Abstract
The macrophage migration inhibition factor (MIF) is a cytokine with multiple biological functions, including the cancer-associated processes, cell cycle deregulation, angiogenesis and metastatization. The present study investigated the expression of MIF and its functionally associated genes (D-DT, CD74, CD44, CXCR2 and CXCR4) in glioblastoma multiforme (GBM). The data were obtained from The Cancer Genome Atlas databank, through the cBioportal web-based utility (cbioportal.org/). A significant increase was observed in the majority of these genes in GBM samples compared with lower grade gliomas, however no significant correlation among the selected genes and the overall survival of the patients was identified. In contrast, the expression of MIF exhibited a trend toward an increase in overall survival and a significant increase of MIF expression was observed in samples of patients who underwent neoadjuvant treatment. In conclusion these data indicate that MIF and its receptors are involved in GBM progression and maintenance. Deciphering the precise biological significance in GBM would favor the adoption of tailored approaches to modulate the function of MIF and its associated genes for the treatment of the disease.
Keywords: macrophage migration inhibitory factor, D-dopachrome tautomerase, glioblastoma, glioma, cluster of differentiation 74
Introduction
Glioblastoma multiforme (GBM) is a glial cancer classified by WHO as a grade IV astrocytoma. Depending on mutation of the isocitrate dehydrogenase (IDH) gene, three main GM subsets can be identified: IDH-wild type, that represent ~90% of the cases, IDH-mutated, generally observed in younger patients with prior lower grade gliomas and NOS (no otherwise specified), where evaluation of the IDH gene cannot be performed (1).
Treatment of first choice is surgery, coupled with chemotherapy and radiotherapy. Since no change in overall survival (OS) has occurred after the introduction in 2005 of the STUPP regimen (radiotherapy or chemotherapy in association with temozolomide), a better understanding of the molecular mechanisms related to GBM proliferation and recurrence could suggest alternative avenues for the development of novel therapies and prognostic markers.
The macrophage migration inhibition factor (MIF) is an uncommon cytokine with multiple biological functions and pleiotropic effects (2,3). MIF binds the HLA class II histocompatibility antigen gamma chain, CD74. The phosphorylation of CD74, following MIF binding, leads to the recruitment of CD44 and the activation of its downstream signaling, through the ERK-MAPK pathway (2,3). MIF is also a non-cognate ligand for the chemokine receptors, CXCR2 and CXCR4. Beside MIF, the MIF superfamily includes the recently identified homolog D-dopachrome tautomerase, D-DT (also known as MIF-2), located on chromosome 22q11.23 (4,5). Similarly to MIF, D-DT possesses enzymatic binding pockets with tautomerase activity for the D-dopachrome and phenylpyruvate substrates. However, the end-products of the dopachrome substrates are different for the two genes. Also, D-DT binds the CD74 ectodomain, with an about 3-fold higher acid dissociation constant and a 11-fold higher dissociation rate as compared to MIF. Interestingly, D-DT lacks the motif that allows MIF binding to the chemokine receptor, CXCR2 (4,5). Studied for its role in immunity, MIF has been primarily identified as an inflammatory mediator involved in the regulation of macrophage activity, chemotaxis, and cytokine secretion and has been indicated as a major player in the pathogenesis of immune-inflammatory and autoimmune diseases including multiple sclerosis, Guillain-Barrè syndrome, type 1 diabetes, systemic lupus erythematosus and rheumatoid arthritis (2,6–9).
However, MIF activity is not limited to the immune system: it can act as a hormone, by exerting glucocorticoid antagonism; as an enzyme, by catalyzing the tautomerization of the D-dopachrome in 5,6-Dihydroxyindole-2-carboxylic Acid (DHICA); as a cell differentiation factor, by activating the Wnt/β-catenin pathway in neurons. MIF is also involved in many tumor processes, including deregulation of cell cycle, angiogenesis and metastasis formation (10).
A significant increase in MIF expression has been observed in several types of cancer, including cervical cancer, breast, prostate, liver, lung cancer, neuroblastoma, colorectal cancer, pancreatic, renal carcinomas and lymphocytic leukemia (10).
There are several biological pathways triggered by MIF that may contribute to tumorigenesis. Indeed, upon endocytosis, MIF interacts with c-Jun activation domain-binding protein-1 (JAB1) and deactivates it. JAB1 is a negative regulator of p27KIP1, that controls cell cycle progression at the G1 phase (11). MIF also promotes tumor growth by activating the MAPK/PI3K/Akt pathways, by inhibiting p53-dependent apoptosis, by promoting angiogenesis via up-regulated secretion of vascular endothelial growth factor (VEGF) (10), and by promoting immune escape via inhibition of Natural Killer cell lysis and recruitment of Myeloid Derived Suppressor Cells (MDSCs). Moreover, MIF favors tumor invasion by facilitating epithelial-to-mesenchymal transition (EMT) (12,13) and by increasing the expression of matrix metalloproteinases (MMPs) (12).
Increasing body of data suggests an important pathogenic role for MIF in the progression of gliomas (14). Immunohistological analysis of GBM samples has shown that MIF strongly accumulates in proximity of necrotic areas and in cancer cells adjacent to the blood vessels (15). It was also shown that MIF immunoreactivity increases with tumor grade (15). Moreover, expression of the CD74 in GBM seems to be involved in the resistance to temozolomide (16). In addition, an association between MIF expression and tumor recurrence and poor prognosis of glioma patients has been reported (17). It has also been reported that MIF enhances autophagy by regulating ROCK1 activity and contributes to the escape of dendritic cell surveillance in GBM (18).
In agreement with these data, a study in primary GBM cells has shown that inhibition of MIF with ISO-1, an inhibitor of its D-dopachrome tautomerase site, reduced the growth rate of primary GBM cells in a dose-dependent manner (19). MiR-608 has also been shown to inhibit the migration and invasion of glioma stem cells by targeting macrophage migration inhibitory factor (20).
The aim of this study was to evaluate the expression of MIF and of its functionally related genes, in different grade gliomas, and to highlight the potential role of MIF in GBM, as biomarker or therapeutic target.
Materials and methods
Characterization of MIF and functionally-related genes in GBM
In order to evaluate the expression levels of MIF and related genes in gliomas, RNA Seq data from the TCGA datasets were downloaded through the cBioportal web-based utility (http://www.cbioportal.org). Selected genes were MIF, D-DT, CD74, CD44, CXCR2, CXCR4 and JAB1. Complete clinical data of the patients were retrieved and only data from primary tumors, with no neoadjuvant therapy prior to excision, were selected. Data were subjected to Kolmogorov-Smirnov test, D'Agostino and Person Omnibus test and Shapiro-Wilk normality test. Accordingly to the results from the normality tests, the Kruskal-Wallis test followed by Dunn's Post Test was applied to assess statistical significance for the differences among cancer types. Overall, this study comprised 153 patients with GBM, 31 with Oligoastrocytoma, 22 with Oligodendroglioma, 15 with Astrocytomas, 29 with Anaplastic Astrocytoma and 16 patients with Anaplastic Oligoastrocytomas.
In order to evaluate how the genes of interest are similar in their expression pattern, a Gene Distance Matrix was built using the Multi Experiment Viewer software (mev.tm4.org). The distance between two genes, which represents the inverse of similarity, was calculated using Pearson correlation. Values can vary from −1 to 1. Correlations near 1 indicate a strong positive correlation, while values closer to −1 indicate a negative correlation.
Identification of genetic aberrations of the genes of interest has been performed on a cohort of 281 and 283 DNA-sequenced GBM and low-grade glioma patients, respectively, as available in the TGCA database.
Correlation between the selected genes and the OS of GBM patients were performed using linear regression analysis and Pearson's test.
Effect of neoadjuvant therapy on MIF expression
Microarray transcriptomic data of 497 tumor samples from patients who underwent tumor excision without neoadjuvant therapy and from 20 patients treated with neoadjuvant therapy before cancer resection were also obtained from the cBioportal web-based utility. No information regarding the type of neoadjuvant therapy has been disclosed. Unpaired Student's t-test was applied to assess statistical significance of the mean difference in MIF levels between the two groups.
Statistical analysis
Expression data are presented as dot plots with line at median value. GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA) was used for all the statistical analysis. Data were subjected to a Kolmogorov-Smirnov test, D'Agostino and Person Omnibus test and Shapiro-Wilk normality test. According to the results from the normality tests, the Kruskal-Wallis test followed by Dunn's post hoc test were applied to assess the statistical significance for the differences among cancer types. The association between the selected genes and the OS of GBM patients was evaluated using linear regression analysis and Pearson's test. An unpaired Student's t-test was applied to assess statistical significance of the mean difference in MIF levels between the group of patients who underwent neoadjuvant therapy and patients without neoadjuvant therapy. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of MIF and its functionally-related genes in GBM
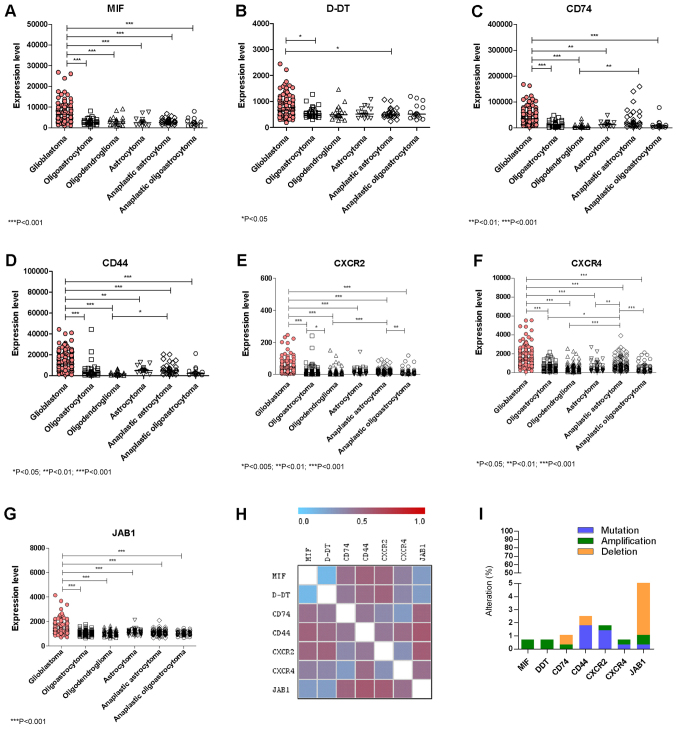
A marked upregulation of MIF expression was observed in GBM as compared to all of the other lower grade Gliomas (Fig. 1A). Similarly to MIF, significant higher levels of D-DT were found in GBM as compared to Oligoastrocytoma and Anaplastic Astrocytoma (Fig. 1B). As regards CD74 and the co-receptor, CD44, significant higher levels were observed in GBM when compared to all of the other lower grade Gliomas (Fig. 1C and D). Along the same lines, the non-cognate receptors CXCR2 and CXCR4 were also significantly overexpressed in GBM samples as compared to all of the other lower grade gliomas (Fig. 1E, F). In addition, in GBM samples significantly higher expression levels of JAB1 were also observed with respect to lower grade gliomas (Fig. 1G). A significant positive correlation in the expression levels was observed between MIF and D-DT (P<0.0001 by Spearman test), CD74 (P=0.0422 by Spearman test), CXCR4 (P<0.0001 by Spearman test) and JAB1 (P<0.0001 by Spearman test) (Fig. 1H).
Figure 1.
Evaluation of the expression levels of (A) MIF, (B) D-DT, (C) CD74, (D) CD44, (E) CXCR2, (F) CXCR4 and (G) JAB1 in glioblastoma and lower grade glioma samples. (H) Gene Distance Matrix was built using the Multi Experiment Viewer software (mev.tm4.org/), in order to evaluate how the genes of interest are similar in their expression pattern. (I) Percentage of genetic aberrations in the genes of interest has been performed on a cohort of 281 and 283 DNA-sequenced glioblastoma and low-grade glioma patients, respectively. Data were retrieved from the The Cancer Genome Atlas dataset through the cBioportal web-based utility (cbioportal.org). *P<0.05, **P<0.01 and ***P<0.001, as indicated. MIF, macrophage migration inhibitory factor; D-DT, D-dopachrome tautomerase; CD, cluster of differentiation; CXCR, C-X-C Motif Chemokine Receptor; JAB1, c-Jun activation domain-binding protein-1.
Characterization of MIF and related genes in GBM
Only a small percentage of samples presented genetic aberrations in the studied genes (Fig. 1I). These aberrations included: 2 cases (over 283 samples) among lower grade gliomas of MIF and D-DT gene amplification; 1 amplification and 2 deletions for CD74 in lower grade glioma samples; 2 cases of CD44 deletion among glioma samples and 2 missense mutations in glioma samples, as well as, 1 missense and 2 nonsense mutations among GBM samples (over 273 samples); 1 case of amplification and 1 missense mutation of the CXCR2 gene in GBM samples, as well as 3 missense mutations among glioma samples; 1 missense mutation of CXCR4 and 1 gene amplification among glioma and GBM samples, respectively; 1 case of missense mutation of JAB1 and 13 cases of deletions among gliomas and 2 deletions among GBM patient (Fig. 1I).
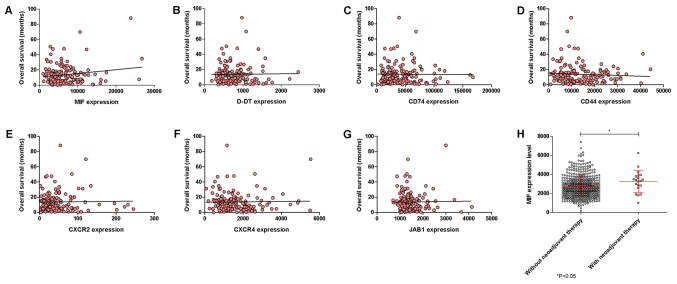
No significant correlations among the selected genes and the OS of the GBM patients could be observed, although a trend of positive correlation was observed for MIF (Fig. 2A-G). Also, neoadjuvant therapy was associated to a moderate, although significant (P=0.0315), increase in MIF expression (Fig. 2H).
Figure 2.
Correlation between overall survival and expression levels (A) MIF, (B) D-DT, (C) CD74, (D) CD44, (E) CXCR2, (F) CXCR4 and (G) JAB1 was performed using linear regression analysis and Pearson's test on glioblastoma samples. (H) Differences in MIF expression levels were calculated in glioblastoma samples from drug-naïve patients and patients who underwent neoadjuvant therapy. Data were retrieved from the The Cancer Genome Atlas dataset through the cBioportal web-based utility (cbioportal.org). *P<0.05, as indicated. MIF, macrophage migration inhibitory factor; D-DT, D-dopachrome tautomerase; CD, cluster of differentiation; CXCR, C-X-C Motif Chemokine Receptor; JAB1, c-Jun activation domain-binding protein-1.
Discussion
The results presented herein partially confirm the previous observations on the modulation of MIF and CD74 in central nervous system (CNS) tumors (15–17) and expand the current knowledge on their most functionally related genes. These data demonstrate that a local over-expression of MIF, D-DT, CD44, CD74, CXCR2, CXCR4 and JAB1 occurs in invasive and malignant forms of GBM, as compared to other lower grade CNS tumors. To the best of our knowledge this is the first paper to demonstrate the overexpression of the MIF related genes, CD44, CXCR2, CXCR4 and JAB1 and D-DT in GBM.
However, in spite of these multiple evidence pointing to a pro-oncogenic role of MIF in GBM, the net biological significance of our present findings is hampered by the observation that the over-expression of MIF does not correlate with a lower OS but rather exhibits an inverse trend to increased OS. In addition, the biological significance of this trend is strengthened by the observation that neoadjuvant therapy is associated to a significant increase in MIF expression. The potential protective role of MIF in GBM may concur with recent findings that CD74 is restricted to microglia/macrophages associated with an M1-polarized immune milieu and prolonged patient survival in gliomas (21). Favoring the beneficial anti-oncogenic role of endogenous MIF in GBM, it is also the observation that bevacizumab resistance in GBM is driven by reduced MIF at the tumor edge, causing proliferative expansion of M2 macrophages, which in turn promotes tumor growth (22).
In contrast, other Authors have reported high levels of CD74 expression in high grade gliomas and have proposed this expression as a mechanism involved in temozolomide resistance (16). Also, Wang and collaborators have shown that in patients with WHO grade III and IV gliomas the survival time was significantly shorter in patients with high expression of MIF or IL-8 in high-grade tumors than those with protein low expression in their tumors (17). However, in multivariate analysis, only histological grade and MIF expression in gliomas were independently associated with survival (17). In particular, the discrepancy of these data with ours can be due to the fact that we analyze MIF expression at the transcriptomic level and not at the protein level. Also, as pointed out by Verjans et al (23), cellular localization of MIF can be associated to different biological effects in cancer. In particular, Verjans et al, have shown that when MIF is localized within breast cancer cells, it acts as a favorable prognostic marker, while it plays a pro-oncogenic role, by promoting breast cancer cells-stroma interactions, when it is localized in the extracellular space (23).
Taken as a whole, these data highlight a complex, pleiotropic and eventually even dichotomous role of MIF in GBM maintenance and progression. The pleiotropism of MIF, as well as of most cytokines, in biological systems is well known. MIF is clearly endowed with pro-inflammatory properties that include induction of nitric oxide, cyclooxygenase 2 (24) and Toll-like receptor 4 (25), the production of pro-inflammatory cytokines (26) and glucocorticoid antagonism (27). Nonetheless, in other settings, MIF acts as anti-inflammatory cytokine that recruits myeloid derived suppressor cells and tumor associated macrophages (28,29) and inhibits CD8+ T- and NK cells-mediated cytotoxicity (30).
The major limitation of our data is that only the transcriptional levels of MIF and related genes have been investigated. Therefore, no direct comparison can be performed between the above mentioned studies and our results. Further analysis on the cellular localization of these proteins will eventually shed light on their role in GBM development, progression and prognosis. Also, it should be pointed out that MIF undergoes post-translational modifications, including carbamylation of the Pro-2; cysteinylation at Cys-60; S-nitrosilation and phosphorylation of Cys-81 and Ser-91 (31), that may alter MIF activity at specific sites, especially where oxidative events occur.
Although the eventual concentration and dose-dependency of the effects of MIF (and D-DT) in the context of GBM remains to be defined, it seems possible to hypothesize that different environmental conditions, the local concentration and/or protein localization may ultimately dictate the effects of MIF in GBM. The exact definition of the precise role of MIF in GBM could lead to important diagnostic and therapeutic achievements in terms of prognosis and identification of biomarkers that could be associated to either beneficial or detrimental effects and predictors of therapeutic responses. In particular, therapeutic approaches based on either MIF-D-DT agonism or antagonisms could be envisaged in a tailored fashion for at least certain subsets of GBM patients, once the protective or pathogenic role of these cytokine homologs is more clearly understood.
Acknowledgements
Not applicable.
Funding
The present study was supported by current research funds 2016 of IRCCS ‘Centro Neurolesi ‘Bonino Pulejo’, Messina-Italy.
Availability of data and materials
All data used in this paper are available for download from the cBioportal website (cbioportal.org).
Authors' contributions
FN and PF conceived and designed the study. MP, EM, MSB and MCP collected and analyzed the data. AB, GC and PB conducted the statistical analysis, interpreted the data and wrote the manuscript. All authors read and revised the manuscript, and approved the final version to be published.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Stosic-Grujicic S, Stojanovic I, Nicoletti F. MIF in autoimmunity and novel therapeutic approaches. Autoimmun Rev. 2009;8:244–249. doi: 10.1016/j.autrev.2008.07.037. [DOI] [PubMed] [Google Scholar]
- 3.Bloom J, Sun S, Al-Abed Y. MIF, a controversial cytokine: A review of structural features, challenges, and opportunities for drug development. Expert Opin Ther Targets. 2016;20:1463–1475. doi: 10.1080/14728222.2016.1251582. [DOI] [PubMed] [Google Scholar]
- 4.Merk M, Zierow S, Leng L, Das R, Du X, Schulte W, Fan J, Lue H, Chen Y, Xiong H, et al. The D-dopachrome tautomerase (DDT) gene product is a cytokine and functional homolog of macrophage migration inhibitory factor (MIF) Proc Natl Acad Sci. 2011;108:E577–E585. doi: 10.1073/pnas.1102941108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merk M, Mitchell RA, Endres S, Bucala R. D-dopachrome tautomerase (D-DT or MIF-2): Doubling the MIF cytokine family. Cytokine. 2012;59:10–17. doi: 10.1016/j.cyto.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connelly KL, Kandane-Rathnayake R, Hoi A, Nikpour M, Morand EF. Association of MIF, but not type I interferon-induced chemokines, with increased disease activity in Asian patients with systemic lupus erythematosus. Sci Rep. 2016;6:29909. doi: 10.1038/srep29909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lang T, Foote A, Lee JP, Morand EF, Harris J. MIF: Implications in the pathoetiology of systemic lupus erythematosus. Front Immunol. 2015;6:577. doi: 10.3389/fimmu.2015.00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benedek G, Meza-Romero R, Jordan K, Zhang Y, Nguyen H, Kent G, Li J, Siu E, Frazer J, Piecychna M, et al. MIF and D-DT are potential disease severity modifiers in male MS subjects. Proc Natl Acad Sci USA. 2017;114:E8421–E8429. doi: 10.1073/pnas.1712288114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicoletti F, Créange A, Orlikowski D, Bolgert F, Mangano K, Metz C, Di Marco R, Al Abed Y. Macrophage migration inhibitory factor (MIF) seems crucially involved in Guillain-Barré syndrome and experimental allergic neuritis. J Neuroimmunol. 2005;168:168–174. doi: 10.1016/j.jneuroim.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 10.Nobre CC, de Araújo JM, Fernandes TA, Cobucci RN, Lanza DC, Andrade VS, Fernandes JV. Macrophage migration inhibitory factor (MIF): Biological activities and relation with cancer. Pathol Oncol Res. 2017;23:235–244. doi: 10.1007/s12253-016-0138-6. [DOI] [PubMed] [Google Scholar]
- 11.Burger-Kentischer A, Finkelmeier D, Thiele M, Schmucker J, Geiger G, Tovar GE, Bernhagen J. Binding of JAB1/CSN5 to MIF is mediated by the MPN domain but is independent of the JAMM motif. FEBS Lett. 2005;579:1693–1701. doi: 10.1016/j.febslet.2005.01.080. [DOI] [PubMed] [Google Scholar]
- 12.Babu SN, Chetal G, Kumar S. Macrophage migration inhibitory factor: A potential marker for cancer diagnosis and therapy. Asian Pac J Cancer Prev. 2012;13:1737–1744. doi: 10.7314/APJCP.2012.13.5.1737. [DOI] [PubMed] [Google Scholar]
- 13.Funamizu N, Hu C, Lacy C, Schetter A, Zhang G, He P, Gaedcke J, Ghadimi MB, Ried T, Yfantis HG, et al. Macrophage migration inhibitory factor induces epithelial to mesenchymal transition, enhances tumor aggressiveness and predicts clinical outcome in resected pancreatic ductal adenocarcinoma. Int J Cancer. 2013;132:785–794. doi: 10.1002/ijc.27736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bach JP, Deuster O, Balzer-Geldsetzer M, Meyer B, Dodel R, Bacher M. The role of macrophage inhibitory factor in tumorigenesis and central nervous system tumors. Cancer. 2009;115:2031–2040. doi: 10.1002/cncr.24245. [DOI] [PubMed] [Google Scholar]
- 15.Bacher M, Schrader J, Thompson N, Kuschela K, Gemsa D, Waeber G, Schlegel J. Up-regulation of macrophage migration inhibitory factor gene and protein expression in glial tumor cells during hypoxic and hypoglycemic stress indicates a critical role for angiogenesis in glioblastoma multiforme. Am J Pathol. 2003;162:11–17. doi: 10.1016/S0002-9440(10)63793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitange GJ, Carlson BL, Schroeder MA, Decker PA, Morlan BW, Wu W, Ballman KV, Giannini C, Sarkaria JN. Expression of CD74 in high grade gliomas: A potential role in temozolomide resistance. J Neurooncol. 2010;100:177–186. doi: 10.1007/s11060-010-0186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang XB, Tian XY, Li Y, Li B, Li Z. Elevated expression of macrophage migration inhibitory factor correlates with tumor recurrence and poor prognosis of patients with gliomas. J Neurooncol. 2012;106:43–51. doi: 10.1007/s11060-011-0640-3. [DOI] [PubMed] [Google Scholar]
- 18.Xu S, Guo X, Gao X, Xue H, Zhang J, Guo X, Qiu W, Zhang P, Li G. Macrophage migration inhibitory factor enhances autophagy by regulating ROCK1 activity and contributes to the escape of dendritic cell surveillance in glioblastoma. Int J Oncol. 2016;49:2105–2115. doi: 10.3892/ijo.2016.3704. [DOI] [PubMed] [Google Scholar]
- 19.Baron N, Deuster O, Noelker C, Stüer C, Strik H, Schaller C, Dodel R, Meyer B, Bacher M. Role of macrophage migration inhibitory factor in primary glioblastoma multiforme cells. J Neurosci Res. 2011;89:711–717. doi: 10.1002/jnr.22595. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Xue Y, Wang P, Zhu J, Ma J. MiR-608 inhibits the migration and invasion of glioma stem cells by targeting macrophage migration inhibitory factor. Oncol Rep. 2016;35:2733–2742. doi: 10.3892/or.2016.4652. [DOI] [PubMed] [Google Scholar]
- 21.Zeiner PS, Preusse C, Blank AE, Zachskorn C, Baumgarten P, Caspary L, Braczynski AK, Weissenberger J, Bratzke H, Reiß S, et al. MIF receptor CD74 is restricted to microglia/macrophages, associated with a M1-polarized immune milieu and prolonged patient survival in gliomas. Brain Pathol. 2015;25:491–504. doi: 10.1111/bpa.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castro BA, Flanigan P, Jahangiri A, Hoffman D, Chen W, Kuang R, De Lay M, Yagnik G, Wagner JR, Mascharak S, et al. Macrophage migration inhibitory factor downregulation: A novel mechanism of resistance to anti-angiogenic therapy. Oncogene. 2017;36:3749–3759. doi: 10.1038/onc.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verjans E, Noetzel E, Bektas N, Schütz AK, Lue H, Lennartz B, Hartmann A, Dahl E, Bernhagen J. Dual role of macrophage migration inhibitory factor (MIF) in human breast cancer. BMC Cancer. 2009;9:230. doi: 10.1186/1471-2407-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jde Rosado D, Rodriguez-Sosa M. Macrophage migration inhibitory factor (MIF): A key player in protozoan infections. Int J Biol Sci. 2011;7:1239–1256. doi: 10.7150/ijbs.7.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gregory JL, Leech MT, David JR, Yang YH, Dacumos A, Hickey MJ. Reduced leukocyte-endothelial cell interactions in the inflamed microcirculation of macrophage migration inhibitory factor-deficient mice. Arthritis Rheum. 2004;50:3023–3034. doi: 10.1002/art.20470. [DOI] [PubMed] [Google Scholar]
- 26.Onodera S, Nishihira J, Koyama Y, Majima T, Aoki Y, Ichiyama H, Ishibashi T, Minami A. Macrophage migration inhibitory factor up-regulates the expression of interleukin-8 messenger RNA in synovial fibroblasts of rheumatoid arthritis patients: common transcriptional regulatory mechanism between interleukin-8 and interleukin-1beta. Arthritis Rheum. 2004;50:1437–1447. doi: 10.1002/art.20190. [DOI] [PubMed] [Google Scholar]
- 27.Fingerle-Rowson G, Koch P, Bikoff R, Lin X, Metz CN, Dhabhar FS, Meinhardt A, Bucala R. Regulation of macrophage migration inhibitory factor expression by glucocorticoids in vivo. Am J Pathol. 2003;162:47–56. doi: 10.1016/S0002-9440(10)63797-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Ye YL, Li MX, Ye SB, Huang WR, Cai TT, He J, Peng JY, Duan TH, Cui J, et al. CXCL2/MIF-CXCR2 signaling promotes the recruitment of myeloid-derived suppressor cells and is correlated with prognosis in bladder cancer. Oncogene. 2017;36:2095–2104. doi: 10.1038/onc.2016.367. [DOI] [PubMed] [Google Scholar]
- 29.Yaddanapudi K, Putty K, Rendon BE, Lamont GJ, Faughn JD, Satoskar A, Lasnik A, Eaton JW, Mitchell RA. Control of tumor-associated macrophage alternative activation by macrophage migration inhibitory factor. J Immunol. 2013;190:2984–2993. doi: 10.4049/jimmunol.1201650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krockenberger M, Dombrowski Y, Weidler C, Ossadnik M, Hönig A, Häusler S, Voigt H, Becker JC, Leng L, Steinle A, et al. Macrophage migration inhibitory factor contributes to the immune escape of ovarian cancer by down-regulating NKG2D. J Immunol. 2008;180:7338–7348. doi: 10.4049/jimmunol.180.11.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schindler L, Dickerhof N, Hampton MB, Bernhagen J. Post-translational regulation of macrophage migration inhibitory factor: Basis for functional fine-tuning. Redox Biol. 2018;15:135–142. doi: 10.1016/j.redox.2017.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used in this paper are available for download from the cBioportal website (cbioportal.org).