Abstract
Lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) are the two major subtypes of lung cancer. To explore mitochondrial respiratory gene expression profiles in LUSC and LUAD, RNA sequencing data from The Cancer Genome Atlas was used for comprehensive analyses to establish the molecular characteristics of LUSC and LUAD. To elucidate expression profiles, subtypes were defined using unsupervised clustering of mitochondrial gene expression data. Differences in nuclear gene expression levels, signaling pathways and tumor microenvironments between subtypes were investigated. The analysis revealed that mitochondrial respiratory genes were generally expressed at lower levels in tumor tissues compared with matched control tissues. The expression of mitochondrially encoded NADH dehydrogenase 5 or 6 was associated with tumor progression in LUAD and LUSC. Patients were clustered into three subgroups based on the expression profile of 13 mitochondrial protein-encoding genes, and patients in Cluster 3 exhibited poor survival rates compared with patients from Cluster 1. Furthermore, this association was also observed in another independent data set. Further analyses of the expression of nuclear-encoded genes in the three clusters revealed the enrichment of several cancer-associated signaling pathways in Cluster 3, particularly the apoptotic signaling pathway, suggesting a potential association between the decreased expression of mitochondrial DNA genes and increased tumor aggressiveness. Furthermore, the analyses of immune cell compositions in the tumor microenvironment detected a significant increase in the proportion of CD4+ T cells and a decrease in the proportion of macrophages in LUAD compared with LUSC (P=0.0000104 and P=0.0000105, respectively). In conclusion, the present study revealed an association between the expression patterns of mitochondrial-encoded genes and lung cancer, which may contribute to novel therapeutic strategies for patients with LUSC and LUAD.
Keywords: lung cancer, mitochondrial DNA, immunotherapy, RNA expression, lung squamous cell carcinoma, lung adenocarcinoma
Introduction
Mitochondrial genome instability and impaired mitochondrial function are hallmarks of cancer progression (1–3). The somatic mutations and copy number variations in mitochondrial genomes have been frequently reported to be associated with the clinical stage of a tumor and notably affect the malignancy progression of lung cancer (4–7) and pan-adenocarcinoma (8). Furthermore, it has been demonstrated that a decrease in mitochondrial DNA (mtDNA) alters mitochondrial gene expression, resulting in a deficiency in oxidative phosphorylation, and this results in the disturbance of cellular functions (6,9). Reznik et al (10,11) reported that the deletion tendency of mtDNA content and the suppression of mitochondrial respiratory gene expression were widely observed in numerous cancer types. Nevertheless, the role of the altered function of the mitochondrial respiratory gene expression remains poorly understood in lung cancer.
Lung cancer is a common cause of cancer-associated mortality globally. Of all patients with lung cancer, <7% of patients survive <10 years across all stages of lung cancer (12). There are two major subtypes in lung cancer: Lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), and these two subtypes differ considerably with regards to their genomic profiles. In LUSC, mutation of the tumor suppressor gene TP53 was observed in 81% of the samples, whereas in LUAD, TP53 mutations were observed in only 46% of tumors, and mutations on other cancer-associated genes, including KRAS proto-oncogene, GTPase, Kelch like ECH associated protein 1 and serine/threonine kinase 11 were commonly observed (13,14). Pan-cancer studies have revealed that the molecular chemistry of carcinogenesis may differ between LUAD and LUSC (13,15–17). The two subtypes are classified into different stages of lung cancer and present distinct expression patterns at different stages (18). Furthermore, the expression pattern of mRNAs and long non-coding RNAs also revealed differences between LUAD and LUSC (19,20). However, the extent to which these differences reflect alteration of the mitochondrial genome and function, and whether such changes contribute to the tumor biology, remain to be elucidated.
In the present study, in order to characterize mitochondrial gene expression profiles in LUSC and LUAD, a comparative study of patients with lung cancer from The Cancer Genome Atlas (TCGA) was conducted. The expression patterns of mtDNA genes were analyzed and changes in the expression level between LUAD and LUSC were compared using high-resolution expression profiles based on RNA sequencing (RNA-seq) data. Analyses of mitochondrial genes in LUAD and LUSC may provide new clues for improving the diagnosis and treatment of patients with LUAD and LUSC.
Materials and methods
Data harvesting
The RNA-seq data was obtained from the Genomic Data Commons Data Portal (GDC; http://portal.gdc.cancer.gov) in April 2017 (21). In brief, a search was performed using the keywords, ‘lung adenocarcinoma’, ‘lung squamous cell carcinoma’, ‘primary tumor and normal tissue’ and ‘gene expression quantification’ in the GDC. The raw gene count data and the fragments per kilobase of transcript per million (FPKM) gene expression data were retrieved. In total, the RNA-seq data of 1,014 primary tumor types (513 LUAD and 501 LUSC) and 108 adjacent normal tissues were downloaded. Of these, 57 LUAD and 49 LUSC profiles included matched normal tissue and tumor gene expression data. The clinical information of the patients, including age, sex and tumor stage according to tumor staging system (22), was collected from the TCGA Firehose pipeline on the Broad Institute website (http://gdac.broadinstitute.org). Additionally, an independent lung cancer microarray dataset (GSE81089) from the Gene Expression Omnibus was used in the present study (23). The dataset contained the expression data from 199 patients with lung cancer, including 108 LUAD, 67 LUSC and 24 from other types of lung cancer.
Relative gene expression pattern
The raw read counts were quantile normalized to allow comparisons between samples. This normalization was performed with a custom script, similar to the GDC/TCGA mRNA expression normalization pipeline. In brief, the raw gene read count was multiplied by a scalar (103) to account for the kilobase units, and then the normalized read count was calculated by dividing the 75th percentile read count value for all genes in each sample. To explore the heterogeneity in expression, a hierarchical clustering algorithms with euclidean distance was applied on the normalized read count, and the number of clusters was manually selected according to a silhouette plot (24). Finally, unpaired student's t-tests were used to compare the expression differences of single genes, and EBseq (25) was adopted to compare the expression profiles between the two groups.
Gene set enrichment analysis (GSEA)
The Java GSEA Desktop v3.0 (July, 2017) application (maintained by the GSEA team) was used to perform gene set enrichment analysis (26). Cancer-associated Kyoto Encyclopedia of Genes and Genomes (27–29), BIOCARTA (30) and Gene Ontology gene sets (31,32) were manually selected for analyses. P-values were estimated by 1,000 permutations.
Immune cell composition
The immune cell composition was determined from the LUAD and LUSC expression profiles by applying CIBERSORT (33) with the LM22 gene signature to identify 22 immune cell types. The immune cell composition analyses were performed with 100 permutations using the default parameters. The results were filtered by a maximum P-value of 0.05.
Statistical analyses
The data are presented as the mean ± standard deviation (SD). P<0.05 was considered to indicate a statistically significant difference. Qualitative variables were compared by Fisher's exact test. The unpaired student's t-test was used for normally distributed data comparison, and the Wilcoxon signed-rank test was used for non-normally distributed data. Two-way analysis of variance test was adopted for comparisons across multiple groups with Tukey's post-hoc test. A log-rank test and Cox's proportional hazards regression analysis were performed with the Survival package for R (34). All statistical analyses were performed in the R software environment (35).
Results
Mitochondrial gene expression patterns in LUAD and LUSC
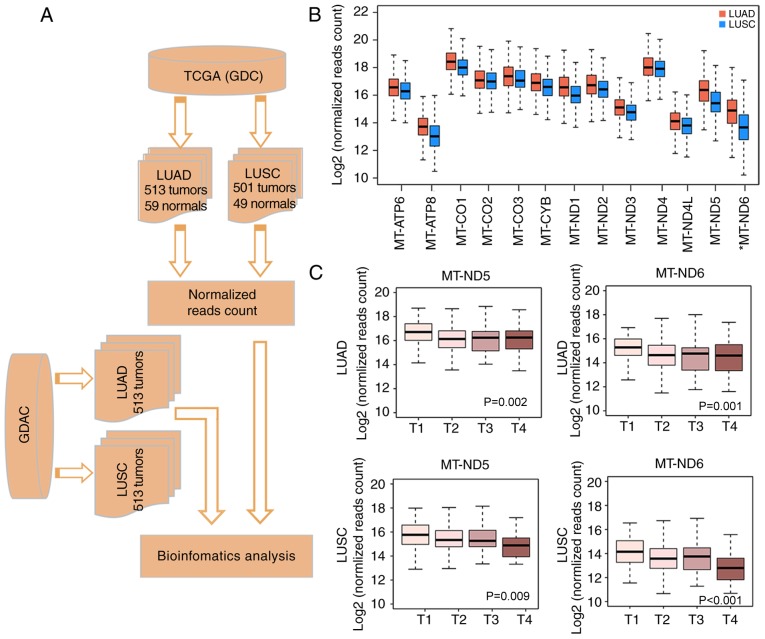
Mitochondrial dysfunction is often identified in cancer (3). For the analysis of mitochondrial gene expression patterns in lung cancer, RNA-seq data was downloaded from TCGA via the GDC (Fig. 1A). In total, 23 expressed mitochondrial genes, including 13 protein-encoding genes and 10 transfer RNA and rRNA genes (median FPKM value>0) were detected in tumor tissues. Genes encoding subunits of the respiratory chain serve a fundamental role in oxidative phosphorylation, which is closely associated with tumorigenesis (36); therefore, the 13 protein-encoding mitochondrial respiratory genes, MT-ATP6, MT-ATP8, MT-CO1, MT-CO2, MT-CO3, MT-CYB, MT-ND1, MT-ND2, MT-ND3, MT-ND4, MT-ND4L, MT-ND5 and MT-ND6 were the focus of the present study. The analyses revealed that their expression was high in normal lung and lung tumor tissues compared with the non-protein encoding mtDNA genes, but when compared with normal lung tissue, a general decreased expression in the tumor tissue was observed.
Figure 1.
Differential mitochondrial gene expression profiles between LUAD and LUSC. (A) Flow chart of protocols in the present study. (B) Comparison of mitochondrial gene expression between LUAD and LUSC. The box plot illustrates that the log2-normalized read counts (y-axis) are decreased in LUSC (blue) compared with LUAD (orange) for the majority of the genes. (C) Association of MT-ND5 and MT-ND6 with tumor stage. The box plot demonstrates the association between the log2-normalized read counts (y-axis) and the tumor stage (x-axis) for MT-ND5 and MT-ND6 in LUAD and LUSC, respectively. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MT-ND, mitochondrially encoded NADH dehydrogenase; TCGA, The Cancer Genome Atlas; GDC, Genomic Data Commons Data Portal; GDAC, Genome Data Analysis Centers.
LUAD and LUSC are the two major types of lung cancer. Previous analyses of genetic alterations revealed a clear difference between LUAD and LUSC (13,14). In the present study, a comparison of mitochondrial gene expression profiles in LUAD and LUSC was performed, and a general trend towards downregulated expression in LUSC compared with LUAD was identified (Fig. 1B). As no significant differences in the expression in the matched normal tissue for these two cancer types were observed, this result indicates the different biological roles of mtDNA expression in these two cancer types.
To investigate the association of mitochondrial gene expression with clinical parameters, the clinical information for the patients was retrieved from the Broad Institute Genome Data Analysis Centers (GDAC) Firehose. The analysis of mitochondrial genes and clinical data identified two genes, mitochondrially encoded NADH dehydrogenase (MT-ND)5 and MT-ND6, which were significantly associated with TNM staging (22) (P=0.002 and P=0.009 for LUAD, respectively; P=0.001 and P<0.001 for LUSC, respectively; Fig. 1C) (36,37). MT-ND5 and MT-ND6 were downregulated with increasing tumor stage (Fig. 1C). This association was observed for LUAD and LUSC. Mutations in MT-ND5 has been previously reported to be associated with elevated reactive oxygen species (ROS) production and the epigenetic regulation of apoptosis in a p53-dependent manner, resulting in the generation of pro-cancerous phenotypes (38), whereas mutated MT-ND6 was reported to be associated with a defective response to hypoxia in human glioma cells (39).
Identification of three subtypes in lung cancer
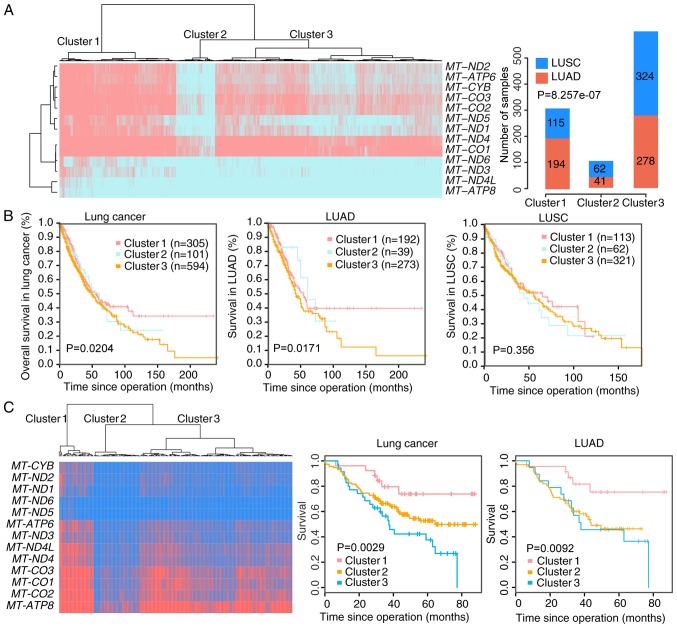
The heterogeneity of lung cancer introduces significant challenges in developing effective treatment strategies. To investigate the heterogeneity in lung cancer, an unsupervised clustering was applied to cluster the 1,014 samples based on the expression of the 13 mtDNA genes. Three clusters were defined using a silhouette plot. The mtDNA gene expression in Cluster 1 was higher compared with the two other clusters. By contrast, these genes were markedly downregulated in Cluster 2 compared with the other two clusters. In Cluster 3, the mean expression differences of each gene were 2.28–2.98 times less compared with that in Cluster 1, and 2.1–3.2 times greater compared with Cluster 2 (Fig. 2A).
Figure 2.
Gene expression profile and prognostic significance of three identified clusters in lung cancer. (A) Gene expression profile of three clusters in lung cancer. Unsupervised clustering of the expression of the 13 mitochondrial protein-coding genes identified 3 clusters of the 1,014 lung cancer tumor expression profiles in the left panel. The total number of LUAD and LUSC profiles in each cluster is illustrated in the right panel as a bar chart. (B) Kaplan-Meier survival curves for Clusters 1, 2 and 3 for overall lung cancer, LUAD and LUSC. (C) Gene expression profile of the three clusters in GSE81089. Unsupervised clustering of the expression of the 13 mitochondrial genes was applied to 175 lung cancer tumor expression profiles. Kaplan-Meier survival curves for Clusters 1, 2 and 3 in overall lung cancer and LUAD were drawn. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
The associations between these clusters and clinical parameters were investigated. The clustering results demonstrated that LUAD tumor types were significantly enriched in Cluster 1 (P<0.0001, Fig. 2A). It was also observed that stage III/IV tumors were more frequent in Cluster 2 (30% in Cluster 2 vs. 20% and 18% in Clusters 1 and 3, respectively) in all patients, and a similar phenomenon was observed in LUSC and LUAD separately (Table I).
Table I.
Association of subtypes and clinical characters in lung cancer (LUAD and LUSC).
| Overall | LUAD | LUSC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cluster 1 | Cluster 2 | Cluster 3 | P-value | Cluster 1 | Cluster 2 | Cluster 3 | P-value | Cluster 1 | Cluster 2 | Cluster 3 | P-value | |
| Samples | 309 | 103 | 602 | 194 | 41 | 278 | 115 | 62 | 324 | |||
| Age (median) | 66.1 | 65.9 | 66.3 | 65.3 | 66.7 | 64.9 | 67.5 | 65.5 | 67.4 | |||
| Sex | ||||||||||||
| Male | 179 (58%) | 69 (67%) | 360 (60%) | 0.27 | 95 (49%) | 21 (51%) | 121 (44%) | 0.39 | 84 (73%) | 48 (77%) | 239 (74%) | 0.83 |
| Female | 130 (42%) | 34 (33%) | 242 (40%) | 99 (51%) | 20 (49%) | 157 (56%) | 31 (27%) | 14 (23%) | 85 (26%) | |||
| Pathological stages | ||||||||||||
| I/II | 246 (80%) | 72 (70%) | 491 (82%) | 0.03 | 153 (80%) | 27 (66%) | 220 (79%) | 0.15 | 93 (81%) | 45 (73%) | 271 (84%) | 0.10 |
| III/IV | 61 (20%) | 31 (30%) | 110 (18%) | 39 (20%) | 14 (34%) | 58 (21%) | 22 (19%) | 17 (27%) | 52 (16%) | |||
To explore the prognostic value of the identified molecular subtypes, a Kaplan-Meier analysis was performed for all patients in the dataset. It was observed that Cluster 3 was significantly associated with a worse survival outcome compared with the other two clusters (P=0.0204), a phenomenon that was validated in another public dataset (Fig. 2B) and remained a significant independent prognostic marker following corrections for age, sex, and clinical stage. Next, the expression data for 175 patients with lung cancer from the published dataset GSE81089 (40), including 108 LUAD and 67 LUSC cases, was analyzed. The survival outcome of Cluster 3 was significantly poorer compared with Cluster 1 in this cohort (P=0.0029; Fig. 2C). This is consistent with the observation that a lower tumor mtDNA copy number was a significant predictor of poor patient survival rate by Reznik et al (10). The association between overall survival rate and the molecular subtypes was not significant in LUSC from this dataset, and therefore further analysis focused on LUAD (Fig. 2B). The downregulation of MT-ND5 or MT-ND6 was associated significantly with a shortened survival time in LUAD. This association persisted when analyzed with Cox's proportional hazards regression analysis (MT-ND5, P=0.08, hazards ratio (HR)=0.76; MT-ND6, P=0.12, HR=0.78) (Table II).
Table II.
The prognostic value of MT-ND5 and MT-ND6 in LUAD.
| MT-ND5 | MT-ND6 | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Gene | 0.761 (0.561~1.03) | 0.080 | 0.787 (0.578~1.07) | 0.129 |
| Age | 1.01 (0.994~1.02) | 0.234 | 1.01 (0.994~1.02) | 0.256 |
| Sex | 1.03 (0.762~1.39) | 0.849 | 1.02 (0.758~1.38) | 0.875 |
| Stage | 2.49 (1.82~3.42) | <0.0001 | 2.46 (1.79~3.38) | <0.0001 |
Signaling pathways and immunological associations with lung cancer subtypes
To further characterize the molecular mechanisms that may potentially contribute to the differences observed between LUSC and LUAD, GSEA (26) was applied to the nuclear gene data to identify the pathways exhibiting aberrant expression patterns potentially associated with tumor progression. The focus was on comparing Cluster 1 and Cluster 3 tumor types, as the prognosis of patients in these two groups differed markedly. It was identified that the expression of genes involved in linoleic acid metabolism was upregulated in Cluster 1 tumor types, whereas Cluster 3 was characterized by the upregulated expression of pathways associated with cancer development. Genes involved in signaling pathways including Fas, apoptosis, protein kinase B, Erb-B2 receptor tyrosine kinase 2, Toll-like receptor signaling and tumor growth factor-β (TGF-β) signaling were highly expressed in Cluster 3 LUAD tumor types (Table III). A previous study suggested that mtDNA deletion may confer specific gene expression in human lung cancer cells (9). Alterations to apoptotic pathways are common in cancer cells and lung tumor types, and have been implicated in lung tumorigenesis (41). Furthermore, defective apoptotic signaling may contribute to chemoresistance and radiotherapy resistance (41,42).
Table III.
Pathways enriched in Cluster 3 specialized for lung adenocarcinoma expression profiles.
| Name of pathwaya | Number of genes | Enrichment score | Normalized enrichment score | NOM P-value | False discover rate Q-value | FWER P-value |
|---|---|---|---|---|---|---|
| Erb-B2 receptor tyrosine kinase 2 signaling | 22 | −0.5588 | −1.8855 | 0.0085 | 0.1091 | 0.1530 |
| Kyoto Encyclopedia of Genes and Genomes | 97 | −0.4611 | −1.9080 | 0.0099 | 0.1237 | 0.1340 |
| Toll-like receptor signaling pathway | ||||||
| Apoptotic signaling (in response to chemical DNA damage) | 22 | −0.4616 | −1.6793 | 0.0064 | 0.1281 | 0.3690 |
| Tumor growth factor-β signaling (which regulates growth and proliferation) | 19 | −0.558 | −1.6585 | 0.0043 | 0.1363 | 0.4000 |
| Fas-mediated apoptotic signaling | 30 | −0.5528 | −1.9988 | 0.0041 | 0.1898 | 0.0820 |
| Protein kinase B signaling pathway | 21 | −0.5857 | −1.9086 | 0.0042 | 0.1840 | 0.1340 |
| KEGG_LINOLEIC_ACID_METABOLISM | 29 | 0.6804 | 1.7514 | 0.0021 | 0.3102 | 0.2640 |
Pathways with a False Discovery Rate ≤0.25 or nominal P-value ≤0.01. NOM, nominal P-value; FWER, family-wise error rate.
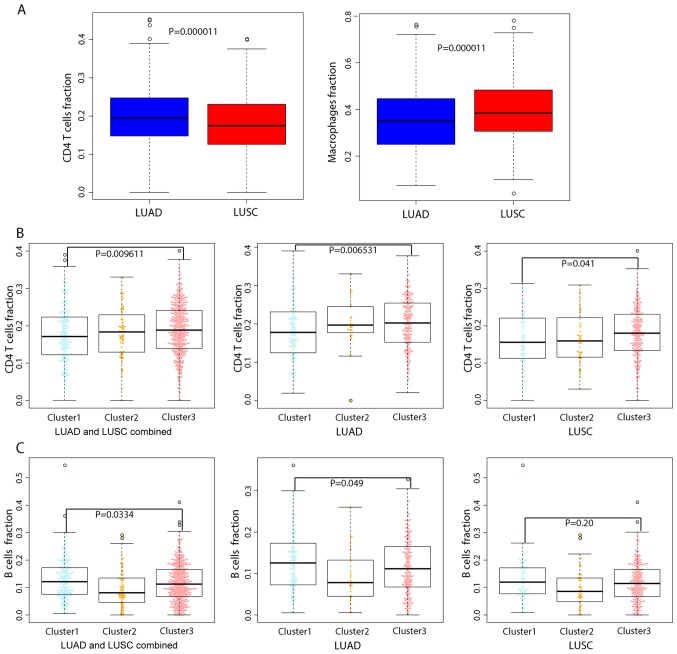
Intra-tumor heterogeneity is associated with leukocyte infiltration. To explore the repertoire of leukocytes in different lung cancer subtypes, a machine-learning approach, CIBERSORT (32), was applied to determine the composition of infiltrating immune cells. The approach evaluated the abundance of 22 distinct immune cell types. It was identified that the proportion of CD4+ T cells was significantly higher in LUAD compared with LUSC, whereas LUSC harbored significantly more macrophages compared with LUAD (P=0.0000104 and P=0.0000105, respectively; Fig. 3A). The proportion of these two leukocyte types also varied considerably across the three clusters; CD4+ T cells and macrophages comprised a significantly larger fraction of cells in Cluster 3 compared with in Cluster 1 (P=0.009611, Fig. 3B). These results are of interest, since CD4+ T cells modulate a wide range of immune responses and are able to differentiate into multiple sublineages in response to tumor antigens (43). Macrophages may be polarized into various subpopulations that exert different effects on the tumor, particularly tumor-associated macrophages (TAMs) which promote tumor proliferation, survival and migration (44). Finally, the analysis demonstrated that the proportion of follicular helper T cells and regulatory T cells, particularly B cells, were significantly reduced in Cluster 2 compared with Cluster 1 and Cluster 3 in LUAD of patients (P=0.0334, Fig. 3C).
Figure 3.
Comparison of the fraction of tumor-infiltrating immune cell types in LUAD and LUSC using nuclear gene mRNA expression profiles. Box plots presenting the evaluated fractions of immune cell types (y-axis) are plotted by cancer subtype (x-axis). (A) Comparison of the evaluated fraction of CD4+ T cells and macrophages between LUAD and LUSC. (B) Comparison of the evaluated fraction of CD4+ T cells across the three lung cancer subtypes. (C) Comparison of the evaluated fraction of B cells across the three lung cancer subtypes. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Discussion
In the present study, the expression profiles of mitochondrial protein-coding genes in lung cancer were analyzed using RNA sequencing data. Briefly, it was identified that the 13 mitochondrial respiratory genes were highly expressed compared with the non-protein encoding mitochondrial genes in normal and tumor tissue, although these genes were widely suppressed in tumor tissue compared with the corresponding normal tissue, emphasizing the potential differences in mitochondrial function in tumor tissue compared with normal lung tissue. Mitochondrial gene expression profiles were examined with a focus on these 13 mitochondrial genes in LUSC and LUAD, the two major subtypes of lung cancer. The expression of these 13 genes was downregulated in LUSC compared with LUAD, consistent with the reported molecular differences between LUAD and LUSC (13,14). In the present study, the expression of MT-ND5 and MT-ND6 was downregulated during tumor progression for LUSC and LUAD, and further analysis also revealed that the low expression of MT-ND5 and MT-ND6 was associated with a shortened survival time in LUAD. Previous studies have suggested that MT-ND5 may serve as a functional target of p53 and B-cell lymphoma-2 proteins to regulate ROS production and cellular invasiveness (38,44). In addition, Ishikawa et al (45) reported in 2008 that MT-ND6 missense mutations contribute to tumor cell metastasis in mouse fibrosarcoma. Furthermore, Yuan et al (46) revealed that nonsense and missense mutations of MT-ND6 gene promote cell migration and invasion in human lung adenocarcinoma. Consistently, the results of the present study indicate that the expression of MT-ND5 and MT-ND6 was significantly lower in tumor types with lymph node infiltration compared with tumor types without lymph node infiltration. However, further studies on MT-ND5 and MT-ND6 in LUAD are required in order to understand their biological functions and mechanisms.
In the present study, a molecular classifier for lung cancer was established based on analysis of the gene expression profiles, including three distinct subtypes (Cluster 1, 2 and 3), which were associated with different clinical and molecular characteristics. The prognostic value of the 13 mitochondrial genes in lung cancer was validated successfully in an independent dataset. In addition, a higher proportion of stage III–IV tumor types (30%) were identified in Cluster 2. Cluster 2 is less well-characterized compared with the other two cluster groups and will require further investigation. It was observed that the mitochondrial genes were expressed at higher levels in Cluster 1 compared with in Cluster 3. Furthermore, Cluster 1 and Cluster 3 were well-differentiated based on survival data analysis. LUAD cases in Cluster 3 exhibited an enriched expression of genes associated with cancer-associated signaling pathways, particularly the apoptotic signaling pathway. Furthermore, through CIBERSORT analysis, it was identified that the proportion of CD4+ T cells and macrophages was significantly higher in Cluster 3 compared with the other two clusters, whereas B cells, follicular helper T cells and regulatory T cells were significantly reduced in Cluster 3 compared with Cluster 1. One previous study indicated that activation of CD4+ T cells indirectly regulate the pulmonary metastasis of mammary carcinomas by enhancing the pro-tumor properties of macrophages (47). Macrophages generally serve a pro-tumoral role, are able to stimulate angiogenesis, and enhance tumor cell invasion, motility and intravasation (48). In lung cancer, TAMs may promote tumor progression by contributing to tumor stroma formation and angiogenesis through their release of platelet-derived growth factor, in conjunction with TGF-β1 production by tumor cells (49). Altogether, these results may help to explain the poor prognosis of patients with LUAD from Cluster 3. Further studies are required to clarify the association between the proportion of these immune cell subsets, and the different types and subsets of lung cancer, to allow the development of more precise immunotherapy against lung cancer.
In the present study, distinct expression profiles of mitochondrial genes were produced for LUSC and LUAD. LUSC may be classified into several subtypes based on nuclear gene expression profiles, including the basal expression, primitive expression, classical expression and secretory expression subtypes (14). Similarly, LUAD includes proximal, proliferative and terminal respiratory unit subtypes (13). Nevertheless, how the mitochondrial gene expression levels affect these subtypes, remains to be fully elucidated. Thus, further comprehensive analyses of mitochondrial and nuclear transcriptomics and genomics data are clearly warranted.
The present study was limited by the lack of information regarding the DNA copy number and does not address the cause of the difference in the expression level of mtDNA encoded genes. The number of copies of mtDNA greatly affect the expression level of mitochondrial genes and this close association between mtDNA copy number and gene expression has been reported by Yuan et al (8) utilizing data from the International Cancer Genome Consortium and Reznik et al (11) using TCGA data. Additionally, certain host genes may also control mitochondrial gene expression (50). Further studies with large lung cancer cohorts will be required to confirm the significance and robustness of the classifications developed in the present study. However, the classifications clearly reflect the inter-heterogeneity of LUSC and LUAD tumor types, and provide a basis for detecting the clinical features of lung cancer or developing therapeutic approaches for the improved management of LUAD and LUSC.
In summary, the present study highlighted: i) The suppression of the expression of MT-ND5 and MT-ND6 respiratory genes in lung cancer, particularly in advanced stages; ii) Clustering-based expression patterns associated with a poor prognosis in LUAD, but not in LUSC; iii) Tumor tissue with a mitochondrial gene expression pattern designated as Cluster 3 exhibited the enhanced expression of genes associated with pathways involved in cancer development and iv) Intra-tumor heterogeneity of the immune cell fraction was associated with the tumor subtypes. It is anticipated that these results will enable biomedical researchers to further explore the mechanisms of lung cancer development and identify novel therapeutic targets and modalities for improving the diagnosis and treatment of patients with lung cancer.
Acknowledgements
The authors would like to thank Dr Chabungbam Orville Singh (Zhejiang University, College of Animal Sciences, China) for revising the manuscript.
Funding
The present study was supported by the Natural Science Foundation of China (grant no. 81672818) and the Guangzhou Science and Technology Plan (grant no. 201604020005). This study was partially supported by the National Natural Science Foundation of China (grant no. 81472553).
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
QG, BR, RL and KK designed the study, interpreted the experimental results and modified the manuscript. NL, JZ and YM analyzed the data. BR drafted the manuscript. All authors have read and approved the final manuscript for publication.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Idaghdour Y, Hodgkinson A. Integrated genomic analysis of mitochondrial RNA processing in human cancers. Genome Med. 2017;9:36. doi: 10.1186/s13073-017-0426-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Choi S, Lee J, Huang YT, Chen F, Zhao Y, Lin X, Neuberg D, Kim J, Christiani DC. Mitochondrial variations in non-small cell lung cancer (NSCLC) survival. Cancer Inform. 2015;14(Suppl 1):S1–S9. doi: 10.4137/CIN.S13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JH, Hwang I, Kang YN, Choi IJ, Kim DK. Genetic characteristics of mitochondrial DNA was associated with colorectal carcinogenesis and its prognosis. PLoS One. 2015;10:e0118612. doi: 10.1371/journal.pone.0118612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuyama W, Nakagawa M, Wakimoto J, Hirotsu Y, Kawabata M, Osame M. Mitochondrial DNA mutation correlates with stage progression and prognosis in non-small cell lung cancer. Hum Mutat. 2003;21:441–443. doi: 10.1002/humu.10196. [DOI] [PubMed] [Google Scholar]
- 5.Dasgupta S, Soudry E, Mukhopadhyay N, Shao C, Yee J, Lam S, Lam W, Zhang W, Gazdar AF, Fisher PB, Sidransky D. Mitochondrial DNA mutations in respiratory complex-I in never-smoker lung cancer patients contribute to lung cancer progression and associated with EGFR gene mutation. J Cell Physiol. 2012;227:2451–2460. doi: 10.1002/jcp.22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu L, Yao X, Shen Y. Altered mitochondrial DNA copy number contributes to human cancer risk: Evidence from an updated meta-analysis. Sci Rep. 2016;6:35859. doi: 10.1038/srep35859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin C, Lee M. Cancer AB001. Low mitochondrial DNA copy number is associated with low mitochondrial DNA integrity and advanced T-status in non- small cell lung cancer. J Thorac Dis. 2017;9 doi: 10.21037/jtd.2017.s001. Suppl 14. [DOI] [Google Scholar]
- 8.Yuan Y, Ju YS, Kim Y, et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. bioRxiv: 161356. 2017 [Google Scholar]
- 9.Magda D, Lecane P, Prescott J, Thiemann P, Ma X, Dranchak PK, Toleno DM, Ramaswamy K, Siegmund KD, Hacia JG. mtDNA depletion confers specific gene expression profiles in human cells grown in culture and in xenograft. BMC Genomics. 2008;9:521. doi: 10.1186/1471-2164-9-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reznik E, Miller ML, Şenbabaoğlu Y, Riaz N, Sarungbam J, Tickoo SK, Al-Ahmadie HA, Lee W, Seshan VE, Hakimi AA, Sander C. Mitochondrial DNA copy number variation across human cancers. Elife. 2016;5:e10769. doi: 10.7554/eLife.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reznik E, Wang Q, La K, Schultz N, Sander C. Mitochondrial respiratory gene expression is suppressed in many cancers. Elife. 2017;6:e21592. doi: 10.7554/eLife.21592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung CHY, Juan HF. Quantitative proteomics in lung cancer. J Biomed Sci. 2017;24:37. doi: 10.1186/s12929-017-0343-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network: Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research Network: Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng X, Chen Z, Farshidfar F, Xu X, Lorenzi PL, Wang Y, Cheng F, Tan L, Mojumdar K, Du D, et al. Molecular characterization and clinical relevance of metabolic expression subtypes in human cancers. Cell Rep. 2018;23(255–269):e4. doi: 10.1016/j.celrep.2018.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, Zhang S. Tumor characterization and stratification by integrated molecular profiles reveals essential pan-cancer features. BMC Genomics. 2015;16:503. doi: 10.1186/s12864-015-1687-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Detterbeck FC, Boffa DJ, Tanoue LT. The new lung cancer staging system. Chest. 2009;136:260–271. doi: 10.1378/chest.08-0978. [DOI] [PubMed] [Google Scholar]
- 19.Chen M, Liu X, Du J, Wang XJ, Xia L. Differentiated regulation of immune-response related genes between LUAD and LUSC subtypes of lung cancers. Oncotarget. 2017;8:133–144. doi: 10.18632/oncotarget.13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, Chen Y, Yang J. LncRNAs are altered in lung squamous cell carcinoma and lung adenocarcinoma. Oncotarget. 2017;8:24275–24291. doi: 10.18632/oncotarget.13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375:1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brierley JD, Gospodarowicz MK, Wittekind C, editors. TNM classification of malignant tumours 978. 8th edition. Wiley-Blackwell; Chichester, West Sussex, UK: 2017. pp. 3241–3244. [Google Scholar]
- 23.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 25.Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, Kendziorski C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacRae SL, Zhang Q, Lemetre C, Seim I, Calder RB, Hoeijmakers J, Suh Y, Gladyshev VN, Seluanov A, Gorbunova V, et al. Comparative analysis of genome maintenance genes in naked mole rat, mouse, and human. Aging Cell. 2015;14:288–291. doi: 10.1111/acel.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latorre-Pellicer A, Moreno-Loshuertos R, Lechuga-Vieco AV, Sánchez-Cabo F, Torroja C, Acín-Pérez R, Calvo E, Aix E, González-Guerra A, Logan A, et al. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature. 2016;535:561–565. doi: 10.1038/nature18618. [DOI] [PubMed] [Google Scholar]
- 31.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carbon S, Dietze H, Lewis SE, et al. Expansion of the gene ontology knowledgebase and resources: The gene ontology consortium. Nucleic Acids Res. 2017;45:331–338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Therneau T. A Package for Survival Analysis in S. version 2.38. 2015. Reference Source. 2016 [Google Scholar]
- 35.Team, R. Core, corp-author. R: A language and environment for statistical computing. 2013 [Google Scholar]
- 36.Lemarie A, Grimm S. Mitochondrial respiratory chain complexes: Apoptosis sensors mutated in cancer? Oncogene. 2011;30:3985–4003. doi: 10.1038/onc.2011.167. [DOI] [PubMed] [Google Scholar]
- 37.Koshikawa N, Akimoto M, Hayashi JI, Nagase H, Takenaga K. Association of predicted pathogenic mutations in mitochondrial ND genes with distant metastasis in NSCLC and colon cancer. Sci Rep. 2017;7:15535. doi: 10.1038/s41598-017-15592-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh RK, Saini S, Verma D, Kalaiarasan P, Bamezai RNK. Mitochondrial ND5 mutation mediated elevated ROS regulates apoptotic pathway epigenetically in a P53 dependent manner for generating pro- cancerous phenotypes. Mitochondrion. 2017;35:35–43. doi: 10.1016/j.mito.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 39.DeHaan C, Habibi-Nazhad B, Yan E, Salloum N, Parliament M, Allalunis-Turner J. Mutation in mitochondrial complex I ND6 subunit is associated with defective response to hypoxia in human glioma cells. Mol Cancer. 2004;3:19. doi: 10.1186/1476-4598-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Djureinovic D, Hallström BM, Horie M, Mattsson JS, La Fleur L, Fagerberg L, Brunnström H, Lindskog C, Madjar K, Rahnenführer J, et al. Profiling cancer testis antigens in non-small-cell lung cancer. JCI Insight. 2016;1:e86837. doi: 10.1172/jci.insight.86837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han SW, Roman J. Targeting apoptotic signaling pathways in human lung cancer. Curr Cancer Drug Targets. 2010;10:566–574. doi: 10.2174/156800910791859461. [DOI] [PubMed] [Google Scholar]
- 42.Viktorsson K, Lewensohn R. Apoptotic signaling pathways in lung cancer. J Thorac Oncol. 2007;2:175–179. doi: 10.1097/JTO.0b013e318031cd78. [DOI] [PubMed] [Google Scholar]
- 43.Hiraoka K, Miyamoto M, Cho Y, Suzuoki M, Oshikiri T, Nakakubo Y, Itoh T, Ohbuchi T, Kondo S, Katoh H. Concurrent infiltration by CD8+ T cells and CD4+ T cells is a favourable prognostic factor in non-small-cell lung carcinoma. Br J Cancer. 2006;94:275–280. doi: 10.1038/sj.bjc.6602934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim EM, Park JK, Hwang SG, Kim WJ, Liu ZG, Kang SW, Um HD. Nuclear and cytoplasmic p53 suppress cell invasion by inhibiting respiratory complex-I activity via Bcl-2 family proteins. Oncotarget. 2014;5:8452–8465. doi: 10.18632/oncotarget.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 46.Yuan Y, Wang W, Li H, Yu Y, Tao J, Huang S, Zeng Z. Nonsense and missense mutation of mitochondrial ND6 gene promotes cell migration and invasion in human lung adenocarcinoma. BMC Cancer. 2015;15:346. doi: 10.1186/s12885-015-1349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM. CD4+T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noy R, Pollard JW. Tumor-associated macrophages: From mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kataki A, Scheid P, Piet M, Marie B, Martinet N, Martinet Y, Vignaud JM. Tumor infiltrating lymphocytes and macrophages have a potential dual role in lung cancer by supporting both host-defense and tumor progression. J Lab Clin Med. 2002;140:320–328. doi: 10.1067/mlc.2002.128317. [DOI] [PubMed] [Google Scholar]
- 50.Pearce SF, Rebelo-Guiomar P, D'Souza AR, Powell CA, Van Haute L, Minczuk M. Regulation of mammalian mitochondrial gene expression: Recent advances. Trends Biochem Sci. 2017;42:625–639. doi: 10.1016/j.tibs.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.