Abstract
Previous studies have demonstrated that the interleukin (IL)-6/ IL-6 receptor (IL-6R) signaling pathway contributes to the pathogenesis of lung cancer. Lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) are the two major pathological subtypes of non-small cell lung cancer (NSCLC). The present study aimed to elucidate the potential clinical prognosis and biological function of IL-6R mRNA expression in LUAD and LUSC. The search term ‘lung cancer’ was used to search through the Gene Expression Omnibus database. Including LUAD and LUSC datasets in The Cancer Genome Atlas database, a total of 8 LUAD and 6 LUSC datasets were included in the present analysis. It was observed that a higher expression level of IL-6R mRNA in tumor tissues was a significant positive prognostic factor for overall survival in LUAD [pooled hazard ratio (HR), 0.48 and P<0.001 for univariate analysis; pooled HR, 0.50 and P<0.001 for multivariate analysis] while there was no similar association in LUSC (pooled HR, 1.59 and P=0.062 for univariate analysis; pooled HR, 1.58 and P=0.079 for multivariate analysis). Correlation analysis revealed that IL-6 and IL-6R were negatively correlated in LUAD and positively correlated in LUSC. IL-6R and its most correlated genes were primarily involved in cell cycle progression in LUAD and primarily involved in tumor angiogenesis, invasion and metastasis in LUSC. These results suggest a possible role of tumoral expression for IL-6R in LUAD, which means it may have potential as a prognostic marker for this type of cancer.
Keywords: lung adenocarcinoma, lung squamous cell carcinoma, IL-6R, overall survival, enrichment analysis
Introduction
Lung cancer is the leading cause of cancer-related deaths in men and the second leading cause of cancer death in women worldwide and presents a serious problem to global health (1). It was estimated that 1.8 million new lung cancer cases and 1.6 million lung cancer related deaths occurred in 2012 worldwide, accounting for approximately 19% of all cancer deaths (2). In recent decades, despite of great research effort in diagnosis and treatment for lung cancer, progress in the treatment is still slow (3). Non-small cell lung cancer (NSCLC) is the major lung cancer, accounts for approximately 85% of lung cancer. Lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) are the major two pathological subtypes of NSCLC and in many ways they are different, such as originate, biological patterns and molecular characteristics (4–6). Related studies on NSCLC should be analyzed separately according to histological type.
Interleukin-6 (IL-6) signaling through IL-6 receptor (IL-6R) regulates cell growth and differentiation and plays an important role in the immune response (7). This signaling pathway can also promote tumor growth which has both pro-inflammatory and anti-inflammatory effect (8). Evidence has shown that the IL-6 signaling pathway contribute to the pathogenesis of NSCLC (9). Several studies have revealed the role of IL-6 in NSCLC and suggested that it promotes tumor growth and survival (10–12). Increased expression of IL-6R in human LUSC-derived cells (HARA-B) has been shown in vitro (13) and in a murine model of brain metastasis (14). Tocilizumab is an anti-IL-6R antibody, upon application, the stimulated growth of HARA-B cells was significantly inhibited and when it was injected to the animal model, the volume of metastatic focus was significantly smaller (14). Studies also have demonstrated that blockade of IL-6R can significantly suppress the proliferation of NSCLC cell and reduce the mRNA levels of IL-6R (15,16). Meanwhile, tocilizumab also seems to be effective for lung cancer cachexia (17,18). However, there was no study investigating the prognostic effect of IL-6R on lung cancer, which is addressed by our current study.
Materials and methods
Search strategy
We searched public databases such as The Cancer Genome Atlas (TCGA) and Gene Expression Ominibus (GEO database; last update by July 03, 2017) using the keywords ‘Lung cancer’. The search strategy is designed as follows: The study type was set as ‘expression profiling by array. The entry type was set as ‘datasets’. The sample size of all selected datasets should be greater or equal to 100. The organism was homo sapiens. Database searching was carried out by two researchers independently.
Data extraction and quality assessment
Data from all eligible datasets were abstracted independently by two authors, using information recorded as follows: First author's surname, publication year, origin of population, sample number, tumor stage, follow-up period and clinic outcome. We separate those microarray datasets into LUAD and LUSC. HRs and 95% CIs were evaluated by Cox's proportional hazards model.
The quality of all eligible studies was assessed according to the Newcastle-Ottawa Quality Assessment Scale (NOS) by two researchers independently (19). The quality scores span from 0 to 9, and higher the score is, higher the quality is.
Statistical analysis
For those public microarray data, gene expression was represented by metric variables. We use Cutoff Finder (http://molpath.charite.de/cutoff) to determine a cutoff point and stratify patients into two groups (20). The range of IL-6R mRNA values for each data and the corresponding cutoff values were listed in Table I. HRs and 95% CIs were calculated to measure the effective prognostic value of expression of IL-6R mRNA in LUAD and LUSC patients. Pooled HRs were carried out using STATA software package (version 12.0; Stata Corp LP, College Station, TX, USA). All P-values were obtained upon two tailed analysis.
Table I.
Cut-off value of IL-6R.
| Datasets | Minimum | P25 | Median | Mean | P75 | Maximum | Cut-off value |
|---|---|---|---|---|---|---|---|
| LUAD | |||||||
| GSE14814 | 4.211 | 4.779 | 5.027 | 5.134 | 5.394 | 6.611 | 4.607 |
| GSE30219 | 5.127 | 6.343 | 6.908 | 7.019 | 7.686 | 9.247 | 7.648 |
| GSE37745 | 3.970 | 7.232 | 7.698 | 7.697 | 8.272 | 10.200 | 7.789 |
| GSE42127 | 3.670 | 4.475 | 4.930 | 4.945 | 5.405 | 7.680 | 5.515 |
| GSE50081 | 5.049 | 7.281 | 7.809 | 7.823 | 8.405 | 10.049 | 7.675 |
| GSE68465 | 31.955 | 246.235 | 361.102 | 437.587 | 543.700 | 1940.570 | 393.500 |
| GSE68571 | −41.850 | 17.188 | 52.725 | 69.762 | 101.213 | 297.650 | 44.020 |
| TCGA | 4.360 | 8.364 | 9.037 | 8.970 | 9.274 | 11.608 | 9.125 |
| LUSC | |||||||
| GSE14814 | 4.324 | 4.644 | 4.912 | 4.909 | 5.076 | 5.871 | 5.072 |
| GSE30219 | 4.232 | 5.430 | 6.004 | 5.946 | 6.453 | 7.988 | 5.741 |
| GSE37745 | 4.523 | 6.395 | 6.992 | 6.851 | 7.458 | 8.770 | 7.637 |
| GSE42127 | 2.830 | 4.100 | 4.390 | 4.456 | 4.730 | 6.260 | 4.115 |
| GSE50081 | 5.475 | 6.681 | 7.144 | 7.125 | 7.421 | 9.009 | 6.678 |
| TCGA | 3.751 | 7.456 | 8.286 | 8.171 | 8.879 | 10.982 | 9.134 |
IL, interleukin; TCGA, The Cancer Genome Atlas; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
In The Cancer Genome Atlas (TCGA) lung adenocarcinoma dataset and TCGA lung squamous cell carcinoma dataset, there were gene expression data in both tumor and normal tissues. We used paired test to compare the differences in IL-6R mRNA expression between tumor and adjacent normal tissues.
For each dataset, we calculated the correlation coefficient between IL-6R and the remaining genes, and then matched the coefficients in all datasets. Genes with absolute correlation coefficient which were greater than 0.4 in half or more publications were extracted. 193 genes in LUAD and 101 genes in LUSC were included in subsequent analysis (Table II).
Table II.
Gene list in LUAD and LUSC.
| Cancer | Gene names |
|---|---|
| LUAD | ABCC3, ADCY9, AKAP13, ALDH2, ALDH5A1, ALOX15B, APLP2, APOH, AQP3, ARHGAP31, ASF1B, ASPM, ATP13A4, AURKB, BFAR, BIRC5, BUB1, BUB1B, C16orf89, C1orf116, C5orf46, CC2, CCNB1, CCNB2, CCND3, CD302, CD81, CDC20, CDC25A, CDC25C, CDC45, CDC6, CDCA2, CDCA3, CDCA5, CDCA8, CDK1, CDKL2, CDKN3, CDT1, CEBPA, CENPM, CENPN, CENPW, CEP55, CHEK1, CISH, CITED2, CKS2, CLIC3, CLIC5, COL4A3, COL4A4, CPM, CRY2, CTDSPL, CTSH, CYP4B1, DEPDC1, DEPDC1B, DJC9, DLC1, DLGAP5, DYNLL1, ECHDC2, ESPL1, EVPL, EXO1, FAM184A, FAM64A, FAM83D, FANCG, FANCI, FBP1, FBXO5, FCGRT, FEN1, FMO5, FNIP2, FOLR1, FOXM1, GALNT11, GAPDH, GGT1, GGTLC1, GINS1, GPD1L, GPR116, GTSE1, H2AFX, HIP1, HJURP, HLF, HNF1B, ICAM4, ICAM5, IL6R, INMT, KIAA0101, KIF11, KIF18B, KIF20A, KIF23, KIF2C, KIF4A, KIFC1, KNTC1, LOC101930052, LRRK2, MAD2L1, MALL, MAMDC2, MELK, MGLL, MGRN1, MIR3658, MIR6883, MKI67, MLLT11, MTHFD2, MUC1, NBEAL1, NCAPG, NCAPH, NDC80, NEK2, NPC2, NRM, NUF2, NUSAP1, OCLN, OIP5, PARM1, PARPBP, PBK, PC, PCDP1, PEBP4, PI4KB, PLA2G1B, PLK1, PLLP, PMVK, POLE2, PON3, PRC1, PRR15L, PSA, PSRC1, PTS, PTTG1, RAD51AP1, RAD54L, RFC4, RRM2, RSE1, RSEH2A, SELENBP1, SFTA1P, SFTPD, SKA1, SKA3, SLC22A3, SLC34A2, SLC41A1, SNX30, SPAG5, SPAG9, SPC25, ST3GAL5, STAT6, STIL, SUSD2, SYNE1, TLR2, TLR5, TMEM163, TMEM243, TMPRSS2, TPX2, TRIP13, TROAP, TTK, TUBA1C, TUBB, TYMS, UBE2C, UBE2T, UHRF1, USP13, ZBTB4, ZNF385B, ZWINT |
| LUSC | ADAP2, ADCY9, ANXA11, APOBR, ARHGEF3, BTK, BUB1B, C1RL, CCR1, CD37, CD4, CD52, CD93, CD97, CEBPD, COQ3, CORO1A, CRTAP, CSE1L, CTSH, DENND2D, DHX9, DOK2, DUT, ECT2, EPAS1, EZH2, FBXO5, FGR, FHOD1, FOS, GIMAP6, HDAC2, HLA-DPB1, HLA-DQB1, IER2, IFITM1, IL2RB, IL6R, ITGAL, JUNB, KIF15, KLF10, LCP2, LRP10, LSM4, LST1, MAST3, MCM6, MOSPD1, MSH6, MSN, MVP, MYO1F, P2RY13, PAGR1, PDCD5, PDE4A, PLEKHO2, PLIN3, POLA1, PPP1CC, PPP1R16B, PRKCB, PTAFR, PTK2B, PTPN6, RAB27A, RAB31, RAD51AP1, RAD54B, RCN2, RHOG, RPL22, S1PR1, SASH3, SIGLEC1, SLA, SLCO2B1, THEMIS2, TMPO, TNFRSF1A, TNFRSF1B, TPP1, TRANK1, TRPV2, TYROBP, VASP, VWF, ZFYVE26, ARHGAP30, ARHGAP9, C1orf162, CARD6, CXCL16, GIMAP1, LOC100129034, LRRCC1, SARNP, TNFAIP8L2, ZNF664 |
LUAD, adenocarcinoma; LUSC, squamous cell carcinoma.
Functional enrichment analysis of genes whose expressions are significantly correlated that of IL-6R was performed to allow the identification of biological processes or functions associated with IL-6R expression. In this study, the Database for Annotation, Visualization and Integrated Discovery (DAVID) was used to analyze gene enrichment and pathway analysis to explore the biological processes of gene enrichment (https://david.ncifcrf.gov/summary.jsp).
Results
Study characteristics
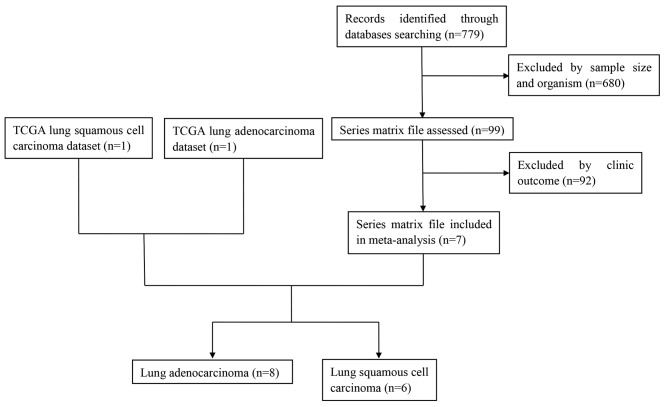
A total of 7 related studies were identified from the GEO database (GSE14814 (21), GSE30219 (22), GSE37745 (23), GSE42127 (24), GSE50081 (25), GSE68465 (26), GSE68571 (27)). Including TCGA lung adenocarcinoma and TCGA lung squamous cell carcinoma datasets, 9 datasets were included in our analysis. In the initial screening, a total of 779 potentially relevant datasets from the GEO database were selected for keyword retrieval. A total of 680 datasets were retrieved after screening sample size and organism. After examination of summary and the clinic outcome of those data, a total of 7 microarray datasets from the GEO database that met the inclusion criteria were included in the present study. Finally, 8 datasets and 6 datasets for adenocarcinoma and squamous cell carcinoma respectively were further analyzed (Fig. 1). Table III showed the baseline characteristics of all included studies. Date of 1,536 LUAD and 739 LUSC patients from Canada, France, UK and USA were included in this analysis.
Figure 1.
Flow diagram of dataset selection process. TCGA, The Cancer Genome Atlas.
Table III.
Baseline characteristics of microarray data.
| Datasets | Author | Year | Country | Median duration, months (range) | Sample size | Stage | Outcome measures | Quality score | Journal | IF | (Refs.) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LUAD | |||||||||||
| GSE14814 | Zhu CQ | 2010 | Canada | 60.84 (0.36–109.08) | 70 | I/II (41/29) | OS | 8 | Journal of clinical oncology | 24.008 | (21) |
| GSE30219 | Rousseaux S | 2013 | France | 68.00 (0–221) | 85 | NR | OS | 7 | Science translational medicine | 16.796 | (22) |
| GSE37745 | Botling J | 2013 | UK | 47.80 (0.20–187.79) | 106 | I/II/III/IV (70/19/13/4) | OS | 8 | Clinical cancer research | 9.619 | (23) |
| GSE42127 | Tang H | 2013 | USA | 45.60 (0–132) | 133 | I/II/III/IV (89/22/20/1) | OS | 7 | Clinical cancer research | 9.619 | (24) |
| GSE50081 | Der SD | 2014 | Canada | 52.44 (1.08–130.56) | 129 | I/II (93/36) | OS | 6 | Journal of thoracic oncology | 6.595 | (25) |
| GSE68465 | Shedden K | 2008 | USA | 47.00 (0.03–204.00) | 442 | NR | OS | 9 | Nature medicine | 29.886 | (26) |
| GSE68571 | Beer DG | 2002 | USA | 29.50 (1.50–110.60) | 86 | I/III (67/19) | OS | 8 | Nature medicine | 29.886 | (27) |
| TCGA | TCGA | 2015 | USA | 15.35 (0.00–223.96) | 485 | I/II/III/IV (262/118/79/25) | OS | 8 | – | – | |
| LUSC | |||||||||||
| GSE14814 | Zhu CQ | 2010 | Canada | 73.56 (8.28–111.48) | 52 | I/II (25/27) | OS | 8 | Journal of clinical oncology | 24.008 | (21) |
| GSE30219 | Rousseaux S | 2013 | France | 59.00 (1–256) | 61 | NR | OS | 7 | Science translational medicine | 16.796 | (22) |
| GSE37745 | Botling J | 2013 | UK | 36.36 (0.20–179.80) | 66 | I/II/III (40/15/11) | OS | 8 | Clinical cancer research | 9.619 | (23) |
| GSE42127 | Tang H | 2013 | USA | 48.00 (0.00–128.40) | 43 | I/II/III (23/10/10) | OS | 7 | Clinical cancer research | 9.619 | (24) |
| GSE50081 | Der SD | 2014 | Canada | 62.16 (5.76–144.48) | 43 | I/II (27/16) | OS | 6 | Journal of thoracic oncology | 6.595 | (25) |
| TCGA | TCGA | 2015 | USA | 17.27 (0.03–174.08) | 474 | I/II/III/IV (232148/84/7) | OS | 8 | – | – |
LUAD, adenocarcinoma; LUSC, squamous cell carcinoma; TCGA, The Cancer Genome Atlas; NR, not report; OS, overall survival; IF, impact factor.
A quality assessment of the eligible datasets included in this meta-analysis has been performed according to Newcastle-Ottawa Quality Assessment Scale (NOS). The quality score span was from 6 to 9 and the mean score was 7.63 for LUAD and the quality score span was from 6 to 8 and the mean score was 7.33 for LUSC. The impact factors of the journals where the studies were published were of high caliber (Table III). Thus, all of those studies were included in following analysis.
Overall survival
Univariate analysis and multivariate analysis were respectively carried out for each dataset. P-values, HRs and 95% CIs of IL-6R mRNA in each article for LUAD and LUSC were shown in Table IV and Figs. 2 and 3.
Table IV.
HR of interleukin-6R mRNA for OS.
| Univariate analysis | Multivariate analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| Datasets | HR | LCI | UCI | P-valuea | HR | LCI | UCI | P-valueb |
| LUAD | ||||||||
| GSE14814c | 0.201 | 0.092 | 0.443 | <0.001 | 0.190 | 0.076 | 0.474 | <0.001 |
| GSE30219d | 0.237 | 0.093 | 0.603 | 0.003 | 0.215 | 0.081 | 0.566 | 0.002 |
| GSE37745c | 0.659 | 0.416 | 1.044 | 0.076 | 0.589 | 0.303 | 1.147 | 0.120 |
| GSE42127c | 0.559 | 0.267 | 1.168 | 0.122 | 0.693 | 0.319 | 1.505 | 0.354 |
| GSE50081e | 0.493 | 0.287 | 0.846 | 0.010 | 0.656 | 0.370 | 1.163 | 0.149 |
| GSE68465f | 0.492 | 0.375 | 0.646 | <0.001 | 0.566 | 0.426 | 0.752 | <0.0001 |
| GSE68571g | 0.492 | 0.217 | 1.117 | 0.089 | 0.602 | 0.256 | 1.418 | 0.246 |
| TCGAh | 0.617 | 0.436 | 0.873 | 0.006 | 0.515 | 0.344 | 0.772 | 0.001 |
| LUSC | ||||||||
| GSE14814i | 1.701 | 0.669 | 4.323 | 0.264 | 1.589 | 0.597 | 4.227 | 0.354 |
| GSE30219d | 1.440 | 0.748 | 2.775 | 0.275 | 1.559 | 0.797 | 3.050 | 0.194 |
| GSE37745c | 2.599 | 1.344 | 5.029 | 0.005 | 4.071 | 1.436 | 11.537 | 0.008 |
| GSE42127c | 9.906 | 1.321 | 74.297 | 0.026 | 8.577 | 1.084 | 67.868 | 0.042 |
| GSE50081j | 0.442 | 0.149 | 1.312 | 0.141 | 0.387 | 0.119 | 1.257 | 0.114 |
| TCGAh | 1.375 | 0.954 | 1.981 | 0.088 | 1.308 | 0.899 | 1.903 | 0.161 |
P<0.05 indicated in bold.
Data were compared using Univaruate Cox's proportional hazards model
data were compared using multivariate Cox's proportional hazards model. Multivariate analysis adjusted variables
age, sex, stage, post-treatment
age, sex, T, N
age, sex, stage, T, N
age, sex, T, N, adjuvant-chemotherapy, adjuvant-radio-therapy
age, sex, tumor size, stage
age, sex, stage, T, N, M
age, stage, post-treatment
age, sex, stage, T. HR, hazard ratio; LCI, lower 95% confidence interval; UCI, upper 95% confidence interval; TCGA, The Cancer Genome Atlas; LUAD, adenocarcinoma; LUSC, squamous cell carcinoma.
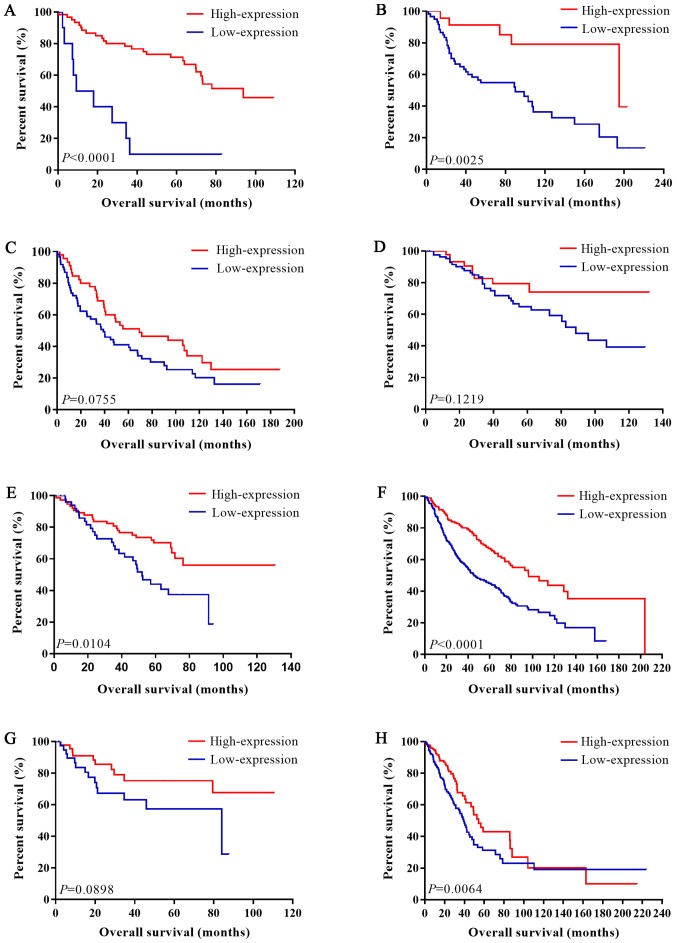
Figure 2.
Association between IL-6R mRNA expression level and overall survival in patients with LUAD. (A) Kaplan-Meier analysis of datasets (A) GSE14814, (B) GSE30219, (C) GSE37745 (D) GSE42127, (E) GSE50081, (F) GSE68465 and (G) GSE68571. (H) Kaplan-Meier analysis of The Cancer Genome Atlas LUAD data. IL, interleukin; LUAD, lung adenocarcinoma.
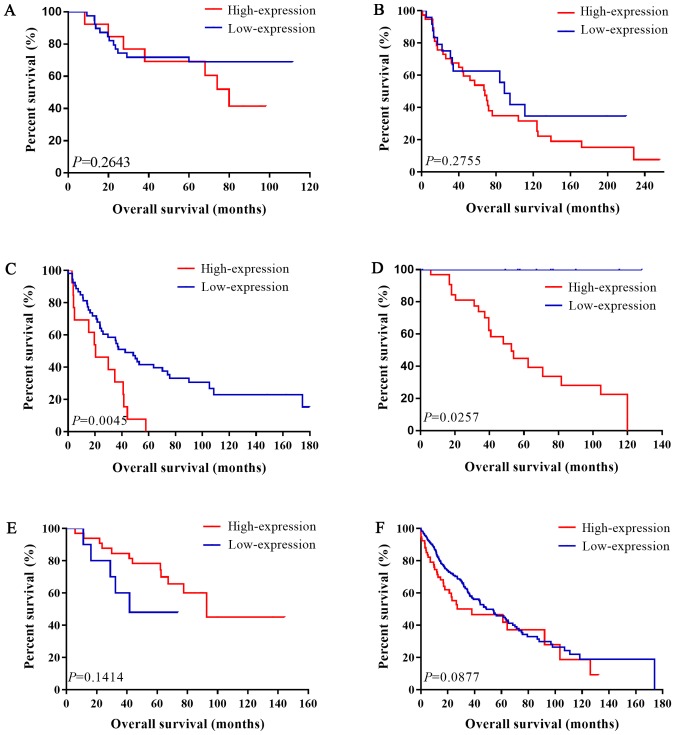
Figure 3.
Association between IL-6R mRNA expression level and overall survival in LUSC patients. Kaplan-Meier analysis of datasets (A) GSE14814, (B) GSE30219, (C) GSE37745, (D) GSE42127 and (E) GSE50081. (F) Kaplan-Meier analysis of The Cancer Genome Atlas LUSC data. LUSC, lung squamous cell carcinoma; IL, interleukin.
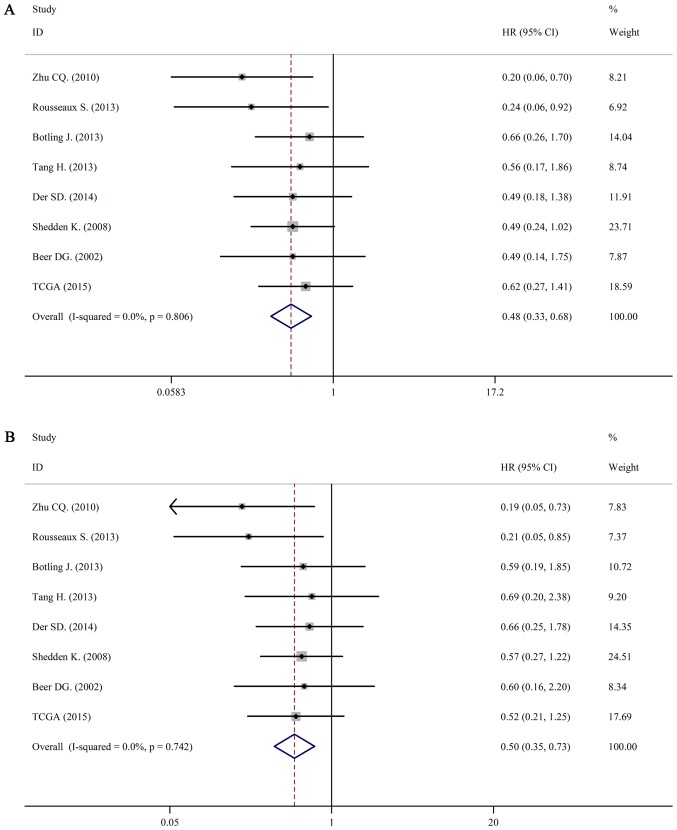
For LUAD, there was no obvious statistical heterogeneity in all of those datasets both in univariate survival analysis and multivariate survival analysis (I2=0.0%, P=0.806; I2=0.0%, P=0.742), a fixed-effects model was used to calculate the pool HRs. Our analysis demonstrated that a higher expression of IL-6R mRNA was significantly associated with better overall survival (OS) (pooled HR=0.50; 95% CI: 0.33–0.68 in univariate analysis; pooled HR=0.50; 95% CI: 0.35–0.73 in multivariate analysis). The forest plots of study-specific HRs for OS were presented in Fig. 4.
Figure 4.
Forest plots of studies evaluating HRs of high IL-6R mRNA expression in lung adenocarcinoma for overall survival. (A) Univariate analysis. HR=0.48, 95% CI: 0.33–0.68. (B) Multivariate analysis. HR=0.50, 95% CI: 0.35–0.73. HR, hazard radio; CI, confidence interval; IL, interleukin.
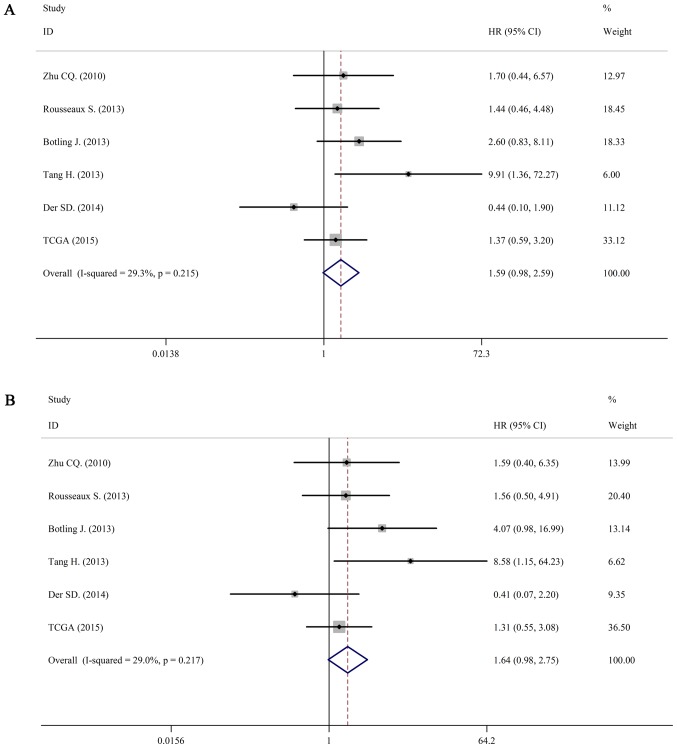
For LUSC, we still used a fixed-effects model to pool the HRs (I2=29.3%, P=0.215; I2=36.5%, P=0.217). Interestingly, there was no association between IL-6 mRNA and OS in patients with LUSC (pooled HR=1.59; 95% CI: 0.98–2.59 in univariate analysis; pooled HR=1.64; 95% CI: 0.98–2.75 in multivariate analysis). The forest plots of study-specific HRs for OS were presented in Fig. 5.
Figure 5.
Forest plots of studies evaluating HRs of high IL-6R mRNA expression in lung squamous cell carcinoma for overall survival. (A) Univariate analysis. HR=1.59, 95% CI: 0.98–2.59. (B) Multivariate analysis. HR=1.64, 95% CI: 0.98–2.75. HR, hazards radio; CI, confidence interval.
Correlation between IL-6 and IL-6R, IL-6R and IL-6ST
The IL-6 receptor is a protein complex consisting of an alpha chain, IL-6R, and IL-6 signal transducer (IL-6ST). Relationship between the mRNA expression of IL-6 and IL-6R, and between IL-6R and IL-6ST were all analyzed in those datasets. We then performed a meta-analysis based on Fisher's z transformation. Interestingly, IL-6 and IL-6R were negatively correlated in LUAD (pooled r=−0.199, P<0.001), while they were positively correlated in LUSC (pooled r=0.288, P=0.001). The correlation coefficient between IL-6R and IL-6ST in LUAD was similar with correlation coefficient in LUSC (pooled r=0.331, P<0.001 in LUAD and pooled r=0.334, P<0.001 in LUSC; Table V).
Table V.
Correlation between IL-6 and IL-6R, IL-6R and IL-6ST.
| IL-6 vs. IL-6R | IL-6R vs. IL-6ST | |||
|---|---|---|---|---|
| Datasets | r | P-value | r | P-value |
| LUAD | ||||
| GSE14814 | −0.345 | 0.004 | 0.141 | 0.245 |
| GSE30219 | −0.515 | <0.001 | 0.506 | <0.001 |
| GSE37745 | −0.107 | 0.272 | 0.294 | 0.002 |
| GSE42127 | 0.041 | 0.642 | 0.243 | 0.005 |
| GSE50081 | −0.207 | 0.019 | 0.363 | <0.001 |
| GSE68465 | −0.166 | 0.001 | 0.271 | <0.001 |
| GSE68571 | −0.194 | 0.073 | 0.191 | 0.079 |
| TCGA | −0.165 | <0.001 | 0.527 | <0.001 |
| Pooled r | −0.199 | <0.001 | 0.331 | <0.001 |
| LUSC | ||||
| GSE14814 | 0.395 | 0.004 | 0.073 | 0.606 |
| GSE30219 | 0.036 | 0.782 | 0.469 | 0.000 |
| GSE37745 | 0.417 | 0.001 | 0.369 | 0.002 |
| GSE42127 | 0.610 | <0.001 | 0.272 | 0.077 |
| GSE50081 | 0.069 | 0.661 | 0.155 | 0.320 |
| TCGA | 0.178 | <0.001 | 0.465 | <0.001 |
| Pooled r | 0.288 | 0.001 | 0.334 | <0.001 |
P<0.05 indicated in bold. Correlations were calculated using Pearson correlation coefficient and pooled correlations were performed using a meta-analysis based on Fisher's z transformation. IL, interleukin; TCGA, The Cancer Genome Atlas; LUAD, adenocarcinoma; LUSC, squamous cell carcinoma.
IL-6R mRNA expression in tumor tissues and adjusted normal tissues
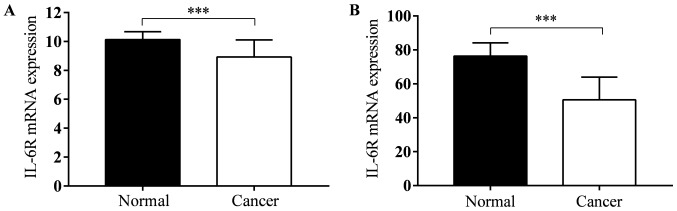
TCGA lung adenocarcinoma and TCGA lung squamous cell carcinoma datasets contains gene expression data both in tumor tissues and normal tissues. There were 57 and 51 pairs in LUAD and LUSC. In both two types of lung cancer, IL-6R mRNA expression level in tumor tissues was less than normal tissues (P<0.001; Fig. 6).
Figure 6.
IL-6R mRNA expression in tumor tissues and adjusted normal tissues for (A) lung adenocarcinoma and (B) lung squamous cell carcinoma. Data are presented as the mean ± standard deviation and were compared using a two-tailed, paired t-test. ***P<0.001. IL, interleukin.
Biological processes and pathway analysis
Functional enrichment analysis was performed on IL-6R and the most related genes (all those genes were shown in Table II). Table VI lists the top five biological processes and pathways enriched in IL-6R correlated genes in LUAD. One of the most significant biological processes is cell division (GO:0051301, P=4.038E-20). Results also showed that those genes enriched in mitotic division. GO:0007067, mitotic nuclear division, GO:0007059, chromosome segregation, GO:0000070, mitotic sister chromatid segregation and GO:0045143, homologous chromosome segregation.
Table VI.
Top 5 biological processes and pathway of interleukin-6R and associated genes in lung adenocarcinoma.
| Analysis | ID | Biological processes | P-value | Genes |
|---|---|---|---|---|
| Biological processes | GO:0051301 | Cell division | 4.038×10−20 | CDK1, CDC6, PSRC1, TPX2, BIRC5, PTTG1, UBE2C, CDC25C, CDC25A, CCNB1, FAM83D, SPC25, NCAPH, CCNB2, CCND3, ZWINT, CDCA2, CKS2, SKA3, SKA1, CDCA5, ASPM, CDCA3 |
| GO:0007067 | Mitotic nuclear division | 3.084×10−14 | CENPN, CDK1, NUF2, PTTG1, CDC25C, CDC25A, FAM83D, SPC25, CCNB2, PLK1, ZWINT, CDCA2, SKA3, CENPW, SKA1, ASPM | |
| GO:0007059 | Chromosome segregation | 1.529×10−09 | SPC25, CENPN, KIF11, OIP5, NEK2, HJURP, SKA3, CENPW, BIRC5, SKA1 | |
| GO:0000070 | Mitotic sister chromatid segregation | 1.186×10−06 | CDCA8, PLK1, NEK2, SPAG5, KIF18B, ESPL1 | |
| GO:0045143 | Homologous chromosome | 5.275×10−06 | PLK1, ESPL1, PTTG1 | |
| Pathway | bta04110 | Cell cycle | 8.584×10−15 | CDK1, CDC6, TTK, CDC20, ESPL1, CHEK1, PTTG1, CDC25C, CDC25A, CCNB1, CDC45, CCNB2, MAD2L1, CCND3, PLK1, BUB1, BUB1B |
| bta04114 | Oocyte meiosis | 2.573×10−07 | CDK1, MAD2L1, LADCY9, PLK1, BUB1, FBXO5, ESPL1, CDC20, PTTG1, CDC25C | |
| bta04914 | Progesterone-mediated oocyte maturation | 3.845×10−06 | CCNB1, CDK1, MAD2L1, CCNB2, LADCY9, PLK1, BUB1, CDC25C, CDC25A | |
| bta04115 | p53 signaling pathway | <0.001 | CCNB1, CDK1, CCNB2, CCND3, RRM2, CHEK1, GTSE1 | |
| bta05166 | HTLV-I infection | 0.008 | MAD2L1, CCND3, POLE2, LADCY9, BUB1B, CDC20, CHEK1, PTTG1 |
The most important pathway in LUAD is cell cycle (bta04110, P=8.584E-15). As with the results of biological processes, pathway analysis also shows that these genes are involved in cell division, eg. oocyte meiosis (bta04114, P=2.573E-07), progesterone-mediated oocyte maturation (bta04914, P=3.845E-06). The other two important pathways are p53 signaling pathway (bta04115, P<0.001) and HTLV-I infection (bta05166, P=0.008).
Table VII lists the top five biological processes and pathways enriched in LUSC. The most significant biological processes are regulation of cell proliferation (GO:0042127, P=0.001) and positive regulation of osteoclast differentiation (GO:0045672, P=0.003). Result also showed that those genes are involved in immune response (GO:0006955, P=0.003). The other two biological processes are trans-membrane receptor protein tyrosine kinase signaling pathway (GO:0007169, P=0.006) and integrin-mediated signaling pathway (GO:0007229, P=0.006).
Table VII.
Top 5 biological processes and pathway of interleukin-6R and associated genes in lung squamous cell carcinoma.
| Analysis | ID | Biological processes | P-value | Genes |
|---|---|---|---|---|
| Biological processes | GO:0042127 | Regulation of cell proliferation | 0.001 | TNFRSF1A, TNFRSF1B, FGR, PTK2B, JUNB, BTK |
| GO:0045672 | Positive regulation of osteoclast differentiation | 0.003 | FOS, KLF10, CCR1 | |
| GO:0006955 | Immune response | 0.003 | TNFRSF1A, TNFRSF1B, CCR1, CD4, CTSH, LCP2 | |
| GO:0007169 | Trans-membrane receptor protein tyrosine kinase signaling pathway | 0.006 | DOK2, FGR, LCP2, BTK | |
| GO:0007229 | Integrin-mediated signaling pathway | 0.006 | ITGAL, FGR, PTK2B, TYROBP | |
| Pathway | ptr04650 | Natural killer cell mediated cytotoxicity | <0.001 | PTPN6, ITGAL, PTK2B, PRKCB, LCP2, TYROBP |
| ptr04380 | Osteoclast differentiation | 0.002 | FOS, TNFRSF1A, JUNB, LCP2, BTK, TYROBP | |
| ptr04611 | Platelet activation | 0.011 | VWF, LADCY9, PPP1CC, LCP2, BTK | |
| ptr04060 | Cytokine-cytokine receptor interaction | 0.015 | TNFRSF1A, TNFRSF1B, IL2RB, CCR1, CXCL16, IL6R | |
| ptr05166 | HTLV-I infection | 0.028 | FOS, ITGAL, TNFRSF1A, IL2RB, LADCY9, BUB1B |
The most important pathway in LUSC is natural killer cell mediated cytotoxicity (ptr04650, P<0.001). And the others are osteoclast differentiation (ptr04380, P=0.002), platelet activation (ptr04611, P=0.011), cytokine-cytokine receptor interaction (ptr04060, P=0.015) and HTLV-I infection (ptr05166, P=0.028).
Discussion
Lung cancer is a serious threat to public health in the world. Cytokines play important roles in tumorigenesis as well as immune surveillance of lung cancer. LUAD and LUSC are two main pathological subtypes of NSCLC. To study the association of various cytokines and their receptors with clinical parameters of NSCLC is an important step for further mechanistic investigations and provides insight into new therapeutic targets. Our study revealed that higher expression levels of IL-6R mRNA in tumor tissues were positively associated with better overall survival in LUAD. These data suggest an antitumoral role of IL-6R signaling.
Recent works have demonstrated that IL-6 signaling pathway plays an important role in the immune response (28). Several studies have demonstrated the pro-tumor effect of IL-6 in NSCLC. Our study revealed a predictive value of IL-6R mRNA expression in LUAD. It suggested that higher IL-6R mRNA was associated with better survival. However, the prognostic value of IL-6R was not shown in LUSC. It is speculated that LUAD and LUSC arise from distinct cells based on the histopathological appearance and gene expression signatures. It is generally accepted that LUAD originates mainly from alveolar epithelial cells and LUSC is possibly derived from basal cells (29,30). LUAD and LUSC undergo distinct developmental processes. Several articles also have showed different result between LUAD and LUSC. One study showed that an increased expression of the embryonic stem cells gene set was associated with overall survival in LUAD. However, there was no correlation in LUSC (31). Meanwhile, other study found that the expression levels of PTN1 genes were associated with survival in LUAD but not LUSC (32). The different results may mainly due to its different cellular origins, developmental stages and tumor microenvironment.
In LUAD, enrichment analysis of IL-6R and its most relevant genes showed that the most significant biological processes were cell division and mitotic division. That means those genes mainly involved in cell cycle progression in LUAD. While in LUSC, the most significant biological processes were regulation of cell proliferation and several signaling pathway. Pathways analysis revealed that those genes were involved in natural killer cell mediated cytotoxicity and platelet activation, meaning that they were mostly involved in tumor angiogenesis, invasion and metastasis. Genes that mostly related with IL-6R and the most significant biological processes were all different in LUAD and LUSC. That means the tumor microenvironment in both cancers were discriminate. This prompts us that studies on LUAD and LUSC should be analyzed separately.
In our report, although there was no statistical significance between IL-6R mRNA expression and OS in LUSC, the lower 95% CI limit of HR (0.98) were very close to 1, showing a trend that IL-6R may be a risk factor for LUSC. Considering the small sample size in each study, further investigations with a larger scale of samples are needed to confirm this result.
IL-6R is a part of the receptor for IL-6 which binds to IL-6 with low affinity, but does not transduce a signal (33). IL-6ST is necessary for this signal activation. Correlation analysis showed that IL-6 and IL-6R were negative correlated in LUAD while they were positive correlated in LUSC, indicating that the higher the expression of IL-6R, the lesser the expression of IL-6 in LUAD and the higher the expression of IL-6 in LUSC. And the pooled correlation coefficient of IL-6R and IL-6ST were positive both in LUAD and LUSC. However, Brooks et al have found that IL-6R protein displayed a positive correlation with IL-6 in LUAD (34). The correlation coefficient of IL-6 and IL-6R in LUAD and LUSC were −0.199 and 0.288, respectively. Both of them were less than 0.3, showing a weak correlation. In our report, the correlation between IL-6 and IL-6R was calculated based on public datasets, showing a possible phenomenon. The possible biological process or interaction between these cytokines should be verified at a tissue, cellular or molecular level by subsequent experimental verification.
In TCGA dataset, we found that IL-6R mRNA expression level in tumor tissues was less than normal tissues in both LUAD and LUSC. Balabko et al have demonstrated the same trend, and they also found that IL-6R mRNA was found significantly induced in the tumoural region of LUAD as compared to LUSC (35). STAT3, a transcription factor downstream of IL-6R, has also been found increased and phosphorylated in LUAD while there was no phosphorylation in LUSC (35). Considering the different effect of IL-6R mRNA in LUAD and LUSC, we deduce that the expression level of IL-6R mRNA can affect the expression level of IL-6 and the activation of downstream pathways and can affect the most important biological processes it involved in.
However, some details need to be further refined. Firstly, this study included only 8 eligible datasets for LUAD and 6 studies for LUSC, which resulted in relatively insufficiency data in the subgroup analyses. Secondly, sample size of LUSC in each study was smaller than LUAD, further articles with a larger scale of samples are needed to confirm the result. Thirdly, these results were calculated based on public datasets and these results should be verified using cells or tumor samples.
In conclusion, our results showed that mRNA levels of IL-6R in LUAD was associated with better prognosis and can potentially be used as a prognostic marker for this cancer. While in LUSC, although there was no statistically significance between IL-6R mRNA and OS in LUSC, taking the small sample size of each dataset, the results should be regarded cautiously. Further prospective studies available of pivotal parameters are needed to verify the prognosis value of IL-6R in LUAD and LUSC patients.
Acknowledgements
Not applicable.
Funding
This study was supported by funding from the National Science and Technology Support Program (grant no. 2015BAI12B12), the National Natural Science Foundation of China (grant nos. 31570877 and 31570908), the Special Funds of Science and Technology of the People's Livelihood Construction Condition of Jiangsu Province (grant no. BL2014034), the Science and Technology Bureau foundation application project of Changzhou (grant no. CJ20159018), the Key R&D Project of Science and Technology Department of Jiangsu Province (grant no. BE2015633) and the Program of Jiangsu Engineering Research Center for Tumor Immunotherapy (grant no. BM2014404).
Availability of data and materials
The datasets analyzed during the current study are available in the Cancer Genome Atlas (cancergenome.nih.gov/) and the Gene Expression Ominibus database (ncbi.nlm.nih.gov/gds/).
Authors' contributions
BL, JJ and YS designed the study and interpreted the results. LL and CY collected the public datasets and reorganized and cleaned the data. QC, CY and BX designed the experiments and helped write the manuscript. QC and BX wrote and organized the manuscript, with editorial input from YS, JJ and BL. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DH, Schiller JH, Bunn PA., Jr Recent clinical advances in lung cancer management. J Clin Oncol. 2014;32:973–982. doi: 10.1200/JCO.2013.53.1228. [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, et al. International association for the study of lung cancer/American thoracic society/European respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhan C, Yan L, Wang L, Sun Y, Wang X, Lin Z, Zhang Y, Shi Y, Jiang W, Wang Q. Identification of immunohistochemical markers for distinguishing lung adenocarcinoma from squamous cell carcinoma. J Thorac Dis. 2015;7:1398–1405. doi: 10.3978/j.issn.2072-1439.2015.07.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kishimoto T, Akira S, Narazaki M, Taga T. Interleukin-6 family of cytokines and gp130. Blood. 1995;86:1243–1254. [PubMed] [Google Scholar]
- 8.Hodge DR, Hurt EM, Farrar WL. The role of IL-6 and STAT3 in inflammation and cancer. Eur J Cancer. 2005;41:2502–2512. doi: 10.1016/j.ejca.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Haura EB, Livingston S, Coppola D. Autocrine interleukin-6/interleukin-6 receptor stimulation in non-small-cell lung cancer. Clin Lung Cancer. 2006;7:273–275. doi: 10.3816/CLC.2006.n.006. [DOI] [PubMed] [Google Scholar]
- 10.Yanagawa H, Sone S, Takahashi Y, Haku T, Yano S, Shinohara T, Ogura T. Serum levels of interleukin 6 in patients with lung cancer. Br J Cancer. 1995;71:1095–1098. doi: 10.1038/bjc.1995.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Songür N, Kuru B, Kalkan F, Ozdilekcan C, Cakmak H, Hizel N. Serum interleukin-6 levels correlate with malnutrition and survival in patients with advanced non-small cell lung cancer. Tumori. 2004;90:196–200. doi: 10.1177/030089160409000207. [DOI] [PubMed] [Google Scholar]
- 12.Chang CH, Hsiao CF, Yeh YM, Chang GC, Tsai YH, Chen YM, Huang MS, Chen HL, Li YJ, Yang PC, et al. Circulating interleukin-6 level is a prognostic marker for survival in advanced nonsmall cell lung cancer patients treated with chemotherapy. Int J Cancer. 2013;132:1977–1985. doi: 10.1002/ijc.27892. [DOI] [PubMed] [Google Scholar]
- 13.Seike T, Fujita K, Yamakawa Y, Kido MA, Takiguchi S, Teramoto N, Iguchi H, Noda M. Interaction between lung cancer cells and astrocytes via specific inflammatory cytokines in the microenvironment of brain metastasis. Clin Exp Metastasis. 2011;28:13–25. doi: 10.1007/s10585-010-9354-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noda M, Yamakawa Y, Matsunaga N, Naoe S, Jodoi T, Yamafuji M, Akimoto N, Teramoto N, Fujita K, Ohdo S, Iguchi H. IL-6 receptor is a possible target against growth of metastasized lung tumor cells in the brain. Int J Mol Sci. 2012;14:515–526. doi: 10.3390/ijms14010515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yi H, Cho HJ, Cho SM, Jo K, Park JA, Kim NH, Amidon GL, Kim JS, Shin HC. Blockade of interleukin-6 receptor suppresses the proliferation of H460 lung cancer stem cells. Int J Oncol. 2012;41:310–316. doi: 10.3892/ijo.2012.1447. [DOI] [PubMed] [Google Scholar]
- 16.Kim NH, Kim SK, Kim DS, Zhang D, Park JA, Yi H, Kim JS, Shin HC. Anti-proliferative action of IL-6R-targeted antibody tocilizumab for non-small cell lung cancer cells. Oncol Lett. 2015;9:2283–2288. doi: 10.3892/ol.2015.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ando K, Takahashi F, Motojima S, Nakashima K, Kaneko N, Hoshi K, Takahashi K. Possible role for tocilizumab, an anti-interleukin-6 receptor antibody, in treating cancer cachexia. J Clin Oncol. 2013;31:e69–e72. doi: 10.1200/JCO.2012.44.2020. [DOI] [PubMed] [Google Scholar]
- 18.Ando K, Takahashi F, Kato M, Kaneko N, Doi T, Ohe Y, Koizumi F, Nishio K, Takahashi K. Tocilizumab, a proposed therapy for the cachexia of Interleukin6-expressing lung cancer. PLoS One. 2014;9:e102436. doi: 10.1371/journal.pone.0102436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 20.Budczies J, Klauschen F, Sinn BV, Győrffy B, Schmitt WD, Darb-Esfahani S, Denkert C. Cutoff Finder: A comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, Thomas RK, Naoki K, Ladd-Acosta C, Liu N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28:4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rousseaux S, Debernardi A, Jacquiau B, Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY, Lantuejoul S, Hainaut P, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. 2013;5:186ra66. doi: 10.1126/scitranslmed.3005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Botling J, Edlund K, Lohr M, Hellwig B, Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Pontén F, et al. Biomarker discovery in non-small cell lung cancer: Integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. 2013;19:194–204. doi: 10.1158/1078-0432.CCR-12-1139. [DOI] [PubMed] [Google Scholar]
- 24.Tang H, Xiao G, Behrens C, Schiller J, Allen J, Chow CW, Suraokar M, Corvalan A, Mao J, White MA, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res. 2013;19:1577–1586. doi: 10.1158/1078-0432.CCR-12-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Der SD, Sykes J, Pintilie M, Zhu CQ, Strumpf D, Liu N, Jurisica I, Shepherd FA, Tsao MS. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. J Thorac Oncol. 2014;9:59–64. doi: 10.1097/JTO.0000000000000042. [DOI] [PubMed] [Google Scholar]
- 26.Director's Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma, corp-author. Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, et al. Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 28.Yu H, Jove R. The STATs of cancer-new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 29.Han X, Li F, Fang Z, Gao Y, Li F, Fang R, Yao S, Sun Y, Li L, Zhang W, et al. Transdifferentiation of lung adenocarcinoma in mice with Lkb1 deficiency to squamous cell carcinoma. Nat Commun. 2014;5:3261. doi: 10.1038/ncomms4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutherland KD, Berns A. Cell of origin of lung cancer. Mol Oncol. 2010;4:397–403. doi: 10.1016/j.molonc.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassan KA, Chen G, Kalemkerian GP, Wicha MS, Beer DG. An embryonic stem cell-like signature identifies poorly differentiated lung adenocarcinoma but not squamous cell carcinoma. Clin Cancer Res. 2009;15:6386–6390. doi: 10.1158/1078-0432.CCR-09-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li P, Zhang L, Yu X, Tong R, Di X, Mao Y, Gao Y, Zhang K, Feng L, Cheng S. Proliferation genes in lung development associated with the prognosis of lung adenocarcinoma but not squamous cell carcinoma. Cancer Sci. 2018;109:308–316. doi: 10.1111/cas.13456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montero-Julian FA. The soluble IL-6 receptors: Serum levels and biological function. Cell Mol Biol (Noisy-le-grand) 2001;47:583–597. [PubMed] [Google Scholar]
- 34.Brooks GD, McLeod L, Alhayyani S, Miller A, Russell PA, Ferlin W, Rose-John S, Ruwanpura S, Jenkins BJ. IL6 trans-signaling promotes KRAS-driven lung carcinogenesis. Cancer Res. 2016;76:866–876. doi: 10.1158/0008-5472.CAN-15-2388. [DOI] [PubMed] [Google Scholar]
- 35.Balabko L, Andreev K, Burmann N, Schubert M, Mathews M, Trufa DI, Reppert S, Rau T, Schicht M, Sirbu H, et al. Increased expression of the Th17-IL-6R/pSTAT3/BATF/RorγT-axis in the tumoural region of adenocarcinoma as compared to squamous cell carcinoma of the lung. Sci Rep. 2014;4:7396. doi: 10.1038/srep07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available in the Cancer Genome Atlas (cancergenome.nih.gov/) and the Gene Expression Ominibus database (ncbi.nlm.nih.gov/gds/).