Abstract
The expression of human transfer ribonucleic acid (tRNA) methyltransferase 9-like (KIAA1456) protein in lung cancer tissue and its effects on certain genes involved in the proliferation, migration and invasion of lung cancer cells were investigated. Immunohistochemistry was applied to stain lung cancer tissue and adjacent tissue sections of 90 lung cancer patients, so as to evaluate the difference in the expression level of KIAA1456 between two tissues. The correlation of KIAA1456 expression with clinicopathological parameters of lung cancer was analyzed. Kaplan-Meier survival curves were used to analyze the relationship between KIAA1456 and postoperative survival in patients with lung cancer. KIAA1456 gene was overexpressed in lung cancer cell lines (A549 and GLC-15), and the influence of KIAA1456 gene on the expression of cyclin D1, neural cadherin (N-cadherin) and epithelial cadherin (E-cadherin) and their involvement in lung cancer cell proliferation, migration and invasion were observed. Compared with that in adjacent tissue, the expression of KIAA1456 in lung cancer tissue was significantly decreased (p<0.05). The low expression of KIAA1456 in lung cancer tissue was clearly associated with pathological tumor (pT) stage, pathological node (pN) stage, tumor-node-metastasis (TNM) stage and pathological stage, but had no correlation with sex, age, tumor size or histology of the patient. KIAA1456 low expression was related with poor prognosis of the lung cancer patient. According to Western blotting, the overexpression of KIAA1456 in lung cancer cells could inhibit the expressions of cyclin D1 and N-cadherin, and promote the expression of E-cadherin. The results show that KIAA1456 expression was low in lung cancer tissue, and was associated with poor prognosis in patients and was an independent prognostic factor in patients with lung cancer. Thus, KIAA1456 can be used as a tumor suppressor gene in lung cancer, suppressing the proliferation, migration and invasion of lung cancer cells.
Keywords: KIAA1456, lung cancer, cyclin D1, N-cadherin, E-cadherin
Introduction
Transfer ribonucleic acids (tRNAs) play a crucial role in protein translation. The abnormal expression of proteins modulating tRNA modification are closely related to diseases such as maternally inherited mitochondrial disease, nervous system disease, tumor and type II diabetes mellitus (1). Human tRNA methyltransferase 9-like (hTRM9L, also known as KIAA1456) protein is a kind of enzyme that catalyzes the swing of tRNA bases (2). The KIAA1456 gene contains two exons, located at the end of chromosome 8. Its protein expression level is the highest in adult brain tissue and relatively low in skeletal muscle, testis and ovary. The content of KIAA1456 in fetal brain tissue is overtly lower than that in adult human brain tissue, and that in cerebellum of adult human brain tissue is the highest (3).
KIAA1456 can catalyze the swing of tRNA bases to make tRNA mature, so as to ensure the normal and orderly translation of proteins. The biological function of KIAA1456 protein is closely correlated with the deoxyribonucleic acid (DNA) damage response. After exogenous overexpression of KIAA1456, the repair capacity of cells to DNA damage is significantly improved, DNA damage in some cells can be completely repaired, but cells with incompletely repaired DNA damage rapidly die (4,5). Previous findings have shown that KIAA1456 is involved in the occurrence and development of tumors, and KIAA1456 protein has evidently lower expression in breast, colon, bladder, cervical and testicular cancer (6). After overexpression of KIAA1456, the migration and invasion abilities and the growth rate of tumor cells are decreased to different extents (7). The abnormal expression of KIAA1456 protein in cells interferes with normal protein translation and DNA damage repair, eventually leading to canceration and malignant proliferation of cells (5). Therefore, considering the important role of KIAA1456 in tRNA modification, it is inferred that KIAA1456 is a potential tumor suppressor gene, and the abnormal decrease or loss of KIAA1456 expression may contribute to tumor formation and development. At present, the expression of KIAA1456 in lung cancer cells is unclear.
In the present study, the expression of KIAA1456 in lung cancer tissue and adjacent tissue were compared, the correlations of KIAA1456 with patient characteristics and clinicopathological stages were analyzed, and the effects of KIAA1456 on lung cancer cell proliferation, migration and invasion were explored preliminarily.
Materials and methods
Lung cancer tissue samples
Human tissue samples (90 pairs of lung cancer and adjacent tissues) were obtained from patients treated in the Pneumology Department, Thoracic Surgery Department and Oncology Department of the First People's Hospital of Xuzhou (Xuzhou, China) from June 2008 to July 2011. These patients were diagnosed according to the American Association for Thoracic Surgery guidelines for lung cancer screening (8), had an average age of 68.26±13.75 years, and included 56 males and 34 females. Postoperative pathological and clinical data were collected from the Department of Pathology and hospital records, and the patients were followed up by the hospital and laboratories. Informed consent of patients was obtained, and the study was approved by the Ethics Review Committee of the First People's Hospital of Xuzhou (Jiangsu, China).
Cells and reagents
293T cells and human lung cancer cell lines (A549 and GLC-15) were purchased from the Shanghai Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% calf serum in a 5% CO2 incubator at 37°C. Mouse anti-human KIAA1456, neural cadherin (N-cadherin), cyclin D1 and epithelial cadherin (E-cadherin) immunoglobulin G (IgG), internal control antibody mouse anti-human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) IgG, and secondary rabbit anti-mouse IgG-horseradish peroxidase (HRP)- and biotin-labeled rabbit anti-mouse IgG were purchased from Abcam (Cambridge, MA, USA). The remaining reagents were purchased from Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd. (Beijing, China).
Immunohistochemistry
Lung cancer tissue and adjacent tissue were collected during surgical procedures from lung cancer patients. The tissues were cut into sections, followed by routine dewaxing and hydration. Then, sections were added with 0.01 mmol/l citrate buffer for antigen retrieval, treated with 3% H2O2 for 20 min, blocked with 10% sheep serum at room temperature for 20 min, and added with mouse anti-human KIAA1456 (1:1,000) for incubation overnight at 4°C. After that, the sections were incubated with a secondary antibody, namely, biotin-labeled rabbit anti-mouse IgG (1:2500), added with diaminobenzidine (DAB) substrate for color development, counterstained with hematoxylin, mounted with neutral resin, dried, and then observed under a microscope (BX-42, Olympus Corporation, Tokyo, Japan).
To determine immunohistochemical results, cells stained in sepia were considered positive cells. ImageJ software (National Institutes of Health, Bethesda, MD, USA) was used to analyze staining intensity. The scoring for staining intensity was: 0 point (negative), 1 point (weakly positive), 2 points (moderately positive), and 3 points (strongly positive). Scoring for percentage of stained cells was: 1 point (1–24%), 2 points (25–49%), 3 points (50–74%), and 4 points (75–100%). After multiplying the above two scores, the following scores 0, 1, 2, 3, 4, 6, 8, 9 and 12 were obtained and further scored was: 0 point, negative; 1–4 points, weakly positive; 6–8 points, moderately positive; 9–12 points, strongly positive. A score <6 points was defined as a low expression of KIAA1456, and a score ≥6 points was defined as a high expression of KIAA1456.
Lentivirus infection
The recombinant lentiviral vector plasmid pLP was co-transfected with 293T cells via the Lipofectamine 2000 reagent and incubated at 37°C for 48 h. The supernatant of the cell medium was collected to obtain lentivirus carrying KIAA1456 gene or empty vector lentivirus solutions. A549 or GLC-15 cells (5×103) were inoculated into a 96-well plate and cultured overnight at 37°C until the rate of cell fusion was 50–60%. The lentivirus pLP carrying KIAA1456 gene [multiplicity of infection (MOI) =4] was added and incubated at 37°C for 24 h. The next day, the medium containing virus was discarded and replaced by DMEM + 10% calf serum medium, followed by 48 h of continuous cultivation. The lentivirus pLP carrying no inserted genes was also used to transfect A549 and GLC-15 cells, used as negative controls.
Western blotting
After 72 h of lentiviral infection of A549 or GLC-15 cells, cells were collected, and added with Tween-20 cell lysis buffer for cell lysis. Then, the supernatant was collected and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After that, protein was transferred onto a polyvinylidene fluoride (PVDF) membrane, followed by blocking with 1% bovine serum albumin (BSA) for 1 h at room temperature. Then, protein was added with mouse polyclonal KIAA1456 antibody (1:500; cat. no. ab68919), N-cadherin (1:500; cat. no. ab98952), cyclin D1 (1:500; cat. no. ab134175) and E-cadherin (1:500; cat. no. ab1416) as well as secondary goat anti-rabbit (HRP) IgG antibody (1:2,000; cat. no. ab6721) and secondary rabbit anti-mouse (HRP) IgG antibody (1:2,000; cat. no. ab6728) for 1 h of incubation at room temperature. All the antibodies were purchased from Abcam. After that, secondary rabbit anti-mouse (HRP) IgG antibody (1:2,000; cat. no. ab6728) was added for 1 h of incubation at room temperature, followed by membrane washing with phosphate-buffered saline (PBS) three times and color development with DAB substrate. GAPDH was used as an internal reference. An Odyssey chemiluminescence instrument was utilized to record the colorimetric results. ImageJ software was used for gray analysis to detect the expression level of the protein of interest.
Statistical analysis
Data were processed using Statistical Product and Service Solutions (SPSS) 17.0 statistical software (SPSS, Inc., Chicago, IL, USA). Measurement data are expressed as mean ± standard deviation, repeated 3 times for each group, and Student's t-test was employed for comparisons among groups. Enumeration data were expressed as cases or percentage, using χ2 test for comparison among groups. The correlations of KIAA1456 expression in lung cancer tissue with the clinicopathological parameters were analyzed via the χ2 test. Survival rate analysis was performed by using Kaplan-Meier survival curves. Threshold of significance was α=0.05, and p<0.05 indicated that the difference was statistically significant.
Results
KIAA1456 expression in lung cancer tissue and adjacent tissue
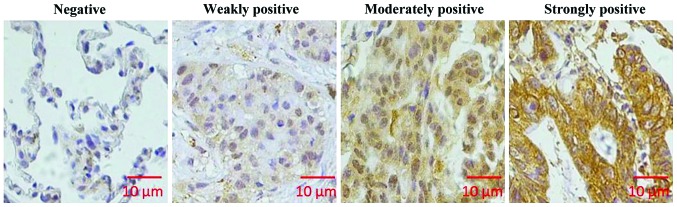
Immunohistochemical analysis was used to analyze the expression of KIAA1456 in lung cancer tissue and adjacent tissue. As shown in Fig. 1, KIAA1456 was mainly expressed in cytoplasm in lung cancer tissue. In 90 cases of lung cancer tissue, the number of tissues with high expression of KIAA1456 accounted for 51.1%, while in adjacent tissue, that accounted for 70%. KIAA1456 expression in lung cancer tissue was distinctly lower than that in adjacent tissue (p<0.05; Table I).
Figure 1.
KIAA1456 staining in lung cancer tissue sections (above, ×200; bottom, ×400). Immunohistochemistry shows that KIAA1456 is mainly expressed in cytoplasm. The above are negative, weakly positive, moderately positive and strongly positive expressions from left to right. KIAA1456, tRNA methyltransferase 9-like protein.
Table I.
Expression of KIAA1456 in lung cancer tissue and adjacent tissue.
| KIAA1456 staining | |||
|---|---|---|---|
| Tissues | n | Low expression (n, %) | High expression (n, %) |
| Lung cancer | 90 | 44 (48.9) | 46 (51.1) |
| Adjacent | 90 | 27 (30) | 63 (70) |
| χ2 | 6.722 | 5.761 | |
| P-value | 0.026 | 0.035 | |
KIAA1456, tRNA methyltransferase 9-like protein.
Correlation of KIAA1456 expression in lung cancer tissue with clinicopathological parameters
The expression of KIAA1456 in lung cancer tissue had no significant correlation with sex, age, tumor size or histological type of the patient (p>0.05), but were significantly correlated with the clinicopathological features of the lung cancer patient, for example, pathological tumor (pT) stage (p<0.01), pathological node (pN) stage (p<0.01), tumor-node-metastasis (TNM) stage (p<0.01) and pathological stage (p<0.01; Table II).
Table II.
Correlation analyses of KIAA1456 expression in lung cancer tissue with clinicopathological parameters.
| KIAA1456 staining | |||||
|---|---|---|---|---|---|
| Clinicopathological parameters | n | Low expression (n, %) | High expression (n, %) | P-valuea | χ2 |
| Sex | 0.118 | 3.271 | |||
| Male | 56 | 29 (51.8) | 27 (48.2) | ||
| Female | 34 | 16 (47.1) | 18 (52.9) | ||
| Age (years) | 0.154 | 2.147 | |||
| ≤60 | 53 | 23 (43.4) | 30 (56.6) | ||
| >60 | 37 | 21 (56.8) | 16 (43.2) | ||
| Max. tumor diameter | 0.125 | 4.158 | |||
| ≤3 cm | 42 | 19 (45.2) | 23 (54.8) | ||
| >3 cm | 48 | 27 (56.2) | 21 (43.8) | ||
| Histology | 0.424 | 3.297 | |||
| Adenocarcinoma | 68 | 32 (47.1) | 36 (52.9) | ||
| Squamous cell carcinoma | 16 | 8 (50) | 8 (50) | ||
| Others | 6 | 4 (66.7) | 2 (33.3) | ||
| pT stage | <0.01 | 6.713 | |||
| pT1 | 27 | 7 (25.9) | 20 (74.1) | ||
| pT2 | 40 | 19 (47.5) | 21 (52.5) | ||
| pT3-pT4 | 23 | 18 (78.3) | 5 (21.7) | ||
| pN stage | <0.01 | 7.291 | |||
| pN0 | 45 | 17 (37.8) | 28 (62.2) | ||
| pN1 | 17 | 7 (41.2) | 10 (58.8) | ||
| pN2-pN3 | 28 | 20 (71.4) | 8 (28.6) | ||
| TNM stage | <0.01 | 6.246 | |||
| I | 33 | 8 (24.2) | 25 (75.8) | ||
| II–III | 57 | 32 (56.1) | 25 (43.9) | ||
| Pathological stage | <0.01 | 6.349 | |||
| G1-G2 | 61 | 23 (37.7) | 38 (62.3) | ||
| G3 | 29 | 21 (72.4) | 8 (27.6) | ||
Chi-square test, p<0.05 suggests that the difference is statistically significant. KIAA1456, tRNA methyltransferase 9-like protein.
Analysis on the relationship between KIAA1456 expression and prognostic survival in patients
To understand the correlation of KIAA1456 expression level with the prognostic survival in patients, Kaplan-Meier survival curves were used to analyze the 5-year survival rates of 90 patients with lung cancer. The results revealed that the overall 5-year survival rate of patients with a high expression of KIAA1456 after operation was obviously higher than that of patients with a low expression of KIAA1456 (p<0.05). The low expression of KIAA1456 was associated with poor postoperative outcomes of patients (Fig. 2).
Figure 2.
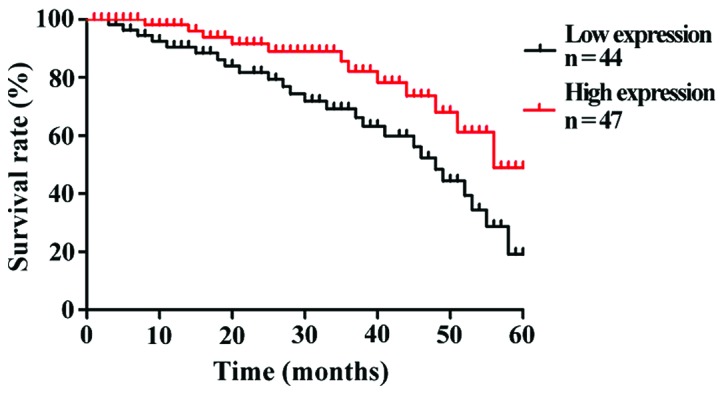
Postoperative survival rates of lung cancer patients with high and low KIAA1456 expression. Lung cancer patients with high KIAA1456 expression have overtly higher overall 5-year survival rates in comparison with those with low expression of KIAA1456 (p<0.05). KIAA1456, tRNA methyltransferase 9-like protein.
Effects of KIAA1456 on lung cancer cell proliferation, migration and invasion
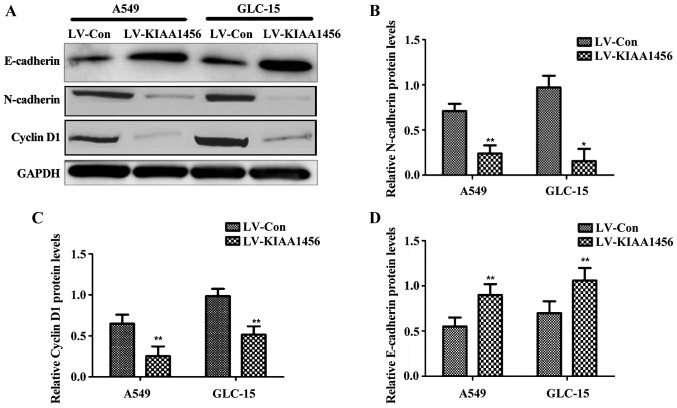
Following the overexpression of KIAA1456 in lung cancer cell lines (A549 and GLC-15), the expression levels of N-cadherin and cyclin D1 were significantly reduced compared with those in control virus-infected cells (p<0.05), while the expression level of E-cadherin was distinctly higher than that in control virus-infected cells (p<0.05; Fig. 3).
Figure 3.
Changes in expression levels of N-cadherin, cyclin D1 and E-cadherin after overexpression of KIAA1456 in A549 and GLC-15 cells. (A) Based on western blotting, after overexpression of KIAA1456, A549 and GLC-15 cells have evidently increased E-cadherin protein expression levels and significantly decreased N-cadherin and cyclin D1 expression levels in comparison with control. (B) N-cadherin expression level; (C) cyclin D1 expression level; and (D) E-cadherin expression level. Compared with LV-Con, *P<0.05; compared with LV-Con, **P<0.01.
Discussion
KIAA1456 is an enzyme catalyzing the swing of tRNA bases, which plays an important role in making tRNA mature with bioactivity. Prasad et al reported that KIAA1456 gene is a potential tumor suppressor gene (9). A study by Begley et al also indicated that KIAA1456 gene is negatively correlated with tumor growth and has significantly decreased expressions in bladder cancer, breast cancer, testicular cancer, colon cancer and cervical cancer (2). There is no research study on KIAA1456 gene in lung cancer. In this study, lung cancer tissue and adjacent tissue were stained by immunohistochemistry to observe whether the expression of KIAA1456 protein was changed in lung cancer tissue. The results showed that the expression level of KIAA1456 in lung cancer tissue was lower than that in adjacent tissue. KIAA1456 expression level was not significantly correlated with sex, age, tumor size and histological type of the patient, but obviously associated with pT stage, pN stage, TNM stage and pathological stage. The overall 5-year survival rate of patients with high KIAA1456 expression after operation was significantly higher than that of patients with low KIAA1456 expression. The low expression of KIAA1456 was related to poor postoperative outcomes of patients.
Cyclin D1, a member of cyclin family, binds cyclin-dependent kinase-4 (CDK-4) proteins to accelerate progression of phase G1-S of cell cycle, thereby promoting tumor cell proliferation (10). Western blot analysis in this study indicated that cyclin D1 protein was clearly decreased when KIAA1456 gene was overexpressed in lung cancer cells, suggesting that KIAA1456 can inhibit the proliferation of lung cancer cells.
Lung cancer is a malignant tumor that has high invasiveness and is easy to metastasize distantly. Epithelial-mesenchymal transition (EMT) promotes the invasion and metastasis of lung cancer cells, which is one of the major causes of high mortality in lung cancer (11). E-cadherin and N-cadherin are the most important markers in the process of EMT. The loss of E-cadherin can lead to the loss of cell-cell junctions, and N-cadherin can increase the motility and invasiveness of tumor cells (12). In this study, when KIAA1456 was overexpressed in lung cancer cells, N-cadherin protein expression was significantly decreased, while E-cadherin protein expression was overtly increased, suggesting that KIAA1456 can reduce lung cancer cell migration, invasion and metastasis.
In conclusion, KIAA1456 protein expression is low in lung cancer tissue. The expression level of KIAA1456 is obviously associated with the clinicopathological features and prognoses of lung cancer. Overexpression of KIAA1456 inhibits the proliferation, migration and invasion of lung cancer cells. KIAA1456 gene can serve as a tumor suppressor gene, and an independent prognostic factor in patients with lung cancer. Studies have shown that the 10-year survival rates of patients with early-stage lung cancer will be up to 75% if lung cancer is diagnosed in the early stage (13). KIAA1456 can act as a new biological indicator for lung cancer detection, which is of great significance for the early diagnosis and targeted therapy of lung cancer.
Acknowledgements
Not applicable.
Funding
This study was supported by the Project of Xuzhou Science and Technology Bureau (project no. KC16SH034), Medical Key Subjects of Jiangsu Province (project no. ZDXKB2016007), Suzhou Clinical Medicine Center (project no. Szzx201502), Key Technology Application Research Project of Suzhou (project no. SS201630).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
SW and XL conceived and designed the experiments, SW wrote the manuscript; JH, SW and YZ performed the experiments; CS and XL analyzed the data of human tissue samples; TL and JY contributed in collecting clinical tissue samples. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Informed consent of patients was obtained, and the study was approved by the Ethics Review Committee of the First People's Hospital of Xuzhou (Xuzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell. 2016;165:1416–1427. doi: 10.1016/j.cell.2016.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begley U, Sosa MS, Avivar-Valderas A, Patil A, Endres L, Estrada Y, Chan CT, Su D, Dedon PC, Aguirre-Ghiso JA, et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-α. EMBO Mol Med. 2013;5:366–383. doi: 10.1002/emmm.201201161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flanagan JM, Healey S, Young J, Whitehall V, Trott DA, Newbold RF, Chenevix-Trench G. Mapping of a candidate colorectal cancer tumor-suppressor gene to a 900-kilobase region on the short arm of chromosome 8. Genes Chromosomes Cancer. 2004;40:247–260. doi: 10.1002/gcc.20039. [DOI] [PubMed] [Google Scholar]
- 4.Shanmugam R, Aklujkar M, Schäfer M, Reinhardt R, Nickel O, Reuter G, Lovley DR, Ehrenhofer-Murray A, Nellen W, Ankri S, et al. The Dnmt2 RNA methyltransferase homolog of Geobacter sulfurreducens specifically methylates tRNA-Glu. Nucleic Acids Res. 2014;42:6487–6496. doi: 10.1093/nar/gku256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbott JA, Francklyn CS, Robey-Bond SM. Transfer RNA and human disease. Front Genet. 2014;5:158. doi: 10.3389/fgene.2014.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng C, Zhang Z, Wu J, Lv Z, Tang J, Xie H, Zhou L, Zheng S. A critical role for ZDHHC2 in metastasis and recurrence in human hepatocellular carcinoma. BioMed Res Int. 2014;2014:832712. doi: 10.1155/2014/832712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaklitsch MT, Jacobson FL, Austin JH, Field JK, Jett JR, Keshavjee S, MacMahon H, Mulshine JL, Munden RF, Salgia R, et al. The American Association for Thoracic Surgery guidelines for lung cancer screening using low-dose computed tomography scans for lung cancer survivors and other high-risk groups. J Thorac Cardiovasc Surg. 2012;144:33–38. doi: 10.1016/j.jtcvs.2012.05.060. [DOI] [PubMed] [Google Scholar]
- 9.Prasad MA, Trybus TM, Wojno KJ, Macoska JA. Homozygous and frequent deletion of proximal 8p sequences in human prostate cancers: Identification of a potential tumor suppressor gene site. Genes Chromosomes Cancer. 1998;23:255–262. doi: 10.1002/(SICI)1098-2264(199811)23:3<255::AID-GCC8>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 10.Bramanti V, Tomassoni D, Bronzi D, Grasso S, Currò M, Avitabile M, Li Volsi G, Renis M, Ientile R, Amenta F, et al. Alpha-lipoic acid modulates GFAP, vimentin, nestin, cyclin D1 and MAP-kinase expression in astroglial cell cultures. Neurochem Res. 2010;35:2070–2077. doi: 10.1007/s11064-010-0256-6. [DOI] [PubMed] [Google Scholar]
- 11.Xiao D, He J. Epithelial mesenchymal transition and lung cancer. J Thorac Dis. 2010;2:154–159. doi: 10.3978/j.issn.2072-1439.2010.02.03.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henschke CI, Yankelevitz DF, Libby DM, Pasmantier MW, Smith JP, Miettinen OS. International Early Lung Cancer Action Program Investigators: Survival of patients with stage I lung cancer detected on CT screening. N Engl J Med. 2006;355:1763–1771. doi: 10.1056/NEJMoa060476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.