Abstract
The present study aimed to investigate microRNA-376a (miR-376a) expression in lymphoma, and to investigate the effect of miR-376a on cell proliferation and apoptosis at cytological and molecular levels. The expression of miR-376a in lymphoma issue and cells was detected by reverse transcription quantitative polymerase chain reaction (RT-qPCR), the expression of forkhead box protein P2 (FOXP2) was detected by RT-qPCR and western blot analysis, and the effect of miR-376a on cell proliferation and apoptosis were studied by an MTT assay and flow cytometry, respectively. Additionally, the expression levels of cyclin D2, cyclin A, cyclin B, apoptosis-associated proteins B-cell lymphoma 2 (Bcl-2) and Bcl-2-associated X protein (Bax) were detected by western blot analysis. Furthermore, the target of miR-376a was predicted and clarified using a dual-luciferase reporter assay. The expression of miR-376a was downregulated and FOXP2 was upregulated in lymphoma tissues and cells. miR-376a overexpression inhibited lymphoma cell proliferation and induced apoptosis by regulating the expression levels of cyclin D2, cyclin A, Bax and Bcl-2. The dual-luciferase reporter assay demonstrated that FOXP2 was a target of miR-376a. miR-376a overexpression induced apoptosis by targeting FOXP2. Overexpression of miR-376a inhibited cell proliferation and induced apoptosis by targeting FOXP2 in lymphoma. Therefore, miR-376a and FOXP2 have the potential for use as biomarkers of lymphoma.
Keywords: lymphoma, microRNA-376a, forkhead box protein P2, cell proliferation, cell apoptosis
Introduction
Lymphoma is a type of blood cell tumor that arises from developing lymphatic cells (1); it represents 3–4% of all types of cancer, making it the seventh most common type of cancer (2). The disease is characterized by painless enlarged lymph nodes (3). There are dozens of subtypes of lymphoma (4); the two primary categories of lymphoma are Hodgkin's lymphoma and non-Hodgkin's lymphoma, with ~90% of lymphoma cases non-Hodgkin lymphoma (5). Recently, in the USA, the 5-year survival rate for non-Hodgkin's lymphoma is 69%, and for the Hodgkin's lymphoma subtypes it is 85% (6). In 2012, there were 566,000 cases of lymphoma and 305,000 incidences of mortality worldwide (7). In order to develop therapies to reduce these numbers, it is important to investigate effective therapeutic strategies for this disease.
MicroRNAs (miRNAs/miRs) are short non-coding RNAs, which have previously been demonstrated to serve an important role in cell proliferation and differentiation (8,9). Notably, miRNAs have important functions in lymphoma biology. For example, miR-155 and miR-34a are crucial to the development of B-cell lymphoma via participation in numerous signaling pathways (nuclear factor κB, phosphoinositide 3-kinase/protein kinase B and transforming growth factor β) fundamental to B-cell development (10). miR-376a has been demonstrated to serve crucial roles in various diseases, including hepatocellular carcinoma (11) and melanoma (12); however, only a small number of studies have assessed its role in lymphoma (13,14). At present, the volume of data concerning the functional role of miRNAs in lymphoma is increasing steadily (15). A previous study by Arribas et al (13) analyzed the miRNA profiles of 31 splenic marginal zone lymphoma cases, and identified that miR-376a was abnormally expressed in lymphoma. However, the exact mechanism of the effect of miR-376a on lymphoma remains unclear.
The present study aimed to investigate miR-376a expression in lymphoma and the associations between aberrantly expressed miR-376a and cell proliferation or apoptosis at the cytological and molecular level. The present study may provide novel insights into the molecular mechanisms underlying lymphoma development.
Patients and methods
Patients and samples
Between January 2015 and February 2016, a total of 29 patients (20 males and 9 females, age range, 24–44 years, mean age, 33.6±10.4 years) who were diagnosed with lymphoma in the Cancer Hospital affiliated to Zhengzhou University, Henan Cancer Hospital (Zhengzhou, China) were enrolled in the present study. Among the 29 patients, 5 cases were Hodgkin's lymphoma and 24 cases were non-Hodgkin's lymphoma. The diagnosis of lymphoma was pathologically defined according to World Health Organization classification of lymphoid neoplasms (16). The pathological lymphoid tissues were collected from patients using resection, extracted and immediately snap-frozen in liquid nitrogen. Additionally, 12 patients (8 males and 4 females, age range, 21–42 years, mean age, 30.8±9.6 years) with benign hyperplastic lymphadenitis were enrolled as controls. All patients provided written informed consent and the present study was approved by the Protection of Human Ethics Committee of the Cancer Hospital affiliated to Zhengzhou University, Henan Cancer Hospital.
Cell culture and transfection
The human mantle cell lymphoma (MCL) JeKo-1 cell line (CRL-3006™) and normal lymphocytes (CRL-2319™), which were obtained from the American Type Culture Collection (Manassas, VA, USA), were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 2 mM L-Glutamine, 50 units penicillin and 50 µg/ml streptomycin (Sigma-Aldrich; Merck KGaA).
Cell transfections were performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. miR-376a mimic, inhibitor and scramble, and small interfering RNA (siRNA) targeting Forkhead box P2 (FOXP2) were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). Cells were transfected with 50 nM miR-376a mimic, 150 nM miR-376a inhibitor and 4 µg si-FOXP2 for 24 h prior to subsequent experimentation. The primer sequences were as follows: miR-376a mimic sense, 5′-GUAGAUUCUCCUUCUAUGAGUA-3′ and anti-sense, 5′-CUCAUAGAAGGAGAAUCUACUU-3′; miR-376a inhibitor 5′-UACUCAUAGAAGGAGAAUCUAC-3′; si-FOXP2 sense, 5′-CGACAGAGACAAUAAGCAACA-3′ and antisense, 5′-UUGCUUAUUGUCUCUGUCGCA-3′.
Cell proliferation assay
Cell proliferation ability was assessed using an MTT assay. At 24 h after transfection, cells were cultured in RPMI-1640 medium supplemented with 10% FBS. JeKo-1 cells were seeded into the 96-well plates at a density of 5×103 cells/well. At various time points (24, 36, 48, 72 and 96 h), cells were centrifuged at 6,000 × g for 5 min at 4°C, and the supernatant was removed. Next, 20 µl 5 mg/ml MTT solution was added into the culture medium and cultured for 4 h at room temperature. Finally, 150 µl dimethyl sulfoxide was added to each well to dissolve the crystals. Following 10 min, the absorbance of cells in each well was observed at 570 nm using an absorption spectrophotometer (Olympus Corporation, Tokyo, Japan). All experiments were conducted independently in triplicate.
Clonogenic assay
A clonogenic assay was performed as described previously (14), with minor modifications. Briefly, 24 h following transfection, JeKo-1 cells were plated in triplicate into the 60-mm tissue culture dishes at a density of 100 cells/dish. After 14 days of culture in RPMI-1640 containing 10% FBS, cells were fixed with 90% methanol for 15 min at room temperature, and stained with the Diff-Quick cell staining kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 20–40 sec at room temperature. Colonies were counted under an inverted microscope (IX83; Olympus Corporation; magnification, ×200), and colonies consisting of at least 30 cells were counted as a single colony.
Apoptosis assay
Cell apoptosis was detected by flow cytometry. The number of apoptotic JeKo-1 cells was determined using an Annexin V-fluorescein isothiocyanate (FITC) cell apoptosis kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. A total of 24 h following cell transfection, JeKo-1 cells were cultured in serum-free RPMI-1640 medium. The cells were harvested and washed 3 times using PBS buffer (pH 7.4), and then resuspended in the staining buffer. Subsequently, 5 µl Annexin-V-FITC and 5 µl propidium iodide (PI) were added into the cells and incubated at room temperature for 10 min. The mixtures were analyzed using a FACScan flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Annexin V-positive and PI-negative cells were considered to be apoptotic cells. The apoptotic cells were analyzed using Cellquest software (version 3.0; BD Biosciences).
Target prediction
Putative target genes of miR-376a were predicted by bioinformatics analysis using TargetScan (version 7; http://www.targetscan.org/vert_71/).
Luciferase reporter analysis
Luciferase reporter plasmid, psiCHECK2, containing the FOXP2 3′-untranslated region (UTR), miR-376a mimic and scramble were produced and synthesized by Sangon Biotech Co., Ltd. Cell transfections were performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. At 48 h after transfection, luciferase activities were measured using the Dual-Luciferase Reporter Assay system (Promega Corporation, Madison, WI, USA). The relative reporter activity was normalized to Renilla luciferase activity.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and JeKo-1 cells using TRIzol® (Life Technologies; Thermo Fisher Scientific, Inc.) and treated with RNase-free DNase I (Promega Corporation). The concentration and purity of isolated RNA were measured with SMA400 UV–VIS (Merinton Instrument, Ltd., Shanghai, China). cDNA was then generated with the PrimerScript 1st Strand cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.). qPCR was performed using a standard SYBR Green I kit (Takara Biotechnology Co., Ltd., Dalian, China). PCR was performed under the following conditions: 1 predenaturation cycle for 1 min at 95°C, 36 cycles at 95°C for 15 sec, 60°C for 30 sec and 72°C for 1 min and a final extension at 72°C for 5 min. U6 and GAPDH were used as the endogenous controls for the normalization of the amount of miRNA and total mRNA. The expression levels were calculated using the 2−ΔΔCt method (17). Sequences of all primers used are summarized in Table I.
Table I.
Primers used for target amplification.
| Gene name | Forward primer, 5′-3′ | Reverse primer, 5′-3′ |
|---|---|---|
| FOXP2 | GAAGACAATGGCATTAAACATGGAGG | GAATAAAGCTCATGAGATTTACCTGTC |
| Cyclin A | GACTGGTTAGTTGAAGTA | CTCCATTCTCAGAACTTG |
| Cyclin B | TAGTTCCATCAGGTATTTGGC | TAGTTCCATCAGGTATTTGGC |
| Cyclin D2 | CTGTGTGCCACCGACTTTAAGTT | GATGGCTGCTCCCACACTTC |
| Bax | CTGAGCTGACCTTGGAGC | GACTCCAGCCACAAAGATG |
| Bcl-2 | CTGGTGGACAACATCGCTCTG | GGTCTGCTGACCTCACTTGTG |
| GAPDH | TGGACTCCACGACGTACTCAG | CGGGAAGCTTGTCATCAATGGAA |
| U6 | GCTTCGGCAGCACATATACTAA | AACGCTTCACGAATTTGCGT |
FOXP2, forkhead box protein P2; Bcl-2, B cell lymphoma 2; Bax, Bcl-2-associated X protein.
Western blot analysis
The protein expression levels of FOXP2, cyclin A, cyclin B, cyclin D2 and GAPDH were determined in total cell extracts from transfected JeKo-1 cells. Total proteins were extracted using radioimmunoprecipitation assay buffer (Sangon Biotech Co., Ltd.) and then the cell lysates were centrifuged at 6,000 × g for 10 min at 4°C and separated using 10% SDS-PAGE. Protein concentration was determined using the bicinchoninic acid assay (Pierce; Thermo Fisher Scientific, Inc.). Then, proteins (20 µg/per lane) were transferred onto the polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA) and probed with the following primary antibodies; FOXP2 (ab16046; dilution 1:1,000), Bcl2 (ab32124; dilution 1:1,000), Bax (ab32503; dilution 1:1,000), cyclin A (ab181591; dilution 1:1,000), cyclin B (ab32053; dilution 1:1,000) and cyclin D2 (ab207604; dilution 1:1,000) and GAPDH (ab8245; dilution 1:5,000) at 4°C overnight. All primary antibodies were purchased fro Abcam (Cambridge, UK). Protein bands were blocked using 5% non-fat dry milk in Tris buffered saline (TBS) supplemented with 0.05% TBS and Tween for 1 h at room temperature. Membranes were then incubated with peroxidase-conjugated goat anti-rabbit (ab6721; dilution 1:5,000; Abcam) for 1 h at room temperature. The immunoreactive protein bands were developed using the enhanced chemiluminescence kit (Thermo Fisher Scientific, Inc.).
Statistical analysis
Statistical analyses were performed using SPSS 19.0 statistical software (IBM Corp., Armonk, NY, USA). Data were expressed as the mean ± standard deviation from 3 independent experiments. One way analysis of variance followed by post hoc Tukey's test was used to calculate the difference >2 groups. Student's t-test was used to calculate the difference between two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-376a is downregulated and FOXP2 is upregulated in lymphoma tissues and cells
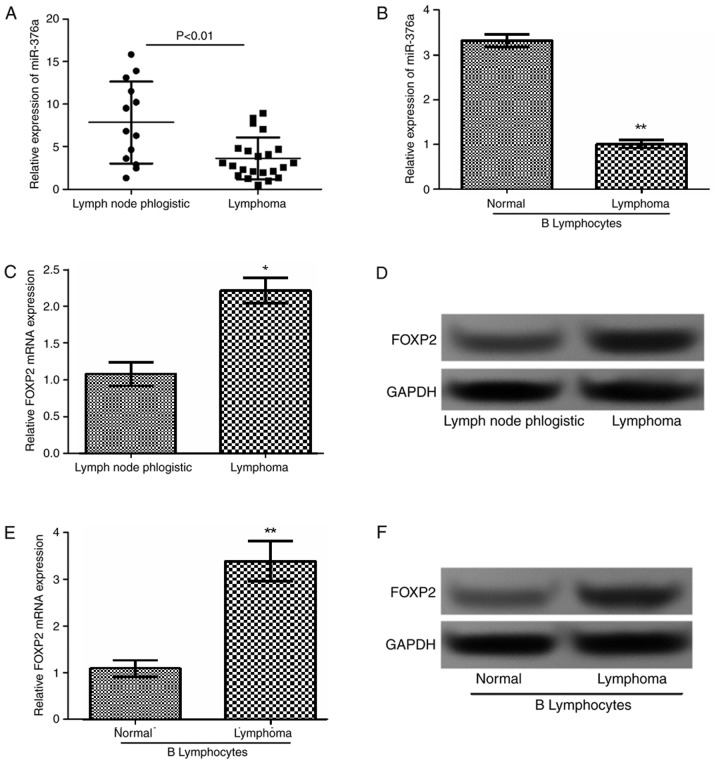
The expression levels of miR-376a and FOXP2 in lymphoma tissues were measured in 29 patients with lymphoma and in lymphoid tissues from 12 patients with benign hyperplastic lymphadenitis by RT-qPCR and western blot analysis. Additionally, miR-376a expression levels were also detected in lymphoma B cells and normal B lymphocytes. As demonstrated in Fig. 1A and B, miR-376a was downregulated in lymphoma tissues and cells compared with the corresponding controls (P<0.01). Additionally, as revealed in Fig. 1C-F, FOXP2 expression in lymphoma tissues and cells was significantly increased compared with that in normal tissues (P<0.01) and cells (P<0.05).
Figure 1.
Expression of miR-376a in lymphoma tissue and cells. The relative expression levels of miR-376a in (A) lymphoma tissues and (B) cell lines detected by RT-qPCR. The relative expression levels of FOXP2 in lymphoma tissues detected by (C) RT-qPCR and (D) western blot analysis. The relative expression levels of FOXP2 in lymphoma cells detected by (E) RT-qPCR and (F) western blot analysis. Data were expressed as mean ± standard deviation. *P<0.05 and **P<0.01 vs. control. miR-376a, microRNA-376a; RT-qPCR, reverse transcription quantitative polymerase chain reaction; FOXP2, forkhead box protein P2.
miR-376a overexpression reduces cell proliferation in B lymphocytes
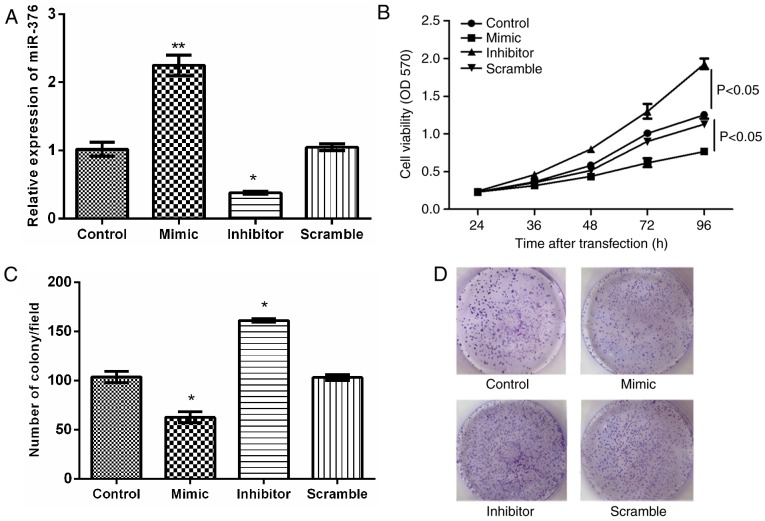
The associations between miR-376a and lymphoma were analyzed at a cytological level. The MCL JeKo-1 cell line was transfected with miR-376a mimic, inhibitor and scramble plasmids. The expression levels of miR-376a in JeKo-1 cells following transfection are depicted in Fig. 2A. To validate the effect of miR-376a on JeKo-1 cell proliferation, MTT and clonogenic assays were used. As indicated in Fig. 2B-D, miR-376a overexpression significantly inhibited cell proliferation and colony formation compared with the control group (P<0.05).
Figure 2.
Effects of miR-376a on cell proliferation. (A) The relative expression level of miR-376a following cell transfection. (B) Cell viability assay following cell transfection using an MTT assay. (C) Clonogenic assay demonstrating the number of colonies/field in each group following cell transfection. (D) Representative images of clonogenic cells. Magnification, ×200. Data were expressed as mean ± standard deviation. *P<0.05 and **P<0.01 vs. control. miR, microRNA; OD, optical density.
miR-376a overexpression induces cell growth arrest by downregulating cyclin D2 and cyclin A
A previous study suggested that cyclin overexpression was a cause of excessive cell proliferation (18). Therefore, the expression levels of cyclin D2, cyclin A and cyclin B were investigated in transfected JeKo-1 cells. As indicated in Fig. 3, the expression levels of cyclin D2 and cyclin A were significantly increased in the miR-376a inhibitor group compared to the scramble group (P<0.05), and significantly decreased in the miR-376a mimic group compared with the control group (P<0.05 and P<0.01, respectively). For the expression of cyclin B, no significant difference was identified among the four groups. This result indicated that miR-376a overexpression could inhibit the proliferation of lymphoma B cells by downregulating expression of cyclin D2 and cyclin A.
Figure 3.

Expression of miR-376a expression on cell cycle-associated mRNA/protein expression. (A) The relative mRNA expression levels of cyclin A, cyclin B and cyclin D2 following cell transfection detected by reverse transcription-quantitative polymerase chain reaction. (B) The protein expression of cyclin A, cyclin B and cyclin D2 following cell transfection, detected by western blot. Data were expressed as mean ± standard deviation. *P<0.05 and **P<0.01 vs. control. miR-376a, microRNA-376a.
miR-376a overexpression induces apoptosis in B lymphocytes
To investigate the effect of miR-376a on apoptosis, flow cytometry was performed. Additionally, the expression levels of apoptosis-associated proteins, including Bcl-2 and Bax in JeKo-1 and transfected JeKo-1 cells were detected by RT-qPCR and western blot analysis. The result of flow cytometry analysis indicated that the overexpression of miR-376a markedly increased the percentage of apoptotic cells, compared with the control group, whereas miR-376a suppression significantly inhibited the percentage of apoptotic cells, compared with the scramble group (both P<0.05; Fig. 4A and B). The expression levels of Bax and Bcl-2 are depicted in Fig. 4C and D. The expression of Bax increased significantly, whereas that of Bcl-2 decreased significantly in the miR-376a mimic group compared with the control group (P<0.01), additionally indicating the pro-apoptotic effect of miR-376a overexpression.
Figure 4.
Effects of miR-376a expression on cell apoptosis. (A) Percentage of apoptotic cells. (B) Representative images of the apoptosis assay following cell transfection using flow cytometry. The relative expression levels of Bax and Bcl-2 following cell transfection detected by (C) reverse transcription-quantitative polymerase chain reaction and (D) western blot analysis. Data were expressed as mean ± standard deviation. *P<0.05 and **P<0.01 vs. control. miR-376a, microRNA-376a; FITC, fluorescein isothiocyanate; PI, propidium iodide; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein.
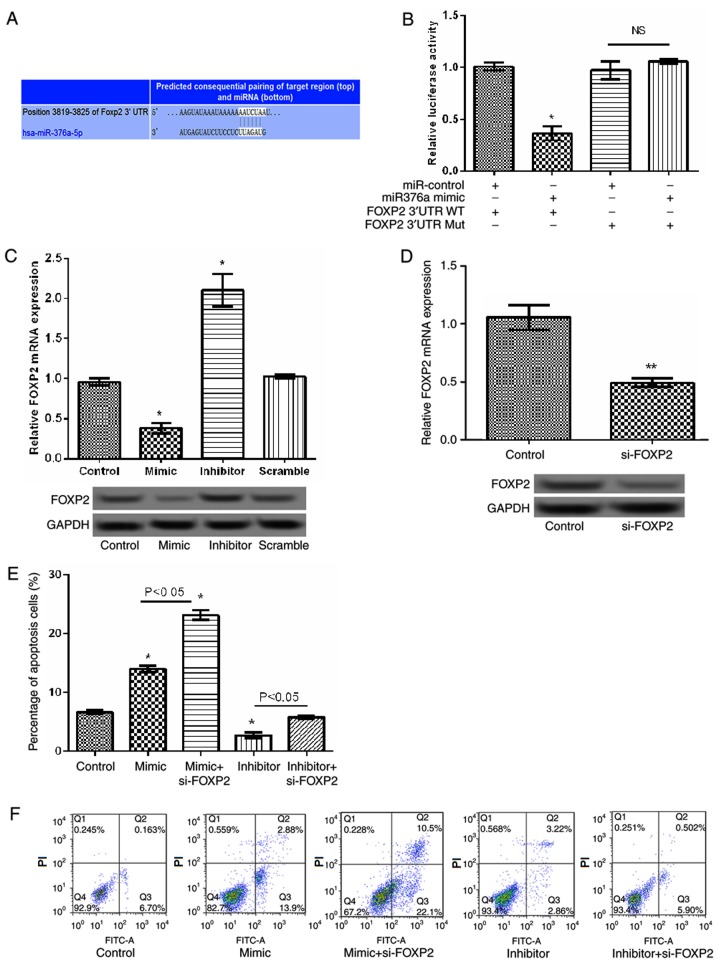
miR-376a overexpression induces apoptosis by targeting FOXP2
FOXP2 was identified to be a potential target gene of miR-376a by bioinformatic prediction (Fig. 5A). Analysis of the relative luciferase activities of FOXP2 identified that the relative luciferase activity of the reporter containing the wild-type 3′-UTR of FOXP2 was significantly reduced in the miR-376a-overexpression group compared with the control (P<0.05; Fig. 5B). Concurrently, RT-qPCR and western blot analysis identified that the relative expression levels of FOXP2 significantly decreased when miR-376a was overexpressed compared with the control group and significantly increased when miR-376a was suppressed compared with the scramble group (both P<0.05; Fig. 5C), indicating that miR-376a may negatively regulate the expression of FOXP2. Subsequently, a siRNA targeting FOXP2 was transfected into JeKo-1 cells. As indicated in Fig. 5D, the expression of FOXP2 decreased significantly in the FOXP2-siRNA group compared with the control group (P<0.01). Additionally, the apoptosis assay identified that, compared with the miR-376a-overexpression group, the percentage of apoptosis cells significantly increased in the FOXP2 siRNA/miR-376a-overexpression group (P<0.05; Fig. 5E and F), indicating that miR-376a overexpression induced apoptosis by targeting FOXP2.
Figure 5.
miR-376a regulated cell apoptosis by targeting FOXP2 in lymphoma. (A) The gene sequences of FOXP2 regulated by miR-376a. (B) The relative luciferase activities in wild-type 3′-UTR of FOXP2 and mutant FOXP2 3′-UTR in transfected cells. (C) The relative mRNA and protein levels of FOXP2 in transfected cells detected by RT-qPCR and western blot analysis. (D) The relative mRNA and protein expression of FOXP2 following cell transfection with si-FOXP2 detected by RT-qPCR and western blot analysis. (E) Percentage of apoptotic cells. (F) Representative images of the apoptosis assay following cell transfection with miR-376a overexpression vectors and si-FOXP2. Data were expressed as mean ± standard deviation (SD). *P<0.05 and **P<0.01 vs. control. miR-376a, microRNA-376a; RT-qPCR, reverse transcription quantitative polymerase chain reaction; FOXP2, forkhead box protein P2; 3′-UTR, 3′-untranslated region; si-FOXP2, small interfering RNA targeting FOXP2; FITC, fluorescein isothiocyanate; PI, propidium iodide; WT, wild type; Mut, mutant; NS, not significant.
Discussion
The present study identified that miR-376a was downregulated in lymphoma tissues and cells compared with the corresponding controls. FOXP2 was a target gene of miR-376a, which was upregulated in lymphoma. Additional experiments identified that miR-376a overexpression inhibited cell proliferation and induced cell apoptosis by regulating FOXP2 expression.
Numerous studies have revealed that miRNAs are potential biomarkers for various types of human cancer (19–21). In the present study, miR-376a was identified to be downregulated in lymphoma, which was in agreement with the results of a previous study by Arribas et al (13). The authors identified that miR-376a was also downregulated in splenic marginal zone lymphoma. miR-376a has also been identified to be downregulated in hepatocellular carcinoma (11). To confirm the biological function of miR-376a in lymphoma, the effects of miR-376a on JeKo-1 cell proliferation and apoptosis were investigated. The results of this investigation demonstrated that miR-376a inhibited JeKo-1 cell proliferation and induced apoptosis. Previous studies have described the roles of miR-376a in different cancer cell lines; for example, Zhang et al (22) suggested that miR-376a was a potential tumor suppressor in human epithelial ovarian cancer. In accordance with previous studies (11,13,22), the findings of the present study indicate that miR-376a suppresses cell proliferation and induced apoptosis in lymphoma.
Previous studies have highlighted the oncogenic nature of abnormal cell cycle in the tumorigenesis (23–25). Cellular proliferation allows for the orderly progression of cells through the cell cycle, which is regulated by certain cell cycle regulators, including cyclins and cyclin-dependent kinases (26,27). Abnormalities in expression levels of these genes/proteins have been demonstrated in several tumor types (28,29). For example, high expression levels of cyclin D2 have been observed in testicular and ovarian tumors (30). The data presented in the present study identified that the expression of cyclin D2 and cyclin A significantly increased when miR-376a was suppressed, whereas they were decreased significantly when miR-376a was overexpressed, additionally indicating that miR-376a suppression may promote cell proliferation via regulating the cell cycle-associated proteins of cyclin D2 and A in lymphoma.
Abnormal or unwanted cells will be removed during organism development (31). The failure of cells to undergo apoptosis is crucial for the initiation and progression of cancer (32). The Bcl-2 family proteins, which exhibit either pro-apoptotic (Bax) or anti-apoptotic (Bcl-2) activities, have been studied extensively due to their importance in the regulation of apoptosis and tumorigenesis (33). In the present study, to investigate the effect of miR-376a on apoptosis, the expression levels of Bax and Bcl-2 were detected in transfected JeKo-1 cells. It was observed that Bax expression increased significantly in the miR-376a mimic group whereas Bcl-2 expression decreased significantly compared with the control group (P<0.01), indicating that miR-376a suppression may reduce the rate of cell apoptosis by regulating Bax and Bcl-2 levels.
FOXP2 is a member of the FOXP family of transcription factors, which has been implicated in regulating language development and developmental neurogenesis in humans (34,35). Notably, this family has also been implicated in various types of human cancer. For example, FOXP1 has been suggested to serve a role in prostate cancer (36), FOXP3 is markedly expressed in lymphoma (37), and FOXP4 is downregulated in kidney cancer (38). The role of FOXP2 in cancer has not been widely studied, although Campbell et al (39) identified that FOXP2 was overexpressed in lymphoma. In concordance with the study by Campbell et al (39), the present study also identified FOXP2 to be overexpressed in lymphoma. Therefore, whether miR-376a was regulated cell apoptosis by targeting FOXP2 was additionally verified by transfecting JeKo-1 cells with miR-376a mimics and FOXP2 siRNA. The data generated by the present study demonstrated that FOXP2 expression was significantly decreased when miR-376a was overexpressed, and significantly increased when miR-376a expression was suppressed, indicating that miR-376a serves a regulatory effect of on FOXP2. Furthermore, quantification of apoptosis identified that FOXP2 suppression combined with miR-376a overexpression exhibited the most marked pro-apoptotic effect on lymphoma cells, indicating that miR-376a overexpression induced cell apoptosis by targeting FOXP2 in lymphoma.
In conclusion, the present study indicates that miR-376a expression is downregulated in lymphoma, whereas that of FOXP2 is upregulated. Overexpression of miR-376a inhibited cell proliferation and induced apoptosis by targeting FOXP2 in lymphoma. Therefore, miR-376a and FOXP2 may be used as biomarkers of lymphoma. However, additional experimental studies will be required to investigate the roles of miR-376a in the progression of lymphoma.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Scott DW, Gascoyne RD. The tumour microenvironment in B cell lymphomas. Nat Rev Cancer. 2014;14:517–534. doi: 10.1038/nrc3774. [DOI] [PubMed] [Google Scholar]
- 2.Marcus EBR, Sweetenham JW, Williams ME. Pathology, diagnosis and treatment. Cambridge University Press; 2013. [DOI] [Google Scholar]
- 3.Kuna T, John O, Kobler P, Filipovićzore I. A finding of diffuse cellular non-Hodgkin lymphoma in the oral cavity-case presentation Dg: Diffuse giant cell Non-Hodgkin lymphoma B-immunophenotype. Acta Stomatol Croatica. 2004;38:279. [Google Scholar]
- 4.Bardia A, Seifter E. In: Johns Hopkins Patients' Guide to Lymphoma. Shockney L, Shapiro GR, editors. Jones and Bartlett; Sudbury, MA, USA: 2011. [Google Scholar]
- 5.Armitage JO, Gascoyne RD, Lunning MA, Cavalli F. Non-Hodgkin lymphoma. Lancet. 2017;390:298–310. doi: 10.1016/S0140-6736(16)32407-2. [DOI] [PubMed] [Google Scholar]
- 6.Surveillance E, Program ER. SEER stat fact sheets: Non-Hodgkin Lymphoma. 2014 [Google Scholar]
- 7.Stewart BW, Wild CP, editors. World Cancer Report 2014. IARC Publications; Lyon, France: 2015. [Google Scholar]
- 8.Paydas S, Acikalin A, Ergin M, Celik H, Yavuz B, Tanriverdi K. Micro-RNA (miRNA) profile in Hodgkin lymphoma: Association between clinical and pathological variables. Med Oncol. 2016;33:34. doi: 10.1007/s12032-016-0749-5. [DOI] [PubMed] [Google Scholar]
- 9.Healy NA, Heneghan HM, Miller N, Osborne CK, Schiff R, Kerin MJ. Systemic mirnas as potential biomarkers for malignancy. Int J Cancer. 2012;131:2215–2222. doi: 10.1002/ijc.27642. [DOI] [PubMed] [Google Scholar]
- 10.Musilova K, Mraz M. MicroRNAs in B-cell lymphomas: How a complex biology gets more complex. Leukemia. 2015;29:1004–1017. doi: 10.1038/leu.2014.351. [DOI] [PubMed] [Google Scholar]
- 11.Zheng Y, Yin L, Chen H, Yang S, Pan C, Lu S, Miao M, Jiao B. miR-376a suppresses proliferation and induces apoptosis in hepatocellular carcinoma. FEBS Lett. 2012;586:2396–2403. doi: 10.1016/j.febslet.2012.05.054. [DOI] [PubMed] [Google Scholar]
- 12.Zehavi L, Avraham R, Barzilai A, Bar-Ilan D, Navon R, Sidi Y, Avni D, Leibowitz-Amit R. Silencing of a large microRNA cluster on human chromosome 14q32 in melanoma: Biological effects of mir-376a and mir-376c on insulin growth factor 1 receptor. Mol Cancer. 2012;11:44. doi: 10.1186/1476-4598-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arribas AJ, Gómez-Abad C, Sánchez-Beato M, Martinez N, Dilisio L, Casado F, Cruz MA, Algara P, Piris MA, Mollejo M. Splenic marginal zone lymphoma: Comprehensive analysis of gene expression and miRNA profiling. Mod Pathol. 2013;26:889–901. doi: 10.1038/modpathol.2012.220. [DOI] [PubMed] [Google Scholar]
- 14.Yu X, Li Z, Liu J. MiRNAs in primary cutaneous lymphomas. Cell Prolif. 2015;48:271–277. doi: 10.1111/cpr.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrie CH. MicroRNAs and lymphomagenesis: A functional review. Br J Haematol. 2013;160:571–581. doi: 10.1111/bjh.12157. [DOI] [PubMed] [Google Scholar]
- 16.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz AD, Jaffe ES. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Sicinski P, Donaher JL, Geng Y, Parker SB, Gardner H, Park MY, Robker RL, Richards JS, McGinnis LK, Biggers JD, et al. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature. 1996;384:470–474. doi: 10.1038/384470a0. [DOI] [PubMed] [Google Scholar]
- 19.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila) 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Ann Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz-Moralli S, Tarrado-Castellarnau M, Miranda A, Cascante M. Targeting cell cycle regulation in cancer therapy. Pharmacol Ther. 2013;138:255–271. doi: 10.1016/j.pharmthera.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 25.Stewart ZA, Westfall MD, Pietenpol JA. Cell-cycle dysregulation and anticancer therapy. Trends Pharmacol Sci. 2003;24:139–145. doi: 10.1016/S0165-6147(03)00026-9. [DOI] [PubMed] [Google Scholar]
- 26.Cordon-Cardo C. Mutations of cell cycle regulators. Biological and clinical implications for human neoplasia. Am J Pathol. 1995;147:545–560. [PMC free article] [PubMed] [Google Scholar]
- 27.Hunter T, Pines J. Cyclins and cancer II: Cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 28.Bartkova J, Lukas J, Strauss M, Bartek J. The PRAD-1/cyclin D1 oncogene product accumulates aberrantly in a subset of colorectal carcinomas. Int J Cancer. 1994;58:568–573. doi: 10.1002/ijc.2910580420. [DOI] [PubMed] [Google Scholar]
- 29.Motokura T, Arnold A. Cyclins and oncogenesis. Biochim Biophys Acta. 1993;1155:63–78. doi: 10.1016/0304-419x(93)90022-5. [DOI] [PubMed] [Google Scholar]
- 30.Surhone LM, Tennoe MT, Henssonow SF, Macromolecule, Acid N. Cyclin D2. 2011 [Google Scholar]
- 31.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 32.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Youle RJ, Strasser A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 34.Tsui D, Vessey JP, Tomita H, Kaplan DR, Miller FD. FoxP2 regulates neurogenesis during embryonic cortical development. J Neurosci. 2013;33:244–258. doi: 10.1523/JNEUROSCI.1665-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vernes SC, Oliver PL, Spiteri E, Lockstone HE, Puliyadi R, Taylor JM, Ho J, Mombereau C, Brewer A, Lowy E, et al. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genetics. 2011;7:e1002145. doi: 10.1371/journal.pgen.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bignone PA, Banham AH. FOXP3+ regulatory T cells as biomarkers in human malignancies. Expert Opin Biol Ther. 2008;8:1897–1920. doi: 10.1517/14712590802494022. [DOI] [PubMed] [Google Scholar]
- 38.Teufel A, Wong EA, Mukhopadhyay M, Malik N, Westphal H. FoxP4, a novel forkhead transcription factor. Biochim Biophys Acta. 2003;1627:147–152. doi: 10.1016/S0167-4781(03)00074-5. [DOI] [PubMed] [Google Scholar]
- 39.Campbell AJ, Lyne L, Brown PJ, Launchbury RJ, Bignone P, Chi J, Roncador G, Lawrie CH, Gatter KC, Kusec R, Banham AH. Aberrant expression of the neuronal transcription factor FOXP2 in neoplastic plasma cells. Br J Haematol. 2010;149:221–230. doi: 10.1111/j.1365-2141.2009.08070.x. [DOI] [PubMed] [Google Scholar]