Abstract
As a mitotic spindle checkpoint gene, baculoviral IAP repeat containing 5 (BIRC5) serves pivotal roles in the development of various types of malignant tumors. In the present study, the expression of BIRC5 in patients with different stages of esophageal squamous cell cancer (ESCC) was investigated. The effect of BIRC5 on the migratory and invasive abilities of different ESCC cell lines was analyzed. Additionally, the effect of BIRC5 on the angiogenesis-associated factor vascular endothelial growth factor, brain-specific angiogenesis inhibitor 1 and methyl-CpG binding domain protein 2 was examined. In addition, the interaction between BIRC5 and the phosphatidylinositol 3-kinase (PI3K)/Akt pathway was analyzed. It was determined that BIRC5 is able to inhibit the migration and invasion of tumor cells, and regulate the expression of angiogenesis-associated factors. In addition, the PI3K/Akt signaling pathway is able to regulate the expression of BIRC5, which affects the development of ESCC.
Keywords: esophageal squamous cell cancer, baculoviral IAP repeat containing 5, phosphatidylinositol 3-kinase/Akt pathway
Introduction
As a type of malignant tumor that develops in the esophagus, esophageal cancer affects ~0.3% of females and 0.8% of males (1). Currently, esophageal cancer is one of the least studied but one of the deadliest types of malignant tumor due to a low survival rate and the extremely aggressive nature of esophageal cancer cells (2,3). The progression of esophageal cancer is a complex process involving various internal and environment factors (1,3). Previous studies have demonstrated that risk factors for esophageal cancer include drinking hot water, poor oral health, smoking, consumption of red meat, alcohol, low socioeconomic status and numerous other factors (3,4). With the development of treatments for malignant tumors, the overall survival rates of patients with a number of different of cancer types have been significantly improved over the past decades (5). However, the treatment of esophageal cancer is unreliable for providing satisfactory outcomes due to an unclear understanding of its pathogenesis (1,6). Therefore, understanding the mechanisms underlying esophageal cancer to improve the treatment of this disease is of great clinical significance.
Mitotic spindle checkpoint signaling, which is essential for normal cell division, has been indicated to be involved in the development of a variety of cancer types (7). A previous study indicated that the alternative reading frame tumor suppressor may maintain mitotic checkpoint fidelity to prevent chromosomal instability (8), indicating the crosstalk between mitotic checkpoint fidelity and tumor development. As a mitotic spindle checkpoint gene, baculoviral IAP repeat containing 5 (BIRC5) was determined to have the ability to regulate cell apoptosis (9). BIRC5 was also indicated to be involved in the onset and development of various types of malignant tumors, including breast (10), colorectal (11) and gastric cancer (12). Therefore, BIRC5 may also serve important roles in the development of esophageal cancer. The level of BIRC5 expression was reported to be increased in esophageal squamous cell carcinoma tissues compared with normal human tissues (13). However, the specific roles of BIRC5 in the pathogenesis of esophageal cancer have not been well studied.
In the present study, expression levels of BIRC5 in patients with different stages of esophageal cancer and in different esophageal cancer cell lines were detected. The effect of BIRC5 on tumor cell migration and invasion was explored. The association between BIRC5 and angiogenesis-associated factor vascular endothelial growth factor (VEGF), brain-specific angiogenesis inhibitor 1 (BAI1) and methyl-CpG binding domain protein 2 (MBD2), and the phosphatidylinositol 3-kinase (PI3K)/Akt pathway was also investigated.
Materials and methods
Patients
A total of 178 patients with esophageal cancer diagnosed via pathological examination and imaging methods, including mini-probe ultrasonography, spiral computed tomography and magnetic resonance imaging were selected at The Fourth Affiliated Hospital of Hebei Medical University (Shijiazhuang, China) from May 2015 to September 2016. The patients included 81 males and 97 females, with an age range of 35–89 years old and a mean age of 54±7.8 years. The inclusion criteria are as follows: i) Diagnosis of malignant esophageal tumor and ii) signing of informed consent. The exclusion criteria are as follows: i) Patients with other types of malignant tumors; ii) patients with severe coagulation dysfunction and iii) patients who refused to sign the informed consent. The present study was approved by the Ethics Committee of The Fourth Affiliated Hospital of Hebei Medical University. According to the pathological examination and imaging results, the patients were divided into different stages of esophageal cancer according to the following criteria, as previously described (14): T0, no detection of primary tumor (n=30); Tis, high-grade dysplasia (n=25); T1, muscularis mucosae, lamina propria or submucosa invaded by the tumor (n=29); T2, muscularis propria invaded by the tumor (n=29); T3, adventitia invaded by the tumor (n=32); T4, adjacent structures invaded by the tumor (n=33). No significant difference in age, sex and other basic information was determined among patients in different stages of esophageal cancer.
Cell lines and cell culture
Homo sapiens esophageal normal cell line Het-1A, esophageal cancer cell line KYSE510 and EC9706 were purchased from American Type Culture Collection (Manassas, VA, USA). All cells were cultured in RPMI-1640 medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) containing 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in an incubator at 37°C with an atmosphere containing 5% CO2. FBS was not added for cells used for treatment with the PI3K/Akt pathway activator SC79 (10 µM).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tumor tissue and in vitro cultured cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and cDNA was synthesized using SuperScript III reverse transcriptase kit (Thermo Fisher Scientific, Inc.). Reverse transcription conditions were: 50 min at 50°C and 5 min at 85°C. SYBR®-Green Real-Time PCR Master Mix (Thermo Fisher Scientific, Inc.) was used to prepare PCR reaction system, and PCR reactions were performed on a CFX96 Touch™ Real-Time PCR Detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The following primers were used in PCR: BIRC5, forward, 5′-TGCCTGGCAGCCCTTTC-3′ and reverse, 5′-CCTCCAAGAAGGGCCAGTTC-3′; VEGF, forward, 5′-ATCTTCAAGCCATCCTGTGTGC-3′ and reverse, 5′-CAAGGCCCACAGGGATTTTC-3′; BAI1, forward, 5′-GCAAACCAAGTTCTGCAACAT-3′ and reverse, 5′-GCAAACCAAGTTCTGCAACAT-3′; MBD2, forward, 5′-CCCTGCTGTTTGGCTTAACACATCTC-3′ and reverse, 5′-TGCGTACTTGCTGTACTCGCTCTTC-3′; GAPDH, forward, 5′-GAGTCAACGGATTTGGTCGT-3′ and reverse, 5′-TTGATTTTGGAGGGATCTCG-3′ for GAPDH. The PCR reaction conditions were: 95°C for 30 sec, followed by 40 cycles of 95°C for 10 sec and 50°C for 45 sec. The data were analyzed using the 2−∆∆Cq method (15), and the relative expression level of each gene was normalized to endogenous control GAPDH.
Small-interfering (si)RNA transfection
BIRC5 siRNA (catalog no. 121294) was purchased from Thermo Fisher Scientific, Inc. Silencer® negative control #1 siRNA (Thermo Fisher Scientific, Inc.) was used as a negative control. Oligofectamine™ reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect 40 nM siRNA into 5×105 cells by incubation with the cells at 37°C in a CO2 incubator for 4 h, followed by incubation in RPMI-1640 medium (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) at 37°C for 48 h prior to harvest.
Transwell migration and invasion assays
Cell migratory ability of each cell line was measured using Transwell cell migration assay (polyester inserts, BD Biosciences, Franklin Lakes, NJ, USA). In brief, ~5×104 cells were transferred to the upper chamber containing RPMI-1640 medium without FCS, while RPMI-1640 medium (Thermo Fisher Scientific, Inc.) containing 20% FCS (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used to fill the lower chamber. The membrane was collected 24 h later, followed by staining for 20 min with 0.5% crystal violet at room temperature. Following staining, the cells on the membrane were counted under a light microscope (magnification, ×20; Olympus Corporation, Tokyo, Japan). Cell invasion assay was performed using the same method except that Matrigel (cat no. 356234; EMD Millipore, Billerica, MA, USA) was used to coat the upper chamber prior to experiment. Each experiment was repeated 3 times to calculate the mean value.
Western blot analysis
Total protein was extracted from cells using Cell Lysis Solutions (Thermo Fisher Scientific, Inc.), and BCA method was used to measure protein concentration. Following denaturation, 30 µg protein (mass per lane) from each sample was subjected to electrophoresis using 10% SDS-PAGE gel, followed by transfer to polyvinylidene fluoride membranes. Following washing with TBS with Tween 20 (TBST) buffer three times for 10 min, the membrane was blocked with 5% skimmed milk for 2 h at room temperature. The membranes were then incubated with corresponding primary antibodies including rabbit anti-BIRC5 polyclonal antibody (dilution, 1:1,000; catalog no. MBS151070; MyBioSource, Inc., San Diego, CA, USA), rabbit anti-VEGF polyclonal antibody (dilution, 1:1,000; catalog no. bs-0279R; Biomathematics and Statistics Scotland, Edinburgh, Scotland), rabbit anti-BAI1 polyclonal antibody (dilution, 1:1,000; catalog no. MBS8242484; MyBioSource, Inc.), rabbit anti-MBD2 polyclonal antibody (dilution, 1:1,000; catalog no. MBS126361; MyBioSource, Inc.), rabbit anti-phospho (p)-Akt polyclonal antibody (dilution, 1:1,000; catalog no. MBS330024; MyBioSource, Inc.) and rabbit anti-GAPDH polyclonal antibody (dilution, 1:1,000; catalog no. MBS9216842; MyBioSource, Inc.) overnight at 4°C. The membrane was then washed with TBST three times (10 min each time), followed by incubation with goat anti-rabbit IgG-horseradish peroxidase secondary antibody (dilution, 1:1,000; catalog no. MBS435036; MyBioSource, Inc.) at room temperature for 2 h. Following washing, chemiluminescence method was used to detect the signal, and the relative expression level of each protein was calculated by normalizing to the endogenous control GAPDH using the ImageJ 1.48 software (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
SPSS statistical software (version 19.0; IBM Corp., Armonk, NY, USA) was used for statistical analysis. All data are expressed as the mean ± standard deviation. Comparisons between two groups of data were performed using an unpaired Student's t-test. Comparisons among multiple groups were performed using one-way analysis of variance, followed by the least significant difference test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression level of BIRC5 mRNA in patients with different stages of esophageal cancer
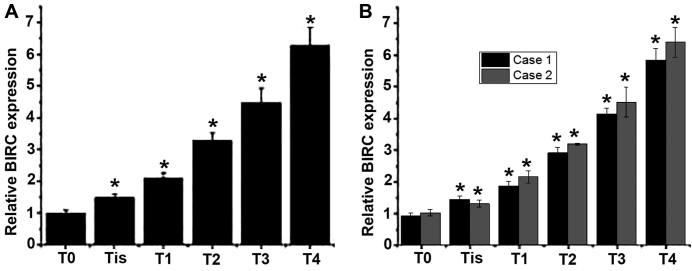
Total RNA was extracted from tumor tissues collected from patients with different stages of esophageal cancer to detect the mRNA expression of BIRC5. As presented in Fig. 1A and B, an increased level of BIRC5 mRNA expression was observed with the progression of esophageal cancer. Those results indicated that BIRC5 may be involved in the development of esophageal cancer.
Figure 1.
Relative expression levels of BIRC5 mRNA in patients with different stages of esophageal cancer. (A) Mean expression level of BIRC5 mRNA in each group. (B) Expression level of BIRC5 mRNA in 2 representative cases of patients in each stage. *P<0.05, compared with the patients with an earlier stage of disease. BIRC, baculoviral IAP repeat containing 5; T0, primary tumor cannot be detected; Tis, high-grade dysplasia; T1, muscularis mucosae, lamina propria or submucosa invaded by the tumor; T2, muscularis propria invaded by the tumor; T3, adventitia invaded by the tumor; T4, adjacent structures invaded by the tumor.
Expression of BIRC5 gene in different cell lines
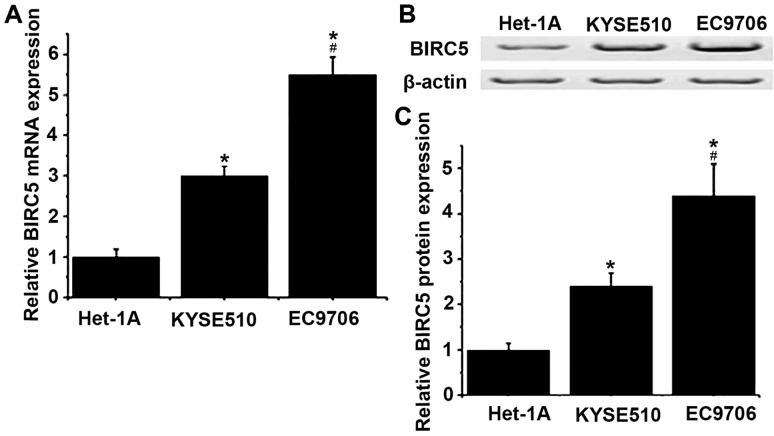
The expression levels of BIRC5 in Homo sapiens normal esophageal cell line Het-1A, and esophageal cancer cell lines KYSE510 and EC9706 were detected by RT-qPCR and western blotting at the level of mRNA and protein, respectively. As presented in Fig. 2, the levels of mRNA and protein of BIRC5 were determined to be significantly increased in KYSE510 and EC9706 cells compared with Het-1A cells (P<0.05). Compared with KYSE510, mRNA and protein expression levels of BIRC5 were significantly increased in EC9706 cells (P<0.05).
Figure 2.
Relative mRNA and protein expression levels of BIRC5 in different cell lines. (A) Relative mRNA expression levels of BIRC5 in different cell lines. (B) Representative results of western blot analysis. (C) Relative protein expression levels of BIRC5 in different cell lines. *P<0.05 vs. Het-1A; #P<0.05 vs. KRSE510. BIRC5, baculoviral IAP repeat containing 5.
Effects of BIRC5 siRNA silencing on the expression levels of angiogenesis-associated factor VEGF, BAI1 and MBD2 proteins in different cell lines
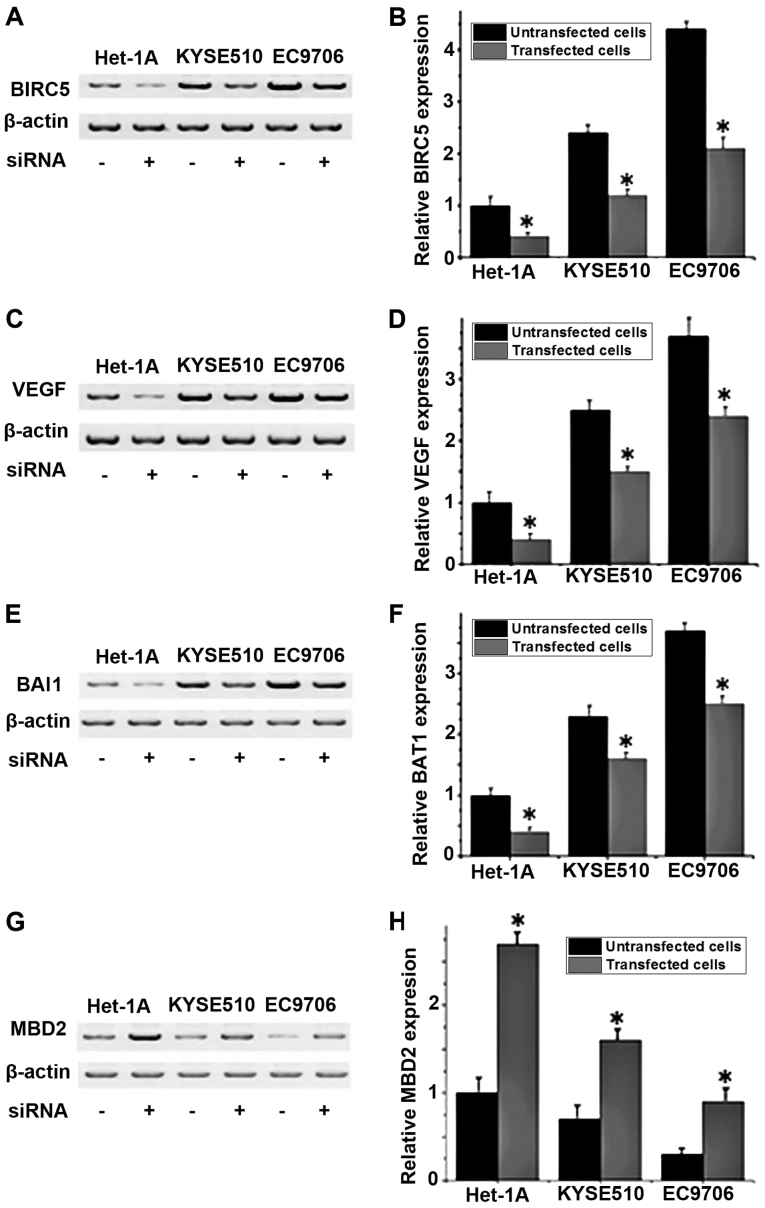
The protein expression levels of VEGF, BAI1 and MBD2 in different cell lines with or without BIRC5 silencing were detected by western blot analysis. No significant differences in expression of those proteins were detected between the control group (untransfected cells) and negative control group (Silencer® Negative Control #1 siRNA; data not shown). As depicted in Fig. 3A and B, the expression level of BIRC5 was significantly decreased in all three cell lines following siRNA transfection compared with untransfected cells (P<0.05), indicating that BIRC5 expression was successfully silenced. Compared with untransfected cells, the levels of VEGF and BAI1 protein expression significantly increased (P<0.05; Fig. 3C-F), and the level of MBD2 protein expression significantly decreased in BIRC5-silenced cells (P<0.05; Fig. 3G and H). These results indicated that the expression level of BIRC5 is positively associated with the expression levels of VEGF and BAI1 and negatively associated with the expression level of MBD2.
Figure 3.
Expression levels of BIRC5, VEGF, BAI1 and MBD2 in different cell lines with or without BIRC5 siRNA. (A) Representative western blot analysis results and (B) relative protein expression level of BIRC5. (C) Representative western blot analysis results and (D) relative protein expression level of VEGF. (E) Representative western blot analysis results and (F) relative protein expression level of BAI1. (G) Representative western blot analysis results and (H) relative protein expression level of MBD2. *P<0.05 vs. untransfected cells. BIRC5, baculoviral IAP repeat containing 5; VEGF, vascular endothelial growth factor; BAI1, brain-specific angiogenesis inhibitor 1; MBD2, methyl-CpG binding domain protein 2; siRNA, small-interfering RNA.
Effects of BIRC5 silencing on migration and invasion in different cell lines
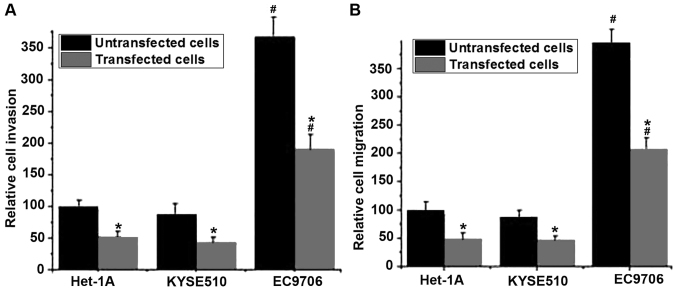
The migratory and invasive abilities of cells in different cell lines were detected using Transwell migration and invasion assays. No significant differences in migratory and invasive abilities of those cell lines were detected between the control group (untransfected cells) and the negative control group (Silencer® negative control #1 siRNA; data not shown). As presented in Fig. 4A, the invasive abilities of the cells were significantly decreased following BIRC5 silencing compared with untransfected cells (P<0.05). Similar results were observed in the cell migration assay. The silencing of BIRC5 was able to significantly reduce the migratory ability of all three cell lines compared with untransfected cells (P<0.05). Those results indicated that the silencing of BIRC5 was able to inhibit the migration and invasion of normal and esophageal cancer cells. In addition, the migratory and invasive abilities of the esophageal cancer cell line EC9706 with or without BIRC5 silencing were significantly higher compared with the esophageal cancer cell line KYSE510 with the same treatment (P<0.05; Fig. 4A and B).
Figure 4.
Relative cell invasion and migration of different cell lines. (A) Relative cell invasion of different cell lines. (B) Relative cell migration of different cell lines. *P<0.05 vs. untransfected cells. #P<0.05 vs. KYSE510 cells administered with the same treatment. BIRC5, baculoviral IAP repeat containing 5; siRNA, small-interfering RNA.
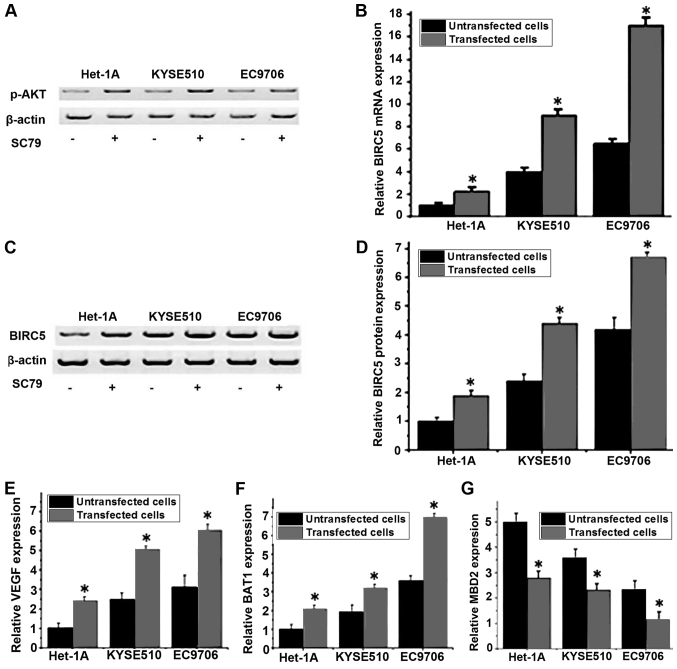
Effect of the PI3K/Akt signaling pathway activator SC79 on the expression level of BIRC5
A previous study has demonstrated that the expression and accumulation of BIRC5 was regulated via the PI3K/Akt pathway (16). The PI3K/Akt pathway activator SC79 was used to treat different cell lines, and the mRNA and protein expression levels of BIRC5 in cells that were treated with or without SC79 were detected by RT-qPCR and western blot analysis, respectively. As presented in Fig. 5A, the expression level of p-Akt was increased in all three cell lines following SC79 treatment, indicating the activation of the PI3K/Akt pathway. As presented in Fig. 5B, compared with untreated cells, the mRNA expression level of BIRC5 was significantly increased in cells that were treated with SC79 in the same cell line (P<0.05). Similar results were detected for the levels of BIRC5 protein expression, where the treatment with SC79 was able to significantly increase the protein expression level of BIRC5 (Fig. 5C and D). Furthermore, RT-qPCR was performed to detect the mRNA expression of VEGF, BAI1 and MBD2 following SC79 treatment. The results indicated that the expression levels of VEGF (Fig. 5E) and BAI1 (Fig. 5F) were significantly increased and the expression level of MBD2 (Fig. 5G) was significantly decreased following treatment with SC79. These results indicated that the activation of the PI3K/Akt signaling pathway may upregulate the mRNA and protein expression level of BIRC5, which in turn promote the expression of VEGF and BAI1 and inhibit the expression of MBD2.
Figure 5.
Relative expression levels of BIRC5 mRNA and protein in different cell lines with or without SC79 treatment. (A) The effect of SC79 treatment on the expression levels of p-AKT in different cell lines. (B) Relative mRNA expression levels of BIRC5 in different cell lines. (C) Representative western blot analysis results of BIRC5. (D) Relative protein expression levels of BIRC5 in different cell lines. (E) The effect of SC79 treatment on the expression levels of VEGF in different cell lines. (F) The effect of SC79 treatment on the expression levels of BAI1 in different cell lines. (G) The effect of BAI1 treatment on the expression levels of MBD2 in different cell lines. *P<0.05 vs. untreated cells. BIRC5, baculoviral IAP repeat containing 5; VEGF, vascular endothelial growth factor; MBD2, methyl-CpG binding domain protein 2; BIRC5, baculoviral IAP repeat containing 5; VEGF, vascular endothelial growth factor; BAI1, brain-specific angiogenesis inhibitor 1; MBD2, methyl-CpG binding domain protein 2; p-Akt, phospho-Akt.
Discussion
BIRC5, also termed survivin, is an anti-apoptosis gene that has been demonstrated to be involved in the progression of a variety of malignant tumor types (17). The expression of BIRC5 was indicated to increase in tumor tissues, compared with normal healthy tissues (12). In a study on breast cancer, Zu et al (18) reported that the expression of BIRC5 was significantly upregulated by the pro-oncogene zinc finger and BTB-domain containing 7A, which in turn promoted the progression of breast cancer. The expression level of BIRC5 was also determined to be significantly increased in colon cancer, and the increased expression of BIRC5 was maintained during the development of tumor (19). A previous study reported that BIRC5 expression level was continuously increased with the development of cervical cancer (20). In a study on esophageal cancer, Kato et al (21) determined that the expression level of BIRC5 was increased to different extents in patients with esophageal cancer, and patients with higher expression levels of BIRC5 were determined to have a reduced survival time compared with those with lower expression levels, indicating that the expression level of BIRC5 may be used as a predictor for the survival of patients.
In the present study, the expression level of BIRC5 was indicated to increase with the progression of esophageal cancer. In addition, the expression level of BIRC5 was also determined to be significantly increased in esophageal cancer cell lines KYSE510 and EC9706, compared with the Homo sapiens normal esophageal cell line Het-1A, which further confirms that BIRC5 may be involved in the development of esophageal cancer. Previous studies have demonstrated that BIRC5 is able to promote the migration and invasion of a number of different types of tumor cells (22). In the present study, the silencing of BIRC5 by siRNA was determined to be able to significantly reduce the migratory and invasive abilities of the normal esophageal and two esophageal cancer cell lines. In addition compared with KYSE510, a significantly higher expression level of BIRC5 mRNA, and increased migratory and invasive abilities were detected in the EC9706 cell line, indicating that the expression level of BIRC5 may be positively associated with the migration and invasion abilities of esophageal cancer cells.
Angiogenesis serves pivotal roles in the development of cancer (23), and anti-angiogenic therapy has been indicated to be a promising strategy in the treatment of various cancer types (24). VEGF and BAI1 are two factors that can promote angiogenesis, while MBD2 can interact with the tumor suppressor gene BAI1 to inhibit its anti-angiogenic function (25). It is understood that BIRC5 serves pivotal roles in angiogenesis during tumor development (26). In a study on glioma, BIRC5 was determined to promote angiogenesis through the interactions with VEGF in vivo and in vitro (26). In the present study, the silencing of BIRC5 by siRNA in three different cell lines were determined to reduce the expression levels of VEGF and BAI1 protein and increase the expression level of MBD2 protein, indicating that the downregulation of BIRC5 may be able to inhibit angiogenesis in esophageal cancer.
The PI3K/AKT signaling pathway, which serves key roles in a variety of physiological and biochemical processes, including cell proliferation, migration and invasion, is usually activated in different types of human cancer (27). Treatment strategies that aimed to inhibit the PI3K/AKT signaling pathway has been frequently used in the treatment of various types of human cancer, including endometrial (28) and colorectal cancer (29). A previous study has demonstrated that PI3K is able to regulate the expression of BIRC5 in the human ovarian cancer cell line OVCAR-3 via the activation of Akt. In the present study, the expression level of BIRC5 was significantly increased following the activation of PI3K/AKT signaling pathway. Furthermore, the expression levels of VEGF and BAI1 were significantly increased and the expression level of MBD2 was significantly decreased following treatment with SC79. Those results indicated that the activation of the PI3K/Akt signaling pathway may upregulate the levels of BIRC5 mRNA and protein expression, which in turn promote the expression of VEGF and BAI1, and inhibit the expression of MBD2.
In conclusion, BIRC5 may be able to promote the development of esophageal cancer via regulating the expression of angiogenesis-associated factor and increasing the migration and invasion of tumor cells. The activation of the PI3K/Akt signaling pathway is able to upregulate the expression level of BIRC5 to participate in development of esophageal cancer.
References
- 1.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 2.Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y, Ueda J, Wei W, Inoue M, Tanaka H. Epidemiology of esophageal cancer in Japan and China. J Epidemiol. 2013;23:233–242. doi: 10.2188/jea.JE20120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598–5606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu M, Zhao JK, Zhang ZF, Han RQ, Yang J, Zhou JY, Wang XS, Zhang XF, Liu AM, van't Veer P, et al. Smoking and alcohol drinking increased the risk of esophageal cancer among Chinese men but not women in a high-risk population. Cancer Causes Control. 2011;22:649–657. doi: 10.1007/s10552-011-9737-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 6.LeVea C. Pathogenesis of Esophageal Cancer [M]/Minimally Invasive Foregut Surgery for Malignancy. Springer International Publishing; 2015. pp. 1–9. [Google Scholar]
- 7.Weaver BA, Cleveland DW. Decoding the links between mitosis, cancer, and chemotherapy: The mitotic checkpoint, adaptation, and cell death. Cancer Cell. 2005;8:7–12. doi: 10.1016/j.ccr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 8.Britigan EM, Wan J, Zasadil LM, Ryan SD, Weaver BA. The ARF tumor suppressor prevents chromosomal instability and ensures mitotic checkpoint fidelity through regulation of Aurora B. Mol Biol Cell. 2014;25:2761–2773. doi: 10.1091/mbc.e14-05-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 10.Gu Y, Li P, Peng F, Zhang M, Zhang Y, Liang H, Zhao W, Qi L, Wang H, Wang C, Guo Z. Autophagy-related prognostic signature for breast cancer. Mol Carcinog. 2016;55:292–299. doi: 10.1002/mc.22278. [DOI] [PubMed] [Google Scholar]
- 11.Wang R, Kang Y, Löhr CV, Fischer KA, Bradford CS, Johnson G, Dashwood WM, Williams DE, Ho E, Dashwood RH. Reciprocal regulation of BMF and BIRC5 (Survivin) linked to Eomes overexpression in colorectal cancer. Cancer Lett. 2016;381:341–348. doi: 10.1016/j.canlet.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim T, Lee I, Kim J, Kang WK. Synergistic effect of simvastatin plus radiation in gastric cancer and colorectal cancer: Implications of BIRC5 and connective tissue growth factor. Int J Radiat Oncol Biol Phys. 2015;93:316–325. doi: 10.1016/j.ijrobp.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 13.Vaĭshlia NA, Zinov'eva MV, Sass AV, Kopantsev EP, Vinogradova TV, Sverdlov ED. Increase of BIRC5 gene expression in non-small cell lung cancer and esophageal squamous cell carcinoma does not correlate with expression of genes SMAC/DIABLO and PML encoding its inhibitors. Mol Biol (Mosk) 2007;42:652–661. (In Russian) [PubMed] [Google Scholar]
- 14.Rice TW, Blackstone EH. Esophageal cancer staging: Past, present, and future. Thorac Surg Clin. 2013;23:461–469. doi: 10.1016/j.thorsurg.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Zhao P, Meng Q, Liu LZ, You YP, Liu N, Jiang BH. Regulation of survivin by PI3K/Akt/p70S6K1 pathway. Biochem Biophys Res Commun. 2010;395:219–224. doi: 10.1016/j.bbrc.2010.03.165. [DOI] [PubMed] [Google Scholar]
- 17.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 18.Zu X, Ma J, Liu H, Liu F, Tan C, Yu L, Wang J, Xie Z, Cao D, Jiang Y. Pro-oncogene Pokemon promotes breast cancer progression by upregulating survivin expression. Breast Cancer Res. 2011;13:R26. doi: 10.1186/bcr2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez JM, Farma JM, Coppola D, Hakam A, Fulp WJ, Chen DT, Siegel EM, Yeatman TJ, Shibata D. Expression of the antiapoptotic protein survivin in colon cancer. Clin Colorectal Cancer. 2011;10:188–193. doi: 10.1016/j.clcc.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu D, Qian J, Yin X, Xiao Q, Wang C, Zeng Y. Expression of PTEN and survivin in cervical cancer: Promising biological markers for early diagnosis and prognostic evaluation. Br J Biomed Sci. 2012;69:143–146. [PubMed] [Google Scholar]
- 21.Kato J, Kuwabara Y, Mitani M, Shinoda N, Sato A, Toyama T, Mitsui A, Nishiwaki T, Moriyama S, Kudo J, Fujii Y. Expression of survivin in esophageal cancer: Correlation with the prognosis and response to chemotherapy. Int J Cancer. 2001;95:92–95. doi: 10.1002/1097-0215(20010320)95:2<92::AID-IJC1016>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 22.Kogo R, How C, Chaudary N, Bruce J, Shi W, Hill RP, Zahedi P, Yip KW, Liu FF. The microRNA-218~Survivin axis regulates migration, invasion, and lymph node metastasis in cervical cancer. Oncotarget. 2015;6:1090–1100. doi: 10.18632/oncotarget.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 24.Shojaei F. Anti-angiogenesis therapy in cancer: Current challenges and future perspectives. Cancer Lett. 2012;320:130–137. doi: 10.1016/j.canlet.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Zhu D, Hunter SB, Vertino PM, Van Meir EG. Overexpression of MBD2 in glioblastoma maintains epigenetic silencing and inhibits the antiangiogenic function of the tumor suppressor gene BAI1. Cancer Res. 2011;71:5859–5870. doi: 10.1158/0008-5472.CAN-11-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang P, Zhen H, Zhang J, Zhang W, Zhang R, Cheng X, Guo G, Mao X, Wang J, Zhang X. Survivin promotes glioma angiogenesis through vascular endothelial growth factor and basic fibroblast growth factor in vitro and in vivo. Mol Carcinog. 2012;51:586–595. doi: 10.1002/mc.20829. [DOI] [PubMed] [Google Scholar]
- 27.Karar J, Maity A. PI3K/AKT/mTOR pathway in angiogenesis. Front Mol Neurosci. 2011;4:51. doi: 10.3389/fnmol.2011.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slomovitz BM, Coleman RL. The PI3K/AKT/mTOR pathway as a therapeutic target in endometrial cancer. Clin Cancer Res. 2012;18:5856–5864. doi: 10.1158/1078-0432.CCR-12-0662. [DOI] [PubMed] [Google Scholar]
- 29.Pandurangan AK. Potential targets for prevention of colorectal cancer: A focus on PI3K/Akt/mTOR and Wnt pathways. Asian Pac J Cancer Prev. 2013;14:2201–2205. doi: 10.7314/APJCP.2013.14.4.2201. [DOI] [PubMed] [Google Scholar]