Abstract
The objective of the present study was to investigate the long non-coding RNA (lncRNA) and mRNA expression profiles that are associated with the invasion and metastasis of papillary thyroid carcinoma (PTC). Transwell invasion assays were used to screen three highly invasive sub-strains of the human PTC IHH4 cell line: IHH4-M1, IHH4-M2 and IHH4-M3. In addition, tumor-bearing nude mice were used to identify the invasive and metastatic capacity of the three sub-strains. Agilent lncRNA microarray chips were used to screen 795 differentially expressed lncRNAs and 788 differentially expressed mRNAs. A total of 10 lncRNAs and 10 mRNAs were randomly selected for RT-qPCR validation to confirm that the results were consistent with the microarray chips, suggesting that the results of the microarray chip analysis were relatively accurate. Gene ontology enrichment-based cluster analysis revealed that the differentially expressed genes were mainly associated with steroid biosynthesis, bioadhesion, intercellular adhesion and other metastasis-associated biological processes. The results of the pathway cluster analysis identified that the differentially expressed genes were associated with tumor metastasis-associated signaling pathways, including the cholesterol metabolic signaling pathway, the sterol regulatory element-binding protein signaling pathway and the integrin signaling pathway, suggesting that lncRNA may regulate PTC metastasis through various signaling pathways. The present study screened and constructed PTC metastasis-associated lncRNA and mRNA expression profiles, and it provides a molecular basis for the future study of high-risk molecular markers of PTC.
Keywords: papillary thyroid carcinoma, metastasis, long non-coding RNA, mRNA
Introduction
Thyroid cancer is the most commonly diagnosed type of endocrine tumor in the USA and its incidence has been rapidly increasing from 1975 to 2014 (1). Thyroid cancer is considered to be among the 10 most common types of malignant tumors (2). According to 2016 data from the National Cancer Institute (National Institutes of Health, Bethesda, MA, USA), the total of newly diagnosed cases of thyroid cancer increased by an average of >5% annually in the USA, and papillary thyroid carcinoma (PTC) was the fastest growing malignancy (3). Although the majority of PTC cases have a good prognosis, with a 10-year survival rate of >90% (1), ~50% of these patients are diagnosed with cervical lymph node metastasis alongside the initial diagnosis of PTC (4). Between 10 and 20% of patients with PTC experience recurrent and distant metastasis (4). Once distant metastasis is identified, the 10-year survival rate of the patients is reduced to ~40% (4).
At present, the main treatment approach for PTC is surgical resection since chemotherapy is often ineffective in patients with PTC (5). However, surgical resection has been the subject of debate among academic researchers in recent years. Certain scientists believe that due to the low degree of PTC malignancy and its slow disease progression, patients with <1 cm papillary thyroid microcarcinoma (PTMC) should be temporarily monitored during follow-up (6). Other scientists have recommended surgical resection and believe that PTMC is not necessarily equivalent to low-risk cancer; indeed, a number of patients with microcarcinomas have very rapid disease progression and early emergence of local and distant metastasis (7). Therefore, in the absence of an effective means of identification, the early diagnosis and treatment of PTC remains necessary. Nevertheless, the underlying molecular mechanisms of PTC onset and progression have not yet been elucidated. Therefore, the study of high-risk molecular events in PTC and the screening of molecular markers for the characterization of high-risk PTC and those associated with PTC metastasis may aid in identifying high-risk individuals at an early stage among numerous patients with PTC to achieve an effective and early diagnosis and prognosis as well as to conduct stratified treatments in patients with important theoretical significance and clinical value.
Long non-coding RNA (lncRNA) is a class of >200 base pair non-coding RNAs, and prior research has identified that lncRNA serves an important function in gene transcription, modification and post-transcriptional regulation (8). Previous studies have identified that lncRNA serves a crucial function in the progression of a number of solid tumors, including gastric, colon, kidney and pancreatic cancer (9–13). LncRNA is involved in tumor suppressor gene and oncogene activity via regulation of the chromosomal structure and the expression of the downstream genes (14). Therefore, lncRNAs are hypothesized to be important molecular markers and potential targets for the diagnosis and treatment of cancer in the future.
The IHH4 cell line has been determined to have a highly metastatic nature (15). The present study screened highly metastatic IHH4 cell sub-strains and used lncRNA and mRNA microarray chips to screen the lncRNA and mRNA expression profiles associated with IHH4 metastasis. The objective of the present study was to provide a molecular basis for the future study of high-risk molecular markers of PTC.
Materials and methods
Cell culture
Since PTMC was defined as PTC with a maximum diameter ≤1 cm, the genetic backgrounds of PTMC and PTC are the same (16), and no specific PTMC cell lines were identified to be available, the human PTC IHH4 cell line was selected for use in the present study. IHH4 cells were derived from a 75-year-old male patient with PTC and purchased from Health Science Research Resources Bank (Osaka, Japan). This cell line was cultured in 1:1 Dulbecco's modified Eagle's medium (DMEM:RPMI-1640 medium containing 10% fetal bovine serum; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The IHH4 cell line was passaged for >60 generations prior to purchase and was stabilized. The IHH4 metastatic tumor model was used in the present study to confirm the characteristics of PTC in metastatic pulmonary tumors.
Screening of highly metastatic thyroid cancer cell sub-strains
A Transwell chamber (8 µm; Corning Incorporated, Corning, NY, USA) was used in the present study to screen the highly metastatic IHH4 cell sub-strains according to previous studies (17–19). A Transwell polycarbonate membrane insert was coated with artificial Matrigel (BD Biosciences, San Jose, CA, USA). IHH4 cells at a density of 2×104 cells were inoculated into each Transwell chamber and incubated at 37°C with 5% CO2 for 24 h. A 1:1 mixture of DMEM:RPMI-1640 medium was added in the upper chamber and a 1:1 mixture of DMEM:RPMI-1640 medium with 10% fetal bovine serum was added in the lower chamber. After washed twice with PBS, the chamber was stained by 0.1% crystal violet for 30 min at room temperature and viewed under a light microscope (×200). Cells that migrated through the Transwell membrane and into the lower chamber were collected to further culture and passage. Sub-strain cell lines collected from the passage were screened repeatedly according to the aforementioned procedures. In order to ensure the reliability of the results, wild-type IHH4 cells were divided into three groups (Group 1, Group 2 and Group 3) and Transwell assays were used to screen the cells of three groups independently. The cells in the upper Transwell chamber that did not migrate through the Matrigel membrane were collected to further culture (DMEM:RPMI-1640 medium containing 10% fetal bovine serum at 37°C with 5% CO2) to obtain the IHH4-C1, IHH4-C2 and IHH4-C3 as the controls. The cells that passed the membrane and were harvested from the lower chamber were cultured for additional screening. Following 15 repeats of screening, three relatively stable cell sub-strains, IHH4-M1, IHH4-M2 and IHH4-M3, were obtained. Furthermore, the proliferation of IHH4-M and IHH4-C cells was detected using a Cell Counting kit-8 (450 nm; Dojindo Molecular Technologies, Inc., Kumamoto, Japan).
In vivo and in vitro metastatic experiments
The animal experiments in the present study have been reviewed and approved by the Animal Ethical and Welfare Committee of Zhejiang Cancer Hospital (Zhejiang, China). The present study used male BALB/c nude mice (purchased from the Research Center of Laboratory Animal Science, Zhejiang Chinese Medical University, Hangzhou, China) weighing 20-25 g at 6–8 weeks of age. Mice were housed in clear polycarbonate cages with controlled environment (20–24°C, 40–60% relative humidity, 12/12 h light/dark cycles and food and water was available ad libitum). A total of 18 nude mice were randomly divided into six groups, namely, the IHH4-C1, IHH4-C2, IHH4-C3, IHH4-M1, IHH4-M2 and IHH4-M3 groups, and were maintained in a specific pathogen-free environment. The IHH4 and IHH4-M cells were dissociated by 0.25% trypsin-EDTA solution (1 ml at 37°C, for 1 min) and harvested by centrifugation (300 × g at 37°C for 3 min) in the logarithmic growth phase and prepared for single-cell suspension. Following centrifugation (300 × g at 37°C, for 3 min) and PBS washing twice (37°C for 15 sec), the number of cells in each group was counted. The cell pellets were resuspended in serum-free RPMI-1640:DMEM culture medium, followed by adjusting the cells at a density of 2×105, which were used to inoculate the nude mice through a caudal vein injection. All the animals in the present study were observed individually once daily and the conditions of the animals were recorded. The duration of the in vivo experiment was 45 days, based on the experience of a preliminary experiment (data not shown). On the 45th day, the tumors in the lungs of the mice were not large. The maximum permitted tumor diameter was 10 mm. Under these circumstances, tumors in the lung may not increase the discomfort or pain, or affect activities of the mice. Therefore, humane endpoints were established that the mice would be humanely sacrificed on the 45th day of experiment or when one of the following conditions was observed in mice according to the Guidance published by The Organization for Economic Co-operation and Development (20): i) Prolonged or irreversible inability to eat or drink, including immobility, obstruction of the oral cavity and missing or abnormal teeth; ii) indication of severe pain, distress or suffering, including fractures, self-induced trauma, abnormal vocalization, abnormal posture or movements, and open wounds or ulcers; iii) rapid or continuing weight loss, e.g., loss of ≥20% body weight over a few days, or gradual but continued weight loss; iv) general decrease in grooming and an abnormal appearance over an extended time period, including rough coat, extensive alopecia, prolonged diarrhea, urine-stained coat, swollen limbs, paralysis, or other central or peripheral nervous disturbances (including convulsions, circling behavior or prostration); v) severe or continuing respiratory distress, including coughing, sneezing, nasal discharge, or bloody nares or mouth; vi) bleeding, anemia or unusual discharges; or vii) microbial infections or other diseases, including those that interfere with the experimental protocol or cause any of the above. In total, 17 mice (94.44%) were humanely sacrificed on the 45th day of the experiment and only one mouse (5.56%) in group IHH4-M3 was humanely sacrificed on the 38th day since tumors formed in the lung and thyroid of the mouse and the tumor in the thyroid had broken out through the skin. All the mice were sacrificed by cervical dislocation in order to collect the lung and thyroid tissues, which were fixed in 10% formalin solution (48 h at 25°C). All the tissues were paraffin-embedded and sectioned into 4-µm thickness for hematoxylin (5 min at 37°C) and eosin (2 min at 37°C) staining to observe the tumor metastasis under a light microscope and captured images was analyzed by NIS-Elements D 3.0 (Nikon Corporation, Tokyo, Japan) with magnification, ×200. The paraffin sections (4-µm thickness) in were placed a 70°C oven for 30 min. As soon as the wax got melt, deparaffinized with xylene and rehydrated. After microwave antigen retrieval performed by 0.01 mol/l sodium citrate buffer (pH=6.0), 5% hydrogen peroxide (H2O2) was used to block the endogenous peroxidase activity (5 min at 37°C). Anti-keratin 19 (CK19) antibody (cat. no. ab52625; Abcam, Cambridge, MA, USA) was used to detect the human-specific CK19 protein content in the tumor cells (1:800 for 1 h at room temperature). Rinsed with PBS buffer three times (5 min each), the sections were incubated with biotinylated goat anti-rabbit IgG-HRP secondary antibody (1:1,000 for 30 min at room temperature cat. no. ab136817, Abcam). The unbound secondary antibody was washed out with PBS buffer (three times, 5 min each). Followed by DAB coloration (2 min at 37°C), nuclear staining with hematoxylin (2 min at 37°C), Scott tapwater bluing (2 min at 37°C) and graded ethanol dehydration (50% ethanol for 30 sec at room temperature; 75% ethanol for 30 sec at room temperature; 80% ethanol for 30 sec at room temperature; 100% ethanol I for 3 min at room temperature and 100% ethanol II for 3 min at room temperature), and sections were sealed with neutral balsam and observed under the light microscope at magnification, ×200 (Nikon Corporation, Tokyo, Japan).
lncRNA chip assays
TRIzol® reagent (Thermo Fisher Scientific, Inc.) was used to extract the total RNA from the IHH4-M and IHH4-C cells. A MirVana miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc.) was used to purify the extracted total RNA. A NanoDrop ND-1000 spectrophotometer was used to detect the OD260/280 ratio to detect RNA purity, followed by 1% agarose gel electrophoresis to examine the RNA integrity (2 µg/loading slot). cDNA labeled with a fluorescent dye (Cy5 and Cy3-dCTP) was produced by Eberwine's linear RNA amplification method (21), and the subsequent enzymatic reaction and procedure was optimized by using CapitalBio cRNA Amplification and Labeling kit (Beijing CapitalBio Technology Co., Inc., Beijing, China) for producing higher yields of labeled cDNA according to the manufacturer's protocol. The labeled cDNA was purified using a PCR NucleoSpin Extract II kit (Macherey-Nagel, Duren, Germany) and resuspended according to the manufacturer's protocol. DNA in hybridization solution was denatured at 95°C for 3 min prior to loading onto a microarray. Arrays were hybridized in an Agilent Hybridization Oven (Agilent Technologies, Inc., Santa Clara, CA, USA) overnight at a rotation speed of 10 × g and a temperature of 42°C and washed with two consecutive solutions (0.2% SDS and 2× SSC at 42°C for 5 min, and 0.2× SSC for 5 min at room temperature). An Agilent Microarray Scanner (Agilent Technologies, Santa Clara, CA, USA) was used to collect the data from the microarray chips. GeneSpring software (version 13.0; Agilent Technologies) was used to collect the data for calibration and quality control. For the selection of the differentially expressed genes, the expression difference ≥2-fold and the P-value of the T-analysis <0.05 was considered as differentially expressed (all the data were Log2 transformed) (22).
Gene function analysis
The differentially expressed genes were submitted to the Database for Annotation, Visualization and Integrated Discovery (version 6.8; http://david.abcc.ncifcrf.gov/), and Gene Ontology (GO; http://www.geneontology.org/) and enrichment-based cluster analysis was used to study the molecular function of the differentially expressed genes. Pathway cluster analysis was used to study the potential functions of the differentially expressed genes in the signaling pathways.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
To further verify the results of the chip screening, 10 lncRNAs (NR_037676.1, ENST00000586841.1, TCONS_00029723, ENST00000510284.1, XR_428320.1, TCONS_00019374, ENST00000504773.1, ENST00000558696.1, ENST00000445300.1 and ENST00000594721.1) and 10 mRNAs [collagen and calcium binding EGF domains 1 (CCBE1), cluster of differentiation (CD)82, frizzled class receptor 4 (FZD4), thyroid receptor-interacting protein 14 (OASL), epithelium-specific ets transcription factor 1 (ELF3), thrombin receptor-like 2 (F2RL2), family with sequence similarity 110 member C (FAM110C), myelodysplasia syndrome-associated protein 1 (MECOM), pappalysin 2 (PAPPA2) and thyroid peroxidase (TPO)] were randomly selected for RT-qPCR. The primers were designed by Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, USA) and the sequences of the primers used in the RT-qPCR are listed in Table I. GAPDH was used as the internal reference for the RT-qPCR to detect the expression of 10 lncRNAs and 10 mRNAs in the IHH4-M1, IHH4-M2, IHH4-M3, IHH4-C1, IHH4-C2 and IHH4-C3 cell sub-strains. The expression levels of target lncRNA/genes were detected using a 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the SYBR Premix Ex Taq II (Takara Bio, Inc., Otsu, Japan). The fold-change of target lncRNA and gene expression intensity was calculated using the 2−ΔΔCq method (23). The thermocycling conditions used in RT-qPCR were as follows: Initial denaturation, 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 34 sec.
Table I.
Primers used for reverse transcription-quantitative polymerase chain reaction.
| lncRNA | mRNA | ||
|---|---|---|---|
| NR_037676.1 | Forward GACTTTGCTGACCCAATGAC | CCBE1 | Forward TGTGCCCACATCTGCATCAA |
| Reverse AGTAGAGACGGGGTTTCACC | Reverse GGCTTTCACCATGTTCTCAGACTTC | ||
| ENST00000586841.1 | Forward TCGTGATCATAACCCCAAAT | CD82 | Forward AGCTGCACTGGTTTCGTGGA |
| Reverse TTACAACAGGGCAGAACCTC | Reverse CAGGACAGAGATGAAACTGCTCTTG | ||
| TCONS_00029723 | Forward TCACCTGAGGTCAGGAGTTC | FZD4 | Forward TGCCAGAACCTCGGCTACAAC |
| Reverse TGATCTCCATAGCCTGCTTC | Reverse GTGCACATTGGCACATAAACAGAAC | ||
| ENST00000510284.1 | Forward CAATTCAAGGCAGCAAGAAG | OASL | Forward GCGGGTGCTGAAGGTAGTCAA |
| Reverse TCGAACTCCTGACCTCAAGT | Reverse GAAGCTGTGGAAACAGCTCAGAAAC | ||
| XR_428320.1 | Forward AACCAGACCTTCTCCTGCTT | ELF3 | Forward GACTTGGTACTGACCCTGAGCAAC |
| Reverse ATGGAGGGGAAAACTAGCTG | Reverse CTTGGTAGCTGATCCAGTCCAGAA | ||
| TCONS_00019374 | Forward CTCATCACCTGGATCATCGT | F2RL2 | Forward CCCTGATTCTCAGGCTAGAAGTGTC |
| Reverse GCAGGTGCATCATCTCTCTC | Reverse GGCATCCCCAGAAGCTCAA | ||
| ENST00000504773.1 | Forward TACAGGCCAGTGTAGGGAAA | FAM110C | Forward CATCATCAAGTGGCTGTACACCTG |
| Reverse GAACCTCTTCCTCCTTCCAG | Reverse GCACACTAGCCAAATGGTCCTG | ||
| ENST00000558696.1 | Forward GCCTCCAAAAGAGACCTTTC | MECOM | Forward GGAATCCAGTGCTATCCAGTCCA |
| Reverse AGTGGCTCATGCCTGTAATC | Reverse GCAGGTTTATAAGGCATTCTGCTCA | ||
| ENST00000445300.1 | Forward GGGTGCCTCCCACTTACTAT | PAPPA2 | Forward GTGCGCAAGACCTGCTTTGA |
| Reverse CAGGACCCCTTTCCTGTTAT | Reverse GTGAGTGACAGCGTCCTTGTCC | ||
| ENST00000594721.1 | Forward TCCCTATGTTGCCTAAGCTG | TPO | Forward AAACACTTGCCTGGCGAACA |
| Reverse GGGGAATTATTTGGAAGACC | Reverse GCTGACTGAAGCCGTCCTCATA | ||
CCBE1, collagen and calcium binding EGF domains 1; CD, cluster of differentiation; FZD4, frizzled class receptor 4; OASL, thyroid receptor-interacting protein 14; ELF3, epithelium-specific ets transcription factor 1; F2RL2, thrombin receptor-like 2; FAM110C, family with sequence similarity 110 member C; MECOM, myelodysplasia syndrome-associated protein 1; PAPPA2, pappalysin 2; TPO, thyroid peroxidase.
Statistical analysis
In the present study the lncRNA and mRNA array data were analyzed for data summarization, normalization and quality control by using the GeneSpring software (version 13.0; Agilent Technologies). An lncRNA or mRNA expression difference of ≥2-fold was defined as differential expression. SPSS software (version 22.0; IBM Corp., Armonk, NY, USA) was used for data analysis. All the data are presented as the mean ± standard deviation and differences between means were analyzed using Student's t-test. Student's t-test was used to examine the difference in expression of genes/lncRNA between IHH4-M and IHH4-C groups. Graphical representations were designed using GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) software. P<0.05 was considered to indicate a statistically significant difference.
Results
Selection of highly invasive IHH4-M cell sub-strains
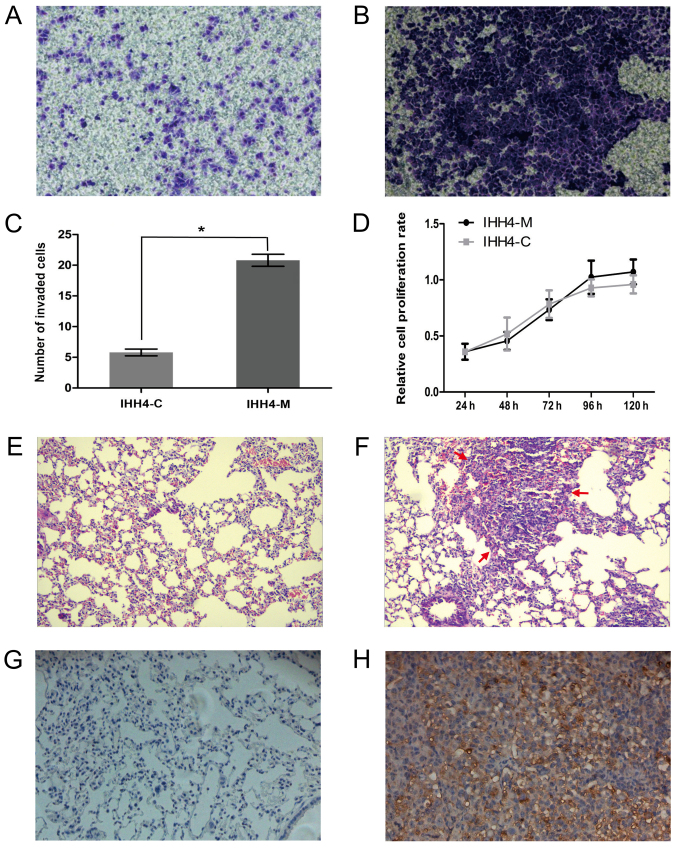
Subsequent to 15 screenings with the Transwell chamber, IHH4 and highly invasive IHH4-M cell sub-strains were successfully obtained and cultured. Transwell invasion assays confirmed that the invasive capacity of the IHH4-M cells was significantly higher than that of the IHH4-C cells (Fig. 1A and B) and the number of cells passed Transwell membrane in IHH4-M group were significantly higher than the IHH4-C group (Fig. 1C). The results of a CCK-8 assay demonstrated that no significant difference between the proliferation of IHH4-M and IHH4-C cells (Fig. 1D; P>0.05). The experimental results of the tumor-bearing nude mice revealed that lung tumor formation in all the mice of the IHH4-M groups was apparent, and only one mouse exhibited thyroid tumor formation, while no lung tumor formation or smaller tumor formation was observed in the mice of the IHH4-C groups compared with the IHH4-M groups (Fig. 1E and F). The results of immunohistochemistry examination demonstrated that all the tumors had a relatively high expression of the human-specific keratin CK19 protein, confirming that the tumor cells from the lungs of nude mice originated from human thyroid tumor cells (Fig. 1G and H). These results confirmed that the IHH4-M cell sub-strains were highly invasive and metastatic in the mice.
Figure 1.
The invasion and metastasis capacity of IHH4-M cells was higher than that of IHH4-C cells. The results of the Transwell assay in (A) IHH4-C and (B) IHH4-M cells (magnification, ×200). (E) No lung tumor formation was identified in the IHH4-C groups (magnification, ×100). (C) The number of cells able to invade through the Matrigel insert was significantly higher in IHH4-M cells, compared with IHH4-C cells. *P<0.001. (D) No significant difference was identified between the proliferation of IHH4-M and IHH4-C cells. (F) Apparent lung tumor formation was identified in the IHH4-M groups (magnification, ×100). CK19 protein expression was relatively low in the (G) IHH4-C groups, compared with higher levels in the (H) IHH4-M groups (magnification, ×200).
Differentially expressed lncRNAs and mRNAs
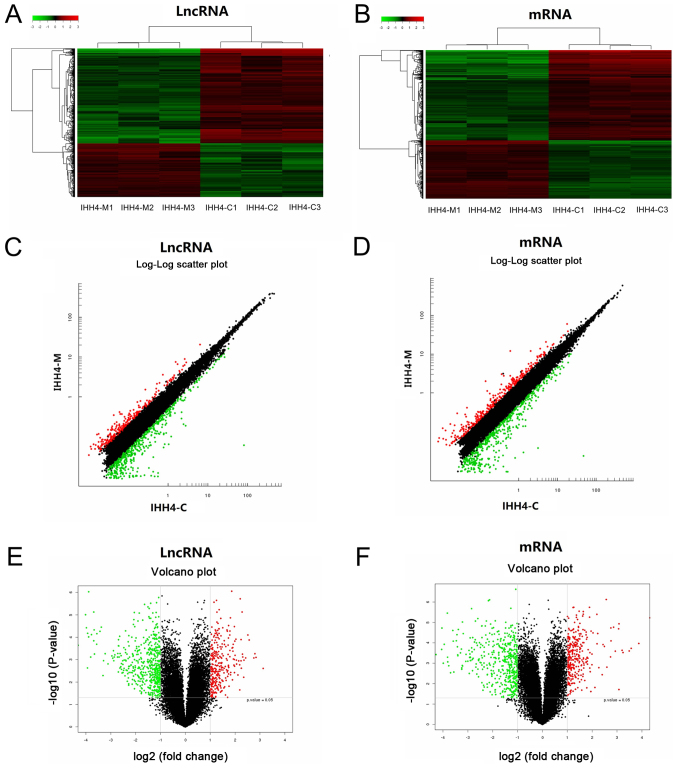
To further clarify the association between PTC metastasis and the relevant lncRNA and mRNA expression, a microarray chip was used to analyze the differential expression profile of the complete genome of lncRNAs and mRNAs in the IHH4-C and IHH4-M cells. It identified that 795 lncRNAs had an expression difference in the IHH4-C and IHH4-M cells; 286 lncRNAs had a high expression in the IHH4-M cells, and 509 lncRNAs had a low expression in the IHH4-M cells. Table II presents the 20 lncRNAs with the largest expression differences; XR_429061.1 had the largest difference among the upregulated lncRNAs, and CDR1AS had the largest difference among the downregulated lncRNAs. Additionally, 788 mRNAs had an expression difference in the IHH4-C and IHH4-M cells; 309 mRNAs had a high expression in the IHH4-M cells and 479 mRNAs had a low expression in the IHH4-M cells. Table III presents the 20 mRNAs with the largest expression differences. Peptidase inhibitor had the highest difference among the upregulated mRNAs, and CDR1 had the highest difference among the downregulated mRNAs. For further cluster analysis, the in vitro samples were divided into two large groups according to the results of arrays. The first group included IHH4-C1, IHH4-C2 and IHH4-C3, and the second group included IHH4-M1, IHH4-M2 and IHH4-M3, indicating that IHH4-M cells had more systematic changes in lncRNA and mRNA expression levels than the IHH4-C cells. It also indicated that these differentially expressed lncRNAs and mRNAs were representative (Fig. 2).
Table II.
The 20 lncRNAs with the largest expression differences.
| Upregulated lncRNAs | Downregulated lncRNAs | ||||
|---|---|---|---|---|---|
| lncRNA | Fold-change | P-value | lncRNA | Fold-change | P-value |
| XR_429061.1 | 8.745 | 2.462×10−3 | CDR1AS | 1,376.528 | 1.131×10−6 |
| ENST00000565748.1 | 7.165 | 7.926×10−4 | NR_104228.1 | 55.781 | 6.841×10−6 |
| ENST00000416341.1 | 6.949 | 5.430×10−4 | ENST00000551210.1 | 48.202 | 6.483×10−5 |
| ENST00000418029.1 | 6.745 | 8.403×10−4 | TCONS_00000982 | 47.740 | 7.044×10−5 |
| XR_245800.1 | 5.921 | 1.327×10−4 | TCONS_00010079 | 36.509 | 9.476×10−6 |
| ENST00000504773.1 | 5.709 | 7.930×10−5 | RNA95223|RNS_305_181 | 29.865 | 2.015×10−3 |
| RNA95432|RNS_514_159 | 5.370 | 8.411×10−4 | ENST00000552046.1 | 23.515 | 2.133×10−5 |
| ENST00000591257.1 | 5.120 | 1.053×10−3 | uc022atw.1 | 20.679 | 1.348×10−4 |
| ENST00000523030.1 | 5.082 | 6.101×10−3 | ENST00000598149.1 | 19.948 | 2.281×10−4 |
| TCONS_00022574 | 4.948 | 7.618×10−4 | TCONS_00017468 | 16.193 | 9.872×10−6 |
| ENST00000567668.1 | 4.911 | 5.678×10−4 | NR_028345.2 | 15.800 | 3.786×10−5 |
| ENST00000569313.1 | 4.863 | 7.475×10−6 | ENST00000445908.1 | 15.342 | 2.350×10−3 |
| uc021wad.1 | 4.757 | 2.416×10−3 | ENST00000473756.1 | 15.283 | 6.667×10−5 |
| TCONS_00022572 | 4.723 | 1.272×10−4 | NR_048567.1 | 14.843 | 9.668×10−5 |
| ENST00000459633.1 | 4.720 | 1.130×10−3 | ENST00000515416.2 | 14.838 | 9.362×10−9 |
| ENST00000553668.1 | 4.612 | 1.927×10−6 | TCONS_00013896 | 14.024 | 7.672×10−3 |
| TCONS_00008360 | 4.556 | 2.244×10−2 | NR_028345.1 | 13.722 | 7.640×10−3 |
| ENST00000453688.1 | 4.540 | 1.702×10−3 | ENST00000456722.1 | 12.883 | 1.440×10−3 |
| TCONS_00010097 | 4.421 | 1.137×10−3 | ENST00000549058.1 | 12.883 | 7.502×10−6 |
| ENST00000469312.2 | 4.328 | 4.838×10−3 | ENST00000555023.1 | 1,376.528 | 1.131×10−6 |
lncRNA, long non-coding RNA.
Table III.
The 20 mRNAs with the largest expression differences.
| Upregulated mRNAs | Downregulated mRNAs | ||||||
|---|---|---|---|---|---|---|---|
| Genbank accession | mRNA | Fold-change | P-value | Genbank accession | mRNA | Fold-change | P-value |
| NM_002638 | PI3 | 20.060 | 5.963×10−6 | NM_004065 | CDR1 | 2,097.717 | 3.607×10−6 |
| NM_014505 | KCNMB4 | 14.685 | 1.076×10−4 | NM_007102 | GUCA2B | 85.694 | 2.864×10−5 |
| NM_006512 | SAA4 | 11.880 | 1.864×10−4 | NR_104228 | LINC00632 | 51.887 | 5.167×10−6 |
| NM_018689 | CEMIP | 11.246 | 2.511×10−4 | NM_138411 | FAM71E1 | 47.223 | 2.703×10−4 |
| NM_020980 | AQP9 | 9.459 | 3.002×10−4 | NM_003022 | SH3BGRL | 41.409 | 1.597×10−4 |
| NM_016529 | ATP8A2 | 9.220 | 2.326×10−4 | NM_080872 | UNC5D | 31.602 | 2.634×10−5 |
| NM_030754 | SAA2 | 8.459 | 1.171×10−5 | NM_001191323 | GREM1 | 21.488 | 1.474×10−3 |
| NM_020641 | EQTN | 8.365 | 2.062×10−5 | NM_024867 | SPEF2 | 19.466 | 3.762×10−4 |
| NM_018689 | CEMIP | 7.628 | 2.568×10−4 | NM_017697 | ESRP1 | 16.618 | 2.114×10−4 |
| NM_001127380 | SAA2 | 7.276 | 1.717×10−5 | NM_001164000 | MECOM | 16.489 | 1.888×10−5 |
| NM_052862 | RCSD1 | 6.766 | 8.038×10−5 | NR_045985 | ALOX15P1 | 15.885 | 1.000×10−3 |
| NM_152637 | METTL7B | 6.744 | 1.867×10−5 | NM_080872 | UNC5D | 15.465 | 1.184×10−4 |
| NM_012193 | FZD4 | 6.680 | 4.910×10−4 | NM_003253 | TIAM1 | 14.180 | 3.511×10−6 |
| NM_004751 | GCNT3 | 6.033 | 1.559×10−3 | NM_014790 | JAKMIP2 | 14.094 | 5.079×10−4 |
| NM_000331 | SAA1 | 5.930 | 7.545×10−7 | NM_020455 | GPR126 | 14.019 | 8.583×10−4 |
| NM_016529 | ATP8A2 | 5.630 | 1.503×10−4 | NM_033071 | SYNE1 | 13.644 | 1.333×10−3 |
| NM_004433 | ELF3 | 5.418 | 1.218×10−4 | NM_014491 | FOXP2 | 12.174 | 2.486×10−5 |
| NM_014228 | SLC6A7 | 4.821 | 1.650×10−3 | NM_032405 | TMPRSS3 | 12.021 | 8.291×10−5 |
| NM_000675 | ADORA2A | 4.673 | 3.503×10−5 | NM_005241 | MECOM | 11.033 | 7.952×10−5 |
| NM_001191055 | ERVV-2 | 4.585 | 1.116 ×10−4 | NM_001033953 | CALCA | 10.850 | 3.999×10−5 |
Figure 2.
Differentially expressed lncRNAs and mRNAs in IHH4-M and IHH4-C cells. Hierarchical clustering analysis of differentially expressed (A) lncRNAs and (B) mRNAs. Red and green colors indicate high and low expression, respectively. In the heat map, columns represent samples and rows represent each mRNA. Log-log scatterplots comparing differentially expressed (C) lncRNAs and (D) mRNAs between IHH4-M and IHH4-C cells. Volcano plots of differentially expressed (E) lncRNAs and (F) mRNAs between IHH4-M and IHH4-C cells. The vertical lines correspond to 2.0-fold up and down and the horizontal line represents a P-value of 0.05. lncRNAs, long non-coding RNAs.
Functional analysis of differentially expressed genes
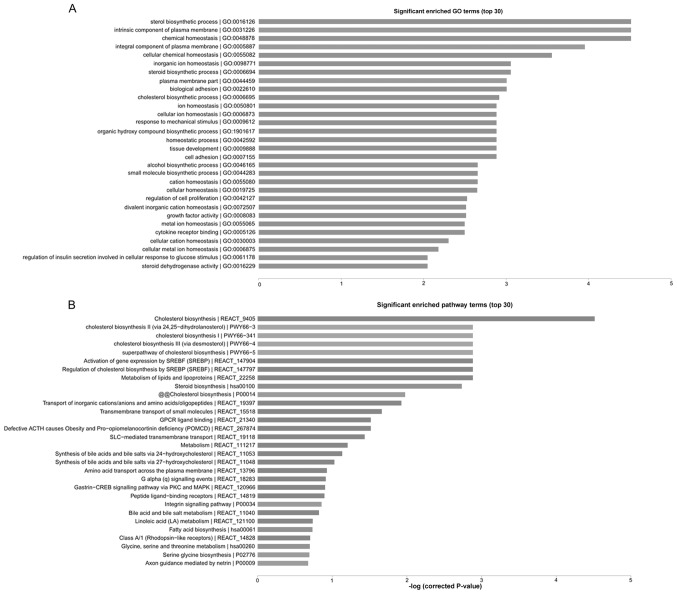
GO cluster analysis and pathway cluster analysis were performed on all the differentially expressed genes to further analyze the function of these genes. The results of GO cluster analysis revealed that the differentially expressed genes were mainly associated with steroid biosynthesis, bioadhesion, intercellular adhesion and other tumor metastasis-associated biological processes (Fig. 3A). The results of pathway cluster analysis revealed that the differentially expressed genes were associated with tumor metastasis-associated signaling pathways, including the cholesterol metabolic signaling pathway, the sterol regulatory element-binding protein signaling pathway and the integrin signaling pathway, suggesting that lncRNAs may regulate PTC metastasis through various signaling pathways (Fig. 3B).
Figure 3.
(A) Enrichment scores of the significantly enriched GO terms for differentially expressed mRNAs. (B) Enrichment scores of the significantly enriched pathway terms for differentially expressed mRNAs. The vertical axis represents the pathway category and the horizontal axis represents the enrichment score [-log10 (P-value)] of the pathway. GO, gene ontology.
RT-qPCR
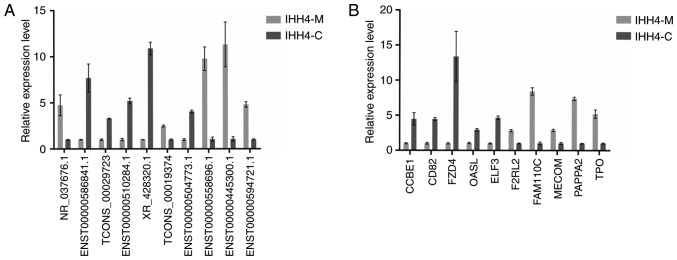
The results of RT-qPCR revealed that the expression of lncRNA ENST00000586841.1, TCONS_00029723, ENST00000510284.1, XR_428320.1, TCONS_00019374, ENST00000504773.1 and ENST00000594721.1 in IHH4-M cells was lower than that in the IHH4-C cells, while the expression of lncRNA NR_037676.1, ENST00000558696.1 and ENST00000445300.1 in IHH4-M cells was higher than that in the IHH4-C cells. Fig. 4A presents the results of RT-qPCR analysis, and the expression of the CCBE1, CD82, FZD4, OASL and ELF3 genes in the IHH4-M cells was lower than that in the IHH4-C cells, while the expression of F2RL2, FAM110C, MECOM, PAPPA2 and TPO in IHH4-M cells was higher than that in the IHH4-C cells. As presented in Fig. 4B, the results were consistent with the findings in the microarray chips, indicating that the chip results were reliable.
Figure 4.
RT-qPCR validation of 10 differentially expressed lncRNAs and mRNAs. (A) RT-qPCR validation of 10 differentially expressed lncRNAs. (B) RT-qPCR validation of 10 differentially expressed mRNAs. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CCBE1, collagen and calcium binding EGF domains 1; CD, cluster of differentiation; FZD4, frizzled class receptor 4; OASL, thyroid receptor-interacting protein 14; ELF3, epithelium-specific ets transcription factor 1; F2RL2, thrombin receptor-like 2; FAM110C, family with sequence similarity 110 member C; MECOM, myelodysplasia syndrome-associated protein 1; PAPPA2, pappalysin 2; TPO, thyroid peroxidase; GO, gene ontology.
Discussion
Previous studies have identified that proto-oncogene B-Raf (BRAF), telomerase reverse transcriptase, phosphatase and tensin homolog, runt-related transcription factor 2, S100 calcium binding protein A4, RAS (NRAS, HRAS and KRAS), Akt serine/threonine kinase 1, ALK receptor tyrosine kinase, RET proto-oncogene, paired box 8/peroxisome proliferator activated receptor γ and galectin 3 genes serve certain functions in local invasion, lymph node metastasis and distant metastasis of PTC (24–27). BRAFV600E has been considered as the most classic gene mutation in PTC and it is associated with lymph node metastasis, recurrence and adenoid tissue invasion of PTC (28). Although some of these markers have been used in the auxiliary diagnosis of thyroid cancer, no breakthrough has been made. Detection of the majority of the markers depends on the acquisition of surgical specimens and therefore cannot facilitate risk stratification in patients preoperatively. This results in a relatively significant challenge in the risk-stratified treatment of PTC. The present study aimed to preliminarily elucidate the role of certain regulatory genes (e.g., lncRNAs) in PTC invasion and metastasis, and to provide a molecular basis for the screening and risk-stratified treatment of high-risk patients with PTC in future studies.
LncRNA was selected as the main research object as it was hypothesized that lncRNA was a more distinctive marker for diagnostic evaluation than traditional molecular markers. LncRNA is abundantly present in the plasma. The detection of lncRNA directly from the patient's plasma for the diagnostic evaluation may effectively avoid the direct stimulation of the primary tumor during examinations (e.g., puncture or biopsy collection), which is more suitable for preoperative diagnosis and risk stratification assessment (29). Furthermore, lncRNA detection depends on RT-qPCR technology, and it requires relatively less time and has a higher sensitivity compared with the semi-quantitative analysis using immunohistochemistry, a traditional detection method of protein molecular markers. Therefore, quantitative lncRNA detection effectively avoids the impact of human factors on the interpretation of the results.
LncRNA has previously been considered as the noise of genomic transcription. However, an increasing number of studies have recognized that lncRNA serves an important function in cell proliferation, differentiation and malignant transformation processes (30,31). Similar to the protein-coding gene, lncRNA may regulate target gene expression and serve a function in oncogene or tumor suppressor gene activity (32). LncRNA research in PTC has been rarely reported. Yoon et al (33) identified that the expression of an lncRNA, non-coding RNA associated with MAP kinase pathway and growth arrest, was downregulated in PTC with a BRAF mutation that was also associated with cell growth arrest. Similarly, BRAF-activated long non-coding RNA expression in PTC cells is significantly higher than that in normal tissues. This overexpression significantly promotes and activates PTC cell proliferation and autophagy (34). The results of these studies suggested that abnormal lncRNA expression in PTC may serve an important function; however, the sample sizes and details of the study designs in the existing literature are limited and require further in-depth study.
To ensure the reliability of the results of the present study, independent cell sub-strains (i.e., IHH4-M1, IHH4-M2 and IHH4-M3) with a high invasive capacity and their corresponding controls were selected to perform chip screening. These three cell sub-strains originated from the IHH4 cell line, but they were screened independently from the beginning of the experiment for 15 generations to ensure a similar genetic background with different invasive and metastatic capacities in the sub-strains. Further application of lncRNA microarray chips to screen the cell sub-strains with differentially expressed lncRNA and mRNA profiles compared with the control cells and the application of GO cluster analysis and pathway cluster analysis for the functional prediction of those lncRNAs and mRNAs revealed that certain genes were associated with bioadhesion, intercellular adhesion and steroid metabolic pathways. This suggested that lncRNAs may regulate PTC metastasis through the regulation of steroid metabolic pathways. To further verify the reliability of the microarray results, 10 differentially expressed lncRNAs and 10 differentially expressed mRNAs were selected and their expression patterns in the IHH4-M and IHH4-C cells was assessed using RT-qPCR. The results were consistent with the microarray, indicating the reliability of the microarray results. In conclusion, abnormal lncRNA expression may serve an important function in the onset and development of PTC. There may be an lncRNA regulatory network in PTC that regulates the downstream target gene expression, thereby affecting thyroid cancer cell proliferation, invasion and metastasis. In the present study, a portion of the lncRNAs associated with PTC metastasis were screened. These lncRNAs may be involved in PTC invasion, metastasis and malignant transformation processes. These data provided an important molecular basis for future screening, stratified diagnosing and treating high-risk patients with PTC in the clinic. Further investigation and study of the mechanism of lncRNA and the cause of abnormal lncRNA expression is required.
However, there are certain limitations to the present study, which require to be expanded upon in further studies. PTC cells were used rather than PTMC cells in the present study due to the lack of PTMC cell lines, resulting in the absence of data on lncRNA and mRNA expression in highly-vs. lowly-invasive PTMC cells. Additionally, a Transwell assay was used rather than in vivo selection to obtain the sub-strains with different invasiveness, as the wild-type IHH4 cells were difficult to develop into a tumor under the skin of the nude mice in preliminary experiments. This may affect the reliability of the results to a certain extent and further studies using in vivo selection are required. Furthermore, Transwell assays and in vivo experiments were used to detect the invasiveness of different sub-strains in the present study. However, certain features, including proliferation and cell death were only preliminarily examined using a CCK-8 assay. This may influence the reliability of the results and additional experiments on proliferation and cell death, including colony formation experiments are required. Immunohistochemical analysis of the 20 genes that were examined was not performed in tissue samples taken from patients with PTMC. The use of tissue samples over cultured cell lines would further validate these data.
Acknowledgements
The authors would like to thank Dr Ke-xin Yin (Second Clinical Medical College, Zhejiang Chinese Medical University, Hangzhou, China) for her technical assistance for cell culture and in vivo experiment support.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81702644), the National Natural Science Foundation of China (grant no. 81672642), the Major Science and Technology Project of Zhejiang Province (grant no. 2015C03027) and the Project of Administration of Traditional Chinese Medicine of Zhejiang Province of China (grant no. 2017ZA037).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MHG made substantial contributions to acquisition of data and analysis and interpretation of data. LHJ made substantial contributions to acquisition of data and analysis of data, and was involved in drafting the manuscript and responsible for the subsequent revision of the manuscript. QLW, ZT and CC provided technical assistance. CMZ and XZ analyzed the data. JWC and ZYZ interpreted the results. XJC made substantial contributions to conception and design, and gave final approval of the version to be published. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The animal experiments in the present study had been reviewed and approved by the Animal Ethical and Welfare Committee of Zhejiang Cancer Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2014. National Cancer Institute; Bethesda, MD: Apr, 2017. based on November 2016 SEER data submission, posted to the SEER web site. [Google Scholar]
- 2.Davies L, Welch HG. Current thyroid cancer trends in the United States. JAMA Otolaryngol Head Neck Surg. 2014;140:317–322. doi: 10.1001/jamaoto.2014.1. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Durante C, Haddy N, Baudin E, Leboulleux S, Hartl D, Travagli JP, Caillou B, Ricard M, Lumbroso JD, De Vathaire F, Schlumberger M. Long-term outcome of 444 patients with distant metastases from papillary and follicular thyroid carcinoma: Benefits and limits of radioiodine therapy. J Clin Endocrinol Metab. 2006;91:2892–2899. doi: 10.1210/jc.2005-2838. [DOI] [PubMed] [Google Scholar]
- 5.American Thyroid Association (ATA) Guidelines Taskforce on Thyroid Nodules and Differentiated Thyroid Cancer, corp-author. Cooper DS, Doherty GM, Haugen BR, Kloos RT, Lee SL, Mandel SJ, Mazzaferri EL, McIver B, Pacini F, et al. Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid. 2009;19:1167–1214. doi: 10.1089/thy.2009.0110. [DOI] [PubMed] [Google Scholar]
- 6.Ito Y, Miyauchi A. Nonoperative management of low-risk differentiated thyroid carcinoma. Curr Opin Oncol. 2015;27:15–20. doi: 10.1097/CCO.0000000000000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wada N, Duh QY, Sugino K, Iwasaki H, Kameyama K, Mimura T, Ito K, Takami H, Takanashi Y. Lymph node metastasis from 259 papillary thyroid microcarcinomas: Frequency, pattern of occurrence and recurrence, and optimal strategy for neck dissection. Ann Surg. 2003;237:399–407. doi: 10.1097/00000658-200303000-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Sun W, Wu Y, Yu X, Liu Y, Song H, Xia T, Xiao B, Guo J. Decreased expression of long noncoding RNA AC096655.1-002 in gastric cancer and its clinical significance. Tumour Biol. 2013;34:2697–2701. doi: 10.1007/s13277-013-0821-0. [DOI] [PubMed] [Google Scholar]
- 10.Sun M, Jin FY, Xia R, Kong R, Li JH, Xu TP, Liu YW, Zhang EB, Liu XH, De W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. doi: 10.1186/1471-2407-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, Jia W. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol. 2013;30:588. doi: 10.1007/s12032-013-0588-6. [DOI] [PubMed] [Google Scholar]
- 12.Qiao HP, Gao WS, Huo JX, Yang ZS. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac J Cancer Prev. 2013;14:1077–1082. doi: 10.7314/APJCP.2013.14.2.1077. [DOI] [PubMed] [Google Scholar]
- 13.Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014;15:2971–2977. doi: 10.7314/APJCP.2014.15.7.2971. [DOI] [PubMed] [Google Scholar]
- 14.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM, Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lv N, Shan Z, Gao Y, Guan H, Fan C, Wang H, Teng W. Twist1 regulates the epithelial-mesenchymal transition via the NF-kB pathway in papillary thyroid carcinoma. Endocrine. 2016;51:469–477. doi: 10.1007/s12020-015-0714-7. [DOI] [PubMed] [Google Scholar]
- 16.Leboulleux S, Tuttle RM, Pacini F, Schlumberger M. Papillary thyroid microcarcinoma: Time to shift from surgery to active surveillance? Lancet Diabetes Endocrinol. 2016;4:933–942. doi: 10.1016/S2213-8587(16)30180-2. [DOI] [PubMed] [Google Scholar]
- 17.Fan YH, Ding J, Nguyen S, Liu XJ, Xu G, Zhou HY, Duan NN, Yang SM, Zern MA, Wu J. Aberrant hedgehog signaling is responsible for the highly invasive behavior of a subpopulation of hepatoma cells. Oncogene. 2016;35:116–124. doi: 10.1038/onc.2015.67. [DOI] [PubMed] [Google Scholar]
- 18.Huang C. Abstract 426: Suppression of cell invasion and migration by inhibitors to ROS level in human lung cancer cells. Cancer Res. 2010;70:426–426. doi: 10.1158/1538-7445.AM10-426. [DOI] [Google Scholar]
- 19.Lu KV, Jong KA, Rajasekaran AK, Cloughesy TF, Mischel PS. Upregulation of tissue inhibitor of metalloproteinases (TIMP)-2 promotes matrix metalloproteinase (MMP)-2 activation and cell invasion in a human glioblastoma cell line. Lab Invest. 2004;84:8–20. doi: 10.1038/labinvest.3700003. [DOI] [PubMed] [Google Scholar]
- 20.William S. Guidance document on the recognition, assessment, and use of clinical signs as humane endpoints for experimental animals used in safety evaluation. OECD Environmental Health and Safety Publications, Series on Testing and Assessment 19; 2000. [Google Scholar]
- 21.Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, Bao W, Fang H, Kawasaki ES, Hager J, Tikhonova IR, et al. Performance comparison of one-color and two-color platforms within the MicroArray quality control (MAQC) project. Nat Biotechnol. 2006;24:1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 22.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Xing M, Haugen BR, Schlumberger M. Progress in molecular-based management of differentiated thyroid cancer. Lancet. 2013;381:1058–1069. doi: 10.1016/S0140-6736(13)60109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu R, Bishop J, Zhu G, Zhang T, Ladenson PW, Xing M. Mortality risk stratification by combining BRAF V600E and TERT promoter mutations in papillary thyroid cancer: Genetic duet of BRAF and TERT promoter mutations in thyroid cancer mortality. JAMA Oncol. 2016 Sep 1; doi: 10.1001/jamaoncol.2016.3288. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 26.Carr FE, Tai PW, Barnum MS, Gillis NE, Evans KG, Taber TH, White JH, Tomczak JA, Jaworski DM, Zaidi SK, et al. Thyroid hormone receptor-β (TRβ) mediates runt-related transcription factor 2 (Run×2) expression in thyroid cancer cells: A novel signaling pathway in thyroid cancer. Endocrinology. 2016;157:3278–3292. doi: 10.1210/en.2015-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joung JY, Kim TH, Jeong DJ, Park SM, Cho YY, Jang HW, Jung YY, Oh YL, Yim HS, Kim YL, et al. Diffuse sclerosing variant of papillary thyroid carcinoma: Major genetic alterations and prognostic implications. Histopathology. 2016;69:45–53. doi: 10.1111/his.12902. [DOI] [PubMed] [Google Scholar]
- 28.Sidaway P. Thyroid cancer: BRAF and/or TERT mutations increase mortality. Nat Rev Clin Oncol. 2016;13:652. doi: 10.1038/nrclinonc.2016.158. [DOI] [PubMed] [Google Scholar]
- 29.Gupta Chandra S, Tripathi Nandan Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int J Cancer. 2017;140:1955–1967. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]
- 30.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eades G, Zhang YS, Li QL, Xia JX, Yao Y, Zhou Q. Long non-coding RNAs in stem cells and cancer. World J Clin Oncol. 2014;5:134–141. doi: 10.5306/wjco.v5.i2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon H, He H, Nagy R, Davuluri R, Suster S, Schoenberg D, Pellegata N, Ade Chapelle L. Identification of a novel noncoding RNA gene, NAMA, that is downregulated in papillary thyroid carcinoma with BRAF mutation and associated with growth arrest. Int J Cancer. 2007;121:767–775. doi: 10.1002/ijc.22701. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Guo Q, Zhao Y, Chen J, Wang S, Hu J, Sun Y. BRAF-activated long non-coding RNA contributes to cell proliferation and activates autophagy in papillary thyroid carcinoma. Oncol Lett. 2014;8:1947–1952. doi: 10.3892/ol.2014.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.