 1 day and analyzed by immunoblotting. (C) N2a cells were transfected with a control shRNA (C) or a Znf179-specific shRNA (shZ) as indicated. After transfection, cells were pre-treated with or without 0.5 μM SAHA for 1 h and treated with 100 µM H2O2 for 1 day. Cell viability was assessed using MTT assays (t-test: *** p < 0.001).
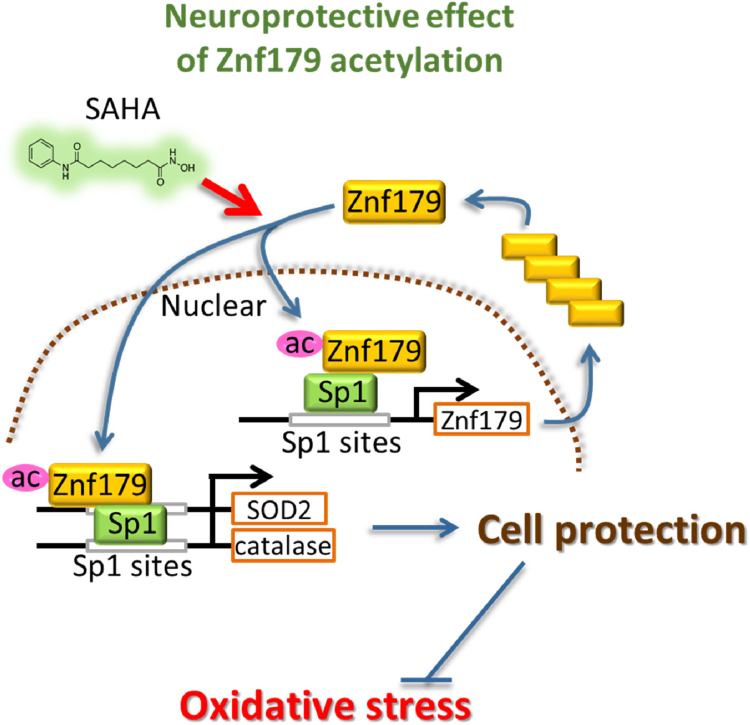
Fig. 6.
SAHA protects cells against oxidative stress via increasing Znf179 acetylation to affect Znf179 autoregulation and expression of its downstream antioxidant enzymes.
4. Discussion
The data obtained in this study demonstrated that SAHA mediated the upregulation of Znf179 and downstream antioxidants in order to play a role in the mitigation of cell death following oxidative stress. First, we showed that peroxide insult induced a dissociation of Znf179 from HDACs, causing an increase in Znf179 acetylation. Second, SAHA-inhibited HDACs activation, which highly elevated levels of Znf179 acetylation and enhanced formation of the Znf179-Sp1 complex at the targeted promoter Sp1 sites. Finally, SAHA induced the positive Znf179 autoregulation and enrichment of antioxidant proteins SOD2, catalase, and Prx3, providing maximal protection against oxidative injury. These findings indicated that HDAC inhibition induced an increase in the neuroprotective function of the Znf179-Sp1 pathway, which may allow for the development of new therapeutic approaches in the prevention or reduction of oxidative damage during degenerative diseases.
Znf179, a novel nuclear–cytoplasmic RING finger protein, is highly expressed in the brain and it regulates multiple genes involved in nervous tissue development, synaptic plasticity, and cellular survival. During the neuronal maturation process, Znf179 is upregulated in order to increase the expression of cell cycle suppressors, promoting an irreversible arrest in the G0/G1 phase of the cell cycle and initiating neuronal differentiation [6]. In addition, a recent study revealed important functions of Znf179 in blood vessel development [20]. In the adult brain, Znf179 can affect membrane trafficking, dendritic spine density, and synaptic plasticity through its endosomal localization mediating endosomal membrane dynamics [21]. Furthermore, our previous studies demonstrated the neuroprotective activity of Znf179 against cell apoptosis in response to oxidative stress [7], [8]. Those results indicate that Znf179 plays critical roles in neuronal development and functions by interacting with different types of protein, including the membrane-bound chaperone Sig-1R [7], E2 ubiquitin-conjugating enzymes [21], transcriptional factors like Sp1 [8], promyelocytic leukemia zinc finger (Plzf) [14], CCAAT/enhancer binding protein delta (CEBPD) [22], and others. While Znf179-mediated cell cycle arrest and cell death/survival is intensely investigated, the regulatory mechanisms of Znf179 interacting with many different types of proteins in response to environmental stress and developmental signals remains poorly understood. Besides phosphorylation, protein acetylation is probably the most abundant and important PTM and it is recognized as a regulatory signal in many cellular processes [9]. In the current study, we examined the human and mouse protein sequences of Znf179 and demonstrated that Znf179 could be acetylated. Furthermore, we found a significant increase in protein acetylation (Fig. 2B) and the transactivation of Znf179 after treatment with H2O2 and an HDAC inhibitor (Fig. 4B and [8]). Since oxidative stress is known to reduce HDAC activity [17], [23], [24], we suggest that HDACs are key players in redox signaling. Moreover, we determined that HDACs like HDAC1 and HDAC6, were regulators upstream of Znf179 and that the inhibition of HDACs by H2O2 or SAHA increased the level of Znf179 acetylation and enhanced formation of the Znf179-Sp1 complex. Therefore, lysine acetylation was a critical regulatory mechanism controlling the interaction of Znf179 with transcriptional factors, which enabled the upregulation of defense factors that target oxidative stress. However, identifying the precise lysine residues in Znf179 marked by acetylation and determining whether lysine acetylation affects Znf179 function in neuronal development require further investigation. In addition, SAHA is a pan HDAC inhibitor, affecting HDAC classes I and II. Although the study demonstrated the occurrence of the complex of Znf179-HDAC1/HDAC6, the HDAC subtypes involved in the Znf179 acetylation and subsequent transcriptional activation of antioxidant enzymes remains unclear. Future studies are thus necessary to identify the HDAC subtypes mediating the SAHA-induced actions shown in this report.
Classical HDACs are comprised of 11 family members and are a family of zinc-dependent enzymes with specific and critical functions in development and tissue homeostasis [25], [26]. In previous studies, increasing evidence has suggested that the dysregulation of HDAC activity is associated with and contributes to neurodegenerative diseases [12]. Generally, an increase in histone deacetylation induced by HDACs results in chromatin compaction, thus attenuating the interactions of transcription factors with target gene promoters and silencing key genes involved in neuroprotection [12], [27]. In addition to histones, HDACs also target many non-histone proteins such as tubulin, p53, p73, retinoblastoma, Hsp90, HIF-1α, steroid receptors, E2F family members, and others, which enables them to regulate several cellular functions associated with metabolism, signaling, and death [12], [28]. In the current study, we demonstrated that HDAC1/HDAC6 were able to interact with Znf179 and inhibited Znf179 activation by catalyzing lysine deacetylation of the protein, but SAHA reversed that effect and induced a defective cellular antioxidant response. In addition to oxidative stress, previous reports have revealed that HDAC inhibition has neuroprotective effects in many brain disorders, including amyotrophic lateral sclerosis (ALS), Alzheimer's disease, Parkinson's disease, Huntington's disease (HD), stroke, spinal cord injury, and traumatic brain injury [12]. Furthermore, it is known that Znf179 expression is reduced in ALS, HD, and Smith-Magenis syndrome [20]. Therefore, the HDAC-inhibitor induced the downregulating function of HDACs and upregulated the expression of Znf179, which may protect neurons against the effects of different types of stress in neurodegenerative diseases.
In conclusion, the results obtained in this study indicated that HDACs represent novel upstream regulators of Znf179 and that protein acetylation plays a positive role in enhancing Znf79 binding to Sp1. Notably, an HDAC inhibitor increased Znf179 acetylation and its expression via an Sp1-dependent autoregulatory loop, enhancing protective action of Znf179 against cellular stress. The results from the study may be useful in the further development of novel treatments of oxidative damage that may occur during neurodegenerative processes.
Acknowledgments
This work was supported by the Ministry of Science and Technology, Taiwan (Grant nos. MOST 104-2923-B-038-002-MY3, MOST 106-2320-B-038-002, and MOST 106-2320-B-038-028) and the Taipei Medical University Hospital, Taiwan (Grant no. 105TMU-TMUH-04).
Acknowledgments
Conflicts of interest
The authors declare no conflicts of interest in this work.
Contributor Information
Wen-Chang Chang, Email: wcchang@tmu.edu.tw.
Jian-Ying Chuang, Email: chuangcy@tmu.edu.tw.
References
- 1.Giorgio M., Trinei M., Migliaccio E., Pelicci P.G. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat. Rev. Mol. Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- 2.Hetz C., Mollereau B. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat. Rev. Neurosci. 2014;15:233–249. doi: 10.1038/nrn3689. [DOI] [PubMed] [Google Scholar]
- 3.Lin M.T., Beal M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 4.Xiong Y., Mahmood A., Chopp M. Animal models of traumatic brain injury. Nat. Rev. Neurosci. 2013;14:128–142. doi: 10.1038/nrn3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nita M., Grzybowski A. The role of the reactive oxygen species and oxidative stress in the pathomechanism of the age-related ocular diseases and other pathologies of the anterior and posterior eye segments in adults. Oxid. Med. Cell. Longev. 2016;2016:3164734. doi: 10.1155/2016/3164734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pao P.C., Huang N.K., Liu Y.W., Yeh S.H., Lin S.T., Hsieh C.P., Huang A.M., Huang H.S., Tseng J.T., Chang W.C., Lee Y.C. A novel RING finger protein, Znf179, modulates cell cycle exit and neuronal differentiation of P19 embryonal carcinoma cells. Cell Death Differ. 2011;18:1791–1804. doi: 10.1038/cdd.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su T.C., Lin S.H., Lee P.T., Yeh S.H., Hsieh T.H., Chou S.Y., Su T.P., Hung J.J., Chang W.C., Lee Y.C., Chuang J.Y. The sigma-1 receptor-zinc finger protein 179 pathway protects against hydrogen peroxide-induced cell injury. Neuropharmacology. 2016;105:1–9. doi: 10.1016/j.neuropharm.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuang J.Y., Kao T.J., Lin S.H., Wu A.C., Lee P.T., Su T.P., Yeh S.H., Lee Y.C., Wu C.C., Chang W.C. Specificity protein 1-zinc finger protein 179 pathway is involved in the attenuation of oxidative stress following brain injury. Redox Biol. 2017;11:135–143. doi: 10.1016/j.redox.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drazic A., Myklebust L.M., Ree R., Arnesen T. The world of protein acetylation. Biochim. Biophys. Acta. 1864;2016:1372–1401. doi: 10.1016/j.bbapap.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Peserico A., Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 2011;2011:371832. doi: 10.1155/2011/371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 12.Chuang D.M., Leng Y., Marinova Z., Kim H.J., Chiu C.T. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32:591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H., Dai X., Qi Y., He Y., Du W., Pang J.J. Histone deacetylases inhibitors in the treatment of retinal degenerative diseases: overview and perspectives. J. Ophthalmol. 2015;2015:250812. doi: 10.1155/2015/250812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin D.Y., Huang C.C., Hsieh Y.T., Lin H.C., Pao P.C., Tsou J.H., Lai C.Y., Hung L.Y., Wang J.M., Chang W.C., Lee Y.C. Analysis of the interaction between Zinc finger protein 179 (Znf179) and promyelocytic leukemia zinc finger (Plzf) J. Biomed. Sci. 2013;20:98. doi: 10.1186/1423-0127-20-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai S.Y., Chuang J.Y., Tsai M.S., Wang X.F., Xi Z.X., Hung J.J., Chang W.C., Bonci A., Su T.P. Sigma-1 receptor mediates cocaine-induced transcriptional regulation by recruiting chromatin-remodeling factors at the nuclear envelope. Proc. Natl. Acad. Sci. USA. 2015;112:E6562–E6570. doi: 10.1073/pnas.1518894112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng W., Wang C., Zhang Y., Xu Y., Zhang S., Liu Z., Xue Y. GPS-PAIL: prediction of lysine acetyltransferase-specific modification sites from protein sequences. Sci. Rep. 2016;6:39787. doi: 10.1038/srep39787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milara J., Navarro A., Almudever P., Lluch J., Morcillo E.J., Cortijo J. Oxidative stress-induced glucocorticoid resistance is prevented by dual PDE3/PDE4 inhibition in human alveolar macrophages. Clin. Exp. Allergy. 2011;41:535–546. doi: 10.1111/j.1365-2222.2011.03715.x. [DOI] [PubMed] [Google Scholar]
- 18.Doetzlhofer A., Rotheneder H., Lagger G., Koranda M., Kurtev V., Brosch G., Wintersberger E., Seiser C. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 1999;19:5504–5511. doi: 10.1128/mcb.19.8.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson T., Tovar C., Yang H., Carvajal D., Vu B.T., Xu Q., Wahl G.M., Heimbrook D.C., Vassilev L.T. Phosphorylation of p53 on key serines is dispensable for transcriptional activation and apoptosis. J. Biol. Chem. 2004;279:53015–53022. doi: 10.1074/jbc.M410233200. [DOI] [PubMed] [Google Scholar]
- 20.Tsou J.H., Yang Y.C., Pao P.C., Lin H.C., Huang N.K., Lin S.T., Hsu K.S., Yeh C.M., Lee K.H., Kuo C.J., Yang D.M., Lin J.H., Chang W.C., Lee Y.C. Important roles of ring finger protein 112 in embryonic vascular development and brain functions. Mol. Neurobiol. 2017;54:2286–2300. doi: 10.1007/s12035-016-9812-7. [DOI] [PubMed] [Google Scholar]
- 21.Lomash R.M., Gu X., Youle R.J., Lu W., Roche K.W. Neurolastin, a dynamin family GTPase, regulates excitatory synapses and spine density. Cell Rep. 2015;12:743–751. doi: 10.1016/j.celrep.2015.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S.M., Lee Y.C., Ko C.Y., Lai M.D., Lin D.Y., Pao P.C., Chi J.Y., Hsiao Y.W., Liu T.L., Wang J.M. Increase of zinc finger protein 179 in response to CCAAT/enhancer binding protein delta conferring an antiapoptotic effect in astrocytes of Alzheimer's disease. Mol. Neurobiol. 2015;51:370–382. doi: 10.1007/s12035-014-8714-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito K., Hanazawa T., Tomita K., Barnes P.J., Adcock I.M. Oxidative stress reduces histone deacetylase 2 activity and enhances IL-8 gene expression: role of tyrosine nitration. Biochem. Biophys. Res. Commun. 2004;315:240–245. doi: 10.1016/j.bbrc.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 24.Ito K., Lim S., Caramori G., Chung K.F., Barnes P.J., Adcock I.M. Cigarette smoking reduces histone deacetylase 2 expression, enhances cytokine expression, and inhibits glucocorticoid actions in alveolar macrophages. Faseb J. 2001;15:1110–1112. [PubMed] [Google Scholar]
- 25.Delcuve G.P., Khan D.H., Davie J.R. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin. Epigenetics. 2012;4:5. doi: 10.1186/1868-7083-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witt O., Deubzer H.E., Milde T., Oehme I. HDAC family: what are the cancer relevant targets? Cancer Lett. 2009;277:8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 27.Pirooznia S.K., Elefant F. Targeting specific HATs for neurodegenerative disease treatment: translating basic biology to therapeutic possibilities. Front. Cell. Neurosci. 2013;7:30. doi: 10.3389/fncel.2013.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ocio E.M., San Miguel J.F. The DAC system and associations with multiple myeloma. Invest. New Drugs. 2010;28(Suppl. 1):S28–S35. doi: 10.1007/s10637-010-9589-x. [DOI] [PMC free article] [PubMed] [Google Scholar]