Abstract
The aim of the present study was to identify potential biomarkers of hepatocellular carcinoma (HCC). Three gene expression profiles of GSE95698, GSE49515 and GSE76427 and a DNA methylation profile of GSE73003 were downloaded from the Gene Expression Omnibus (GEO) database, each comprising data regarding HCC and control tissue samples. The differentially expressed genes (DEGs) between the HCC group and the control group were identified using the limma software package. The Database for Annotation, Visualization and Integrated Discovery (DAVID) was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of the overlapping DEGs. The PPI network of the overlapping DEGs was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins. A total of 41 DEGs were identified in HCC the group compared with control group. The overlapping DEGs were enriched in 11 GO terms and 3 KEGG pathways. A total of 6,349 DMSs were identified, and 6 of the differentially expressed genes were also differentially methylated [Denticleless protein homolog (DTL), Dual specificity phosphatase 1 (DUSP1), Eomesodermin, Endothelial cell specific molecule 1, Nuclear factor κ-light-chain gene enhancer of activated B cells inhibitor, α (NFKBIA) and suppressor of cytokine signaling 2 (SOCS2)]. The present study suggested that DTL, DUSP1, NFKBIA and SOCS2 may be potential biomarkers of HCC, and the tumor protein ‘p53 signaling’, ‘forkhead box O1’ signaling and ‘metabolic’ pathways may serve roles in the pathogenesis of HCC.
Keywords: hepatocellular carcinoma, pathogenesis, gene expression profile, DNA methylation profile
Introduction
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer in adults, the fifth most common type of cancer and the third leading cause of cancer-associated mortality worldwide (1). HCC is closely associated with chronic viral hepatitis infection and exposure to toxins, including alcohol and aflatoxin (2). Other diseases can markedly increase the risk of HCC, including chronic liver inflammation, hemochromatosis and α1-antitrypsin deficiency (3,4). Chronic infections of hepatitis B and/or C contribute to the development of HCC by repeatedly causing the immune system to attack the liver cells, and consumption of large amounts of alcohol can have a similar effect (5). The prevalence of aflatoxin and hepatitis B has caused relatively high rates of HCC in a number of developing countries (6,7). Aflatoxin is produced by certain Aspergillus fungus species, and is a carcinogen known to contribute to the carcinogenesis of HCC (6). Tumors can develop following a mutation to cellular machinery, causing the cell to replicate at a higher rate and/or the avoidance of apoptosis (8). Numerous genes have been reported to serve roles in HCC carcinogenesis, which may be exploitable for the development of effective prevention and treatment regimens for HCC (9,10). Downregulated expression of human PMS1 homolog 2, mismatch repair system component (hPMS2) provides a growth advantage and stimulates proliferation of HCC cells, and it is believed that hPMS2 is involved at an early stage of HCC (11). Knockdown of DLC1 has been demonstrated to aid MYC in the induction of hepatoblast transformation in vitro, and in the development of HCC in vivo (12). Another study demonstrated that the ‘cell cycle’ pathway and cell division cycle 25a may be crucial in the pathogenesis and progression of HCC (13). In addition, hypermethylation of DNA has been implicated as an early event in hepatocarcinogenesis (14). A mate-analysis study provided empirical evidence that abnormal suppressor of cytokine signaling 1 (SOCS1) promoter-methylation may contribute to the pathogenesis of HCC (15). Retinol metabolism genes and serine hydroxymethyltransferase 1 have also been indicated to be epigenetically regulated through promoter DNA methylation in alcohol-associated HCC (16). However, our incomplete understanding of the molecular and cellular mechanisms that drive HCC limits the available therapeutic options. In the present study, three gene expression profiles and a DNA methylation profile were jointly analyzed in order to identify biomarkers of HCC.
Materials and methods
Gene expression and DNA methylation profiles
The gene expression profiles, GSE95698, GSE49515 (17) and GSE76427, and DNA methylation prolife, GSE73003, were downloaded from the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/). A total of 3 human HCC tissue samples (HCC group) and their matching paracancerous tissue samples (control group) were selected from GSE95698, and the genes were detected using the Agilent-039494 SurePrint G3 Human GE V2 8×60 K Microarray 039381 platform. Data regarding 20 peripheral blood samples from 10 HCC patients (HCC group) and 10 healthy people (control group) were selected from the GSE49515 profile, and the genes were detected with Affymetrix Human Genome U133 Plus 2.0 Array. A total of 115 HCC tissue samples (HCC group) and 52 paracancerous tissue samples (control group) were selected from GSE76427, which were detected using an Illumina HumanHT-12 V4.0 expression beadchip. A total of 20 HCC tissue samples (HCC group) and 20 non-tumor tissue samples (control group) were selected from GSE73003, which were detected using an Illumina HumanMethylation27 BeadChip (Human Methylation27_270596_V1.2).
Data processing and differential analysis
For the gene expression profiles of GSE95698, GSE49515 and GSE76427, the raw data were obtained and normalized using the preprocess Core function package (version 3.5; http://www.bioconductor.org/packages/release/bioc/html/preprocessCore.html). If multiple probes corresponded to the same gene, the average expression of these probes was used as the expression value of the gene. Subsequently, the differentially expressed genes (DEGs) in the HCC group compared with control group for the 3 sets of gene expression profiles were identified using the limma software package (version 3.18.13; http://www.bioconductor.org/packages/2.13/bioc/html/limma.html). P<0.05 and |log (fold-change)|>0.5 were used as the DEG threshold criteria. Thus, 3 sets of DEGs were obtained and the overlapping DEGs were selected for further study. For the DNA methylation profile of GSE73003, the preprocess Core function package (version 3.5) was used for normalization, and the differentially methylated sites (DMSs) were identified with the β-distribution test and t-test using a threshold of P<0.05. Overlaps between the overlapping DEGs and the differentially methylated sites were further studied.
Functional- and pathway-enrichment analyses of the overlapping DEGs
The Database for Annotation, Visualization and Integrated Discovery (DAVID; version 6.8; http://david.ncifcrf.gov/) is a widely used web-based tool for functional and pathway enrichment analyses (18). In the present study, it was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the overlapping DEGs. The GO terms and KEGG pathways with P<0.05 were selected.
Construction of the protein-protein interaction (PPI) network
Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org) is a biological database and web resource used to identify known and predicted PPIs (19). In the present study, the PPI network of the overlapping DEGs was constructed using STRING (version 10.0) and visualized using Cytoscape software (version 3.5.1; http://www.cytoscape.org/download.php). Significant nodes were selected using a threshold score of >500 (obtained via STRING).
Patients
A total of 5 HCC patients (3 male and 2 female; age range, 32–65 years old; mean age 52.5 years old) were accepted in the Second People's Hospital of Tianjin (Tianjin, China) between November 2016 and May 2017. Tumor and paracancerous tissue specimens were collected. All patients signed informed consent prior to enrollment in the study, and all procedures were conducted under the ethical approval of the Second People's Hospital of Tianjin and the Chinese national research committee.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR and methylation-specific PCR (MSP) were performed to detect the mRNA expression level and methylation status of Denticleless protein homolog (DTL), Dual specificity phosphatase 1 (DUSP1), Nuclear factor k-light-chain gene enhancer of activated B cells inhibitor, a (NFKBIA) and SOCS2. The total RNA was extracted using TRIzol (Invitrogen; Scientific, Inc., Waltham, MA, USA). The PrimeScript® 1st Stand cDNA Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China), the SYBR® Premix Ex Taq™ kit (Takara Bio, Inc., Otsu, Japan) and the Applied Biosystems™ QuantStudio™ 5 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) were used to conduct RT-PCR, according to the manufacturer's instructions. The reaction conditions of reverse transcription were as follows: 30°C for 10 min, 42°C for 60 min, and 95°C for 5 min. The EZ-DNA Methylation-Gold kit™ (Zymo Research Corp., Irvine, CA, USA) was used to conduct MSP, according to the manufacturer's instructions. All primers were designed and synthesized by Takara Biomedical Technology Co., Ltd. (Beijing, China), and their sequences are listed in Table I. β-actin was used as an internal control. The thermocycling conditions were as follows: 95°C for 5 min; followed by 40 cycles of 95°C for 15 sec, 60°C for 30 sec, and 72°C for 35 sec; and a final 5 min at 72°C extension. The 2−ΔΔCt method was used to calculate the relative expression value of the target gene (20).
Table I.
Primer sequences.
| Gene | Primer sequences (5′-3′) | Size (bp) |
|---|---|---|
| DTL (mRNA) | 490 | |
| Forward | CCTCTGTCCGATCCTCCAAA | |
| Reverse | AAAGATTTTCAGTCCCGCGG | |
| DTL (methyl) | 152 | |
| Forward | TTTTTTGTTCGATTTTTTAAAGACG | |
| Reverse | TAATAACCCTACAACTCTACCCGAA | |
| DUSP1 (mRNA) | 490 | |
| Forward | AGAATGTTCCTGACTCGGCA | |
| Reverse | AAGCAAAATCCAATCCCGGG | |
| DUSP1 (methyl) | 142 | |
| Forward | AGGCGAAAATATATAAGTTAAGCGA | |
| Reverse | AATACCGAATCAAAAACATTCTACG | |
| NFKBIA (mRNA) | 560 | |
| Forward | AGCGATGGGGTCTCACTATG | |
| Reverse | TCCAACAGCTTAGGTCAGGG | |
| NFKBIA (methyl) | 147 | |
| Forward | ATTTTAGTTTTTTAAGTAGGTGCGA | |
| Reverse | ATAAAACAAAAAAATCACTTACGTT | |
| SOCS2 (mRNA) | 630 | |
| Forward | GCTTGGGGTTAAATGGTGCA | |
| Reverse | AAGGGATGGGGCTCTTTCTC | |
| SOCS2 (methyl) | 181 | |
| Forward | GTATTGATTTTAAGGAAGGACGC | |
| Reverse | CCTACGAAAATAACTCCTCCG | |
| β-actin (mRNA) | 308 | |
| Forward | CATCTCTTGCTCGAAGTCCA | |
| Reverse | ATCATGTTTGAGACCTTCAACA | |
| β-actin (methyl) | 110 | |
| Forward | ATTATTATTGGTAATGAGCGGTTTC | |
| Reverse | TTCATAATAAAATTAAATATAATTTCGTA |
RT-qPCR, reverse transcription-quantitative polymerase chain reaction; MSP, methylation-specific polymerase chain reaction; methyl, methylation; DTL, denticleless protein homolog; DUSP1, dual specificity phosphatase 1; NFKBIA, nuclear factor of k light chain gene enhancer in B cells inhibitor, α; SOCS2, suppressor of cytokine signaling 2.
Statistical analysis
SPSS (version 17; SPSS Inc., Chicago, IL, USA) was used for all statistical analyses, and data are expressed as the mean ± standard deviation. Student's t-test was used to compare 2 groups. P<0.05 was considered to indicate a statistically significant difference.
Results
DEGs and DMSs
A total of 1,402 (562 upregulated and 840 downregulated), 2,049 (859 upregulated and 1,190 downregulated) and 2,159 (1,339 upregulated and 820 downregulated) DEGs were separately identified in the HCC group compared with control group in GSE95698, GSE49515 and GSE76427, respectively. A total of 41 overlaps were identified among all 3 sets of DEGs (Fig. 1; Table II). Among these, 8 genes were upregulated simultaneously [CCNB1, CEP55, DTL, Endothelial cell specific molecule 1 (ESM1), NEU1, RRM2, UHRF1 and VPS72], and 14 genes were downregulated simultaneously (CCL21, CYP1A1, DAO, DUSP1, GABARAPL1, GADD45B, HSD11B1, NFIL3, NFKBIA, NR4A2, NSUN6, RHOB, SLC27A2 and TRIB1) in the 3 sets of DEGs.
Figure 1.
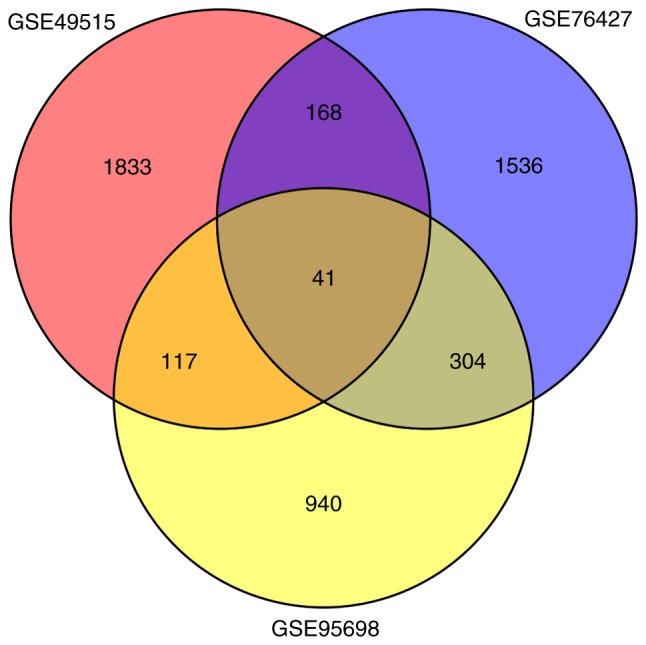
The Venn diagram illustrating the 3 sets of differentially expressed genes.
Table II.
Overlapping differentially expressed genes in the hepatocellular carcinoma group compared with the control group in GSE95698, GSE49515 and GSE76427.
| A, GSE49515 | ||
|---|---|---|
| Genes | Mean lgFC | P-value |
| CCNB1 | 0.80 | 2.09×10−3 |
| CEP55 | 1.14 | 1.25×10−3 |
| DTL | 0.83 | 9.88×10−3 |
| ESM1 | 0.53 | 4.88×10−3 |
| NEU1 | 0.78 | 2.31×10−7 |
| RRM2 | 0.56 | 2.41×10−2 |
| UHRF1 | 1.04 | 7.11×10−4 |
| VPS72 | 0.65 | 7.69×10−5 |
| CCL21 | −0.60 | 8.37×10−5 |
| CYP1A1 | −0.57 | 1.40×10−4 |
| DAO | −3.93 | 1.53×10−8 |
| DUSP1 | −1.11 | 1.64×10−5 |
| GABARAPL1 | −0.73 | 3.31×10−5 |
| GADD45B | −0.66 | 9.85×10−7 |
| HSD11B1 | −0.90 | 6.32×10−3 |
| NFIL3 | −1.66 | 1.04×10−6 |
| NFKBIA | −0.78 | 4.47×10−3 |
| NR4A2 | −2.5 | 2.49×10−6 |
| NSUN6 | −0.6 | 4.80×10−3 |
| RHOB | −0.88 | 1.13×10−5 |
| SLC27A2 | −1.43 | 1.49×10−4 |
| TRIB1 | −0.53 | 1.26×10−3 |
| ABHD6 | 1.02 | 1.20×10−7 |
| AKR7A3 | 0.92 | 1.65×10−5 |
| ALDH8A1 | 0.85 | 1.12×10−2 |
| ANG | 1.51 | 1.13×10−5 |
| C11orf24 | 0.58 | 2.66×10−4 |
| CLEC1B | 2.09 | 3.86×10−3 |
| CYP2J2 | 0.52 | 1.12×10−2 |
| EOMES | 0.64 | 4.07×10−2 |
| GHR | 1.62 | 1.67×10−5 |
| GZMK | 0.67 | 4.42×10−3 |
| ITPRIPL2 | 0.55 | 1.58×10−3 |
| PDK4 | 0.61 | 4.69×10−3 |
| RARRES3 | 1.98 | 1.48×10−4 |
| SESTD1 | −0.54 | 1.81×10−4 |
| SOCS2 | 0.85 | 1.26×10−5 |
| ST3GAL6 | 0.56 | 5.16×10−3 |
| TMEM45A | 1.28 | 3.71×10−2 |
| UGP2 | 0.86 | 8.84×10−6 |
| XAF1 | 0.81 | 1.55×10−2 |
| B, GSE76427 | ||
| Genes | Mean lgFC | P-value |
| CCNB1 | 0.62 | 1.77×10−13 |
| CEP55 | 0.75 | 3.33×10−12 |
| DTL | 0.60 | 3.92×10−10 |
| ESM1 | 1.13 | 8.87×10−13 |
| NEU1 | 0.71 | 5.27×10−11 |
| RRM2 | 0.51 | 1.55×10−13 |
| UHRF1 | 0.59 | 1.34×10−12 |
| VPS72 | 0.56 | 3.47×10−17 |
| CCL21 | −0.81 | 1.01×10−4 |
| CYP1A1 | −0.72 | 1.69×10−2 |
| DAO | −0.56 | 7.49×10−6 |
| DUSP1 | −0.77 | 1.65×10−5 |
| GABARAPL1 | −1.27 | 3.43×10−17 |
| GADD45B | −1.40 | 2.45×10−15 |
| HSD11B1 | −0.59 | 1.83×10−2 |
| NFIL3 | −0.53 | 1.39×10−6 |
| NFKBIA | −0.63 | 4.06×10−11 |
| NR4A2 | −0.51 | 9.59×10−4 |
| NSUN6 | −0.89 | 1.34×10−17 |
| RHOB | −1.09 | 1.25×10−10 |
| SLC27A2 | −0.83 | 3.90×10−7 |
| TRIB1 | −0.99 | 1.79×10−12 |
| ABHD6 | −0.87 | 3.63×10−16 |
| AKR7A3 | −1.04 | 1.15×10−8 |
| ALDH8A1 | −1.12 | 2.44×10−16 |
| ANG | −0.70 | 4.79×10−6 |
| C11orf24 | −0.74 | 3.27×10−12 |
| CLEC1B | −3.49 | 2.37×10−32 |
| CYP2J2 | −0.88 | 3.49×10−10 |
| EOMES | −0.68 | 1.94×10−7 |
| GHR | −2.04 | 5.96×10−22 |
| GZMK | −0.58 | 7.91×10−4 |
| ITPRIPL2 | −0.55 | 5.94×10−6 |
| PDK4 | −0.97 | 5.40×10−7 |
| RARRES3 | −0.55 | 4.11×10−5 |
| SESTD1 | 0.78 | 4.46×10−18 |
| SOCS2 | −1.42 | 6.75×10−16 |
| ST3GAL6 | −1.31 | 4.97×10−17 |
| TMEM45A | −0.88 | 1.55×10−5 |
| UGP2 | −0.74 | 1.02×10−13 |
| XAF1 | −0.89 | 2.72×10−8 |
| C, GSE95698 | ||
| Genes | Mean lgFC | P-value |
| CCNB1 | 2.29 | 1.92×10−2 |
| CEP55 | 2.00 | 2.17×10−2 |
| DTL | 3.32 | 1.93×10−3 |
| ESM1 | 6.12 | 1.75×10−3 |
| NEU1 | 1.18 | 2.25×10−2 |
| RRM2 | 2.23 | 4.49×10−2 |
| UHRF1 | 2.63 | 1.79×10−2 |
| VPS72 | 1.16 | 2.32×10−2 |
| CCL21 | −2.05 | 4.34×10−2 |
| CYP1A1 | −5.69 | 6.24×10−5 |
| DAO | −4.92 | 4.83×10−2 |
| DUSP1 | −1.66 | 1.55×10−2 |
| GABARAPL1 | −1.64 | 1.07×10−2 |
| GADD45B | −2.17 | 2.79×10−3 |
| HSD11B1 | −3.30 | 1.10×10−2 |
| NFIL3 | −1.04 | 3.03×10−2 |
| NFKBIA | −0.90 | 4.79×10−2 |
| NR4A2 | −2.76 | 9.34×10−3 |
| NSUN6 | −1.85 | 2.99×10−2 |
| RHOB | −1.94 | 1.13×10−2 |
| SLC27A2 | −3.15 | 4.95×10−2 |
| TRIB1 | −1.88 | 1.74×10−2 |
| ABHD6 | −2.22 | 9.50×10−3 |
| AKR7A3 | −2.81 | 2.15×10−2 |
| ALDH8A1 | −3.97 | 3.41×10−2 |
| ANG | −2.44 | 3.99×10−2 |
| C11orf24 | −1.00 | 4.95×10−2 |
| CLEC1B | −7.55 | 6.77×10−5 |
| CYP2J2 | −3.32 | 2.29×10−2 |
| EOMES | −3.02 | 6.40×10−3 |
| GHR | −4.4 | 2.30×10−2 |
| GZMK | −2.42 | 2.89×10−2 |
| ITPRIPL2 | 0.99 | 3.91×10−2 |
| PDK4 | −2.39 | 4.18×10−2 |
| RARRES3 | −1.76 | 9.60×10−3 |
| SESTD1 | 1.88 | 3.61×10−2 |
| SOCS2 | −2.21 | 2.07×10−2 |
| ST3GAL6 | −2.76 | 2.32×10−3 |
| TMEM45A | −2.25 | 1.37×10−2 |
| UGP2 | −1.42 | 3.12×10−2 |
| XAF1 | −1.51 | 1.79×10−2 |
FC, fold change.
For GSE73003, a total of 6,349 (2,854 upregulated and 3,495 downregulated) DMSs were identified in the HCC group compared with the control group, and the top 40 most-significant DMSs are listed in Table III. A total of 12 genes were among the 41 overlapping DEGs and the differentially methylated genes, and they are presented in Fig. 2. The expressions of DTL, DUSP1, eomesodermin (EOMES), ESM1, NFKBIA and SOCS2 were negatively with their methylation levels.
Table III.
The top 40 most significant differentially methylated sites in the hepatocellular carcinoma group compared with the control group.
| Gene | Regulation | P-value | Db-value |
|---|---|---|---|
| LYPD3 | Up | 1.13×10−11 | 0.26 |
| FLJ21159 | Up | 1.70×10−11 | 0.26 |
| ZNF154 | Up | 2.94×10−11 | 0.52 |
| ZNF540 | Up | 7.96×10−11 | 0.33 |
| GPR25 | Up | 8.68×10−11 | 0.20 |
| MS4A3 | Down | 9.61×10−11 | −0.23 |
| LDHB | Up | 1.62×10−10 | 0.44 |
| ALOX12 | Up | 1.65×10−10 | 0.12 |
| PRKG2 | Down | 1.99×10−10 | −0.31 |
| FLJ25773 | Down | 2.10×10−10 | −0.24 |
| PKDREJ | Up | 2.14×10−10 | 0.17 |
| TBC1D1 | Up | 2.47×10−10 | 0.13 |
| SLC39A12 | Down | 2.94×10−10 | −0.30 |
| SERHL | Up | 2.98×10−10 | 0.26 |
| C6orf206 | Up | 3.19×10−10 | 0.32 |
| C11orf2 | Up | 3.22×10−10 | 0.06 |
| CCDC37 | Up | 3.39×10−10 | 0.32 |
| LILRA1 | Down | 3.79×10−10 | −0.21 |
| OR51B4 | Down | 4.22×10−10 | −0.27 |
| HDAC1 | Down | 4.30×10−10 | −0.05 |
| XLF | Down | 5.08×10−10 | −0.04 |
| KCTD4 | Up | 5.57×10−10 | 0.24 |
| INA | Up | 5.58×10−10 | 0.34 |
| ABHD9 | Up | 6.00×10−10 | 0.42 |
| AQP6 | Down | 6.06×10−10 | −0.07 |
| ANKRD33 | Up | 6.90×10−10 | 0.19 |
| TSPYL5 | Up | 9.71×10−10 | 0.26 |
| RPS6KC1 | Down | 1.19×10−9 | −0.07 |
| FLJ13149 | Down | 1.33×10−9 | −0.06 |
| FCAR | Down | 1.38×10−9 | −0.22 |
| CD1B | Down | 1.40×10−9 | −0.23 |
| KLK2 | Down | 1.50×10−9 | −0.21 |
| CDH18 | Down | 1.50×10−9 | −0.34 |
| BOLL | Up | 1.67×10−9 | 0.24 |
| SGNE1 | Up | 1.67×10−9 | 0.25 |
| CYP11B1 | Down | 1.75×10−9 | −0.38 |
| S100A8 | Down | 1.92×10−9 | −0.21 |
| MAPKAP1 | Down | 2.01×10−9 | −0.06 |
| QTRT1 | Down | 2.21×10−9 | −0.09 |
| KLK9 | Down | 2.32×10−9 | −0.20 |
| FLJ46481 | Down | 2.35×10−9 | −0.24 |
Figure 2.
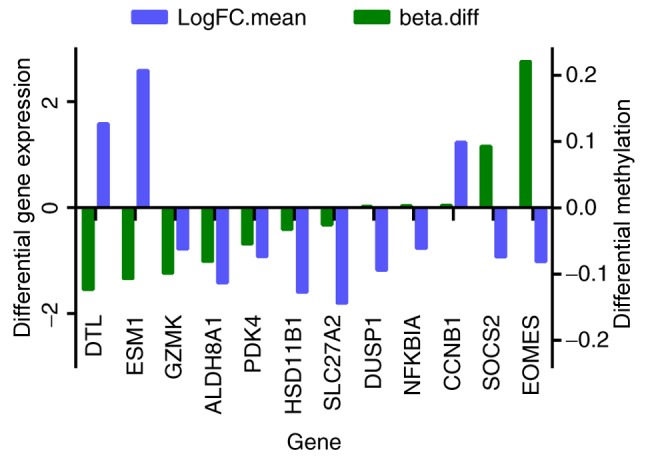
In total, 12 overlapping genes were differentially expressed and methylated in the HCC group compared with control. Beta.diff, fold-change in gene methylation level; LogFC.mean, fold-change in gene differential expression; HCC, hepatocellular carcinoma; DTL, denticleless E3 ubiquitin protein ligase; ESM1, endothelial cell specific molecule 1; GZMK, granzyme K; ALDH8A1, aldehyde dehydrogenase 8 family, member A1; PDK4, pyruvate dehydrogenase kinase 4; HSD11B1, hydroxysteroid 11-beta dehydrogenase 1; SLC27A2, solute carrier family 27 member 2; DUSP1, dual specificity phosphatase 1; NFKBIA, nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, α; CCNB1, cyclin B1; SOCS2, suppressor of cytokine signaling 2; EOMES, eomesodermin.
Enriched GO terms and KEGG pathways
The overlapping DEGs were enriched in 11 GO terms and 3 KEGG pathways, listed in Tables IV and V, respectively.
Table IV.
Enriched GO terms of the overlapping differentially expressed genes.
| Category | Term | Number of enriched genes | P-value |
|---|---|---|---|
| BP | GO:0044255: Cellular lipid metabolic process | 8 | <0.01 |
| BP | GO:0008610: Lipid biosynthetic process | 5 | 0.01 |
| BP | GO:0071310: Cellular response to organic substance | 8 | 0.03 |
| BP | GO:1901701: Cellular response to oxygen-containing compound | 5 | 0.04 |
| BP | GO:0043436: Oxoacid metabolic process | 5 | 0.04 |
| BP | GO:0031668: Cellular response to extracellular stimulus | 3 | 0.04 |
| BP | GO:0044242: Cellular lipid catabolic process | 3 | 0.04 |
| BP | GO:0006082: Organic acid metabolic process | 5 | 0.05 |
| CC | GO:0043231: Intracellular membrane-bounded organelle | 23 | 0.05 |
| MF | GO:0016712: Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen | 2 | <0.01 |
| MF | GO:0020037: Heme binding | 2 | 0.03 |
GO, gene ontology; BP, biological process; CC, cellular components; MF, molecular function.
Table V.
Enriched Kyoto Encyclopedia of Genes and Genomes pathways of the overlapping differentially expressed genes.
| Term | Number of enriched genes | P-value | Genes |
|---|---|---|---|
| cfa04115: P53 signaling pathway | 3 | 0.012539936 | CCNB1, RRM2, GADD45B |
| cfa04068: Foxo signaling pathway | 3 | 0.046310836 | CCNB1, GABARAPL1, GADD45B |
| cfa01100: Metabolic pathways | 7 | 0.081506099 | CYP2J2, CYP1A1, RRM2, ST3GAL6, HSD11B1, DAO, UGP2 |
The PPI network
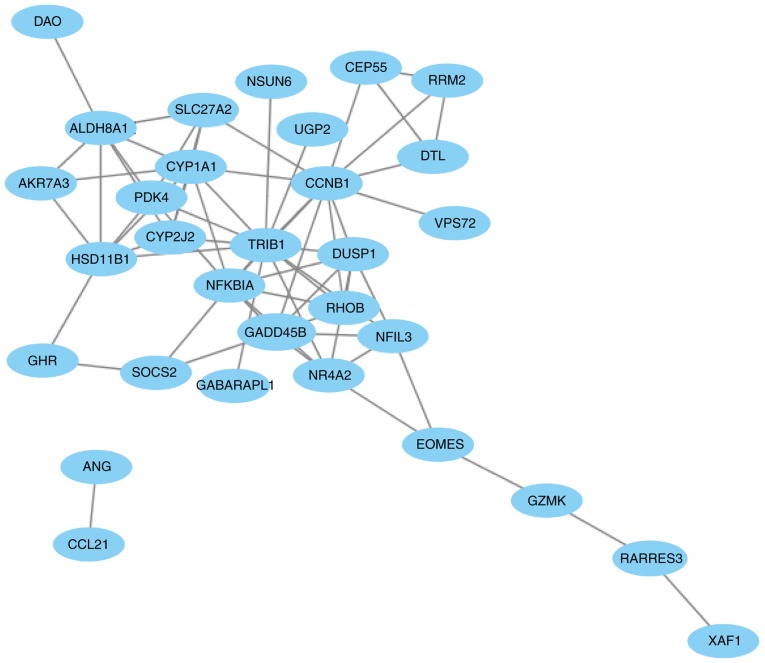
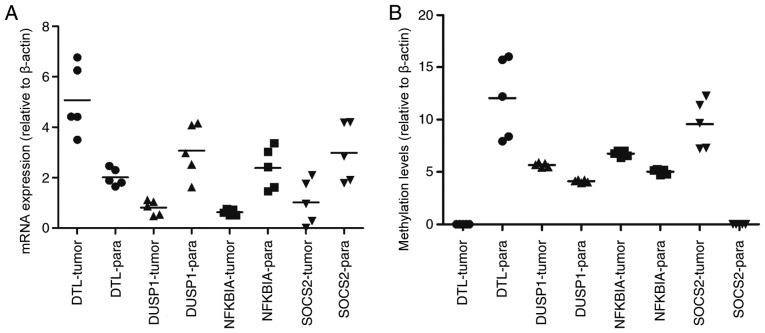
The PPI network of the 41 overlapping DEGs, including 66 interaction pairs, is presented in Fig. 3. Furthermore, the PPI network of nodes with scores >500 is presented in Fig. 4, including DTL, DUSP1, NFKBIA and SOCS2. DTL was differentially expressed (upregulated) and differentially methylated (hypomethylated). DUSP1, NFKBIA and SOCS2 were differentially expressed (downregulated) and differentially methylated (upregulated). The results of RT-qPCR and MSP are presented in Fig. 5. The mRNA expression of DTL was significantly increased in HCC tissue compared with paracancerous samples, and the mRNA expression of DUSP1, NFKBIA and SOCS2 was significantly decreased (P<0.05). Furthermore, the opposite effect on the methylation levels of these genes was observed (P<0.05). These results were consistent with those of the bioinformatics differential expression analysis.
Figure 3.
The protein-protein interaction network of the 41 overlapping differentially expressed genes.
Figure 4.
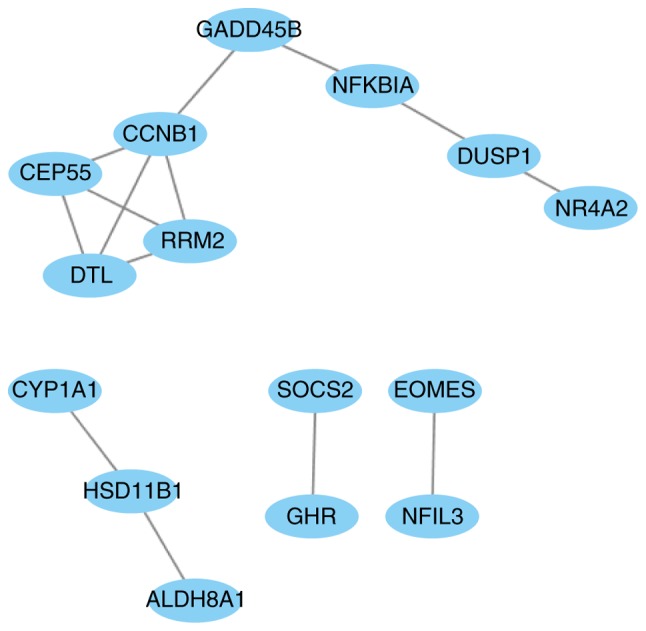
The protein-protein interaction network based on nodes scoring >500 in Search Tool for the Retrieval of Interacting Genes (version 10).
Figure 5.
Clinical verification of mRNA expression and methylation levels of DTL, DUSP1, NFKBIA and SOCS2 in HCC tumor and paracancerous samples. (A) Comparison of tumor vs. paracancerous mRNA expression levels: DTL, P=0.0007; DUSP1, P=0.0268; NFKBIA, P=0.0008, and SOCS2, P=0.0091. (B) Comparison of tumor vs. paracancerous methylation levels: DTL, P<0.0001; DUSP1, P<0.0001; NFKBIA, P<0.0001, and SOCS2, P<0.0001. -tumor, measured in tumor samples, -para measured in paracancerous samples.
Discussion
Recent advances have significantly improved our understanding of the molecular pathogenesis of HCC and its complex genetic landscape (21–23). The integration of multiple profiling data may provide additional insight into the molecular mechanisms of HCC. In the present study, 41 genes were simultaneously differentially expressed in 3 expression profiles, and they were enriched in 3 KEGG pathways: ‘p53 signaling’, ‘Foxo signaling’ and ‘metabolic’ pathways (Table V). The ‘p53 signaling’ pathway influences a myriad of diverse cellular processes, and p53 has been suggested to be activated in >50% human cancer types (24). The ‘p53 signaling’ pathway has been associated cancer occurrence and in mediating the response to cancer therapies (25). Kirstein and Vogel (26) summarized the pathological mechanisms of HCC carcinogenesis and progression. It was demonstrated that the most frequently identified mutations in HCC led to inactivation of p53. Cisplatin has been effectively used in the treatment of HCC, and p53 signaling is a potential target of cisplatin treatment (27). Paired box 5 (PAX5) has been suggested to be a functional tumor suppressor involved in HCC through direct regulation of the p53 signaling pathway, and Ras association domain family member 10 has been demonstrated to suppress human HCC growth by activating p53 signaling (28,29). The tyrosine kinase inhibitor, genistein, has been demonstrated to induce apoptosis and cell cycle arrest in HepG2 cells by activating the p53 signaling pathway (30). The forkhead box O1 (Foxo) signaling pathway participates in the regulation of multiple biological processes, including cellular responses to oxidative stress, cellular apoptosis and cell proliferation (31–33). It was reported that juglanthraquinone C could induce the apoptosis of HCC cells by activating the Foxo signaling pathway (34). PS341 (Bortezomib) was the first proteasome-inhibitor drug to be approved in clinical treatment for multiple myeloma, functioning by mediating targeted therapy against HCC through the Foxo3 signaling pathway (35). A previous study reported that Foxo-inhibition resulted in the myeloid maturation and death of acute myeloid leukemia cells. Leukemic cells resistant to Foxo-inhibition responded to JUN N-terminal kinase inhibition (36). Metabolic pathways mainly comprise metabolism of substance and energy, involved in a variety growth and development processes, and the occurrence and development of disease. Lu et al (37) hypothesized that blocking the cholesterol metabolic pathway may have potential therapeutic applications for patients with HCC. Björnsson (38) explored the central metabolic pathway in HCC using a differential rank conservation algorithm, revealing the involvement of ‘fatty acid metabolism’. Therefore, ‘p53 signaling’, ‘Foxo signaling’ and ‘metabolic’ pathways may serve roles in the pathogenesis of HCC, and may guide future research into the development of novel HCC therapies.
Combination analysis of the 41 overlapping DEGs revealed that the expression and methylation of 6 genes was downregulated in HCC (DTL, DUSP1, EOMES, ESM1, NFKBIA and SOCS2). In order to further investigate the association between these genes, a PPI network was constructed. DTL, DUSP1, NFKBIA and SOCS2 were nodes a score >500 (Fig. 4). Denticleless protein homolog (DTL) is a cell cycle-regulated nuclear and centrosome protein a protein (39). It was confirmed to be a critical target of miR-215, and DTL-knockdown by siRNA resulted in enhanced G2-arrest, p53 and p21 induction, and reduced cell proliferation of osteosarcoma and colon cancer cells (39). A previous study reported that overexpression of DTL promoted proliferation of tumor cells and was associated with a malignant outcome in esophageal squamous cell carcinoma (40). Another study reported that gastric carcinoma patients with DTL-overexpressing tumors had a relatively poor overall survival rate compared with those not exhibiting DTL expression (P=0.0498) and disease-free survival rate (P=0.0324) (41). DUSP1 has been demonstrated to be involved in cell cycle inhibition, apoptosis and senescence (42). A South Korean study revealed that DUSP1 functioned as a tumor suppressor during hepatocarcinogenesis, and that the DUSP1 expression was associated with the activation of p53 (43). Hao et al (44) revealed that disruption of a positive regulatory loop between DUSP1 and p53 promoted HCC development and progression. Wei et al (45) found that miR-101 inhibited macrophage-induced growth of HCC tumors by regulating tumor growth factor-β secretion via targeting DUSP1. NFKBIA inhibits NF-κB by forming a heterodimer with NF-κB, and preventing its translocation to the nucleus (46). A previous study investigation revealed the distribution frequency of the NFKBIA genotype and haplotype polymorphisms between HCC and control specimens (47). It has been reported that the expression of NFKBIA is decreased in liver cancer tissue compared with control tissue, and that negative NFKBIA expression predicted poor prognosis in patients with primary HCC (48). A nested case-control study in Shanghai (China) suggested that genetic variants of NFKB1 influenced liver cancer-susceptibility in the Chinese population (49). SOCS2 has been reported to inhibit tumor metastasis (50). The expression of SOCS2 has also been demonstrated to be markedly reduced in HCC and associated with aggressive tumor progression and poor prognosis in patients with HCC (51). Although ESM1 and EOMES were differentially expressed and methylated in the HCC group compared with control group, the differences were non-significant (P=0.061, 0.053, respectively) and they were minimally involved in the PPI networks. Therefore, we hypothesize that ESM1 and EOMES do not serve important roles in the pathogenesis.
In summary, DTL, DUSP1, NFKBIA and SOCS2 were closely associated with HCC, and may provide useful insight for developing the treatment and prognosis of HCC. In the present study, it was confirmed that DTL expression was upregulated in HCC, and DUSP1, NFKBIA and SOCS2 were downregulated, and the opposite effects on their methylation levels were observed (Fig. 5). We hypothesize that DTL, DUSP1, NFKBIA and SOCS2 may be involved in HCC carcinogenesis. In conclusion, the present study identified potential biomarkers of HCC, including DTL, DUSP1, NFKBIA and SOCS2. Furthermore, the ‘p53 signaling’, ‘Foxo signaling’ and ‘metabolic’ pathways may be involved in the carcinogenesis of HCC, and may provide insight into the development of novel HCC therapies. Future work will involve detection of these potential biomarkers in large clinical samples by immunohistochemistry (IHC).
Acknowledgements
Not applicable.
Funding
The present study was supported by the Tianjin Clinical Research Center for Organ Transplantation (grant no. 15ZXLCSY00070) and the Tianjin Municipal Science and Technology Commission (grant no. 17ZXMFSY00040).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
All authors had access to the data and were responsible for the concept and design of the study, data analysis/interpretation and critical revision and approval of the article. CM and WJ were responsible for data collection. WJ was responsible for drafting the article. CM and XS were responsible for statistical analyses.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen KW, Ou TM, Hsu CW, Horng CT, Lee CC, Tsai YY, Tsai CC, Liou YS, Yang CC, Hsueh CW, Kuo WH. Current systemic treatment of hepatocellular carcinoma: A review of the literature. World J Hepatol. 2015;7:1412–1420. doi: 10.4254/wjh.v7.i10.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sinn DH, Lee J, Goo J, Kim K, Gwak GY, Paik YH, Choi MS, Lee JH, Koh KC, Yoo BC, Paik SW. Hepatocellular carcinoma risk in chronic hepatitis B virus-infected compensated cirrhosis patients with low viral load. Hepatology. 2015;62:694–701. doi: 10.1002/hep.27889. [DOI] [PubMed] [Google Scholar]
- 3.Debes JD, Chan AJ, Balderramo D, Kikuchi L, Ballerga Gonzalez E, Prieto JE, Tapias M, Idrovo V, Davalos MB, Cairo F, et al. Hepatocellular carcinoma in South America: Evaluation of risk factors, demographics, and therapy. Liver Int. 2018;38:136–143. doi: 10.1111/liv.13502. [DOI] [PubMed] [Google Scholar]
- 4.Petrick JL, Campbell PT, Koshiol J, Thistle JE, Andreotti G, Beane-Freeman LE, Buring JE, Chan AT, Chong DQ, Doody MM, et al. Abstract 3007: Tobacco smoking, alcohol use and risk of hepatocellular carcinoma and intrahepatic cholangiocarcinoma: The Liver Cancer Pooling Project. Cancer Res. 2017;77 doi: 10.1158/1538-7445.AM2017-3007. [DOI] [Google Scholar]
- 5.Zampino R, Pisaturo MA, Cirillo G, Marrone A, Macera M, Rinaldi L, Stanzione M, Durante-Mangoni E, Gentile I, Sagnelli E, et al. Hepatocellular carcinoma in chronic HBV-HCV co-infection is correlated to fibrosis and disease duration. Ann Hepatol. 2015;14:75–82. [PubMed] [Google Scholar]
- 6.Hamid AS, Tesfamariam IG, Zhang Y, Zhang ZG. Aflatoxin B1-induced hepatocellular carcinoma in developing countries: Geographical distribution, mechanism of action and prevention. Oncol Lett. 2013;5:1087–1092. doi: 10.3892/ol.2013.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alavian SM, Haghbin H. Relative Importance of Hepatitis B and C viruses in hepatocellular carcinoma in EMRO countries and the Middle East: A systematic review. Hepat Mon. 2016;16:e35106. doi: 10.5812/hepatmon.35106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balan S, Finnigan J, Bhardwaj N. Dendritic cell strategies for eliciting mutation-derived tumor antigen responses in patients. Cancer J. 2017;23:131–137. doi: 10.1097/PPO.0000000000000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y, Li L, Xiang X, Wang H, Cai W, Xie J, Han Y, Bao S, Xie Q. Three common functional polymorphisms in microRNA encoding genes in the susceptibility to hepatocellular carcinoma: A systematic review and meta-analysis. Gene. 2013;527:584–593. doi: 10.1016/j.gene.2013.05.085. [DOI] [PubMed] [Google Scholar]
- 10.Lyragonzález I, Floresfong LE, Gonzálezgarcía I, Medina-Preciado D, Armendáriz-Borunda J. Adenoviral gene therapy in hepatocellular carcinoma: A review. Hepatol Int. 2013;7:48–58. doi: 10.1007/s12072-012-9367-2. [DOI] [PubMed] [Google Scholar]
- 11.Zekri AR, Sabry GM, Bahnassy AA, Shalaby KA, Abdel-Wahabh SA, Zakaria S. Mismatch repair genes (hMLH1, hPMS1, hPMS2, GTBP/hMSH6, hMSH2) in the pathogenesis of hepatocellular carcinoma. World J Gastroenterol. 2005;11:3020–3026. doi: 10.3748/wjg.v11.i20.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimonjic DB, Popescu NC. Role of DLC1 tumor suppressor gene and MYC oncogene in pathogenesis of human hepatocellular carcinoma: Potential prospects for combined targeted therapeutics (review) Int J Oncol. 2012;41:393–406. doi: 10.3892/ijo.2012.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, Sun W, Tang Y, Zhu L, Li Y, Ou C, Yang C, Su J, Luo C, Hu Y, Cao J. Identification of key genes in hepatocellular carcinoma and validation of the candidate gene, cdc25a, using gene set enrichment analysis, meta-analysis and cross-species comparison. Mol Med Rep. 2016;13:1172–1178. doi: 10.3892/mmr.2015.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis-A Comprehensive Study of loss of heterozygosity and microsatellite instability at 39 Loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32:970–979. doi: 10.1053/jhep.2000.19797. [DOI] [PubMed] [Google Scholar]
- 15.Zhao RC, Zhou J, He JY, Wei YG, Qin Y, Li B. Aberrant promoter methylation of SOCS-1 gene may contribute to the pathogenesis of hepatocellular carcinoma: A meta-analysis. J Buon. 2016;21:142–151. [PubMed] [Google Scholar]
- 16.Udali S, Guarini P, Ruzzenente A, Ferrarini A, Guglielmi A, Lotto V, Tononi P, Pattini P, Moruzzi S, Campagnaro T, et al. DNA methylation and gene expression profiles show novel regulatory pathways in hepatocellular carcinoma. Clin Epigenetics. 2015;7:43. doi: 10.1186/s13148-015-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmerli L, Hou BH, Tsai CH, Jakab G, Mauch-Mani B, Somerville S. The xenobiotic beta-aminobutyric acid enhances Arabidopsis thermotolerance. Plant J. 2008;53:144–156. doi: 10.1111/j.1365-313X.2007.03343.x. [DOI] [PubMed] [Google Scholar]
- 18.Tiezzi F, Parker-Gaddis KL, Cole JB, Clay JS, Maltecca C. A genome-wide association study for clinical mastitis in first parity US Holstein cows using single-step approach and genomic matrix re-weighting procedure. PLoS One. 2015 Feb 6;10(2):e0114919. doi: 10.1371/journal.pone.0114919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Jayachandran M. An updated portrait of pathogenesis, molecular markers and signaling pathways of hepatocellular carcinoma. Curr Pharm Design. 2017;23:2356–2365. doi: 10.2174/1381612823666170329124852. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Yang Z, Du F, Yang Q, Hou J, Yan X, Geng Y, Zhao Y, Wang H. Molecular mechanisms of pathogenesis in hepatocellular carcinoma revealed by RNA-sequencing. Mol Med Rep. 2017;16:6674–6682. doi: 10.3892/mmr.2017.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Qian Z, Li F, Li J, Lu Y. Integrative analysis of microarray data to reveal regulation patterns in the pathogenesis of hepatocellular carcinoma. Gut Liver. 2017;11:112–120. doi: 10.5009/gnl16063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stegh AH. Targeting the p53 signaling pathway in cancer therapy-the promises, challenges and perils. Expert Opin Ther Targets. 2012;16:67–83. doi: 10.1517/14728222.2011.643299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grochola LF, Zeronmedina J, Mériaux S, Bond GL. Single-nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb Perspect Biol. 2010;2:a001032. doi: 10.1101/cshperspect.a001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirstein MM, Vogel A. The pathogenesis of hepatocellular carcinoma. Dig Dis. 2014;32:545–553. doi: 10.1159/000360499. [DOI] [PubMed] [Google Scholar]
- 27.Wang P, Cui J, Wen J, Guo Y, Zhang L, Chen X. Cisplatin induces HepG2 cell cycle arrest through targeting specific long noncoding RNAs and the p53 signaling pathway. Oncol Lett. 2016;12:4605–4612. doi: 10.3892/ol.2016.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W, Li X, Chu ES, Go MY, Xu L, Zhao G, Li L, Dai N, Si J, Tao Q, et al. Paired box gene 5 is a novel tumor suppressor in hepatocellular carcinoma through interaction with p53 signaling pathway. Hepatology. 2011;53:843–853. doi: 10.1002/hep.24124. [DOI] [PubMed] [Google Scholar]
- 29.Jin Y, Cao B, Zhang M, Zhan Q, Herman JG, Yu M, Guo M. RASSF10 suppresses hepatocellular carcinoma growth by activating P53 signaling and methylation of RASSF10 is a docetaxel resistant marker. Genes Cancer. 2015;6:231–240. doi: 10.18632/genesandcancer.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong X, Liu PQ, Liang JX, Yun FU. Genistein induces apoptosis through upregulation of p53 signaling pathway. J Trop Med. 9:893–896. 910. [Google Scholar]
- 31.Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villén J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, Bonni A. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 32.Da NI, Xue S, Lian F, Wang WJ. Platelet-derived growth factor stimulates vascular smooth muscle cell proliferation through AKT-FoxO signaling pathway. Mol Cardiol China. 2011 [Google Scholar]
- 33.Huang P, Zhou Z, Wang H, Wei Q, Zhang L, Zhou X, Hutz RJ, Shi F. Effect of the IGF-1/PTEN/Akt/FoxO signaling pathway on the development and healing of water immersion and restraint stress-induced gastric ulcers in rats. Int J Mol Med. 2012;30:650–658. doi: 10.3892/ijmm.2012.1041. [DOI] [PubMed] [Google Scholar]
- 34.Hou YQ, Yao Y, Bao YL, Song ZB, Yang C, Gao XL, Zhang WJ, Sun LG, Yu CL, Huang YX, et al. Juglanthraquinone C induces intracellular ROS increase and apoptosis by activating the Akt/Foxo signal pathway in HCC cells. Oxid Med Cell Longev. 2016;2016:4941623. doi: 10.1155/2016/4941623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z, Liu S, Zhu M, Zhang H, Wang J, Xu Q, Lin K, Zhou X, Tao M, Li C, Zhu H. PS341 inhibits hepatocellular and colorectal cancer cells through the FOXO3/CTNNB1 signaling pathway. Sci Rep. 2016;6:22090. doi: 10.1038/srep22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sykes SM, Lane SW, Bullinger L, Kalaitzidis D, Yusuf R, Saez B, Ferraro F, Mercier F, Singh H, Brumme KM, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146:697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu M, Hu XH, Li Q, Xiong Y, Hu GJ, Xu JJ, Zhao XN, Wei XX, Chang CC, Liu YK, et al. A specific cholesterol metabolic pathway is established in a subset of HCCs for tumor growth. J Mol Cell Biol. 2013;5:404–415. doi: 10.1093/jmcb/mjt039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Björnsson E. The differential rank conservation algorithm (DIRAC) reveals deregulation of central metabolic pathways in hepatocellular carcinoma. 2014 [Google Scholar]
- 39.Song B, Wang Y, Titmus MA, Botchkina G, Formentini A, Kornmann M, Ju J. Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol Cancer. 2010;9:96. doi: 10.1186/1476-4598-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawaguchi T, Komatsu S, Ichikawa D, Hirajima S, Kosuga T, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Otsuji E. Overexpression of denticleless E3 ubiquitin protein ligase homolog (DTL) is related to tumor cell proliferation and malignant outcome in esophageal squamous cell carcinoma. J Am Coll Surgeons. 2015;221(Suppl 2):e132–e133. doi: 10.1016/j.jamcollsurg.2015.08.255. [DOI] [Google Scholar]
- 41.Kobayashi H, Komatsu S, Ichikawa D, Kawaguchi T, Hirajima S, Miyamae M, Okajima W, Ohashi T, Kosuga T, Konishi H, et al. Overexpression of denticleless E3 ubiquitin protein ligase homolog (DTL) is related to poor outcome in gastric carcinoma. Oncotarget. 2015;6:36615–36624. doi: 10.18632/oncotarget.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gang L, Qun L, Liu WD, Li YS, Xu YZ, Yuan DT. MicroRNA-34a promotes cell cycle arrest and apoptosis and suppresses cell adhesion by targeting DUSP1 in osteosarcoma. Am J Transl Res. 2017;9:5388–5399. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Kato I, Maita H, Takahashi-Niki K, Saito Y, Noguchi N, Iguchi-Ariga SM, Ariga H. Oxidized DJ-1 inhibits p53 by sequestering p53 from promoters in a DNA-binding affinity-dependent manner. Mol Cell Biol. 2013;33:340–359. doi: 10.1128/MCB.01350-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hao PP, Li H, Lee MJ, Wang YP, Kim JH, Yu GR, Lee SY, Leem SH, Jang KY, Kim DG. Disruption of a regulatory loop between DUSP1 and p53 contributes to hepatocellular carcinoma development and progression. J Hepatol. 2015;62:1278–1286. doi: 10.1016/j.jhep.2014.12.033. [DOI] [PubMed] [Google Scholar]
- 45.Wei X, Tang C, Lu X, Liu R, Zhou M, He D, Zheng D, Sun C, Wu Z. MiR-101 targets DUSP1 to regulate the TGF-β secretion in sorafenib inhibits macrophage-induced growth of hepatocarcinoma. Oncotarget. 2015;6:18389–18405. doi: 10.18632/oncotarget.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramos-Marquès E, Zambrano S, Tiérrez A, Bianchi ME, Agresti A, García-Del Portillo F. Single-cell analyses reveal an attenuated NF-κB response in the Salmonella-infected fibroblast. Virulence. 2016;8:719–740. doi: 10.1080/21505594.2016.1229727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng CW, Su JL, Lin CW, Su CW, Shih CH, Yang SF, Chien MH. Effects of NFKB1 and NFKBIA gene polymorphisms on hepatocellular carcinoma susceptibility and clinicopathological features. PLoS One. 2013;8:e56130. doi: 10.1371/journal.pone.0056130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang WJ, Ren G, Qi JD, Liu SH, Liang J. Expression changes of NFKBIA in hepatocellular carcinoma and its significance. Shandong Medical Journal. 2015;22:16–19. [Google Scholar]
- 49.Gao J, Xu HL, Gao S, Zhang W, Tan YT, Rothman N, Purdue M, Gao YT, Zheng W, Shu XO, Xiang YB. Genetic polymorphism of NFKB1 and NFKBIA genes and liver cancer risk: A nested case-control study in Shanghai, China. BMJ Open. 2014;4:e004427. doi: 10.1136/bmjopen-2013-004427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cui M, Ji S, Hou J, Fang T, Wang X, Ge C, Zhao F, Chen T, Xie H, Cui Y, et al. The suppressor of cytokine signaling 2 (SOCS2) inhibits tumor metastasis in hepatocellular carcinoma. Tumour Biol. 2016;37:13521–13531. doi: 10.1007/s13277-016-5215-7. [DOI] [PubMed] [Google Scholar]
- 51.Qiu X, Zheng J, Guo X, Gao X, Liu H, Tu Y, Zhang Y. Reduced expression of SOCS2 and SOCS6 in hepatocellular carcinoma correlates with aggressive tumor progression and poor prognosis. Mol Cell Biochem. 2013;378:99–106. doi: 10.1007/s11010-013-1599-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.