Abstract
MicroRNAs (miRNAs) have been proven to have important effects on the proliferation and metastasis of multiple cancers, including hepatocellular carcinoma (HCC). In the present study, our aim was to explore the biological function of miR-106b in HCC cell proliferation and metastasis. qPCR analysis showed that miR-106b was expressed at higher levels, while disabled homolog 2 (DAB2) was expressed at lower levels in HCC tissues and cells. Moreover, the aberrant miR-106b expression in HCC affected the cell proliferative and migratory ability by MTT and Transwell assay. DAB2 was identified as a specific target of miR-106b in HCC by luciferase reporter assay and regression analysis showed a negative correlation between DAB2 and miR-106b expression. In addition, DAB2 may attenuate the miR-106b promotion effect on HCC cell proliferation and migration. In short, miR-106b may promote HCC cell proliferation and migration by targeting DAB2.
Keywords: miR-106b, proliferation, migration, HCC, DAB2
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors and can be divided into two major categories: Primary and secondary. Primary liver malignancy is a malignant tumor which life-threatening and has a high mortality rate in China (1,2). At present, the major treatments for HCC are surgical resection, chemotherapy, radiation therapy and gene therapy (3,4). Although there are obvious improvements in the diagnostic approach and treatment of HCC, the curative ratio remains low. Thus, identifying treatment strategies and gaining an understanding of the underlying mechanism of HCC progression is required.
miRNAs inhibit the expression of protein coding genes by partially binding the 3-UTR mRNA (5). Mounting evidence has shown that miRNA maladjustment is involved in the progression of many tumor diseases (6–8), including breast, prostate and lung cancers as well as HCC. For instance, miR-196b upregulated HCC cell invasion and migration by binding to the 3-UTR of FOXP2 (9). Xue and Tian concluded that the miR-429/RAB23 axis provided a potential target for treating HCC (10). miR-3613 and miR-1271 affected cell proliferation and cycle of HCC via their target genes (11,12).
The deregulated expression of miR-106b plays an abnormal role in regulating the development of various cancers (13,14). miR-106b was found to be expressed at a lower level in osteosarcoma cells and target HMGA2 to inhibit the cell progression (15). However, Zhang et al provided evidence that miR-106b was upregulated in colorectal cancer and promoted cell invasion and migration by targeting DLC1 (16). Furthermore, a recent study has shown that in the progression of cervical cancer, DAB2 was confirmed as a target of miR-106b (17). Another recent study showed that miRNA-106b expression was markedly increased and regulated HCC development by targeting mRNA (18,19). However, whether miR-106b targeted DAB2 in the regulation of HCC progression has yet to be reported.
Disabled homolog 2 (DAB2) is a member of the disable gene family. DAB2/DOC-2 has been proven to function as a new tumor suppressor that plays an important role in the occurrence and development of tumors (20,21). A previous study reported that DAB2 was downregulated in ovarian cancer (21). Subsequently, a lower DAB2 expression was detected and cell development was regulated in various types of cancer, including breast (22), prostate (23), and non-small lung cancer (24), nasopharyngeal (25) and esophageal squamous cell carcinoma (26). Previous findings have shown that DAB2 expression is decreased in HCC cells (27,28) and regulates the progression of HCC. However, the biological mechanism of DAB2 in HCC cells regulated by miR-106b has not been reported yet.
In the present study, to the best of our knowledge, we showed for the first time that, DAB2 acted as a specific target of miR-106b and confirmed the promotion effect of miR-106b in regulating HCC cell proliferation and migration. miR-18a was overexpressed in HCC, whereas DAB2 was expressed at a lower level in HCC and miR-106b expression was negatively associated with DAB2 expression. The data suggested that miR-106b may promote HCC cell viability and migration via inhibiting DAB2. This mechanism provides a therapeutic strategy for treating HCC.
Materials and methods
Specimens and cells culture
We collected 50 HCC samples from patients who underwent complete surgery at The Third People's Hospital of Qingdao (Qingdao, China) from July, 2013 to September, 2017. The samples were immediately placed in a liquid nitrogen tank and stored in a refrigerator at −80°C. Written informed consent was signed by all the patients and the study was approved by the Ethics Committee of The Third People's Hospital of Qingdao.
We purchased the HCC cell lines (Hep3B, Huh7 and Bel-7402) and normal liver cell L02 from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). The cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 µg/ml). Subsequently, the cells were maintained in an incubator at 37°C under a 5% CO2 atmosphere.
qPCR
Total RNA was isolated from HCC tissues and cells by using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). miR-106b expression was detected by miRNA First-Strand Synthesis and miRNA Quantitation kits (Takara Biotechnology Co., Ltd., Dalian, China). DAB2 was examined by CellAmp™ Direct Prep kit for RT-PCR and Protein Analysis (Takara Biotechnology Co., Ltd.). Primer sequences used were: miR-106b-F: CTTCCTGTCATAAAGTGCTGAC AGTGCAGATCTGCAGTCTGGAGTTTCA, miR-106b-R: TGACAGGAAGTAAAGTGCTGACAGTGCAGATCGAGA TCTTGGGCCTCT. DAB2-F: GTAGAAACAAGTGCAACC AATGG, DAB2-R: GCCTTTGAACCTTGCTAAGAGA. U6-F: CTCGCTTCGGCAGCACA. U6-R: AACGCTTCAC GAATTTGCGT. GAPDH-F: TGGTATCGTGGAAGGA CTC, GAPDH-R: AGTAGAGGCAGGGATGATG. U6 and GAPDH were used as the internal references to standardize the miR-106b and DAB2 relative expression respectively. The 2−ΔΔCq method was used to detect the differential expression of miR-106b and DAB2.
Western blot analysis
After transfection for 48 h, RIPA lysis containing proteinase inhibitors (Beyotime Institute of Biotechnology, Haimen, China) and phenylmethylsulfonyl fluoride were used to extract the total protein from HCC cells. The protein concentrations were tested with the BCA protein assay kit (Beyotime Institute of Biotechnology). The total proteins (50 µg) were added into the SDS-PAGE gels and performed electrophoresis at 60 V when bromophenol blue ran out from the bottom. The proteins were then transferred to nitrocellulose filter (NC) membranes. Then, skimmed milk (5-10%) was used to block the proteins on the membranes at room temperature for 2 h. Subsequently, the membranes were incubated with the primary antibody: Rabbit polyclonal anti-DAB2 (ab76253; 1:1,000; Abcam, Cambridge, MA, USA) were added in to incubate the samples at 4°C, and then horseradish peroxidase-conjugated (HRP, 1:10,000). GAPDH primary antibody (5174P; 1:5,000; Cell Signaling Technology, Inc., Danvers, MA, USA) was chosen as the internal reference. After being washed three times with 1X TBST, they were incubated with secondary antibody goat anti-rabbit IgG-HRP (sc-2,004; 1:3,000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) at room temperature for 2 h. Protein bands were detected using the chemiluminescence method (ECL; Millipore, Billerica, MA, USA).
Cell transfection
We purchased miR-106b mimic and inhibitor from GenePharma Co., Ltd. (Suzhou, China) and DAB2 vector from Shanghai Genechem Co., Ltd. (Shanghai, China). Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect miR-106b mimic, miR-106b inhibitor, DAB2 vector or both DAB2 vector and miR-106b mimic, respectively, into Hep3B cells following the manufacturers protocol.
MTT assay
An MTT assay was carried out to examine cell viability to determine HCC cell proliferation. The cells (5×103 cells/ml) treated with different transfection were planted in 96-well plates and incubated for 48 h at 37°C with 5% CO2. Then, we added MTT reagent (20 ml) to each well at 0, 1, 2, 3 and 4 days followed by incubation for another 4 h. Dimethyl sulfoxide (150 ml) was added to dissolve the crystallization. The absorbance value of cells was measured at 490 nm using enzyme-linked immunoassay.
Transwell assay
Cell migration was performed using the Transwell assay. A Transwell chamber with 8 µm pore size polycarbonic membrane (CoStar Group, Inc., Washington, DC, USA) was placed into the 24-well plates to separate the upper and lower chambers. After transfection for 48 h, the cells were added into the upper chamber coated with gelatin and DMEM medium (600 ml) supplemented with 10% fetal bovine serum was seeded in the lower chamber. Then, the cells in each well were incubated at 37°C for 24 h. Cells migrating from the upper to the lower chamber were fixed with 90% ethanol. Then, 0.05% crystal violet was added to stain the migrated cells for 15 min. Finally, cotton swab was used to gently scrape off the cells that did not migrate. Images of the migration cells were captured under a microscope (BX53; Olympus Corporation, Tokyo, Japan).
Luciferase assay
Relative luciferase ability was performed using the recombinant pMIR-reporter luciferase vector (Guangzhou RiboBio Co., Ltd., Guangzhou, China). A site-directed mutagenesis kit (cat. no. 210518; Agilent Technologies, Inc., Santa Clara, CA, USA) was carried out to introduce the site-directed mutagenesis into the miR-106b binding site of DAB2 mRNA. The wild-type and mut-type miR-106b putative targets on DAB2 3-UTR mRNA were cloned into the downstream of pGL3 luciferase vector (Promega Corporation, Madison, WI, USA). The HCC cells were transfected with miR-106b mimic using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The Dual Luciferase Assay (Promega Corporation) was subsequently used to analyze the luciferase activity values.
Statistical analysis
All the experiments were repeated in triplicate. Data are expressed as mean ± SD. SPSS 19.0 software (SPSS, Inc., Chicago, IL, USA). One-way ANOVA was used to perform the statistical analyses. The data were evaluated using the Students t-test or Tukeys post-hoc test, with statistically significant difference considered as P<0.05. Correlation between mRNA and miRNA was estimated using the Spearmans correlation method.
Results
miR-106b is overexpressed in HCC
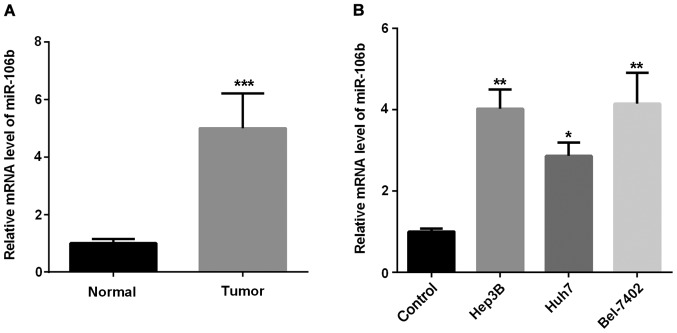
To understand the role miR-106b played in HCC progression, we investigated miR-106b expression in HCC tissues and cells. Firstly, we examined the expression of miR-106b in HCC tissues. As shown in Fig. 1A, miR-106 expression was markedly higher in HCC tissues than that in the adjacent normal tissues. Secondly, we examined miR-106b expression in three HCC cell lines. As shown in Fig. 1B, miR-106b expression in HCC cells increased in different degrees compared with the normal cells. Thus, miR-106b may function as a tumor promoter in regulating HCC development.
Figure 1.
Increased expression of miR-106b in HCC. (A) Relative miR-106b mRNA expression tested in HCC and normal tissues by qPCR (***P<0.001 vs. normal). (B) Relative miR-106b mRNA expression tested in three HCC cell lines and normal L02 cells (*P<0.05, **P<0.01 vs. L02). HCC, hepatocellular carcinoma.
miR-106b promotes HCC cell proliferation and migration
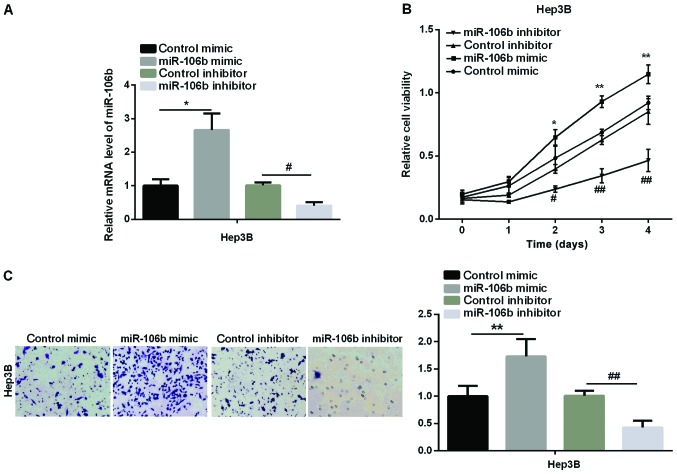
We then examined the effect of miR-106b on HCC cell proliferation and migration. MTT and Transwell assays were performed to test the cell viability and relative cell migration in an HCC cell line (Hep3B). Firstly, we transfected Hep3B with miR-106b mimic or miR-106b inhibitor to overexpress or silence miR-106b. The transfection efficiency was detected by qPCR. As shown in Fig. 2A, miR-106b expression increased markedly in the miR-106b mimic group, but significantly decreased in the miR-106b inhibitor group. Secondly, MTT assay showed that the relative cell viability was obviously higher in Hep3B after treatment with miR-106b mimic, whereas it was lower in miR-106b inhibitor than in the control group (Fig. 2B). Finally, we used Transwell assay to detect the change in HCC cell migration. The results in Fig. 2C revealed that relative cell migration was significantly increased after the overexpression of miR-106b, while it was inhibited by miR-106b silencing. The above results clearly showed that miR-106b promotes the HCC development by enhancing cell proliferation and migration due to its high-level expression in HCC.
Figure 2.
The promotion effect of miR-106b in HCC cell proliferation and migration. (A) Relative miR-106b expression checked by qPCR in Hep3B cell line after treatment with miR-106b mimic or inhibitor for 48 h. (B) Relative cell viability tested using MTT assays in Hep3B cell line after treatment with miR-106b mimic or inhibitor at 0, 1, 2, 3 and 4 days. (C) Relative cell migration tested using Transwell assay after treatment with miR-106b mimic or inhibitor for 48 h in Hep3B cell line (*P<0.05, **P<0.01 vs. control mimic; #P<0.05, ##P<0.01 vs. control inhibitor). HCC, hepatocellular carcinoma.
miR-106b specifically targets DAB2 in HCC
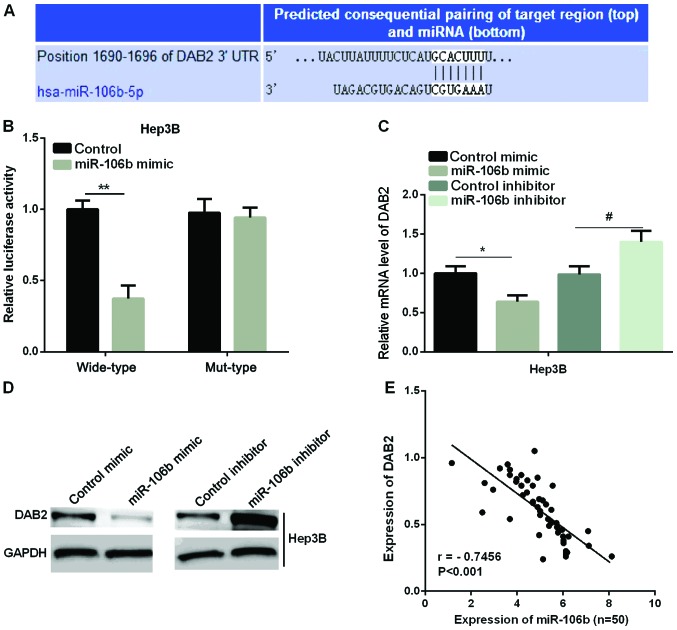
Previous studies have confirmed that DAB2 acts as a tumor suppressor in HCC (28). We predicted that DAB2 was a target of miR-106b in the regulation of HCC cells. We first used TargetScanHuman 7.1 to validate this prediction. The binding sites of DAB2 and miR-106b are shown in Fig. 3A. A luciferase reporter assay was to detect the luciferase ability in two HCC cell lines to further determine the accuracy of this prediction. It was found that the luciferase activity in the miR-106b mimic group was significantly reduced compared with the control group in wild-type, whereas there were no effects in mut-type in Hep3B (Fig. 3B). Furthermore, we examined the mRNA and protein expression of DAB2 after the overexpression or knockdown of miR-106b in the Hep3B cell line. As shown in Fig. 3C and D, both the miR-106b mRNA and protein level were significantly reduced by the miR-106b mimic, while it was increased by miR-106b inhibitor in Hep3B cell line. Regression analysis showed that DAB2 expression and miR-106b expression were negatively correlated in HCC tissues (Fig. 3E).
Figure 3.
Confirmation DAB2 as the target of miR-106b in HCC. (A) Prediction of the binding site of miR-106b with DAB2. (B) Detection of luciferase activities in Hep3B cell line after transfection with DAB2-3-UTR-wild (wild-type) or DAB2-3-UTR-mutant (mut-type). (C and D) Relative DAB2 mRNA and protein expression tested using qPCR and western blot analysis in Hep3B cell line (*P<0.05 vs. control mimic; #P<0.05; **P<0.01 vs. control inhibitor). (E) The negative correlation between DAB2 and miR-106b expression in HCC tissues (r=−0.7456, P<0.001). HCC, hepatocellular carcinoma.
miR-106b upregulates HCC progression by targeting DAB2
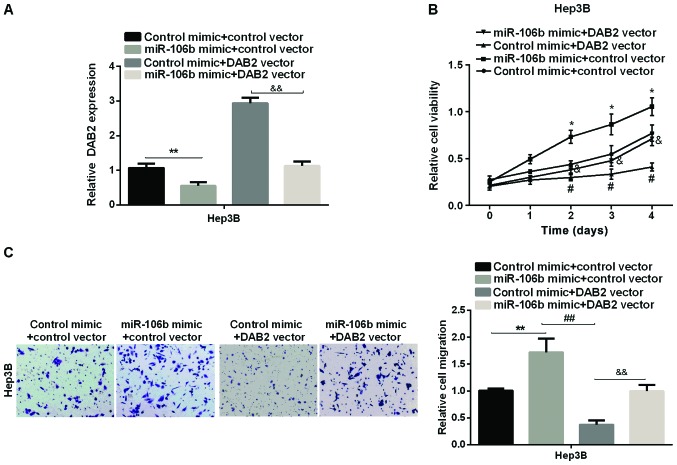
We first used the DAB2 vector to overexpress DAB2 in Hep3B cell line (Fig. 4A) to investigate whether DAB2 is the downstream mediator of miR-106b in promoting HCC proliferation and migration. Subsequently, the effect of DAB2 on relative cell viability and cell migration regulated by miR-106 was investigated by MTT and Transwell assay. The results showed that the overexpression of DAB2 reduced viability and migration of HCC cells, and DAB2 may reverse the miR-106b-promoting effect on HCC cell proliferation (Fig. 4B). The relative cell migration of HCC, which was increased by miR-106b, was supressed by DAB2 re-expression in Hep3B cell line (Fig. 4C). The data suggested that miR-106b promoted HCC cell proliferation and migration via targeting DAB2.
Figure 4.
Promotion effect of miR-106b in HCC cell proliferation and migration via regulating DAB2. (A) Examination of DAB2 mRNA levels in Hep3B cell line after overexpression of DAB2. (B) Detection of cell viability after it was transfected with miR-106b mimic, DAB2 vector, or both miR-106b mimic and DAB2 vector in Hep3B cell line (*P<0.05 vs. control mimic + control vector; #P<0.05 vs. control mimic + control vector; &P<0.05 vs. miR-106b mimic + control vector). (C) Detection of cell migration after transfection with miR-106b mimic, DAB2 vector, or both miR-106b mimic and DAB2 vector in Hep3B cell line (**P<0.01 vs. control mimic + control vector; ##P<0.01 vs. control mimic + control vector; &&P<0.01 vs. miR-106b mimic + control vector). HCC, hepatocellular carcinoma.
Discussion
Many studies have reported that the miRNAs were abnormally expressed in HCC patients, such as miR-196b, miR-429, miR-3613 and miR-1271 (9–12). Although the abnormal expression of miRNAs in HCC was regarded as a cause of HCC, we need to further explore the potential mechanism of the impact of miRNA on the progression of HCC. Previous findings showed that miRNA-106b expression is obviously increased in HCC and may provide a new biomarker for the early diagnosis of HCC (18). Those findings are in line with those of our study showing that miR-106b was overexpressed in HCC tissues and cells. However, the underlying mechanism of miR-106 in regulating HCC progression remains unclear.
Mounting evidence indicates that malignant tumor cell proliferation, migration and invasion are the main causes of human tumors (29). Previous findings have shown that the different expression of miRNAs has different effects on HCC cell development. It was proven that miR-765 mimic may make the number of HCC cells increase by regulating INPP4B (30). However, miR-340 may suppress HCC cell proliferation by upregulating JAK1 as its expression was downregulated in HCC (31). In the present study, we revealed that the higher expression of miR-106 promoted HCC cell proliferation and migration, whereas the lower expression of miR-106 repressed HCC cell development.
DAB2 is involved in the development of multiple cancers, including prostate, non-small lung and breast cancers (22–24). In HCC, a lower expression of DAB2 was detected in HCC and inhibiting DAB2 may promote HCC cell proliferation and invasion (28). Furthermore, loss of DAB2 induced a significantly higher apoptosis in HCC (32). Findings of those studies are in agreement with our results, which showed DAB2 expression was lower in HCC and upregulation of DAB2 inhibited the suppression effect on HCC cell proliferation and migration ability.
In summary, miR-106b expression was higher while that of DAB2 was lower in HCC and they were negatively correlated. To the best of our knowledge, these are the first findings that DAB2 was a direct target of miR-106b in regulating the progression of HCC and DAB2 may partially reverse the promotion effect of miR-106b in HCC, indicating that the miR-106b/DAB2 axis has a potential for use in HCC diagnosis and therapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors contributions
CS conceived and designed this study. XY and QJ performed the assay and interpreted the results. XS was involved in drafting the manuscript and revising it critically for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of The Third People's Hospital of Qingdao (Qingdao, China). Patients who participated in this study had complete clinical data. Signed written informed consents were obtained from the patients and/or guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Inokawa Y, Inaoka K, Sonohara F, Hayashi M, Kanda M, Nomoto S. Molecular alterations in the carcinogenesis and progression of hepatocellular carcinoma: Tumor factors and background liver factors. Oncol Lett. 2016;12:3662–3668. doi: 10.3892/ol.2016.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernández-Rodríguez CM, Gutiérrez-García ML. Prevention of hepatocellular carcinoma in patients with chronic hepatitis B. World J Gastrointest Pharmacol Ther. 2014;5:175–182. doi: 10.4292/wjgpt.v5.i3.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, Man W, Guo H, Yang P. Clinical study of transcatheter arterial chemoembolization combined with microwave ablation in the treatment of advanced hepatocellular carcinoma. J Cancer Res Ther. 2016;12(Supplement):C217–C220. doi: 10.4103/0973-1482.200598. [DOI] [PubMed] [Google Scholar]
- 4.Buendia MA, Neuveut C. Hepatocellular carcinoma. Cold Spring Harb Perspect Med. 2015;5:a021444. doi: 10.1101/cshperspect.a021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ, Jr, Lazo JS, Wang Z, Zhang L, Yu J. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 2009;69:8157–8165. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Bulkowska M, Rybicka A, Senses KM, Ulewicz K, Witt K, Szymanska J, Taciak B, Klopfleisch R, Hellmén E, Dolka I, et al. MicroRNA expression patterns in canine mammary cancer show significant differences between metastatic and non-metastatic tumours. BMC Cancer. 2017;17:728. doi: 10.1186/s12885-017-3751-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Z, Lin X, Tian M, Chang W. microRNA 196b promotes cell migration and invasion by targeting FOXP2 in hepatocellular carcinoma. Oncol Rep. 2018;39:731–738. doi: 10.3892/or.2017.6130. [DOI] [PubMed] [Google Scholar]
- 10.Xue H, Tian GY. MiR-429 regulates the metastasis and EMT of HCC cells through targeting RAB23. Arch Biochem Biophys. 2018;637:48–55. doi: 10.1016/j.abb.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Qin A, Zhu J, Liu X, Zeng D, Gu M, Lv C. MicroRNA-1271 inhibits cellular proliferation of hepatocellular carcinoma. Oncol Lett. 2017;14:6783–6788. doi: 10.3892/ol.2017.7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang D, Liu E, Kang J, Yang X, Liu H. MiR-3613-3p affects cell proliferation and cell cycle in hepatocellular carcinoma. Oncotarget. 2017;8:93014–93028. doi: 10.18632/oncotarget.21745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu S, Qin X, Chen T, Zhou L, Xu X, Feng J. MicroRNA-106b-5p regulates cisplatin chemosensitivity by targeting polycystic kidney disease-2 in non-small-cell lung cancer. Anticancer Drugs. 2017;28:852–860. doi: 10.1097/CAD.0000000000000524. [DOI] [PubMed] [Google Scholar]
- 14.Arias Sosa LA, Cuspoca Orduz AF, Bernal Gómez BM. Deregulation of microRNAs in gastric cancer: Up regulation by miR-21 and miR-106. Rev Gastroenterol Peru. 2017;37:65–70. (In Spanish) [PubMed] [Google Scholar]
- 15.He QY, Wang GC, Zhang H, Tong DK, Ding C, Liu K, Ji F, Zhu X, Yang S. miR-106a-5p suppresses the proliferation, migration, and invasion of osteosarcoma cells by targeting HMGA2. DNA Cell Biol. 2016;35:506–520. doi: 10.1089/dna.2015.3121. [DOI] [PubMed] [Google Scholar]
- 16.Zhang GJ, Li JS, Zhou H, Xiao HX, Li Y, Zhou T. MicroRNA-106b promotes colorectal cancer cell migration and invasion by directly targeting DLC1. J Exp Clin Cancer Res. 2015;34:73. doi: 10.1186/s13046-015-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piao J, You K, Guo Y, Zhang Y, Li Z, Geng L. Substrate stiffness affects epithelial-mesenchymal transition of cervical cancer cells through miR-106b and its target protein DAB2. Int J Oncol. 2017;50:2033–2042. doi: 10.3892/ijo.2017.3978. [DOI] [PubMed] [Google Scholar]
- 18.Samal J, Kandpal M, Vivekanandan P. HBeAg-induced miR-106b promotes cell growth by targeting the retinoblastoma gene. Sci Rep. 2017;7:14371. doi: 10.1038/s41598-017-14652-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu C, Shi L, Chen W, Fang P, Li J, Jin L, Pan Z, Pan C. MiR-106b inhibitors sensitize TRAIL-induced apoptosis in hepatocellular carcinoma through increase of death receptor 4. Oncotarget. 2017;8:41921–41931. doi: 10.18632/oncotarget.16707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albertsen HM, Smith SA, Melis R, Williams B, Holik P, Stevens J, White R. Sequence, genomic structure, and chromosomal assignment of human DOC-2. Genomics. 1996;33:207–213. doi: 10.1006/geno.1996.0185. [DOI] [PubMed] [Google Scholar]
- 21.Mok SC, Chan WY, Wong KK, Cheung KK, Lau CC, Ng SW, Baldini A, Colitti CV, Rock CO, Berkowitz RS. DOC-2, a candidate tumor suppressor gene in human epithelial ovarian cancer. Oncogene. 1998;16:2381–2387. doi: 10.1038/sj.onc.1201769. [DOI] [PubMed] [Google Scholar]
- 22.Xu S, Zhu J, Wu Z. Loss of Dab2 expression in breast cancer cells impairs their ability to deplete TGF-β and induce Tregs development via TGF-β. PLoS One. 2014;9:e91709. doi: 10.1371/journal.pone.0091709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Y, Zhang Y, Jiang L, Zhang M, Chen Z, Liu D, Huang Q. Disabled homolog 2 is required for migration and invasion of prostate cancer cells. Front Med. 2015;9:312–321. doi: 10.1007/s11684-015-0401-3. [DOI] [PubMed] [Google Scholar]
- 24.Li C, Chen J, Chen T, Xu Z, Xu C, Ding C, Wang Y, Lei Z, Zhang HT, Zhao J. Aberrant Hypermethylation at Sites −86 to 226 of DAB2 Gene in Non-Small Cell Lung Cancer. Am J Med Sci. 2015;349:425–431. doi: 10.1097/MAJ.0000000000000436. [DOI] [PubMed] [Google Scholar]
- 25.Xu YF, Mao YP, Li YQ, Ren XY, He QM, Tang XR, Sun Y, Liu N, Ma J. MicroRNA-93 promotes cell growth and invasion in nasopharyngeal carcinoma by targeting disabled homolog-2. Cancer Lett. 2015;363:146–155. doi: 10.1016/j.canlet.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 26.Li C, Ding C, Chen T, Chen J, Xu Z, Lei Z, Xu C, Zhao J. Micro ribonucleic acid-93 promotes proliferation and migration of esophageal squamous cell carcinoma by targeting disabled 2. Thorac Cancer. 2015;6:524–533. doi: 10.1111/1759-7714.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA, Schroeder I, Factor VM, Thorgeirsson SS. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Li N, Li X, Zhao W, Qiao Y, Liang L, Ding Y. Low expression of DAB2IP contributes to malignant development and poor prognosis in hepatocellular carcinoma. J Gastroenterol Hepatol. 2012;27:1117–1125. doi: 10.1111/j.1440-1746.2011.07049.x. [DOI] [PubMed] [Google Scholar]
- 29.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Xie BH, He X, Hua RX, Zhang B, Tan GS, Xiong SQ, Liu LS, Chen W, Yang JY, Wang XN, et al. Mir-765 promotes cell proliferation by downregulating INPP4B expression in human hepatocellular carcinoma. Cancer Biomark. 2016;16:405–413. doi: 10.3233/CBM-160579. [DOI] [PubMed] [Google Scholar]
- 31.Yuan J, Ji H, Xiao F, Lin Z, Zhao X, Wang Z, Zhao J, Lu J. MicroRNA-340 inhibits the proliferation and invasion of hepatocellular carcinoma cells by targeting JAK1. Biochem Biophys Res Commun. 2017;483:578–584. doi: 10.1016/j.bbrc.2016.12.102. [DOI] [PubMed] [Google Scholar]
- 32.Calvisi DF, Pinna F, Pellegrino R, Sanna V, Sini M, Daino L, Simile MM, De Miglio MR, Frau M, Tomasi ML, et al. Ras-driven proliferation and apoptosis signaling during rat liver carcinogenesis is under genetic control. Int J Cancer. 2008;123:2057–2064. doi: 10.1002/ijc.23720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.