Abstract
Poor prognosis and chemotherapy tolerance are the main obstacles encountered in the treatment of cholangiocarcinoma. Chloroquine (CQ), an antimalarial agent, is able to induce sustained endoplasmic reticulum (ER) stress by functioning as an autophagy inhibitor. The present study indicated that CQ had the ability to induce apoptosis in QBC939 cholangiocarcinoma cells. Furthermore, using western blotting, Hoechst staining and flow cytometry, it was demonstrated that CQ induced the apoptosis of QBC939 cholangiocarcinoma cells. Analysis by a polymerase chain reaction (PCR) array and confirmation via quantitative PCR technology indicated that the expression levels of growth arrest and DNA damage 153 [C/EBP homologous protein (CHOP)], a key molecule involved in ER stress-induced apoptosis, and its downstream death receptors were increased following CQ stimulation. It was considered that the upregulation of CHOP may mediate CQ-induced extrinsic pathways and autophagy-dependent apoptosis; therefore, the role of autophagy in cholangiocarcinoma treatment was elucidated based on the data demonstrating that CQ regulates the ER-autophagy network in tumor cells. Furthermore, it was considered that CQ may become a novel and effective strategy for the treatment of cholangiocarcinoma.
Keywords: chloroquine, cholangiocarcinoma, autophagy, endoplasmic reticulum stress
Introduction
Hepatocellular carcinoma and cholangiocarcinoma are the two most common cancer types in the hepatobiliary system (1). Extrahepatic cholangiocarcinoma is a malignancy that originates from the confluence of right and left hepatic ducts to the distal end of the common bile duct (2–4). Clinically, the treatment options include surgery, radiotherapy and chemotherapy; however, the prognosis remains poor following these treatments (5). Recent studies have indicated that autophagy is able to protect cells by degrading misfolded proteins, which may shed new light on the treatment of cholangiocarcinoma (6,7).
Chloroquine (CQ), which is frequently used clinically as an antimalarial agent, is a classic inhibitor of autophagy that blocks the binding of autophagosomes to lysosomes by altering the acidic environment of lysosomes, resulting in the accumulation of a large number of degraded proteins in cells (8). The inhibition of autophagy by CQ results in the accumulation of a large number of damaged proteins in the cytoplasm and induces endoplasmic reticulum (ER) stress (9). Persistent ER stress eventually results in cell death (10,11). A study confirmed that CQ is able to induce tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis of glioma cells (12). Our previous experiments demonstrated that cisplatin increases the ER stress and autophagy level in HeLa cells, whereas blocking autophagy significantly increased the chemosensitivity of HeLa cells to cisplatin (13); therefore, CQ may serve an antitumor role by regulating the ER-autophagy network, although the specific mechanism is currently unclear.
Growth arrest and DNA damage 153 [C/EBP homologous protein (CHOP)] is considered to be one of the most important molecules mediating ER stress-induced apoptosis (14). Activated CHOP exerts a pro-apoptotic effect by upregulating the expression of B-cell lymphoma-2 (Bcl-2)-associated X (Bax), a pro-apoptotic member of the Bcl-2 family (15,16). Additionally, CHOP promotes the transcription of death receptor 5 (DR5), also known as TRAIL-receptor, and activates the caspase-8-dependent extrinsic apoptosis pathway (17). Cells may be protected from ER stress-induced apoptosis by inhibition of CHOP expression. Furthermore, a previous study indicated that CHOP-deficient mice were more tolerant to tunicamycin (an inducer of endoplasmic reticulum stress) (18); therefore, CHOP may serve a key role in targeted cell apoptosis induced by ER stress. Further studies have demonstrated the involvement of an autophagy-dependent death-inducing signaling complex known as intracellular death-inducing signaling complex (iDISC) in the ER stress-apoptosis signaling pathway. iDISC is associated with the autophagy substrate p62 and initiates apoptosis through activation of caspase-8 (19,20). CHOP may be the core molecule associated with multiple ER stress-induced apoptotic pathways. By investigating the regulatory mechanisms of the autophagy inhibitor CQ underlying ER stress and apoptosis, the role of autophagy-ER stress in antitumor therapy may be further elaborated, potentially indicating the development of novel targets.
In the present study, the autophagy inhibitor CQ was used to regulate the ER-autophagy network, in order to examine whether autophagy provides data on novel treatments of cholangiocarcinoma. Additionally, by elucidating the mechanism by which CQ exerts cytotoxicity against the cholangiocarcinoma cell line QBC939, CQ was considered a novel and effective strategy for the treatment of cholangiocarcinoma.
Materials and methods
Materials
CQ diphosphate salt was purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany) and was dissolved in water to produce a 10 mM stock solution.
Hoechst 33342 and MTT were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). Anti-PARP (sc-7150), anti-Bak (sc-832), anti-Bax (sc-4239), and anti-HSPA5 (sc-13968) antibodies were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Anti-LC3 (3868), anti-CHOP (2895), anti-Caspase-3 (9662), anti-Cleaved-Caspase-3 (9664) and anti-β-actin (3700) were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Anti-p62 (ab109012) was purchased from Abcam (Cambridge, UK). Horseradish peroxidase-conjugated goat anti-rabbit (H+L) immunoglobulin (Ig)G (SA00001-2) and goat anti-mouse (H+L) IgG (SA00001-1) secondary antibodies were purchased from Proteintech (Chicago, IL).
Cell lines and culture
The human cholangiocarcinoma cell line QBC939 was purchased from America Tissue Culture Collection (Manassas, VA, USA) and cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA), 100 U/ml penicillin, and 100 mg/ml streptomycin (complete medium) in a humidified cell culture incubator with an atmosphere containing 5% CO2 at 37°C.
MTT assay
Cellular viability was assessed using MTT assays. Briefly, cells were seeded in 96-well plates at a density of 1×104 cells/well in 100 µl complete medium at 37°C in an atmosphere containing with 5% CO2 and exposed to different doses of CQ (0, 1.5, 3.1, 6.2, 12.5, 25, 50, 100 and 200 µM) for 24 h. At the end of the treatment 10 µl 10 mg/ml MTT reagent (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in PBS was added to each well and incubated at 37°C for a further 4 h. Any formazan crystals that formed were dissolved in 150 µl dimethyl sulfoxide and absorbance was recorded at a wavelength of 490 nm. Each treatment was repeated in five separate wells.
Analysis of apoptosis by Hoechst 3324 staining
Apoptotic morphological alterations in nuclear chromatin were detected by Hoechst 3324 staining. Briefly, QBC939 cells were cultured in 24-well plates and treated with 50 µM CQ for 12 and 24 h, with the untreated group used as a control. Cells were washed with ice-cold PBS three times and fixed with 4% (w/v) paraformaldehyde at 37°C for 15 min. The plates were then incubated with 10 M Hoechst 33258 staining solution for 10 min at 37°C and the cells were visualized under a fluorescence microscope (magnification, ×10) (IX-71; Olympus Corporation, Tokyo, Japan).
Western blot analysis
Subsequent treating with 50 µM CQ for 6, 12 and 24 h, with the untreated group used as a control, cells were collected and lysed with radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China). The lysate was agitated with ultrasound for 20 sec, and placed on ice for 45 min. The lysate was then centrifuged at 3,170 × g for 5 min at 4°C and the precipitate was discarded. Briefly, following quantification of protein in each sample with the Bio-Rad protein reagent (Bio-Rad Laboratories, Inc., Hercules, CA, USA), 40 µg total protein per well was separated by 12% SDS-PAGE and transferred onto an Immun-Blot polyvinylidene fluoride membrane (Bio-Rad Laboratories, Inc.). The membranes were blocked in Tris buffered saline containing 5% (w/v) non-fat dry milk at room temperature for 1 h, and then incubated with specific primary antibodies overnight at 4°C. Following washing with PBS Tween-20 (0.1%, v/v) for three times, membranes were incubated with horseradish-peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at room temperature for 1 h. Membranes were then incubated in enhanced chemiluminescent reagents (Thermo Fisher Scientific, Inc.) and images were captured by Syngene Bio Imaging (Synoptics, Ltd., Cambridge, UK). Densitometric quantitation of bands was performed using Syngene Bio Imaging tools (version 1.2.2.0; Synoptics, Ltd., Cambridge, UK).
Quantitative polymerase chain reaction (qPCR)
Cells were harvested and total RNA was extracted with TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). qPCR was performed using SYBR®-Green Real-Time PCR Master Mix (Takara Biotechnology Co., Ltd., Dalian, China) with relevant primers using the MX300P instrument (Agilent Technologies, Inc., Santa Clara, CA, USA), according to the manufacturer's protocol in the following conditions: 94.0°C for 30 sec, 40 cycles of 94.0°C for 5 sec and 60.0°C for 30 sec. A melting curve was detected between 60 and 94°C to confirm the PCR product of interest. The primer sequences used are as follows: TNF receptor superfamily member 10c (DCR1), 5′-CCTCCTTGCTTCCCATGTAC-3′ (forward), and 5′-CTTCAACACACTGGTATCATC-3′ (reverse); TNF receptor superfamily member 10d (DCR2), 5′-TCACGTCCTTCACGAGTTCC-3′ (forward), and 5′-CTCCGACACCCTTCAGCTTC-3′ (reverse); TNF receptor superfamily member 25 (DR3), 5′-CCTCAATGTGCCAGGCTCTT-3′ (forward), and 5′-ATGACGGCACGCTCACACT-3′ (reverse); TNF receptor superfamily member 10a (DR4); 5′-CTCGCTGTCCACTTTCGTCTC-3′ (forward), and 5′-GTACCAGCTCTGACCACATC-3′ (reverse); TNF receptor superfamily member 10b (DR5), 5′-AAGACCCTTGTGCTCGTTGT-3′ (forward), and 5′-AGGCGGACACAATCCCTCTG-3′ (reverse); tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), 5′-GAGCTGAAGCAGATGCAGGAC-3′ (forward), and 5′-TGACGGAGTTGCCACTTGACT-3′ (reverse); TNF receptor 2 (TNFR2), 5′-GCCCCACCAGATCTGCAACGTG-3′ (forward), and 5′-TGAGGCACCTTGGCTTCTCTC-3′ (reverse); and Fas cell surface death receptor (FAS), 5′-TGTAGATTGTGTGATGAAGG-3′ (forward), and 5′-GATCCCATGTTCACATTTGG-3′ (reverse). Analysis of relative gene expression data using qPCR and the 2−ΔΔCq method (21).
Caspase-8 activity
Caspase-8 activity was measured using the Caspase-Glo 8 assay kit (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocols. Each experiment was performed in quintuplicate.
Flow cytometry
Apoptosis was determined by an Annexin-V and PI kit (cat. no. 556547; BD Biosciences, San Jose, CA, USA). Cells were seeded in 6-well plates overnight at a density of 5×105 cells/well. Following exposure to 50 µM CQ for 12 and 24 h, the untreated group was used as a control, cells were trypsinized at 37°C for 5 min and incubated with PI and Annexin V-FITC for 15 min at 37°C. The samples were analyzed using a BD Accuri C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). All experiments were performed in triplicate.
RT2 Profiler PCR Array System
The Human Unfolded Protein Response RT2 Profiler™ PCR Array (SABiosciences-Qiagen, Hilden, Germany) profiles the expression of 84 key genes involved in unfolded protein accumulation in the ER. Total cellular RNA was extracted from cultured cells according to the manufacturer's instructions. Single-stranded cDNA was obtained by reverse transcription of 1 µg of total RNA using the SABiosciences RT2 First Strand kit. qPCR was performed using Applied Biosystems 7300 Fast with SYBR Green Fluorophore. The reactions were carried out using an RT2 SYBR-Green Mastermix. cDNA was used as template and cycling parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Fluorescence intensities were analyzed using the manufacturer's software, and relative quantification was calculated using the 2−ΔΔCq method. Change of expression of the 84 genes was shown by heat imaging. GAPDH was used as a reference gene.
Immunoprecipitation assay
Cells were lysed in NP40 lysis buffer. Equal amounts of lysates were immunoprecipitated with 2 µg of the p62-linkage Specific Polyubiquitin antibody overnight at 4°C. A total of 25 µl protein A and G agarose (Beyotime Institute of Biotechnology) were used in each sample. Beads were washed with PBS three times with 1 ml each. The eluted proteins were examined by using western blotting as described above.
Statistical analysis
Data are expressed as the mean ± standard deviation. SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA) was used for analysis. All experiments were repeated at least three times. The statistical significance of the difference between groups was assessed using one-way analysis of variance with Least-Significant-Difference post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Autophagy inhibitor CQ induces apoptosis of QBC939 cells
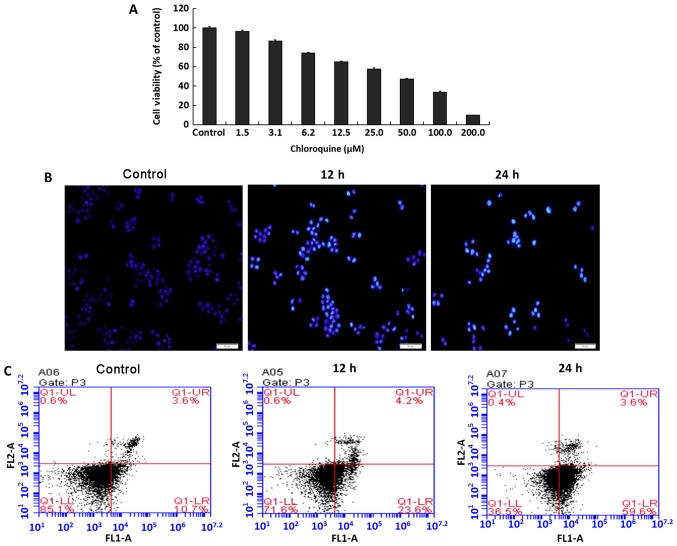
CQ has been frequently used as an autophagy inhibitor. QBC939 cells died following treatment with different doses of CQ (Fig. 1A), and the half-maximal inhibitory concentration (IC50) was 53.01 µM (for the convenience of calculation and statistics, 50 µM was used as the IC50 dose). However, the exact mechanism underlying cell death caused by CQ remains unclear. Staining the cells with Hoechst 33342 indicated typical apoptotic changes, including nuclear fragmentation (Fig. 1B). The data produced by flow cytometry were consistent with the fluorescence results, where the apoptosis rate of cells gradually increased proportional to increasing CQ treatment time (Fig. 1C).
Figure 1.
Autophagy inhibitor CQ induces apoptosis of QBC939 cells. (A) QBC939 cells were treated with various doses of CQ for 24 h and cell viability was determined by an MTT assay. Data are presented as mean ± standard deviation, n=3. (B) Cells were treated with 50 µM CQ for 12 and 24 h and then stained with Hoechst 33258. Cell morphology was observed by fluorescence microscopy (scale bar, 50 µm). The apoptosis ratio of QBC939 cells was detected by flow cytometric analysis after treatment with 50 µM CQ for 12 and 24 h. CQ, chloroquine.
Autophagy inhibitor CQ initiates the iDISC-associated apoptosis pathway
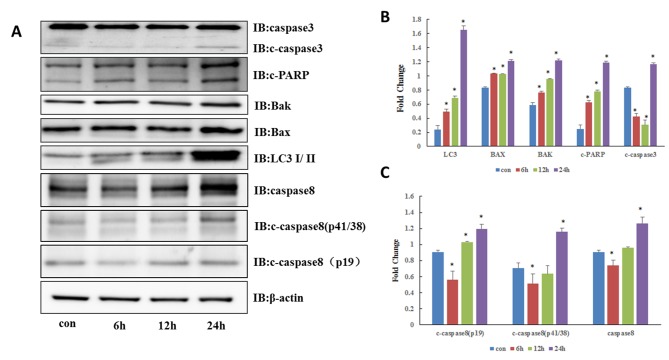
To verify the hypothesis that CQ induces the apoptosis of QBC939 cells, western blotting was performed to detect the expression of apoptosis-associated proteins. As depicted in Fig. 2A and B, the expression of caspase-3 increased following CQ treatment for 6, 12 and 24 h (P<0.05). Accordingly, the level of poly(ADP-ribose) polymerase (PARP) cleavage also increased (P<0.05). Additionally, 50 µM CQ resulted in the notable accumulation of microtubules associated protein light chain 3 II (P<0.05) and blocked the binding of autophagosomes to lysosomes, resulting in the accumulation of autophagosomes in the cytoplasm. CQ also promoted the expression of pro-apoptotic Bcl-2 family proteins Bax and Bcl-2 antagonist/killer (Bak) (P<0.05) and accelerated apoptosis; therefore, CQ results in the apoptosis of QBC939 cells.
Figure 2.
Autophagy inhibitor CQ initiates the intracellular death-inducing signaling complex-associated apoptosis pathway. (A) Expression of apoptosis-associated proteins and caspase-8 was analyzed by western blotting in QBC939 cells treated with CQ (50 µM). (B) Quantification of LC3, Bak, Bax, c-caspase3 and c-PARP. (C) Quantification of c-caspase8 and c-caspase8. *P<0.05 vs. control. C, cleaved; LC3, microtubules associated protein light chain 3; Bax, B-cell lymphoma 2-associated X; Bak, B-cell lymphoma 2 antagonist/killer; PARP, poly(ADP-ribose) polymerase; CQ, chloroquine.
Caspase-8 is a key protease in the induction of the extrinsic pathway of apoptosis (10). Due to the aforementioned results indicating that CQ-induced apoptosis may be associated with the death receptor-mediated extrinsic apoptotic pathway, the effect of CQ on caspase-8 was determined. As depicted in Fig. 2A and C, activation of caspase-8 significantly increased throughout CQ treatment 12 and 24 h (P<0.05) indicating that caspase-8 may be involved in CQ-induced apoptosis.
Autophagy inhibitor CQ induces ER stress in QBC939 cells
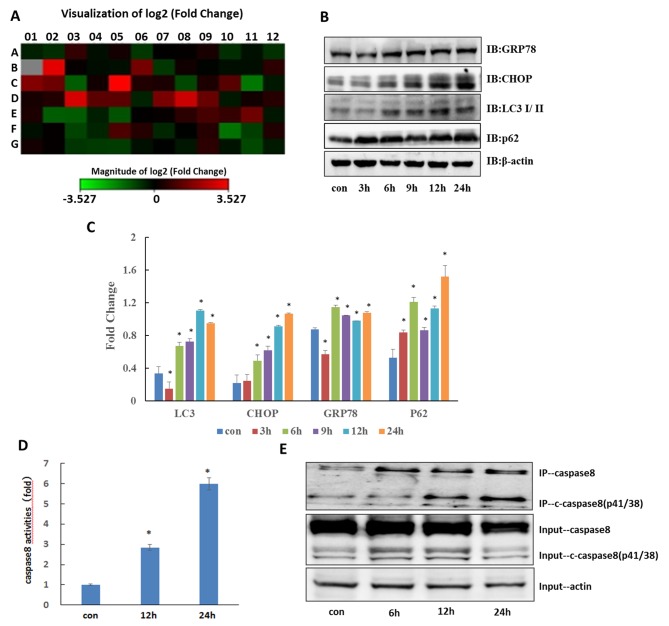
Previous studies have demonstrated that ER stress may be a cause of apoptosis (6,12–14); therefore, in order to examine whether ER stress serves a key role in CQ-induced apoptosis, 84 ER stress-associated genes were screened using a PCR Array (Table I). The results depicted in Fig. 3A demonstrated significant changes in the expression of numerous ER stress-associated genes. Compared with the control, the expression levels of HSPA5 (DNA damage inducible transcript 3 and heat shock protein family A (Hsp70) member 5) were verified to be significantly increased (Table II; P<0.05), and also demonstrated that the protein expression of CHOP and HSPA5 were significantly increased (Fig. 3B and C; P<0.05); therefore, CQ induced ER stress in QBC939 cells. Particularly, CHOP, as a downstream molecule, may serve a key role in the mechanism of CQ-induced apoptosis.
Table I.
84 ER stress-associated genes in PCR array.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | AMFR | ATF4 | ATF6 | ATF6B | ATXN3 | BAX | CALR | CANX | CCT4 | CCT7 | CEBPB | CREB3 |
| B | CREB3L3 | DDIT3 | DERL1 | DERL2 | DNAJB2 | DNAJB9 | DNAJC10 | DNAJC3 | DNAJC4 | EDEM1 | EDEM3 | EIF2A |
| C | EIF2AK3 | ERN1 | ERN2 | ERO1L | ERO1LB | ERP44 | FBXO6 | GANAB | GANC | HERPUD1 | HSPA1B | HSPA1L |
| D | HSPA2 | HSPA4 | HSPA4L | HSPA5 | HSPH1 | HTRA2 | HTRA4 | INSIG1 | INSIG2 | MANF | MAPK10 | MAPK8 |
| E | MAPK9 | MBTPS1 | MBTPS2 | NPLOC4 | NUCB1 | OS9 | PDIA3 | PFDN2 | PFDN5 | PPIA | PPP1R15A | PRKCSH |
| F | RNF139 | RNF5 | RPN1 | SCAP | SEC62 | SEC63 | SEL1L | SELS | SERP1 | SIL1 | SREBF1 | SREBF2 |
| G | SYVN1 | TCP1 | TOR1A | UBE2G2 | UBE2J2 | UBXN4 | UFD1L | UGGT1 | UGGT2 | USP14 | VCP | XBP1 |
| H | ACTB | B2M | GAPDH | HPRT1 | RPLP0 | HGDC | RTC | RTC | RTC | PPC | PPC | PPC |
Figure 3.
Autophagy inhibitor CQ induces ER stress in QBC939 cells. (A) QBC939 cells were treated with 50 µM CQ for 6 h and the expression of 84 ER stress-associated genes was examined using a Human Unfolded Protein Response PCR Array. Changes are presented as heat images where green indicates downregulation and red indicates upregulation. Grey represents no value obtained. Data are derived from three experiments. (B) Cells were treated with CQ and expression of HSPA5, CHOP, LC3 and p62 was analyzed by western blotting. (C) Results of quantitative analysis of Fig. 3B. *P<0.05 vs. control. (D) Cells were analyzed using a caspase-8 activity assay. Data are presented as mean ± standard deviation, n=3. *P<0.05 vs. control. (E) QBC939 cells were treated with 50 e-8 activity assay. Data are presented as mean ± indicates upregulation ubiquitination, translocation anti-caspase 8 antibody. HSPA5, heat shock protein family A (Hsp70) member 5; CHOP, C/EDP homologous protein; LC3, microtubules associated protein light chain 3; CQ, chloroquine.
Table II.
Upregulated genes in the polymerase chain reaction array.
| Gene | CHOP | EIF2AK3 | ERN1 | GADD34 | HSPA5 |
|---|---|---|---|---|---|
| Fold-change | 9.1219 | 3.5365 | 2.8352 | 2.8954 | 2.3864 |
CHOP, C/EDP homologous protein; EIF2AK3, eukaryotic translation initiation factor 2 α kinase 3; ERN1, endoplasmic reticulum to nucleus signaling 1; GADD34, growth arrest and DNA damage inducible 34; HSPA5, heat shock protein family A (Hsp70) member 5.
As an autophagy inhibitor, CQ can induce the accumulation of the autophagy substrate p62. Johnson et al demonstrated that ER stress can induce mTORC1-overactive cells to produce an autophagy-dependent death-inducing signal complex iDISC and promotes the establishment of such a death platform, a protein complex that binds to multiple proteins during apoptosis, by facilitating the binding between p62 and caspase-8 (10). In the present study, CQ promoted the accumulation of p62 protein and activation of caspase-8 in QBC939 cells (Fig. 3B and C; P<0.05); therefore, any interaction between these two factors was investigated. Immunoprecipitation experiments demonstrated that the binding of caspase-8 with p62 significantly increased following CQ treatment (Fig. 3D and E; P<0.05); therefore, it was considered that an autophagy-dependent apoptosis mechanism was involved in CQ-induced apoptosis.
Autophagy inhibitor CQ promotes the high expression of multiple death receptors downstream of CHOP
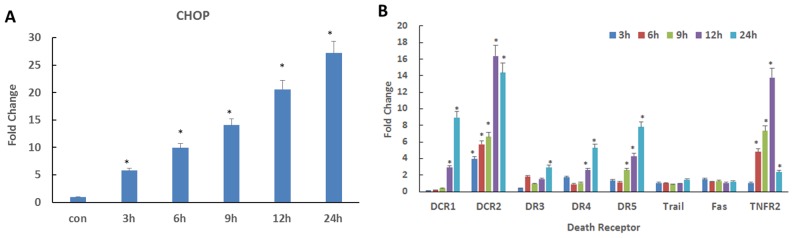
Previous studies demonstrated that CHOP, as a transcription factor, has the ability to bind to a variety of death receptor promoters, promoting their transcriptional expression (22–24). In the present study, CQ induced ER stress in QBC939 cells and promoted the expression of CHOP at mRNA and protein levels (Figs. 3A and 4A; P<0.05). To determine whether the continuous activation of CHOP resulted in the expression of downstream death receptor genes, qPCR analysis of multiple death receptor genes downstream of CHOP was performed. As depicted in Fig. 4B, the gene level expression of DCR1, DCR2, DR3, DR4, DR5 and TNFR2 significantly increased during CQ treatment (P<0.05); however, there was no significant change in the gene expression of TRAIL and FAS. Therefore, CQ may initiate death receptor pathway-dependent apoptosis by inducing CHOP, a key molecule associated with ER stress.
Figure 4.
Autophagy inhibitor CQ promotes high expression of multiple DRs downstream of CHOP. (A) mRNA expression of CHOP in QBC939 cells following treatment with 50 µM CQ *P<0.05 vs. control. (B) Quantitative polymerase chain reaction analysis of the mRNA expression of DR genes in QBC939 cells treated with 50 µM CQ. *P<0.05 vs. control. DR, death receptor; CHOP, C/EDP homologous protein; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; FAS, Fas cell surface death receptor; TNFR2, TNF receptor 2; CQ, chloroquine; DCR1, TNF receptor superfamily member 10c; DCR2, TNF receptor superfamily member 10d. DR3, TNF receptor superfamily member 25; DR4, TNF receptor superfamily member 10a; DR5, TNF receptor superfamily member 10b.
Discussion
CQ was initially used as an antimalarial drug, but previous studies demonstrated that CQ has inhibitory effects on a number of tumor types (25–27). Further investigations into the antitumor mechanism underlying CQ demonstrated that CQ was able to inhibit the growth of lung cancer cells by inducing the expression of prostate apoptosis response-4 and p53 (28). In bile duct carcinoma CCKS1 and HuCCT1 cells, CQ increases cell invasion by suppressing the autophagy-associated gene autophagy and beclin 1 regulator 1 (29). Furthermore, CQ can increase the sensitivity of cholangiocarcinoma QBC939 cells to cisplatin by upregulating the mitochondrial level of reactive oxygen species (30). The present study demonstrated that CQ reduced the viability of cholangiocarcinoma QBC939 cells. The cells exhibited notable apoptotic changes, including nuclear fragmentation and the increased activation of apoptotic protein caspase-3 and its substrate PARP, and markedly increased the apoptotic rate, indicating that the inhibitory effect of CQ on QBC939 cholangiocarcinoma cells was achieved by inducing apoptosis. Apoptosis is generally considered to primarily involve the extrinsic apoptotic pathway and mitochondrial apoptosis pathway (12,31,32). In the present study, the expression of pro-apoptotic Bcl-2 family proteins Bax and Bak was significantly increased following CQ treatment of cholangiocarcinoma QBC939 cells, and the protein expression and activity of caspase-8, which is associated with the extrinsic apoptotic pathway, was also significantly increased. Therefore, CQ may not trigger apoptosis through a single pathway but rather through the co-activation of multiple apoptotic pathways.
Inhibition of autophagy by CQ results in the accumulation of large quantities of protein in the cytoplasm, whcih cannot be degraded and will inevitably result in further cytotoxic effects, including ER stress (31,33). Whilst a low stress level assists in maintaining the normal functions of cells, excessively high ER stress induces cell death. CHOP is an important effector of ER stress (34). Activated CHOP, as a transcription factor, performs numerous functions, including suppressing Bcl-2, stimulating DR5, activating caspases, integrating mitochondrial events and amplifying death signals (35). In models where ER stress induced the apoptosis of colon cancer HCT116 cells and prostate cancer DU145 cells, autophagy may reduce cell vacuoles, alleviate the ER stress and protect cells from death (36–38). In the present study, the inhibition of autophagy in cholangiocarcinoma QBC939 cells by CQ significantly increased the expression of two key proteins in ER stress GRP78 and CHOP, particularly the latter, indicating the occurrence of ER stress. CHOP, as a transcription factor, has the ability to bind to a variety of death receptor promoters and thus directly induce cell death.
The expression of classic death receptor genes including DCR1, DCR2, DCR3, DCR4, DCR5 and TNFR2 was significantly increased in the present experimental model. Thus, CQ may initiate the extrinsic apoptosis pathway. CQ blocks autophagy during autolysosome formation, resulting in the accumulation of the p62 protein. p62 and caspase-8 co-existed in QBC939 cells and their levels increased following CQ treatment. One previous study demonstrated that tunicamycin and thapsigargin (two classic inducers of endoplasmic reticulum stress) are able to induce ER stress-autophagy-dependent activation of caspase-8 in cells deficient for caspase-9 or Bax/Bak (39). CQ treatment results in an increase in the remaining autophagosomes in the cytoplasm. These autophagosomes include a variety of autophagy-associated proteins, including p62, which form iDISC, or other known stress-inducible complexes or stressors, with caspase-8 (40,41). It is speculated that CQ induces the establishment of a novel ER stress-associated death platform in QBC939 cells, which results in autophagy-dependent apoptosis.
In the present study, the cytotoxic mechanism underlying the autophagy inhibitor CQ in cholangiocarcinoma QBC939 cells was elucidated through in vitro experiments. ER stress and autophagy inhibition occur during the stimulation of cells by CQ, resulting in intrinsic, extrinsic and autophagy-dependent apoptosis. By investigating the role of CQ in the promotion of apoptosis through regulation of the ER-autophagy network, novel insights into the mechanism underlying CQ in antitumor therapy were produced. Numerous studies have indicated that conventional agents used to treat other diseases have antitumor effects, including blocking autophagy, a process termed drug re-positioning (42–45). The present study indicated that CQ may function synergistically with traditional chemotherapeutics to increase apoptosis of cancer cells by autophagy inhibition; however, there are a number of limitations of the present study. For example, only one cholangiocarcinoma cell line was used in the present study, and as a result it lacks persuasiveness. Additionally, the association between the upregulation of p53 and CHOP was not investigated. Future studies will need to use other cell lines in order to verify the mechanism underlying CQ and to justify the clinical application of CQ in the chemotherapy of cholangiocarcinoma. Additionally, the role of p53 in the CQ-mediated increase of CHOP expression and apoptosis of human cholangiocarcinoma cells will be investigated, and experiments involving RNA interference-mediated reduction in CHOP expression will be conducted in order to verify whether the effects of CQ treatment are mediated by CHOP. Furthermore, the aim of future studies will be to determine how the dose used to treat QBC939 cells compares with the physiological dose administered to humans. Due to there being no exact drug dose used clinically at present, this is important for future studies to investigate.
Acknowledgments
Not applicable.
Funding
National Natural Science Foundation of China (nos. 81672948, 81772794, 81572927 and 81501982). Jilin Provincial Research Foundation for the Development of Science and Technology Projects (nos. 20170623021TC and 20160414005GH). Jilin Provincial Education Department Foundation for ‘The 13th five-year’ Science and Technology Project (nos. JJKH20170825KJ and JJKH20170834KJ). Jilin University Bethune Plan B Projects (no. 2015222).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SLK, LYP and LYH made substantial contributions to design. XYN, YXY, LJL, WY, GR, ZJJ and ZLC were involved in performing the experiments, acquiring and analyzing associated articles and data. JBX was a major contributor in designing research program, analyzing data and writing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Likhitrattanapisal S, Tipanee J, Janvilisri T. Meta-analysis of gene expression profiles identifies differential biomarkers for hepatocellular carcinoma and cholangiocarcinoma. Tumour Biol. 2016;37:12755–12766. doi: 10.1007/s13277-016-5186-8. [DOI] [PubMed] [Google Scholar]
- 2.Blechacz B, Komuta M, Roskams T, Gores GJ. Clinical diagnosis and staging of cholangiocarcinoma. Nat Rev Gastroenterol Hepatol. 2011;8:512–522. doi: 10.1038/nrgastro.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliveira IS, Kilcoyne A, Everett JM, Mino-Kenudson M, Harisinghani MG, Ganesan K. Cholangiocarcinoma: Classification, diagnosis, staging, imaging features, and management. Abdom Radiol (NY) 2017;42:1637–1649. doi: 10.1007/s00261-017-1094-7. [DOI] [PubMed] [Google Scholar]
- 4.Matull WR, Khan SA, Pereira SP. Re: Impact of classification of hilar cholangiocarcinomas (Klatskin tumors) on the incidence of intra- and extrahepatic cholangiocarcinoma in the United States. J Natl Cancer Inst. 2007;99:407–408. doi: 10.1093/jnci/djk069. [DOI] [PubMed] [Google Scholar]
- 5.Roskams T. Liver stem cells and their implication in hepatocellular and cholangiocarcinoma. Oncogene. 2006;25:3818–3822. doi: 10.1038/sj.onc.1209558. [DOI] [PubMed] [Google Scholar]
- 6.Carew JS, Medina EC, Esquivel JA, II, Mahalingam D, Swords R, Kelly K, Zhang H, Huang P, Mita AC, Mita MM, et al. Autophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulation. J Cell Mol Med. 2010;14:2448–2459. doi: 10.1111/j.1582-4934.2009.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thorburn A, Thamm DH, Gustafson DL. Autophagy and cancer therapy. Mol Pharmacol. 2014;85:830–838. doi: 10.1124/mol.114.091850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mushtaque M, Shahjahan Reemergence of chloroquine (CQ) analogs as multi-targeting antimalarial agents: A review. Eur J Med Chem. 2015;90:280–295. doi: 10.1016/j.ejmech.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 9.Fernández A, Ordóñez R, Reiter RJ, González-Gallego J, Mauriz JL. Melatonin and endoplasmic reticulum stress: Relation to autophagy and apoptosis. J Pineal Res. 2015;59:292–307. doi: 10.1111/jpi.12264. [DOI] [PubMed] [Google Scholar]
- 10.Johnson CE, Hunt DK, Wiltshire M, Herbert TP, Sampson JR, Errington RJ, Davies DM, Tee AR. Endoplasmic reticulum stress and cell death in mTORC1-overactive cells is induced by nelfinavir andenhanced by chloroquine. Mol Oncol. 2015;9:675–688. doi: 10.1016/j.molonc.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Høyer-Hansen M, Jäättelä M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 12.Park EJ, Min KJ, Choi KS, Kubatka P, Kruzliak P, Kim DE, Kwon TK. Chloroquine enhances TRAIL-mediated apoptosis through up-regulation of DR5 by stabilization of mRNA and protein in cancer cells. Sci Rep. 2016;6:22921. doi: 10.1038/srep22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Li D, Zeng L, Wang C, Zhang L, Wang Y, Yu Y, Liu S, Li Z. Proteasome inhibitor lactacystin enhances cisplatin cytotoxicity by increasing endoplasmic reticulum stress-associated apoptosis in HeLa cells. Mol Med Rep. 2015;11:189–195. doi: 10.3892/mmr.2014.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiribau CB, Gaccioli F, Huang CC, Yuan CL, Hatzoglou M. Molecular symbiosis of CHOP and C/EBP beta isoform LIP contributes to endoplasmic reticulum stress-induced apoptosi. Mol Cell Biol. 2010;30:3722–3731. doi: 10.1128/MCB.01507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teske BF, Fusakio ME, Zhou D, Shan J, McClintick JN, Kilberg MS, Wek RC. CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol Biol Cell. 2013;24:2477–2490. doi: 10.1091/mbc.e13-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moon DO, Park SY, Choi YH, Ahn JS, Kim GY. Guggulsterone sensitizes hepatoma cells to TRAIL-induced apoptosis through the induction of CHOP-dependent DR5: Involvement of ROS-dependent ER-stress. Biochem Pharmacol. 2011;82:1641–1650. doi: 10.1016/j.bcp.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 18.Chen CM, Wu CT, Chiang CK, Liao BW, Liu SH. C/EBP homologous protein (CHOP) deficiency aggravates hippocampal cell apoptosis and impairs memory performance. PLoS One. 2012;7:e40801. doi: 10.1371/journal.pone.0040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iurlaro R, Muñoz-Pinedo C. Cell death induced by endoplasmic reticulum stress. FEBS J. 2016;283:2640–2652. doi: 10.1111/febs.13598. [DOI] [PubMed] [Google Scholar]
- 20.Young MM, Takahashi Y, Khan O, Park S, Hori T, Yun J, Sharma AK, Amin S, Hu CD, Zhang J, et al. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem. 2012;287:12455–12468. doi: 10.1074/jbc.M111.309104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Wijdeven RH, Pang B, Assaraf YG, Neefjes J. Old drugs, novel ways out: Drug resistance toward cytotoxic chemotherapeutics. Drug Resist Updat. 2016;28:65–81. doi: 10.1016/j.drup.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 23.He H, Ke R, Lin H, Ying Y, Liu D, Luo Z. Metformin, an old drug, brings a new era to cancer therapy. Cancer J. 2015;21:70–74. doi: 10.1097/PPO.0000000000000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mudduluru G, Walther W, Kobelt D, Dahlmann M, Treese C, Assaraf YG, Stein U. Repositioning of drugs for intervention in tumor progression and metastasis: Old drugs for new targets. Drug Resist Updat. 2016;26:10–27. doi: 10.1016/j.drup.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Sung B, Prasad S, Ravindran J, Yadav VR, Aggarwal BB. Capsazepine, a TRPV1 antagonist, sensitizes colorectal cancer cells to apoptosis by TRAIL through ROS-JNK-CHOP-mediated upregulation of death receptors. Free Radic Biol Med. 2012;53:1977–1987. doi: 10.1016/j.freeradbiomed.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He L, Jang JH, Choi HG, Lee SM, Nan MH, Jeong SJ, Dong Z, Kwon YT, Lee KS, Lee KW, et al. Oligomycin A enhances apoptotic effect of TRAIL through CHOP-mediated death receptor 5 expression. Mol Carcinog. 2013;52:85–93. doi: 10.1002/mc.21831. [DOI] [PubMed] [Google Scholar]
- 27.Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P, Ashkenazi A. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345:98–101. doi: 10.1126/science.1254312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burikhanov R, Hebbar N, Noothi SK, Shukla N, Sledziona J, Araujo N, Kudrimoti M, Wang QJ, Watt DS, Welch DR, et al. Chloroquine-inducible par-4 secretion is essential for tumor cell apoptosis and inhibition of metastasis. Cell Rep. 2017;18:508–519. doi: 10.1016/j.celrep.2016.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nitta T, Sato Y, Ren XS, Harada K, Sasaki M, Hirano S, Nakanuma Y. Autophagy may promote carcinoma cell invasion and correlate with poor prognosis in cholangiocarcinoma. Int J Clin Exp Pathol. 2014;7:4913–4921. [PMC free article] [PubMed] [Google Scholar]
- 30.Qu X, Sheng J, Shen L, Su J, Xu Y, Xie Q, Wu Y, Zhang X, Sun L. Autophagy inhibitor chloroquine increases sensitivity to cisplatin in QBC939 cholangiocarcinoma cells by mitochondrial ROS. PLoS One. 2017;12:e0173712. doi: 10.1371/journal.pone.0173712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura T, Takabatake Y, Takahashi A, Isaka Y. Chloroquine in cancer therapy: A double-edged sword of autophagy. Cancer Res. 2013;73:3–7. doi: 10.1158/0008-5472.CAN-12-2464. [DOI] [PubMed] [Google Scholar]
- 32.Vlahopoulos S, Critselis E, Voutsas IF, Perez SA, Moschovi M, Baxevanis CN, Chrousos GP. New use for old drugs? Prospective targets of chloroquines in cancer therapy. Curr Drug Targets. 2014;15:843–851. doi: 10.2174/1389450115666140714121514. [DOI] [PubMed] [Google Scholar]
- 33.Rashid HO, Yadav RK, Kim HR, Chae HJ. ER stress: Autophagy induction, inhibition and selection. Autophagy. 2015;11:1956–1977. doi: 10.1080/15548627.2015.1091141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malhi H, Kaufman RJ. Endoplasmic reticulum stress in liver disease. J Hepatol. 2011;54:795–809. doi: 10.1016/j.jhep.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trivedi R, Maurya R, Mishra DP. Medicarpin, a legume phytoalexin sensitizes myeloid leukemia cells to TRAIL-induced apoptosis through the induction of DR5 and activation of the ROS-JNK-CHOP pathway. Cell Death Dis. 2014;5:e1465. doi: 10.1038/cddis.2014.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukhopadhyay S, Panda PK, Sinha N, Das DN, Bhutia SK. Autophagy and apoptosis: Where do they meet? Apoptosis. 2014;19:555–566. doi: 10.1007/s10495-014-0967-2. [DOI] [PubMed] [Google Scholar]
- 37.Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: The interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gordy C, He YW. The crosstalk between autophagy and apoptosis: Where does this lead? Protein Cell. 2012;3:17–27. doi: 10.1007/s13238-011-1127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang S, Okamoto K, Yu C, Sinicrope FA. p62/sequestosome-1 up-regulation promotes ABT-263-induced caspase-8 aggregation/activation on the autophagosome. J Biol Chem. 2013;288:33654–33666. doi: 10.1074/jbc.M113.518134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomar D, Prajapati P, Sripada L, Singh K, Singh R, Singh AK, Singh R. TRIM13 regulates caspase-8 ubiquitination, translocation to autophagosomes and activation during ER stress induced cell death. Biochim Biophys Acta. 2013;1833:3134–3144. doi: 10.1016/j.bbamcr.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 41.Kantari C, Walczak H. Caspase-8 and bid: Caught in the act between death receptors and mitochondria. Biochim Biophys Acta. 2011;1813:558–563. doi: 10.1016/j.bbamcr.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 42.Huang H, Xie H, Pan Y, Zheng K, Xia Y, Chen W. Plumbagin triggers ER stress-mediated apoptosis in prostate cancer cells via induction of ROS. Cell Physiol Biochem. 2018;45:267–280. doi: 10.1159/000486773. [DOI] [PubMed] [Google Scholar]
- 43.Mathur A, Elmageed Abd ZY, Liu X, Kostochka ML, Zhang H, Abdel-Mageed AB, Mondal D. Subverting ER-stress towards apoptosis by nelfinavir and curcumin coexposure augments docetaxel efficacy in castration resistant prostate cancer cells. PLoS One. 2014;9:e103109. doi: 10.1371/journal.pone.0103109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khan I, Paul S, Jakhar R, Bhardwaj M, Han J, Kang SC. Novel quercetin derivative TEF induces ER stress and mitochondria-mediated apoptosis in human colon cancer HCT-116 cells. Biomed Pharmacother. 2016;84:789–799. doi: 10.1016/j.biopha.2016.09.094. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida GJ. Therapeutic strategies of drug repositioning targeting autophagy to induce cancer cell death: From pathophysiology to treatment. J Hematol Oncol. 2017;10:67. doi: 10.1186/s13045-017-0436-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.