Abstract
Long non-coding RNA (lncRNA) activated by TGF-β (ATB) has been reported to be widely expressed in different types of cancer; however, the function of ATB in lung cancer remains unclear. In order to elucidate the role of ATB in lung cancer, reverse transcription-quantitative polymerase chain reaction was used to detect the expression of ATB in tumor tissues and corresponding non-tumor lung tissues from 32 patients with lung cancer. Furthermore, the association between the expression of ATB and clinical characteristics was investigated. Cell proliferation was assessed using a cell counting kit-8 assay and cell migration was assessed using a wound healing assays. Epithelial-mesenchymal-transition and mitogen-activated protein kinase signaling pathway activity was examined using western blotting. It was demonstrated that ATB was highly expressed in lung cancer tissues compared with noncancerous tissues, and associated with tumor size and metastasis. It was also demonstrated that ATB was highly expressed in the lung cancer cell lines, A549 and HCC827, compared with the HBE-1 cell line. Suppression of ATB significantly inhibited the proliferation and migratory rate of lung cancer cells. The protein expression levels of p38, E-cadherin and N-cadherin were altered by suppression of ATB expression. Overall, the present study demonstrated that ATB may promote the development of lung cancer.
Keywords: long non-coding RNA activated by TGF-β, lung cancer, p38
Introduction
Lung cancer is the leading cause of cancer-associated mortality globally, and is characterized by uncontrolled proliferation and metastasis of lung cancer cells (1,2). Although progress has been made in lung cancer research, the outcome of patients with lung cancer remains poor, and only 17% of patients survive >5 years after diagnosis (1,2). Proliferation and metastasis are the primary causes of mortality in patients with lung cancer (3), which emphasizes the requirement for an improved understanding of lung cancer carcinogenesis.
Long non-coding RNAs (lncRNAs) are >200 nucleotides in length and regulate gene expression at transcriptional or post-transcriptional levels in various biological processes, including cancer development. Liu et al (4) reported that lncRNA regulator of reprogramming (ROR) shares the binding site of miR-145 with POU class 5 homeobox 1 (OCT4) mRNA. Suppression of lncRNA ROR expression in prostate cancer cells may lead to increased expression of microRNA (miR)-145, thus preventing cell proliferation by decreasing OCT4 expression. Wang et al (5) demonstrated that lncRNA urothelial cancer associated 1 (UCA1) increased the metastatic ability of gastric cancer cells by promoting G protein-coupled receptor kinase 2 ubiquitination and degradation, suggesting that lncRNA UCA1 may function as a mediator of protein ubiquitination and may be a target for gastric cancer therapy. Chen et al (6) reported that lncRNA X inactive specific transcript (XIST) was able to bind miR-200-3p, promoting zinc finger E-box binding homeobox (ZEB)1 expression, a tumor suppressor gene, suggesting that lncRNA XIST may be used as a prognostic biomarker for patients with CRC. Wang et al (7) demonstrated that lncRNA HOXD cluster antisense RNA 1 was highly expressed in hepatocellular carcinoma, competitively binding to miR-130a-3p to increase the expression of SRY-box 4, and subsequently promoting metastasis of hepatocellular carcinoma. Numerous reports have indicated lncRNAs participate in tumor genesis and progression, including that of lung cancer (8,9). However, the detailed molecular mechanisms of lncRNA activity in lung cancer require further study.
LncRNA activated by TGF-β (ATB) was first identified in hepatocellular carcinoma, in which it upregulated ZEB1 and ZEB2 expression by competitively binding the miR-200 family, thus inducing epithelial-mesenchymal-transition (EMT) and invasion of cancer cells (10). Recently, it has been demonstrated that ATB promotes malignancy in other types of cancer, including breast cancer (11), glioma (12) and colon cancer (13). However, the function of ATB in lung cancer remains unclear.
The present study aimed to investigate the role of ATB in lung cancer. It was demonstrated that ATB was highly expressed in lung cancer tissue, and that knockdown of ATB in lung cancer cell lines inhibited the proliferation and metastasis of lung cancer cells. Finally, the potential mechanism of ATB in lung cancer was investigated.
Materials and methods
Clinical sample collection and preparation
A total of 32 lung cancer tissue specimens and adjacent non-cancerous tissue specimens were collected from patients (21 male and 11 female; the age ranged from 47 to 73 years, with a mean age of 63.3 years) from Southwest Hospital in (Chongqing, China) between January 2015 and December 2016. None of these patients had received radiotherapy or chemotherapy prior to tissue collection. All of the tissues were washed with the RNase-free PBS and frozen in liquid nitrogen at −196°C. Written informed consent was obtained from all patients and the experimental protocol was approved by the Institutional Board of the Southwest Hospital.
Cell lines and cell culture
The human lung cancer cell lines, H1299, H358, HCC827 and A549, and the normal human bronchial epithelial cell line, HBE-1, were obtained from the Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences (Shanghai, China). H358, HCC827, H1299 and A549 cells were maintained and grown in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and HBE cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare, Chicago, IL, USA), 100 U/ml penicillin and 100 µg/ml streptomycin in an atmosphere of 5% CO2 at 37°C.
Cell Counting Kit-8 (CCK-8) assay
The target cells were seeded in 96-well plates at a density of 2×103 cells/well in a 200-µl suspension. A total of 10 µl Cell Counting kit-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) in 100 µl RPMI-1640 medium was added to each well, and cultured for 2 h at 37°C. The absorbance was measured at 450 nm. A total of five repeats/group were performed, three times independently.
Cell transfection
The cells (A549 and HCC827) were plated in a 6-well plate. Serum-free culture DMEM medium was added 2 h before transfection at 70–90% confluency. The suppression plasmid (pGPU6/GFP/Neo) of lncRNA ATB was synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The short hairpin RNA (shRNA) sequences in the plasmid vectors were as follows: ATB shRNA, TATGGCCTAGATTACCTTTCCATT; negative control (NC), UUCUCCGAACGUGUCACGUTT. Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) was used to transfect cells according to the manufacturer's protocol. A total of 1×105 cells were seed in each well; with a transfection time of 6 h, then the supernatant was discarded and DMEM medium containing 10% serum was added into the well.
Wound-healing assay
The cells (A549 and HCC827) were seeded into 6-well plates at a density of 4×105 cells/well, and cultured to 80% confluence. The cell monolayer was scratched with a 10 µl pipette tip to in a straight line and then washed with PBS. Then, the cells were cultured in serum-free RPMI-1640 medium for 24 h at 37°C. The wound width was then inspected and imaged to analyze the ability of cell migration. The experiment was repeated three times. The migration ability was measured by as the migration rate (MR): (D0-D1)/D0. D0 being the width of the wound at 0 h and D1 being the width of the wound at 24 h. The MR was measured at five different points of each wound.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from cultured cells and clinical tissues was extracted using RNAsio Plus (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocol. The RNA was reverse transcribed into cDNA using PrimeScript™ RT reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. The quantitative analysis of lncRNA ATB expression was performed using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) using 2 µl cDNA, 1 µl forward primer, 1 µl reverse primer, 0.4 µl ROX (from SYBR Premix Ex Taq), 5.6 µl RNase-free water and 10 µl SYBR mix. The thermocycling conditions were as follows: 10 min at 95°C; and then 40 cycles of the following three steps: 10 sec at 95°C, 30 sec at 60°C, and 20 sec at 72°C. GAPDH was used as an internal control to quantify lncRNA-ATB expression. The primer sequences were as follows: lncRNA-ATB forward, 5′-CTTCACCAGCACCCAGAGA-3′ and reverse, 5′-AAGACAGAAAAACAGTTCCGAGTC-3′; GADPH forward, 5′-GGTGAAGGTCGGAGTCAACG-3′ and reverse, 5′-CAAAGTTGTCATGGATGHACC-3′). Expression was quantified using the 2−ΔΔCq method (14).
Western blotting
The target cells were washed three times with PBS, and total protein was extracted using radioimmunoprecipitation assay buffer containing the protein inhibitor, phenylmethylsulfonyl fluoride (Beyotime Institute of Biotechnology, Haimen, China). The concentration of protein was assessed using a BCA assay. A total of 40 µg protein was loaded for 10% SDS-PAGE, then transferred onto Immobilon-P polyvinylidene fluoride membranes (Merck KGaA, Darmstadt, Germany). The membranes were then blocked with 5% non-fat milk for 1 h at room temperature. This was followed by incubation of the following primary antibodies at 4°C overnight: Anti-E-cadherin (dilution, 1:1,000; cat. no. 14472; anti-mouse), anti-p38 (dilution, 1:1,000; cat. no. 9212; anti-rabbit), anti-N-cadherin (dilution, 1:1,000; cat. no. 14215; anti-mouse) (all from Cell Signaling Technology, Inc., Danvers, MA, USA). The membranes were washed using Tris-buffered saline with 0.1% Tween 20 for 10 min at room temperature, then incubated with secondary antibodies [Goat anti-Mouse IgG (H+L) Cross-Adsorbed secondary antibody, dilution, 1:1,000; cat. no. A1607; goat anti-mouse; and Goat anti-Rabbit IgG (H+L) Cross-Adsorbed secondary antibody, dilution 1:1,000; cat. no. A16104; goat anti-rabbit; both from Thermo Fisher Scientific, Inc.] for 2 h at room temperature. The protein blots were visualized using an ECL chemiluminescent detection system (Thermo Fisher Scientific, Inc.) and the protein bands were quantified with Quantity One software (version 4.62; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
All data are expressed as the mean ± standard deviation, and all experiments were repeated ≥3 times. The difference between two groups was analyzed by the Student's t-test, and one-way analysis of variance (ANOVA) followed by post-hoc Brown-Forsythe test, to validate ANOVA, was used to analyzed data from >2 groups. Survival analysis was performed using the Kaplan-Meier method, and the log-rank test was used to assess the statistical significance. P<0.05 was considered to indicate a statistically significant difference.
Results
LncRNA ATB is highly expressed in lung cancer tissue and cancer cell lines
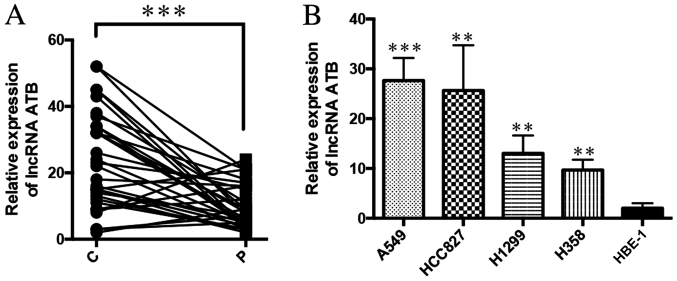
To investigate the role of ATB in lung cancer, the expression of ATB in lung cancer tissue (n=32) was analyzed by RT-qPCR, which revealed that ATB was significantly increased in the lung cancer tissue compared non-cancerous adjacent tissues (Fig. 1A). The expression of ATB was also analyzed in lung cancer cell lines, H358, HCC827, H1299 and A549, and the normal lung epithelial cell line, HBE-1. It was demonstrated that ATB was also highly expressed in lung cancer cell lines compared with HBE-1 cells (Fig. 1B). Among the four lung cancer cell lines, ATB was most highly expressed in A549 and HCC827, which were, therefore, selected for use in the following experiments. These results indicated that ATB was highly expressed in lung cancer, but its function remained unclear.
Figure 1.
LncRNA ATB is highly expressed in lung cancer tissues and cancer cell lines. (A) The expression of ATB in lung cancer tissue and paracancerous tissue was detected by RT-qPCR. ***P<0.0001; n=32. (B) RT-qPCR was used to detect the expression of ATB in lung cancer cell line and normal lung epithelial cell line HBE-1, from left to right, P=0.0007, P=0.003, P=0.007, P=0.0045. **P<0.001, ***P<0.0001 vs. HBE-1. LncRNA, long non-coding RNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; C, lung cancer tissue; P, paracancerous tissue.
ATB expression indicates a poor prognosis of patients with lung cancer
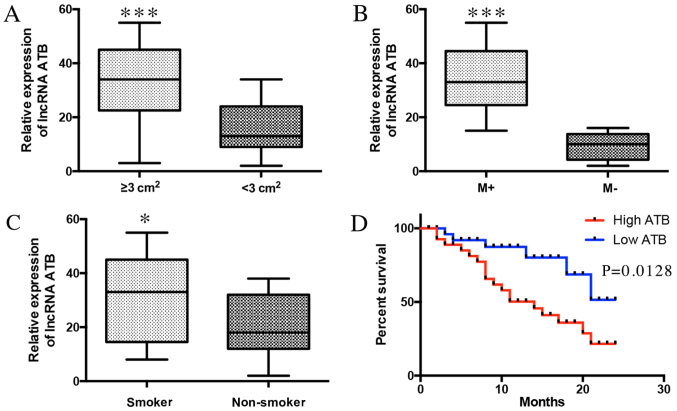
To evaluate the clinical significance of ATB in lung cancer, the associations between ATB and clinicopathological characteristics were investigated. ATB was positively associated with tumor size (Fig. 2A) and lymph node metastasis (Fig. 2B). ATB was also associated with smoking history (Fig. 2C). Additionally, it was demonstrated that high expression of ATB was negatively associated with the survival time of lung cancer patients (Fig. 2D). These results indicate that the expression of ATB was associated with poor prognosis of patients with lung cancer, and positively associated with lung cancer progression.
Figure 2.
LncRNA ATB indicates a poor prognosis of patients with lung cancer. (A) The association between ATB expression and tumor size, P=0.0003. (B) The association between ATB expression and lymph node metastasis, P=0.0002. (C) The association between ATB expression and smoking history of lung cancer patients, P=0.0295. (D) Association between patient survival time (log-rank test) and expression of ATB, P=0.0128. *P<0.05, ***P<0.0001. LncRNA, long non-coding RNA; M+, lymph node metastasis, M-, no lymph node metastasis.
Knockdown of ATB inhibits proliferation and metastasis of lung cancer cell in vitro
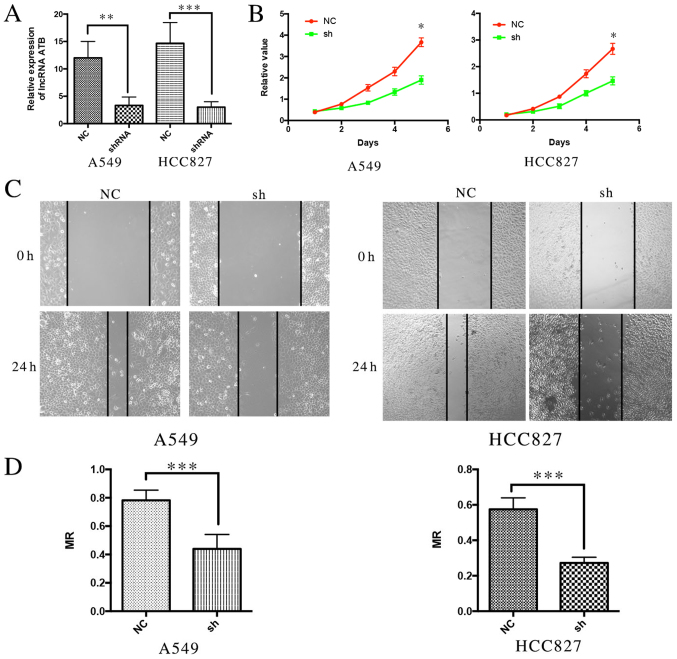
To further investigate the biological function of ATB in lung cancer cell lines, ATB knockdown was performed using shRNA (Fig. 3A). It was demonstrated that suppression of ATB expression significantly inhibited the proliferation of A549 and H358 cells compared with the respective NC groups (Fig. 3B), suggesting that ATB promotes cell growth. A wound healing assay was performed to analyze the effect of ATB on the migration of lung cancer cells. It was demonstrated that silencing ATB significantly inhibited the migratory ability of the two lung cancer cell lines compared with their controls (Fig. 3C and D). These results indicate that knockdown of ATB may inhibit the proliferation and metastasis of lung cancer cells.
Figure 3.
Knockdown of lncRNA ATB inhibits proliferation and metastasis of lung cancer cells in vitro. (A) The expression of ATB in lung cancer cell lines following ATB was knocked down, P=0.0086, P=0.0067. (B) Cell Counting kit-8 assays demonstrated that the proliferation ability of lung cancer cell lines was reduced following ATB knockdown, P=0.022, P=0.013. (C) Wound Healing assays demonstrated that the metastasis ability of lung cancer cell lines was reduced following ATB knockdown. (D) The MR indicated the migration ability of lung cancer cells following ATB knockdown. P=0.003, P=0.002. *P<0.05, **P<0.001, ***P<0.0001. LncRNA, long non-coding RNA; NC, negative control; sh/shRNA, short hairpin RNA; MR, migration rate.
Suppression of ATB downregulates p38 and N-cadherin protein expression
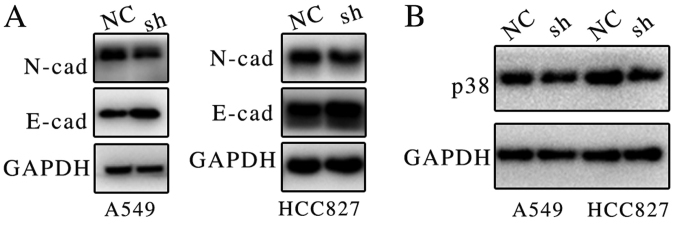
Previous results suggested that ATB may be associated with proliferation and metastasis (12,13); however, the molecular mechanism remained unclear. Previous studies have indicated that ATB may bind members of the miR-200 family, and induce EMT and invasion in different types of cancer (10,11). It was demonstrated in the present study that the suppression of ATB expression markedly increased E-cadherin and decreased N-cadherin protein expression in both lung cancer cell lines (Fig. 4A).
Figure 4.
Knockdown of lncRNA ATB leads to downregulation of p38 and N-cadherin expression. (A) The protein expression levels of N-cadherin and E-cadherin were detected by western blotting in lung cancer cells with and without ATB knockdown. (B) The protein expression level of p38 in lung cancer cells with and without ATB knockdown was detected by western blotting. LncRNA, long non-coding RNA; NC, negative control; sh, short hairpin RNA; N-cad, N-cadherin; E-cad, E-cadherin.
A recent study demonstrated that miR-200 may inhibit p38 mitogen-activated protein kinase (MAPK) signaling pathway via MAPK14, thus promoting cell death (14). The present study indicates that ATB may promote the expression of p38. It was demonstrated that suppression of ATB markedly inhibited the expression of p38 (Fig. 4B). Activation of p38 MAPK signaling may promote proliferation in lung cancer (15). It is hypothesized that ATB may promote proliferation and metastasis of lung cancer via regulation of E-cadherin and p38 expression.
Discussion
Previous research has suggested that lncRNAs regulate the progression of different types of cancer (11–13). Ke et al (16) demonstrated that lncRNA ATB was highly expressed in lung cancer tissue, and that downregulation of ATB was able to promote cell apoptosis, viability, migration and invasion. However, the molecular mechanism of ATB in lung cancer has remained unclear. Therefore, the present study aimed to the influence of ATB expression on the proliferation and metastasis of lung cancer, and its underlying mechanism. The results demonstrate that ATB expression was significantly increased in lung cancer tissues and cell lines, compared with normal controls. The expression of ATB was also significantly associated with poor prognosis of patients with lung cancer. Suppression of ATB expression may inhibit the proliferation and metastasis of lung cancer cells. It was also indicated that this function may be executed via E-cadherin and p38.
ATB was reported to be activated by transforming growth factor-β (TGF-β) in liver cancer (10). In the present study, it was demonstrated that ATB was highly expressed in 32 lung cancer tissues, and significantly associated with tumor size and lymph node metastasis. These results indicated that ATB was highly expressed in lung cancer. It is well established that smoking is a key risk factor in lung cancer (17). Checa et al (18) demonstrated that cigarette smoke enhanced the expression of TGF-β in alveolar epithelial cells, and Islas-Vazquez et al (19) demonstrated that cigarette smoke increased the expression of TGF-β in lung adenocarcinoma. The present study suggests that high expression of ATB may be associated with smoking history in patients with lung cancer.
Previous studies have suggested that ATB may target the miR-200 family to promote cancer development (10,12,20). A previous study demonstrated that miR-200 inhibits p38 expression in inducing cell death (21). The present study demonstrated that suppression of ATB expression markedly downregulated the protein expression level of p38. It has been reported that p38 MAPK signaling is important in regulating proliferation of lung cancer cells (22). Taken together, these results suggest that ATB may affect proliferation of lung cancer via p38.
It has been previously suggested that ATB may affect the migration of cancer by regulating EMT (11,13). Yue et al (13) demonstrated that ATB suppressed E-cadherin expression, thus promoting EMT in colon cancer progression. In addition, Shi et al (11) reported that ATB promoted trastuzumab resistance and EMT in breast cancer. Similarly, in the present study, it was demonstrated that suppression of ATB increased the expression of E-cadherin, while decreasing that of N-cadherin, suggesting that ATB may influence the migratory ability of lung cancer via EMT.
Overall, it was demonstrated that ATB was highly expressed in lung cancer tissues and cells, and was associated with tumor size, lymph node metastasis and smoking history, indicating that lncRNA ATB could be used as a potential treatment target for lung cancer. In vitro studies demonstrated that suppression of ATB causes significant inhibition of proliferation and migration of lung cancer cells, which may occur via p38 and EMT regulation; however, the mechanism underlying lncRNA ATB regulating p38 and EMT requires further study.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LW performed data analyses and wrote the manuscript. TW and PH contributed significantly in data analyses. JLZ and WW conceived and designed the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all patients and the experimental protocol was approved by the Institutional Board of the Southwest Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hao Y, Yang X, Zhang D, Luo J, Chen R. Long noncoding RNA LINC01186, regulated by TGF-β/SMAD3, inhibits migration and invasion through Epithelial-Mesenchymal-Transition in lung cancer. Gene. 2017;608:1–12. doi: 10.1016/j.gene.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Sang H, Liu H, Xiong P, Zhu M. Long non-coding RNA functions in lung cancer. Tumour Biol. 2015;36:4027–4037. doi: 10.1007/s13277-015-3449-4. [DOI] [PubMed] [Google Scholar]
- 4.Liu T, Chi H, Chen J, Chen C, Huang Y, Xi H, Xue J, Si Y. Curcumin suppresses proliferation and in vitro invasion of human prostate cancer stem cells by ceRNA effect of miR-145 and lncRNA-ROR. Gene. 2017;631:29–38. doi: 10.1016/j.gene.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Wang ZQ, He CY, Hu L, Shi HP, Li JF, Gu QL, Su LP, Liu BY, Li C, Zhu Z. Long noncoding RNA UCA1 promotes tumour metastasis by inducing GRK2 degradation in gastric cancer. Cancer Lett. 2017;408:10–21. doi: 10.1016/j.canlet.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 6.Chen DL, Chen LZ, Lu YX, Zhang DS, Zeng ZL, Pan ZZ, Huang P, Wang FH, Li YH, Ju HQ, Xu RH. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017;8:e3011. doi: 10.1038/cddis.2017.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, Deng X, Jin H, Wang N, Wang C, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Z, Wang J, Wang S, Chang H, Zhang T, Qu J. LncRNA CCAT2 promotes tumorigenesis by over-expressed Pokemon in non-small cell lung cancer. Biomed Pharmacother. 2017;87:692–697. doi: 10.1016/j.biopha.2016.12.122. [DOI] [PubMed] [Google Scholar]
- 9.Roth A, Diederichs S. Long noncoding RNAs in lung cancer. Curr Top Microbiol Immunol. 2016;394:57–110. doi: 10.1007/82_2015_444. [DOI] [PubMed] [Google Scholar]
- 10.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–63. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma CC, Xiong Z, Zhu GN, Wang C, Zong G, Wang HL, Bian EB, Zhao B. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J Exp Clin Cancer Res. 2016;35:90. doi: 10.1186/s13046-016-0367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu F, Peng Z, Yan D. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol. 2016;31:595–603. doi: 10.1111/jgh.13206. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Khan GJ, Gao Y, Gu M, Wang L, Khan S, Naeem F, Yousef BA, Roy D, Semukunzi H, Yuan S, Sun L. TGF-β1 causes EMT by regulating N-Acetyl Glucosaminyl transferases via downregulation of non muscle Myosin II-A through JNK/P38/PI3K pathway in lung cancer. Curr Cancer Drug Targets. 2018;18:209–219. doi: 10.2174/1568009617666170807120304. [DOI] [PubMed] [Google Scholar]
- 16.Ke L, Xu SB, Wang J, Jiang XL, Xu MQ. High expression of long non-coding RNA ATB indicates a poor prognosis and regulates cell proliferation and metastasis in non-small cell lung cancer. Clin Transl Oncol. 2017;19:599–605. doi: 10.1007/s12094-016-1572-3. [DOI] [PubMed] [Google Scholar]
- 17.van der Aalst CM, Ten Haaf K, de Koning HJ. Lung cancer screening: Latest developments and unanswered questions. Lancet Respir Med. 2016;4:749–761. doi: 10.1016/S2213-2600(16)30200-4. [DOI] [PubMed] [Google Scholar]
- 18.Checa M, Hagood JS, Velazquez-Cruz R, Ruiz V, Garcia-De-Alba C, Rangel-Escareno C, Urrea F, Becerril C, Montano M, Garcia-Trejo S, et al. Cigarette smoke enhances the expression of profibrotic molecules in alveolar epithelial cells. PLoS One. 2016;11:e0150383. doi: 10.1371/journal.pone.0150383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Islas-Vazquez L, Prado-Garcia H, Aguilar-Cazares D, Meneses-Flores M, Galicia-Velasco M, Romero-Garcia S, Camacho-Mendoza C, Lopez-Gonzalez JS. LAP TGF-Beta subset of CD4(+)CD25(+)CD127(−) Treg cells is increased and overexpresses LAP TGF-Beta in lung adenocarcinoma patients. Biomed Res Int. 2015;2015:430943. doi: 10.1155/2015/430943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han F, Wang C, Wang Y, Zhang L. Long noncoding RNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200s. Am J Cancer Res. 2017;7:770–783. [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao Y, Yan W, Lu L, Wang Y, Lu W, Cao Y, Cai W. p38/p53/miR-200a-3p feedback loop promotes oxidative stress-mediated liver cell death. Cell Cycle. 2015;14:1548–1558. doi: 10.1080/15384101.2015.1026491. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Chen T, Gong W, Tian H, Wang H, Chu S, Ma J, Yang H, Cheng J, Liu M, Li X, Jiang C. Fibroblast growth factor 18 promotes proliferation and migration of H460 cells via the ERK and p38 signaling pathways. Oncol Rep. 2017;37:1235–1242. doi: 10.3892/or.2016.5301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.