Abstract
Alpha-1 antitrypsin (AAT) deficiency–related emphysema is the fourth leading indication for lung transplant. Chymotrypsin-like elastase 1 (Cela1) is a digestive protease that is expressed during lung development in association with regions of elastin remodeling, exhibits stretch-dependent expression during lung regeneration, and binds lung elastin in a stretch-dependent manner. AAT covalently neutralizes Cela1 in vitro. We sought to determine the role of Cela1 in postnatal lung physiology, whether it interacted with AAT in vivo, and to detect any effects it may have in the context of AAT deficiency. The lungs of Cela1−/− mice had aberrant lung elastin structure and higher elastance as assessed with the flexiVent system. On the basis of in situ zymography with ex vivo lung stretch, Cela1 was solely responsible for stretch-inducible lung elastase activity. By mass spectrometry, Cela1 degraded mature elastin similarly to pancreatic elastase. Cela1 promoter and protein sequences were phylogenetically distinct in the placental mammal lineage, suggesting an adaptive role for lung-expressed Cela1 in this clade. A 6-week antisense oligonucleotide mouse model of AAT deficiency resulted in emphysema with increased Cela1 mRNA and reduction of approximately 70 kD Cela1, consistent with covalent binding of Cela1 by AAT. Cela1−/− mice were completely protected against emphysema in this model. Cela1 was increased in human AAT-deficient emphysema. Cela1 is important in physiologic and pathologic stretch-dependent remodeling processes in the postnatal lung. AAT is an important regulator of this process. Our findings provide proof of concept for the development of anti-Cela1 therapies to prevent and/or treat AAT-deficient emphysema.
Keywords: emphysema, lung development, lung matrix remodeling, alpha-1 antitrypsin, elastin
Clinical Relevance
Chymotrypsin-like elastase 1 (Cela1) is a digestive protease with increased expression in lung epithelial cells and macrophages during development and post-pneumonectomy lung regeneration. Expression is increased by lung stretch, and Cela1 is covalently bound by alpha-1 antitrypsin (AAT) in vitro. In the present study, we show that postnatal lung remodeling by Cela1 reduces postnatal lung elastance, that Cela1 is entirely responsible for stretch-inducible lung elastase activity, that AAT covalently binds Cela1 in vivo, and that Cela1 is required for emphysema in an antisense oligonucleotide model of AAT deficiency. Cela1 expression increases in both murine and human AAT-deficient emphysema. Phylogenetic analysis demonstrates divergence of Cela1 from other Cela homologs in placental mammals, suggesting adoption of a nondigestive role after a gene duplication event. Cela1 reduces postnatal lung elastance in the mammalian lung, but it promotes emphysema in the absence of its cognate antiprotease: AAT.
Alpha-1 antitrypsin (AAT) deficiency–related lung disease (AAT-RLD) is an underdiagnosed progressive emphysema that develops in the fourth and fifth decades of life, and it is the fourth leading indication for lung transplant (1, 2). AAT, like all serpins, neutralizes target proteases via protease cleavage of a specific reactive center loop sequence that triggers covalent binding of the serpin to the protease (3). In AAT, this reaction exposes an FVFLM motif, permitting uptake of the complex by endothelial cells (4, 5). Unopposed neutrophil elastase, protease 3, cathepsin G, and matrix metalloproteinases (MMPs) have all been implicated in AAT-RLD pathogenesis (6–8); however, the standard of care—intravenous AAT replacement therapy—only modestly slows emphysema progression in this disease (9). One major hurdle to understanding AAT-RLD is the genetic heterogeneity and variable penetrance (3). The lack of a genuine animal model of AAT-RLD has hampered progress in defining the pathogenesis and natural course of this disease (10).
Chymotrypsin-like elastase 1 (Cela1) is a pancreatic protease that we and others have noted to be expressed in lung epithelial, intestinal, and immune cells (11–13). Cela1 expression increases during post-pneumonectomy compensatory lung growth in a stretch-dependent manner (14), and Cela1-positive cells colocalize to regions of elastin remodeling during lung development (12). Cela1 binds lung elastin with stretch-dependent binding kinetics, and it is covalently bound by AAT in vitro (14), as was previously reported for Cela2a (15). Although these observations suggest that Cela1 might play an important role in lung structure, function, and disease, this has yet to be tested in animal models.
We developed a biologically faithful model of AAT deficiency using a previously published antisense silencing oligonucleotide (16) and created a Cela1−/− mouse using CRISPR/Cas9 gene editing to test whether Cela1 is important in developmental and pathological lung matrix remodeling. Using these tools, we show that Cela1 is entirely responsible for the stretch-inducible lung elastase activity seen in wild-type mouse lung (17) and that Cela1 is important in elastin fiber organization and function. Cela1 covalently interacts with AAT in vivo and is increased in human AAT-deficient emphysema. Cela1−/− mice are completely protected from antisense oligonucleotide–mediated AAT-deficient emphysema. Our findings demonstrate a role for Cela1 in normal postnatal lung matrix remodeling and implicate Cela1 in AAT-RLD pathogenesis.
Methods
Detailed information on the methods is provided in the Supplementary Materials and Methods section of the data supplement.
Human Lung Experiments
Human tissues were used under an exemption from the Cincinnati Children’s Hospital Institutional Review Board (2016-9641).
Animal Experiments
Animal use and care
All animal use was approved by the Cincinnati Children’s Hospital Institutional Animal Care and Use Committee. C57BL/6 mice were housed in a pathogen-free facility with access to chow and water ad libitum. For each experiment, numbers of male and female mice were roughly equivalent.
Cela1−/− mouse
CRISPR/Cas9 technology was used to make Cela1−/− mice using the guide RNAs listed in Table E1 in the data supplement (18), with validation and genotyping as described in the data supplement.
Anti-AAT oligonucleotide model of AAT-RLD
We extended the duration of a previously published antisense oligonucleotide mouse model of AAT deficiency (16) from 3 to 6 weeks with validation by liver AAT mRNA and serum AAT protein concentrations.
Lung Tissue Analysis
Western blot analysis
Lung protein was quantified by Western blot using a previously validated guinea pig anti-Cela1 antibody (12), antitropoelastin antibody (Abcam), and anti–β-actin (Abcam) antibodies.
Proximity ligation in situ hybridization
According to the methods of Nagendran and colleagues (19), the probes and oligonucleotides listed in Table E2 were used for proximity ligation in situ hybridization of human lung specimens with antibody-based immunofluorescence costaining for alveolar epithelial type II (ATII) cells and macrophage markers, as detailed in the data supplement.
Morphometry
Mean linear intercept (20) and airspace diameter distribution (21) were determined by using previously published methods, with further description provided in the data supplement. Morphometric quantification of tile-scanned, Hart-stained, and antibody-labeled lung sections was performed using a computerized threshold identification of positive objects, as outlined in the data supplement.
Stretch-inducible lung elastase assay
The stretch-inducible elastase assay of live lung sections was performed as previously published (14, 17), except that biaxial stretch was applied instead of uniaxial stretch, as detailed in the data supplement.
Recombinant Cela1 and Elastin Proteolysis
Recombinant proteins
Cela1 with and without propeptide was synthesized using commercial (full-length) and synthesized (truncated) plasmids.
Analysis of Cela1 elastin digests
The digested fragments of recombinant human tropoelastin and mature elastin after incubation with full-length Cela1 were analyzed by nano-HPLC–nano-electrospray ionization quadrupole-quadrupole time-of-flight mass spectrometry, as detailed in the data supplement.
Phylogeny
Ensembl protein and promoter sequences for Cela1, Cela2a, Cela2b, Cela3a, and Cela3b were used to construct phylogenetic trees using ClustalW and Megalign Pro (DNAStar).
Statistical Analysis
Parametric data were evaluated with Student’s t test or one-way ANOVA with Tukey’s post hoc comparison, and nonparametric data were evaluated with the Mann-Whitney U test or ANOVA on ranks with Dunn’s post hoc comparison test. P values less than 0.05 were considered significant. Parametric data are presented as bar charts with error bars representing the SEM, and nonparametric data are presented as box plots with whiskers representing 5th- and 95th-percentile values.
Results
Cela1 Regulates Lung Elastin Structure and Function
Cela1 mRNA concentrations peak during the alveolar stage of lung development (12); lung elastin remodeling is maximal during this period (17); and Cela1-postive cells are located in close proximity to this remodeling (12). To determine the role of Cela1 in lung development, we used CRISPR/Cas9 gene editing (18) to create a Cela1−/− mouse on the C57BL/6 background. Cela1 mRNA was absent from the pancreas of these mice, and there were no gross developmental or behavioral differences between wild-type and Cela1−/− animals (Figures E1A–E1D).
Analysis of these mice identified a role for Cela1 in lung structure and function. Cela1−/− mice underwent alveolar septation; however, septal tip elastin was less discrete, elastin fibers were increased in airspace walls, and airspace size was reduced in Cela1−/− animals (Figures 1A–1C, E1E, and E1F). Lung wet weight to dry weight ratio was increased in Cela1−/− mice (Figure E1G). Thus, Cela1 appeared to have a role in distal lung remodeling during the alveolar stage of lung development.
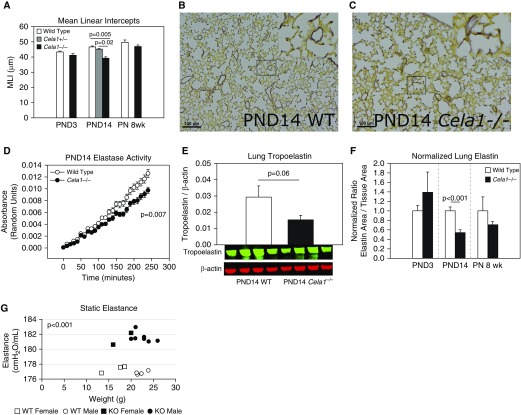
Figure 1.
Role of Chymotrypsin-like elastase 1 (Cela1) in postnatal lung elastin remodeling. (A) The mean linear intercepts (MLI) of Cela1−/− Postnatal Day (PND)14 lungs (alveolar stage of lung development; n = 9) were 18% smaller than wild type (WT, n = 6; Cela1+/−, n = 5), with a trend toward smaller intercepts at PND3 (saccular stage of lung development; WT, n = 8; Cela1−/−, n = 7) and 8 weeks postnatally (PN) (adult lung; WT and Cela1−/−, n = 8 per group). (B) PND14 WT lungs demonstrated the typical localization of dense elastin bands to septal tips. Scale bars: 100 μm. (C) Cela1−/− lungs had less dense septal tip elastin bands and more diffuse elastin fibers distributed throughout distal airspace walls. (D) The lung homogenates of Cela1−/− PND14 mice had less elastase activity than WT lung homogenates (n = 6 per group). (E) Western blot of PND14 lung homogenates indicated that Cela1−/− mouse lungs had approximately half the soluble tropoelastin of WT lungs. (F) Morphometric quantification of total lung elastin demonstrated a 45% reduction in lung elastin at PND14 and a 29% reduction at 8 weeks PN. To account for differences in lung density, values were normalized to tissue area, but similar findings were present when normalized to lung area. (G) As assessed using the flexiVent system, 8 weeks PN Cela1−/− mouse lungs (n = 9) were more elastic (i.e., more stiff) than WT lungs (n = 7). These differences were not age or sex dependent. KO = knockout.
We next sought to assess the impact of Cela1 on lung elastin and lung elastin remodeling. We previously reported that lung elastase activity is maximal at Postnatal Day (PND)14 (17). The elastase activity of Cela1−/− lung homogenate at this age was reduced by 25% compared with wild type (Figure 1D). We therefore expected to see excessive lung elastin in Cela1−/− lungs; however, at PND14, Cela1−/− mouse lungs had less elastin; largely lacked discrete bands at the tips of secondary alveolar septae; and had more diffuse, thinner elastin fibers throughout the alveolar walls (Figures 1B and 1C). Cela1−/− lungs contained half as much tropoelastin as wild type (Figure 1E), which we confirmed by morphometric analysis. This reduced lung elastin was noted in adult lung, but not in PND3 lung (Figure 1F), consistent with a role for Cela1 in the alveolar stage of lung development. There were modestly reduced quantities of many elastin-associated gene mRNAs in PND14 lung (Figure E1H), without any differences in total lung collagen (Figures E1I–E1K). Cela1 is an important contributor to elastin remodeling during alveolar development, although the relationship between reduced elastin remodeling in alveolar walls and reduced elastogenesis and septal tip elastin deposition is unclear.
Because proper elastic fiber density and localization are critical for normal lung dynamics, we used the flexiVent system (SCIREQ) to quantify the impact of these differences on lung dynamics in animals at 8 weeks postnatally. Cela1−/− mice had significantly higher elastance lungs than wild-type mice (Figures 1F and 2). The key findings derived from these experiments are that 1) Cela1 contributes to lung elastin remodeling during development by regulating elastin fiber localization, and 2) Cela1 reduces postnatal lung elastance, likely secondary to this effect.
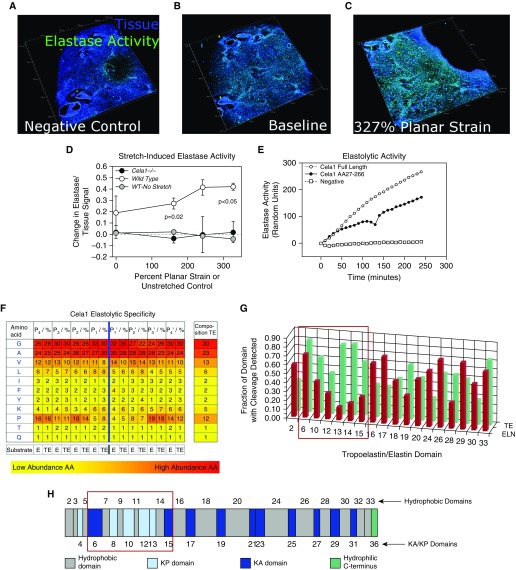
Figure 2.
Elastolytic profile of Cela1. (A) Live lung sections of 12-week-old mice were inflated with gelatin, then 200-μm sections were adhered to the silicone insert of a three-dimensional printed confocal lung-stretching device (see Figure E3), and the gelatin was removed. Lung tissue was defined by autofluorescence, and elastase activity was determined using a soluble elastin substrate conjugated to a quenched fluorophore. A three-dimensional image of a lung section is shown, with each major tick mark representing 500 μm. (B) After application of the zymography substrate, there was an appreciable increase in the elastase signal. A representative WT lung section is shown. (C) The same lung section is shown at the maximal strain with an increase in the elastase signal. (D) The stretch-induced lung elastase activity of WT (n = 3) and Cela1−/− (n = 2) lung sections was compared. WT lung not exposed to stretch but incubated with substrate for an equivalent time did not demonstrate increased elastase signal. Cela1−/− lung that was stretched lacked the inducible elastase activity observed in WT lung. Comparisons were carried out using one-way ANOVA. (E) As assessed using a plate-based fluorometric elastase assay, full-length Cela1 demonstrated elastolytic activity comparable to that of Cela1 without signaling and propeptide. (F) Full-length recombinant Cela1 was incubated with soluble human tropoelastin and human skin elastin, and degradation products were analyzed to determine proteolytic specificity. Whereas Cela1 had a propensity for hydrophobic residues, the only amino acid that was nonpreferred in regions adjacent to cleavage sites was proline. However, proline was preferred at P4-P2 and P3′-P4′ sites, suggesting that the conformational turn induced by proline residues at these locations enhanced elastin–Cela1 interaction. (G) Although there was no major difference in the amino acid residues cleaved by Cela1, mature elastin had substantially fewer cleavage sites in domains 6–15 (red box). (H) Although there was no major difference in the hydrophobicity of these domains (ivory color, domains numbered above simplified tropoelastin protein sequence) compared with others, these domains contain lysine-proline (KP) cross-linking sites (light blue color). Because soluble tropoelastin lacks these cross-links, they likely account for the difference in elastin degradation between mature elastin and soluble tropoelastin. AA = amino acid; E, ELN = elastin; KA = lysine-alanine; TE = tropoelastin.
Characterization of the Cela1 Elastolytic Profile
Because we previously demonstrated that Cela1 binds to lung elastin with stretch-dependent binding kinetics (14), we sought to characterize the Cela1 elastolytic profile more fully. To determine stretch-dependent elastase activity (17), we used a biaxial live lung–stretching elastin device (Figure E3) and live sectioned lung with fluorescent elastin in situ zymography. As previously reported (14), stretch doubled the elastase activity of wild-type lung, but there was no increase in the elastase activity of wild-type lung incubated with substrate for an equivalent time. However, Cela1−/− lung completely lacked stretch-inducible elastase activity (Figures 2A–2D). At least in this ex vivo murine lung model, Cela1 mediated stretch-inducible lung elastase activity.
Cela family members and related chymotrypsin proteases are synthesized as zymogens and activated in the intestinal lumen after interaction with pepsin. However, the protease that would accomplish this task in the lung was unclear. We therefore synthesized recombinant Cela1 with and without its inhibitory propeptide to confirm that Cela1 was indeed a zymogen. Interestingly, full-length Cela1 (with its signaling peptide and propeptide intact) had a level of elastolytic activity comparable to that of truncated Cela1 (Figure 2E). Thus, Cela1 synthesized in the lung may not require exogenous activation by another protease, as is typically required for proteases in this class.
Lastly, we wished to understand the specificity with which Cela1 cleaves tropoelastin and elastin. In the lung, translated tropoelastin forms coacervate droplets that are directed to microfibrils and formed into elastin fibers coordinately with a range of microfibril-associated proteins (22). Tropoelastin monomers on adjacent microfibrils are cross-linked at oxidized and deaminated lysine residues, increasing fiber strength (23). Porcine pancreatic elastase, which is composed largely of Cela family members, cleaves elastin and tropoelastin in a nondiscriminatory manner (24). Using mass spectrometry to analyze elastin degradation products of full-length Cela1, both mature elastin and recombinant tropoelastin without substantial amino acid specificity apart from, preferring regions with prolines outside of the P1-P2′ positions (Figures 2F and E4). Thus, Cela1 has a proteolytic profile similar to that of pancreatic elastase. However, when we compared the degradation of recombinant tropoelastin (un-cross-linked) and mature elastin, we observed a substantial difference in degradation with regard to the domains cleaved. Whereas the fraction of amino acids with documented cleavage sites was relatively consistent across tropoelastin domains, cleavage of domains 6–15 was reduced in elastin compared with tropoelastin (Figure 2G). These domains involve or are adjacent to lysine- and proline-containing cross-linking domains 4, 8, 10, 12, and 13 (Figure 2H). This suggests that steric hindrance of these cross-links denies Cela1 substrate access, accounting for the differential degradation of mature elastin and recombinant tropoelastin. Taken together, these findings indicate that full-length Cela1 cleaves elastin in a manner similar to other Cela family members and that it alone is responsible for stretch-inducible elastase activity in the lung.
Evolutionarily Conserved Role for Cela1 in Reducing Distal Lung Elastance in Placental Mammals
We next sought to understand why Cela1, a digestive protease, was expressed in the lung, hypothesizing that its role in reducing postnatal lung compliance may have conferred an evolutionary advantage to animals that breathe by expansion of gas-exchanging surfaces. To define the evolutionary conservation of Cela1, we constructed phylogenetic trees of Cela protein and promoter sequences. Whereas mammalian Cela2 and Cela3 protein sequences were interspersed within the different nonmammalian Cela sequences, mammalian Cela1 protein sequences were phylogenetically distinct (Figure 3A). Similarly, mammalian Cela1 promoter sequences were phylogenetically unique from mammalian Cela2, Cela3, and all nonmammalian Cela sequences (Figure 3B). In comparing the two phylogenetic trees, we found that the only mammals with Cela1 protein and promoter sequences distant from the other mammalian Cela1 sequences (and thus interspersed with all other Cela proteases) were the platypus (Ornithorhynchus anatinus) and the Tasmanian devil (Sarcophilus harrisii). Thus, placental mammal Cela1 has diverged from mammalian Cela2 and Cela3, which have remained more similar to nonmammalian Cela proteases.
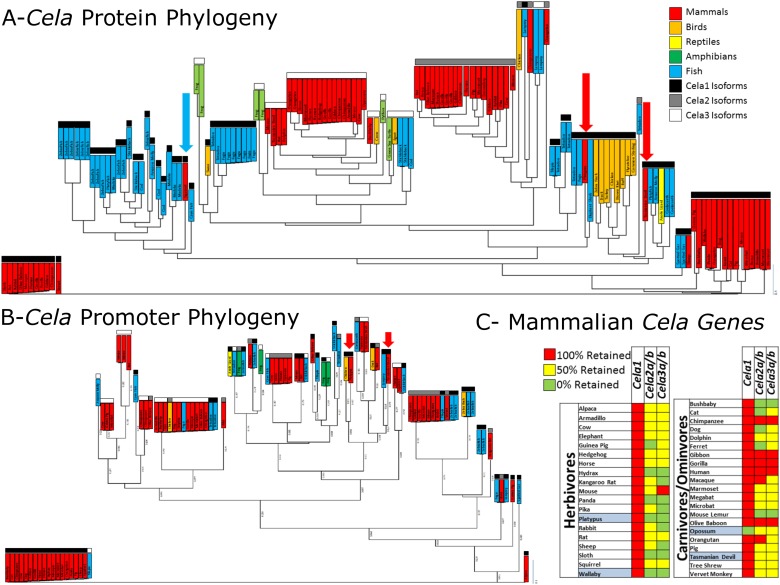
Figure 3.
Unique role for Cela1 in the placental mammalian lineage. (A) Cela1 protein sequences were analyzed with clustering of mammalian Cela1 protein sequences away from mammalian Cela2 and Cela3 and nonmammalian Cela1, Cela2, and Cela3. Common names are listed. Different vertebrate classes are color coded, and sequences for Cela1, Cela2, and Cela3 are coded in black, gray, and white, respectively. Only two mammalian Cela1 protein sequences failed to cluster with the other mammalian Cela1 sequences: the platypus and the Tasmanian devil (red arrows). The rabbit Cela1 protein sequence was likely incorrect because it included introns in its protein sequence, but it was retained for completeness (blue arrow). (B) Phylogenetic tree of Ensembl 200-bp promoter sequences for all Cela family members. Like the pattern observed with protein sequences, mammalian Cela2 and Cela3 promoter sequences were similar to nonmammalian Cela1, Cela2, and Cela3 sequences, but mammalian Cela1 sequences were less similar. Again, the Tasmanian devil and platypus Cela1 sequences failed to cluster with other mammalian Cela1 sequences. (C) Using the Ensembl and Entrez Gene databases, the absence or presence of the Cela1 gene (green or red, respectively) or the presence of zero, one, or two Cela2 and Cela3 genes (red, yellow, and green, respectively) were plotted in a simplified heat map. Annotated mammals were segregated by diet, and nonplacental mammals are highlighted in blue. Except for the opossum, all annotated mammals had retention of the Cela1 gene, whereas herbivores had more frequent loss of Cela2 and Cela3 homologs. Primates had higher retention of both Cela2 and Cela3 homologs.
Gene duplication events result in two genes: a parent gene, which remains relatively stable, and a daughter gene (homolog), which undergoes a higher rate of substitution. This homolog may provide an evolutionary advantage and be retained, or it may fail to provide such an advantage and be lost (25). If homologs retain the same function, they are termed orthologs, and if they adopt a new function, they are termed paralogs. To determine whether there were differences in the retention of Cela1, Cela2a, Cela2b, Cela3a, and Cela3b homologs over the course of mammalian evolution, we used the Ensembl and Entrez Gene databases to identify retained and lost Cela genes. Cela1 was retained in 39 of 40 mammals, being absent only in opossum (Monodelphis domestica), a marsupial. Ten of 19 herbivores had loss of all Cela2 and Cela3 homologs, whereas only 2 of 21 carnivores/omnivores demonstrated such loss (Figure 3C). These two omnivores were the bushbaby (Otolemur garnettii) and the mouse lemur (Microcebus murinus), both of which consume mostly fruits and insects. Most primates had retention of all four Cela2 and Cela3 homologs. These data suggest that Cela2 and Cela3 are orthologs and redundantly fulfill a digestive role, but that in the placental mammal lineage, Cela1 is a paralog with a nondigestive role in placental mammals.
Anti-AAT Oligonucleotide Model of AAT-RLD
We next investigated whether AAT was an important regulator of Cela1-mediated remodeling. The rationale for these experiments was our previous observation that Cela1 was covalently bound by AAT in cell culture supernatant but not in cell lysate (14).
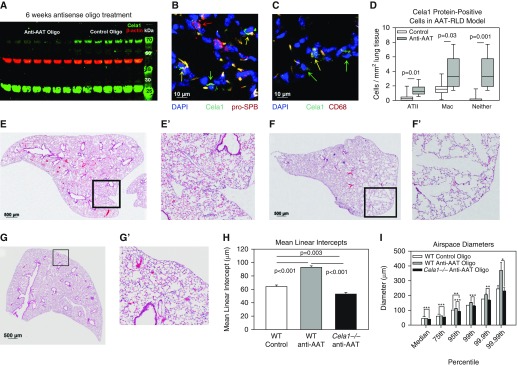
To develop a model of AAT-RLD, we used a previously published antisense oligonucleotide targeting all five murine AAT isoforms but extended treatment from 3 to 6 weeks (16). Once-weekly administration of 100 mg/kg anti-AAT oligonucleotide resulted in a greater than 99% decrease in liver AAT mRNA concentration (Figure 4A) and a 77% reduction in serum protein concentration (Figure 4B) 8 days after the fifth dose of antisense oligonucleotide. Because antisense oligonucleotide therapy has been associated with significant inflammation, we quantified lung neutrophil infiltration and inflammatory gene mRNAs in untreated, control oligonucleotide–treated, and anti-AAT oligonucleotide–treated lungs. Lung myeloperoxidase activity, a measure of lung neutrophil infiltration, was slightly elevated in anti-AAT oligonucleotide–treated lungs compared with untreated lungs (P = 0.8), but it was increased threefold in control oligonucleotide–treated lungs (P < 0.001) (Figure 4C). Whereas there were no differences in the mRNAs of IL6, IL8, or TNFα between control and anti-AAT oligonucleotide, there was a nonsignificant increase in these mRNAs compared with untreated lungs. There were no differences in the macrophage chemoattractant protein 1 (MCP1, CCL2) mRNA in all three groups. The mRNAs of Toll-like receptor 7 (TLR7) effectors IFNα and IFNβ were elevated in anti-AAT oligonucleotide–treated lungs compared with control, but these anti-AAT concentrations were comparable to those in untreated lungs, making the significance of this finding unclear (Figure 4D). We next quantified the mRNAs of proteases previously shown to be important in AAT-RLD (6–8). The mRNAs of neutrophil elastase (ELANE), protease 3, cathepsin G, MMP12, and MMP14 were not elevated in anti-AAT oligonucleotide– versus control oligonucleotide–treated lungs (Figure 4E). However, Cela1 mRNA was increased eightfold (P < 0.001, Mann-Whitney U test). Taken together, these data demonstrate that anti-AAT oligonucleotide administration efficiently reduces liver AAT synthesis, that anti-AAT oligonucleotide does not trigger significant neutrophil infiltration in the lung, and that anti-AAT oligonucleotide does not increase the mRNAs of classically associated proteases, but that it does increase Cela1 mRNA.
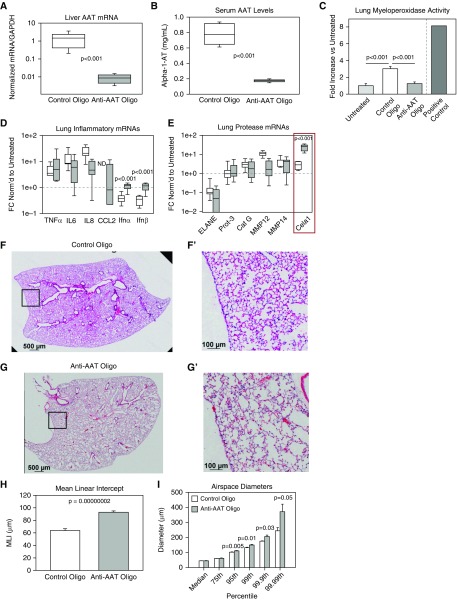
Figure 4.
Mouse model of alpha-1 antitrypsin (AAT) deficiency–related lung disease (AAT-RLD). (A) Compared with control oligonucleotide, anti-AAT oligonucleotide administration reduced liver AAT concentrations by more than 99% 8 days after administration of the final dose. Note the logarithmic scale and nonparametric data (n = 8 per group, comparison by Mann-Whitney U test). (B) Serum AAT concentrations were reduced in these same animals. (C) Lung myeloperoxidase activity, a measure of neutrophil infiltration, was elevated in control oligonucleotide–treated lungs, but not anti-AAT oligonucleotide–treated lungs, compared with untreated lungs (n = 5). Lung from an animal subjected to intestinal ischemia reperfusion in a previous study (42) was used as a positive control. (D) Left lung inflammatory mRNAs were no different after treatment with anti-AAT and control oligonucleotide. Values were normalized to untreated adult control lung, and samples with undetectable mRNA were assigned a value of zero for statistical analysis. IFN-α and IFN-β mRNA concentrations were elevated in anti-AAT oligonucleotide–treated lungs but comparable to untreated control lungs. P < 0.001 (n = 8 per group). (E) The mRNAs of many genes associated with AAT-RLD were no different in control oligonucleotide–treated and anti-AAT oligonucleotide–treated lungs, but Cela1 mRNA concentrations were increased eightfold. (F and G) A representative lower lung lobe of a wild-type mouse treated with (F) control oligonucleotide and (G) anti-AAT oligonucleotide (E′, F′, and G′ are higher magnification images of respective boxed regions). Scale bars: 500 μm (F and G) and 100 μm (F′ and G′). (H) As assessed by MLI, anti-AAT oligonucleotide treatment (n = 8) significantly increased airspace size compared with control oligonucleotide–treated mice (n = 7) (Student’s t test). (I) Whereas median and 75th percentile alveolar diameters were no different in anti-AAT oligonucleotide– and control oligonucleotide–treated lungs, diameters were significantly larger at percentiles beyond the 95th (Student’s t test), indicating that only the largest airspaces were contributing to the measured difference in MLI. Cat G = cathepsin G; ELANE = neutrophil elastase; FC = fold change; ND = not detected; Prot-3 = protease 3.
The lungs of anti-AAT oligonucleotide–treated mice demonstrated emphysematous changes reminiscent of AAT-RLD (Figures 4F and 4G), with a near-doubling of mean linear intercept (26) (Figure 4H). We further characterized this emphysema using a computerized morphometry program (21) that we modified to increase sampling (Figure E5 and other data supplement material). Using this technique, we determined that the difference in mean linear intercept was driven by enlargement of airspaces in the 95th percentile of anti-AAT oligonucleotide–treated mice (i.e., airspace sizes were comparable below the 95th percentile) (Figure 4I). These data demonstrate that anti-AAT oligonucleotide administration to mice is a natural and faithful model of AAT-RLD and that Cela1 expression increases in this model.
Role for Cela1 in AAT-Deficient Emphysema
Because Cela1 mRNA was increased in this mouse model of AAT-RLD, we sought to determine whether Cela1 was playing an important role in emphysema pathogenesis. In the lung, Cela1 exists in both low (∼28 kD) and high (∼70 kD) molecular weight forms (12). In vitro this higher-molecular-weight species is a covalent Cela1 and AAT product (14). Western blot analysis of control and anti-AAT oligonucleotide–treated lungs showed a 77% reduction in high-molecular-weight Cela1 (P < 0.001, Student’s t test) (Figure 5A), demonstrating that Cela1 is covalently bound by AAT in vivo. Although there was no difference in the 28 kD Cela1 species by Western blot analysis, quantitative image analysis revealed that the total number of Cela1-positive cells was increased in anti-AAT oligonucleotide–treated mice 3.9-fold (P = 0.002, Student’s t test), with 3- and 2.4-fold increases in Cela1+ ATII cells and macrophages, respectively, but a 13-fold increase in cells expressing neither marker (Figures E6 and 5B–5D).
Figure 5.
Cela1 in a murine model of AAT-deficient emphysema. (A) We previously reported a higher-than-expected molecular weight for Cela1 in mouse lung (1) and that Cela1 formed a covalent complex with AAT in vitro (2), likely accounting for this observation. A 77% reduction in this high-molecular-weight form of Cela1 was observed in anti-AAT oligonucleotide–treated lungs compared with control oligonucleotide–treated lungs (P < 0.001, Student’s t test). Little difference was noted in the quantity of native molecular weight Cela1. (B) Cela1-positive cell quantification was performed by tile scan of Cela1, prosurfactant protein B (pro-SPB), and CD68 costaining, as outlined in Figure E6. A confocal image of an anti-AAT oligonucleotide section is shown, demonstrating single-positive alveolar epithelial type II cells (ATII; red arrows), single-positive Cela1 cells (green arrow), and double-positive cells (yellow arrows). Scale bar: 10 μm. (C) Same section showing a macrophage (Mac) with Cela1 protein (yellow arrow) and nonmacrophage Cela1-positive cells (green arrows). Scale bar: 10 μm. (D) Quantification of Cela1-positive cells in control (n = 7) and anti-AAT oligonucleotide (n = 8) right lower lobe sections demonstrating increases in the number of Cela1-positive cells in the AAT-RLD model. (E) Control oligonucleotide–treated middle lobe. (F) Anti-AAT oligonucleotide–treated middle lobe. (G) Cela1−/− mice treated with anti-AAT oligonucleotide did not demonstrate any evidence of airspace injury or emphysema after 6 weeks of treatment. Scale bars: 500 μm (E, F, and G). (H) The airspaces of Cela1−/− mice treated with anti-AAT oligonucleotide (n = 7) were smaller than wild-type (WT) mice treated with either anti-AAT or control oligonucleotide. This smaller airspace size is consistent with the developmental findings outlined in Figure 1. Analysis was performed by one-way ANOVA with post hoc comparison using Dunn’s method. (I) In comparing airspace diameter percentiles, we found that the Cela1−/− anti-AAT oligonucleotide–treated mice had an airspace diameter distribution comparable to that of WT control oligonucleotide–treated mice. *P < 0.05; **P < 0.01; ***P < 0.001.
To test whether Cela1 was important in the development of emphysema in AAT deficiency, we treated Cela1−/− mice with anti-AAT oligonucleotide. In contrast to observations in wild-type mice, treatment of Cela1−/− mice for 6 weeks with anti-AAT oligonucleotide did not result in any appreciable lung injury or evidence of emphysema (Figures 5E–5G). On the contrary, anti-AAT oligonucleotide–treated Cela1−/− airspaces were even smaller than those in control oligonucleotide–treated wild-type mice (Figures 5H and 5I). This decrease is consistent with the findings in untreated mice (Figure 1A), but it is more pronounced in this experiment because of open-chest inflation versus previous closed-chest inflation. Thus, we found that in this mouse model of AAT deficiency, Cela1 is required for emphysema.
Cela1 in Normal Human Lung and in AAT-RLD
To demonstrate the relevance of our findings in human AAT-RLD, we evaluated the expression of Cela1 in human lung and determined whether its expression was increased in AAT-RLD. To do so, we obtained lung tissue from an adult organ donor and immunostained paraffin-embedded lung specimens from explants of individuals with AAT deficiency undergoing lung transplant, similar explants from individuals with confirmed AAT sufficiency, and control specimens from individuals undergoing lung nodule resection with the surrounding tissue considered normal lung.
Cela1 PCR and sequencing from normal human lung confirmed the presence of Cela1 mRNA (Figure E7). These findings are in agreement with published single-cell mRNA sequencing data showing Cela1 mRNA in lung epithelial cells during development and later increasing in lung immune cells (13, 27).
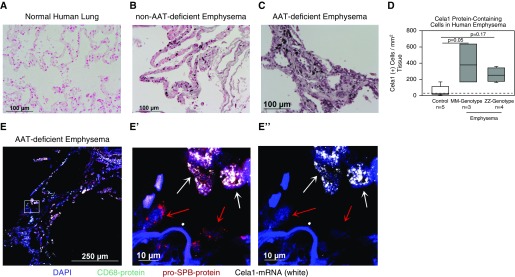
To determine whether Cela1 expression was increased in human AAT-RLD, we performed immunohistochemistry and immunofluorescence staining and imaging on control AAT-sufficient and AAT-deficient emphysema sections. Immunohistochemistry showed that Cela1-positive cells were largely absent from control lung (Figure 6A) but were abundant in the lungs of individuals with emphysema with AAT sufficiency (MM genotype Figure 6B) as well as in individuals with ZZ genotype AAT deficiency (Figure 6C). Morphometric quantification confirmed these findings (Figure 6D). Proximity in situ hybridization for Cela1 mRNA demonstrated increased Cela1 mRNA–containing cells in both AAT-sufficient and AAT-deficient emphysema, with most Cela1-expressing cells being ATII cells (Figure 6E). Our findings demonstrate increased numbers of Cela1-expressing cells in both AAT-deficient and AAT-sufficient emphysema, and they support a central role for Cela1 in the pathogenesis of AAT-RLD.
Figure 6.
Cela1 in human emphysema. (A) Immunohistochemistry for Cela1 in normal human lung revealed very few Cela1 protein–containing cells. (B) A specimen obtained from a patient without AAT deficiency (MM genotype) undergoing lung transplant revealed scattered Cela1 protein–containing cells throughout the alveolar interstitium. (C) Examination of a lung specimen from an individual with AAT deficiency (ZZ genotype) undergoing lung transplant identified Cela1 protein–containing cells in regions of airspace destruction. (D) Quantification of Cela1 protein–containing cells in control, AAT-sufficient (MM genotype), and AAT-deficient (ZZ genotype) patients identified a trend toward an increased number of Cela1-positive cells in AAT-sufficient and AAT-deficient emphysema. Only two of five control lungs had numbers of Cela1-containing cells above the number observed in secondary-alone control (dashed line). (E) A separate ZZ genotype AAT-deficient emphysema specimen with staining for the macrophage marker CD68 (green) and pro-SPB (red) human Cela1 mRNA by proximity ligation in situ hybridization (white) and DAPI (blue). Scattered Cela1 mRNA–containing cells are seen in this region of airspace destruction. E′ is a magnification of the boxed area in E demonstrating ATII cells with (white arrows) and without (red arrows) Cela1 mRNA. E″ displays magnification of only DAPI and Cela1 proximity ligation in situ hybridization signals. Scale bars: 100 μm (A, B, and C), 250 μm (E), and 10 μm (E′ and E").
Discussion
We have demonstrated, for the first time to our knowledge, a role for Cela1 in lung matrix remodeling in development and disease. We also have provided phylogenetic evidence that Cela1 was adapted for a nondigestive role in placental mammals. We have shown the in vivo interaction of AAT with Cela1, and we have provided murine loss-of-function and associative human data suggesting that Cela1 may be important in the pathogenesis of AAT-deficient emphysema. We conceptualize that Cela1 acts to improve respiratory efficiency by remodeling the distal lung in a stretch-dependent manner (14) and that this reduced lung elastance conferred a selective advantage in the placental mammal lineage. This remodeling is regulated by AAT, and in the absence of its antiprotease, excessive remodeling can lead to a progressive emphysema.
Cela1 is potentially an accessible and specific target for the treatment of AAT-RLD. Neutrophil and/or macrophage depletion (28, 29) partially prevents emphysema in cigarette smoke models, supporting the role of these cells in early alveolar injury, and deletion of specific myeloid-derived proteases, such as neutrophil elastase (7), cathepsin G (30), proteinase 3 (30), and MMP12 (31), has been shown to have similar partial protection in cigarette smoke models of emphysema. None of these mediators were increased after antisense oligonucleotide–induced AAT depletion, suggesting that the pathogenesis of AAT-RLD is distinct from that of AAT-sufficient emphysema—a hypothesis that is supported by differing ages of presentation, histology, and lobar patterning of airspace destruction (3). Given the stretch-dependent biology of lung elastin remodeling (17) and Cela1 expression and activity (14), targeting Cela1 in AAT deficiency may prevent emphysema. Whether anti-Cela1 therapies can halt or reverse emphysema progression in already established lung disease will be addressed in future studies.
We have characterized a novel mouse model of AAT-RLD that overcomes many of the limitations of previous models. Whereas humans have one AAT gene (Sepina1), different mouse strains have up to five (Serpina1a–Serpina1e) (10). Furthermore, homozygous deletion of Serpina1a was reported to be embryonic lethal (32). The “pallid” mouse has a mutation in the pallidin gene that reduces AAT secretion and develops emphysema at 12 months of age (33, 34). However, the “pallid” mouse exhibits multiple other phenotypic and functional anomalies (10, 34, 35). Mice with knock-in of the human Z-type AAT gene (PiZ mice) have normal concentrations of AAT and no lung phenotype (36). The anti-AAT oligonucleotide model of AAT-RLD is a genuine, specific, efficient, and natural model of human AAT-RLD.
This study leaves several important questions unanswered. First, how does the lack of stretch-dependent remodeling activity impair the formation of the elastin band at alveolar septal tips? Clearly, Cela1 is not an important regulator of overall lung elastin content, because lung elastin was decreased and collagen was unchanged in Cela1−/− mice. Although we hypothesize that Cela1 remodels matrix in a targeted manner to alter local tissue mechanics and that mechanosensitive matrix synthesis accounts for observed differences in overall matrix content (37–39), we can provide little support for this conjecture. Second, how can it be that this relatively aggressive protease does not exist in the lung as a zymogen? Whether Cela1 is complexed to a reversible inhibitory protein in the intracellular and/or extracellular space, is contained in vesicles, and/or can activate itself needs to be investigated in future studies. Third, and relatedly, how is it that neutralization of Cela1 by AAT is critical to maintaining lung integrity when the majority of lung Cela1 exists in its unbound state? Answering how Cela1 exists in different cellular compartments should provide insight into this important question. Last, how is the stretch-dependent expression of Cela1 transcriptionally regulated?
The reader should consider several limitations of our study. First, our model is based on a classical protease/antiprotease model of AAT-RLD; however, AAT has immunomodulatory effects (40, 41), and we did not extensively investigate the impact of Cela1 on lung inflammation or immune status. Second, we cannot say for certain that the increased lung elastance of Cela1−/− mice did not confer some degree of protection against anti-AAT oligonucleotide therapy. Either conditional deletion of Cela1 after lung morphogenesis is complete or specific anti-Cela1 therapy would be required to overcome this study limitation. Last, there may have been increased TLR7-mediated inflammation between the control and anti-AAT oligonucleotide groups, although the concentrations of IFNα and IFNβ were comparable to untreated lung.
In summary, we present phylogenetic, physiologic, and biochemical data demonstrating that Cela1 is an evolutionarily conserved lung-remodeling enzyme with an important role in postnatal lung matrix remodeling. In the absence of AAT, continued lung remodeling leads to the emphysema characteristic of AAT-RLD. The therapeutic potential of Cela1 targeting needs to be investigated in follow-up studies.
Acknowledgments
Acknowledgment
The authors thank the following individuals for their assistance with this project:
John Matthew Kofron and the CCRF Confocal Imaging Core for assistance with image acquisition and analysis; Monica Delay and the CCRF Flow Cytometry Core for assistance with flow cytometry experiment design and analysis; Xie Hurang, formerly of the CCRF Mouse Transgenic Core and now at the Michigan State Mouse Transgenic Core, for assistance with CRISPR guide RNA design and validation as well as with Cela1−/− mouse creation; Satish Madala of the Cincinnati Children’s Hospital Medical Center Division of Pulmonary Medicine for assistance with flexiVent experiments; Jason Gokey of the CCHMC Division of Pulmonary Biology for assisting with proximity ligation in situ hybridization experiments; Basilia Zingarelli and Giovanna Piraino of the CCHMC Division of Critical Care Medicine for assisting with myeloperoxidase assay and providing positive control tissue; and The Alpha-1 Foundation and the University of Cincinnati Pathology Department for providing AAT-RLD, emphysema, and control lung specimens.
Footnotes
Supported by the Cincinnati Children’s Research Foundation (CCRF) Procter Fellowship Award (B.M.V.), the Parker B. Francis Fellowship Award (B.M.V.), Alpha-1 Foundation research grant 498262 (B.M.V.), National Heart, Lung, and Blood Institute grant K08HL131261 (B.M.V.), German Research Foundation (DFG) grant HE 6190/1-2 (A.H.), the LEO Foundation Center for Cutaneous Drug Delivery (grant 2016-11-01 [A.H.]), and Fraunhofer Attract program grant 069-608203 (C.E.H.S.).
Author Contributions: R.J.: developed Cela1−/− mice and designed and performed most animal and lung histological experiments; A.H.: performed elastin degradation analysis and provided insight into the nature of Cela-mediated proteolysis; Q.F.: identified gain of function in full-length Cela1 and improved the live lung–sectioning technique; S.G.: performed validation and preliminary experiments of anti–alpha-1 antitrypsin (AAT) and control oligonucleotides; B.M.: developed the antisense oligonucleotide therapy used in AAT silencing; C.E.H.S.: designed mass spectrometric experiments and analyzed elastin degradation data; A.S.W.: identified connections between cross-linking domains and differential degradation of soluble and mature elastin; M.B.: designed and constructed the three-dimensional printed confocal lung-stretching device; H.P.: wrote the MATLAB airspace morphometry program; and B.M.V.: identified the evolutionary relationship between Cela isoforms and conceptualized, coordinated, and executed the project.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2017-0361OC on February 8, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Whitson BA, Hayes D., Jr Indications and outcomes in adult lung transplantation. J Thorac Dis. 2014;6:1018–1023. doi: 10.3978/j.issn.2072-1439.2014.07.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silverman EK, Weiss ST, Drazen JM, Chapman HA, Carey V, Campbell EJ, et al. Gender-related differences in severe, early-onset chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2000;162:2152–2158. doi: 10.1164/ajrccm.162.6.2003112. [DOI] [PubMed] [Google Scholar]
- 3.Stoller JK, Aboussouan LS. A review of α1-antitrypsin deficiency. Am J Respir Crit Care Med. 2012;185:246–259. doi: 10.1164/rccm.201108-1428CI. [DOI] [PubMed] [Google Scholar]
- 4.Joslin G, Fallon RJ, Bullock J, Adams SP, Perlmutter DH. The SEC receptor recognizes a pentapeptide neodomain of alpha 1-antitrypsin-protease complexes. J Biol Chem. 1991;266:11282–11288. [PubMed] [Google Scholar]
- 5.Sohrab S, Petrusca DN, Lockett AD, Schweitzer KS, Rush NI, Gu Y, et al. Mechanism of alpha-1 antitrypsin endocytosis by lung endothelium. FASEB J. 2009;23:3149–3158. doi: 10.1096/fj.09-129304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandhaus RA, Turino G. Neutrophil elastase-mediated lung disease. COPD. 2013;10:60–63. doi: 10.3109/15412555.2013.764403. [DOI] [PubMed] [Google Scholar]
- 7.Shapiro SD, Goldstein NM, Houghton AM, Kobayashi DK, Kelley D, Belaaouaj A. Neutrophil elastase contributes to cigarette smoke-induced emphysema in mice. Am J Pathol. 2003;163:2329–2335. doi: 10.1016/S0002-9440(10)63589-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Löffek S, Schilling O, Franzke CW. Series “matrix metalloproteinases in lung health and disease”: biological role of matrix metalloproteinases: a critical balance. Eur Respir J. 2011;38:191–208. doi: 10.1183/09031936.00146510. [DOI] [PubMed] [Google Scholar]
- 9.Gøtzsche PC, Johansen HK. Intravenous alpha-1 antitrypsin augmentation therapy for treating patients with alpha-1 antitrypsin deficiency and lung disease. Cochrane Database Syst Rev. 2016;9:CD007851. doi: 10.1002/14651858.CD007851.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni K, Serban KA, Batra C, Petrache I. Alpha-1 antitrypsin investigations using animal models of emphysema. Ann Am Thorac Soc. 2016;13(Suppl 4):S311–S316. doi: 10.1513/AnnalsATS.201510-675KV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bird AD, Tan KH, Olsson PF, Zieba M, Flecknoe SJ, Liddicoat DR, et al. Identification of glucocorticoid-regulated genes that control cell proliferation during murine respiratory development. J Physiol. 2007;585:187–201. doi: 10.1113/jphysiol.2007.136796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, Young SM, Varisco BM. Dynamic expression of chymotrypsin-like elastase 1 over the course of murine lung development. Am J Physiol Lung Cell Mol Physiol. 2014;306:L1104–L1116. doi: 10.1152/ajplung.00126.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Y, Guo M, Whitsett JA, Xu Y. ‘LungGENS’: a web-based tool for mapping single-cell gene expression in the developing lung. Thorax. 2015;70:1092–1094. doi: 10.1136/thoraxjnl-2015-207035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi R, Liu S, Brown MD, Young SM, Batie M, Kofron JM, et al. Stretch regulates expression and binding of chymotrypsin-like elastase 1 in the postnatal lung. FASEB J. 2016;30:590–600. doi: 10.1096/fj.15-277350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szepessy E, Sahin-Tóth M. Inactivity of recombinant ELA2B provides a new example of evolutionary elastase silencing in humans. Pancreatology. 2006;6:117–122. doi: 10.1159/000090031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo S, Booten SL, Watt A, Alvarado L, Freier SM, Teckman JH, et al. Using antisense technology to develop a novel therapy for α-1 antitrypsin deficient (AATD) liver disease and to model AATD lung disease. Rare Dis. 2014;2:e28511. doi: 10.4161/rdis.28511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young SM, Liu S, Joshi R, Batie MR, Kofron M, Guo J, et al. Localization and stretch-dependence of lung elastase activity in development and compensatory growth. J Appl Physiol (1985) 2015;118:921–931. doi: 10.1152/japplphysiol.00954.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Z, Ren M, Wang Z, Zhang B, Rong YS, Jiao R, et al. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics. 2013;195:289–291. doi: 10.1534/genetics.113.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagendran M, Riordan DP, Harbury PB, Desai TJ. Automated cell-type classification in intact tissues by single-cell molecular profiling. Elife. 2017;7:e30510. doi: 10.7554/eLife.30510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knudsen L, Weibel ER, Gundersen HJ, Weinstein FV, Ochs M. Assessment of air space size characteristics by intercept (chord) measurement: an accurate and efficient stereological approach. J Appl Physiol (1985) 2010;108:412–421. doi: 10.1152/japplphysiol.01100.2009. [DOI] [PubMed] [Google Scholar]
- 21.Parameswaran H, Majumdar A, Ito S, Alencar AM, Suki B. Quantitative characterization of airspace enlargement in emphysema. J Appl Physiol (1985) 2006;100:186–193. doi: 10.1152/japplphysiol.00424.2005. [DOI] [PubMed] [Google Scholar]
- 22.Yeo GC, Keeley FW, Weiss AS. Coacervation of tropoelastin. Adv Colloid Interface Sci. 2011;167:94–103. doi: 10.1016/j.cis.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Zhao BH, Zhou JH. Decreased expression of elastin, fibulin-5 and lysyl oxidase-like 1 in the uterosacral ligaments of postmenopausal women with pelvic organ prolapse. J Obstet Gynaecol Res. 2012;38:925–931. doi: 10.1111/j.1447-0756.2011.01814.x. [DOI] [PubMed] [Google Scholar]
- 24.Getie M, Schmelzer CE, Neubert RH. Characterization of peptides resulting from digestion of human skin elastin with elastase. Proteins. 2005;61:649–657. doi: 10.1002/prot.20643. [DOI] [PubMed] [Google Scholar]
- 25.Pegueroles C, Laurie S, Albà MM. Accelerated evolution after gene duplication: a time-dependent process affecting just one copy. Mol Biol Evol. 2013;30:1830–1842. doi: 10.1093/molbev/mst083. [DOI] [PubMed] [Google Scholar]
- 26.Dunnill MS. Quantitative methods in the study of pulmonary pathology. Thorax. 1962;17:320–328. doi: 10.1136/thx.17.4.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du Y, Kitzmiller JA, Sridharan A, Perl AK, Bridges JP, Misra RS, et al. Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax. 2017;72:481–484. doi: 10.1136/thoraxjnl-2016-209598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ofulue AF, Ko M. Effects of depletion of neutrophils or macrophages on development of cigarette smoke-induced emphysema. Am J Physiol. 1999;277:L97–L105. doi: 10.1152/ajplung.1999.277.1.L97. [DOI] [PubMed] [Google Scholar]
- 29.Hautamaki RD, Kobayashi DK, Senior RM, Shapiro SD. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. Science. 1997;277:2002–2004. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- 30.Guyot N, Wartelle J, Malleret L, Todorov AA, Devouassoux G, Pacheco Y, et al. Unopposed cathepsin G, neutrophil elastase, and proteinase 3 cause severe lung damage and emphysema. Am J Pathol. 2014;184:2197–2210. doi: 10.1016/j.ajpath.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 31.Haq I, Lowrey GE, Kalsheker N, Johnson SR. Matrix metalloproteinase-12 (MMP-12) SNP affects MMP activity, lung macrophage infiltration and protects against emphysema in COPD. Thorax. 2011;66:970–976. doi: 10.1136/thx.2011.159087. [DOI] [PubMed] [Google Scholar]
- 32.Wang D, Wang W, Dawkins P, Paterson T, Kalsheker N, Sallenave JM, et al. Deletion of Serpina1a, a murine α1-antitrypsin ortholog, results in embryonic lethality. Exp Lung Res. 2011;37:291–300. doi: 10.3109/01902148.2011.554599. [DOI] [PubMed] [Google Scholar]
- 33.Martorana PA, Brand T, Gardi C, van Even P, de Santi MM, Calzoni P, et al. The pallid mouse: a model of genetic alpha 1-antitrypsin deficiency. Lab Invest. 1993;68:233–241. [PubMed] [Google Scholar]
- 34.Mao GF, Goldfinger LE, Fan DC, Lambert MP, Jalagadugula G, Freishtat R, et al. Dysregulation of PLDN (pallidin) is a mechanism for platelet dense granule deficiency in RUNX1 haplodeficiency. J Thromb Haemost. 2017;15:792–801. doi: 10.1111/jth.13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida M, Sakiyama S, Kenzaki K, Toba H, Uyama K, Takehisa M, et al. Functional evaluation of pallid mice with genetic emphysema. Lab Invest. 2009;89:760–768. doi: 10.1038/labinvest.2009.34. [DOI] [PubMed] [Google Scholar]
- 36.Carlson JA, Rogers BB, Sifers RN, Finegold MJ, Clift SM, DeMayo FJ, et al. Accumulation of PiZ α1-antitrypsin causes liver damage in transgenic mice. J Clin Invest. 1989;83:1183–1190. doi: 10.1172/JCI113999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanazawa T, Furumatsu T, Hachioji M, Oohashi T, Ninomiya Y, Ozaki T. Mechanical stretch enhances COL2A1 expression on chromatin by inducing SOX9 nuclear translocalization in inner meniscus cells. J Orthop Res. 2012;30:468–474. doi: 10.1002/jor.21528. [DOI] [PubMed] [Google Scholar]
- 38.Papachristou DJ, Papachroni KK, Basdra EK, Papavassiliou AG. Signaling networks and transcription factors regulating mechanotransduction in bone. Bioessays. 2009;31:794–804. doi: 10.1002/bies.200800223. [DOI] [PubMed] [Google Scholar]
- 39.Tschumperlin DJ. Matrix, mesenchyme, and mechanotransduction. Ann Am Thorac Soc. 2015;12(Suppl 1):S24–S29. doi: 10.1513/AnnalsATS.201407-320MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jonigk D, Al-Omari M, Maegel L, Müller M, Izykowski N, Hong J, et al. Anti-inflammatory and immunomodulatory properties of α1-antitrypsin without inhibition of elastase. Proc Natl Acad Sci USA. 2013;110:15007–15012. doi: 10.1073/pnas.1309648110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cosio MG, Bazzan E, Rigobello C, Tinè M, Turato G, Baraldo S, et al. Alpha-1 antitrypsin deficiency: beyond the protease/antiprotease paradigm. Ann Am Thorac Soc. 2016;13(Suppl 4):S305–S310. doi: 10.1513/AnnalsATS.201510-671KV. [DOI] [PubMed] [Google Scholar]
- 42.Daly MC, Atkinson SJ, Varisco BM, Klingbeil L, Hake P, Lahni P, et al. Role of matrix metalloproteinase-8 as a mediator of injury in intestinal ischemia and reperfusion. FASEB J. 2016;30:3453–3460. doi: 10.1096/fj.201600242R. [DOI] [PMC free article] [PubMed] [Google Scholar]