Abstract
Differentiation of mesenchymal stromal/stem cells (MSCs) involves a series of molecular signals and gene transcription events required for attaining cell lineage commitment. Modulation of the actin cytoskeleton using cytochalasin D (CytoD) drives osteogenesis at early timepoints in bone marrow-derived MSCs and also initiates a robust osteogenic differentiation program in adipose tissue-derived MSCs. To understand the molecular basis for these pronounced effects on osteogenic differentiation, we investigated global changes in gene expression in CytoD-treated murine and human MSCs by high-resolution RNA-sequencing (RNA-seq) analysis. A three-way bioinformatic comparison between human adipose tissue-derived MSCs (hAMSCs), human bone marrow-derived MSCs (hBMSCs), and mouse bone marrow-derived MSCs (mBMSCs) revealed significant upregulation of genes linked to extracellular matrix organization, cell adhesion and bone metabolism. As anticipated, the activation of these differentiation-related genes is accompanied by a downregulation of nuclear and cell cycle-related genes presumably reflecting cytostatic effects of CytoD. We also identified eight novel CytoD activated genes—VGLL4, ARHGAP24, KLHL24, RCBTB2, BDH2, SCARF2, ACAD10, HEPH—which are commonly upregulated across the two species and tissue sources of our MSC samples. We selected the Hippo pathway-related VGLL4 gene, which encodes the transcriptional co-factor Vestigial-like 4, for further study because this pathway is linked to osteogenesis. VGLL4 small interfering RNA depletion reduces mineralization of hAMSCs during CytoD-induced osteogenic differentiation. Together, our RNA-seq analyses suggest that while the stimulatory effects of CytoD on osteogenesis are pleiotropic and depend on the biological state of the cell type, a small group of genes including VGLL4 may contribute to MSC commitment toward the bone lineage.
Keywords: : osteogenesis, mesenchymal stromal cell, stem cell, cytochalasin D, cell signaling, bone, osteoblast
Introduction
The osteogenic potential of mesenchymal stromal/stem cells (MSCs) renders them promising biological tools for skeletal regenerative therapies and bone tissue repair [1,2]. Bone anabolic agents, including growth factors and epigenetic drugs, are considered for bone tissue regeneration [3–6]. We recently showed that the cytoskeletal modifying drug cytochalasin D (CytoD), which is a secondary metabolite derived from molds, improves the osteogenic capacity of adipose-derived MSCs [7]. Previously, CytoD has also been shown to promote osteogenesis in mouse bone marrow-derived MSCs (mBMSCs) and human bone marrow-derived MSCs (hBMSCs) by influencing MSC lineage commitment via intranuclear actin transport and structure within the nucleus [8,9]. While CytoD initiates a robust osteogenic program in MSCs, the underlying regulatory mechanisms activated during CytoD-dependent inhibition of cytosolic actin polymerization remain unexplored.
To understand the principal signaling pathways that are stimulated by CytoD, we applied next generation RNA sequencing (RNA-seq) to examine gene expression profiles that may account for the bone anabolic properties of CytoD at the transcriptome level. Our group has previously utilized RNA-seq to decipher the molecular pathways and key genes involved in cellular responses and phenotypic changes in musculoskeletal cells and stem cell types, as well at the tissue level [3–5,10–14]. Here, we have utilized the RNA-seq platform to identify novel genes involved in CytoD-mediated osteogenesis of MSCs. Our analysis included both adipose-derived and bone marrow-derived MSCs that may biologically differ in their osteogenic potential, as well as MSCs from different donors to account for individual variation.
In this study, we established two main findings based on comparative transcriptomic analyses of messenger RNA (mRNA) isolated from either bone marrow-derived or adipose-derived MSCs across multiple donors of either human or murine origin. First, the effects of CytoD are highly dependent on the biological state of the sample. For example, CytoD stimulated expression profiles vary with human donor, mammalian species and specific MSC type from different tissue sources. Second, notwithstanding the variability of expression profiles associated with CytoD, we were able to identify a distinct signature of eight relatively unexplored genes that are consistently stimulated by CytoD under diverse biological conditions, regardless of tissue origin, species, culture condition or donor. The latter finding indicates that this unique subset of genes, which includes the Hippo-pathway related gene VGLL4, may mediate the osteogenic response of CytoD.
Materials and Methods
Isolation and culture of MSCs
MSCs from human adipose tissue were isolated from the stromal vascular fraction of the fat tissue obtained from healthy donors in an outpatient clinic after informed consent as described previously [3]. Procedures for harvesting and expansion of these cells in platelet lysate were reviewed and approved by the Mayo Clinic Institutional Review Board. hBMSCs were derived from commercially purchased bone marrow mononuclear cells that were sourced from bone marrow aspirates (cat. no. 2M-125; Lonza, Walkersville, MD). Lonza representatives obtained permission for their use in research applications by informed consent or legal authorization. Mouse MSCs were collected from the marrow of 8- to 12-week-old wild-type mouse femurs and isolated by plastic adherence [15] in compliance with approved Institutional Animal Care and Use Committee (IACUC) protocols (University of North Carolina IACUC protocol ID# 18-054.0 JR).
Human adipose tissue-derived MSCs (hAMSCs) were maintained in advanced minimal essential medium (MEM) supplemented with 5% platelet lysate (PL-Max; Mill Creek Life Sciences, Rochester, MN) [16]. Bone marrow MSCs from either human or mouse sources were maintained in MEM containing 10% fetal bovine serum and 100 μg/mL penicillin/streptomycin. For experiments, cells from passages 4–6 were plated at a density of 10,000 cells per square centimeter in six-well culture plates (Fisher) in MEM and cultured for 1 day before application of treatments. MSCs from human donors from passage 4 were seeded at a density of 5,000 cells per cm2. After 24 h, cells were treated with or without CytoD (0.1 μg/mL) in maintenance media.
RNA-sequencing
Next generation RNA-seq was performed using hAMSCs, hBMSCs, and mBMSCs. MSC cultures were each treated in triplicate for 24 h with or without CytoD. Biological triplicates for each of the three sample types (n = 3) underwent RNA-seq to yield technically robust datasets. RNA-seq analysis of MSCs was performed as described previously [3] using a well-established standard biochemical protocol and institutionally standardized bioinformatics pipeline with oligo dT purified mRNA and indexed complementary DNAs (cDNAs) (standard TruSeq Kits 12-Set A and 12-Set B) on the Illumina platform (Illumina's RTA version 1.17.21.3). Raw reads were mapped using standard pipeline (Bioinformatics Core standard tool) comprising MAPRSeq v.1.2.1 analysis, alignment with TopHat 2.0.6, and gene counting with the HTSeq application, as well as normalization and expression analysis using edgeR. Gene expression is reported in reads per kilobase pair per million mapped reads (RPKM).
RNA-interference
Transfection of hAMSCs was performed with small interfering RNA (siRNA) (50 nM) in serum-free Opti-MEM overnight before replacing the medium and adding reagents for control or treatment groups. VGLL4 siRNA and scrambled non-silencing RNAs were purchased from Dharmacon (GE Healthcare) and transfection was performed as per the protocol recommended by the manufacturer. Total RNA was isolated at days 2 and 5 posttransfection and reverse transcribed to cDNA using SuperScript III First-Strand Synthesis System (Invitrogen). Gene expression was quantified by real-time polymerase chain reaction PCR using 10 ng cDNA per reaction using the QuantiTect SYBR Green PCR Kit and a CFX384 Real-Time qPCR System (Bio-Rad, Hercules, CA). Gene expression was quantified using 2−ΔΔCt method and is represented as relative expression units of VGLL4 normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
Real-time PCR
To evaluate gene expression of commonly upregulated genes with CytoD, hAMSCs were seeded at 3,000 cells per cm2 and allowed to adhere overnight. The following day, cells were treated with 0.1 μg/mL CytoD or vehicle (DMSO or dimethyl sulfoxide). After 24 h, cells were lysed with Trizol for isolation of total RNA. Primers were designed for VGLL4, KLHL24, RCBTB2, SCARF2, ARHGAP24, ACAD10, HEPH, and BDH2. cDNA preparation and gene expression analysis were performed as described above in the RNA-interference section.
Osteogenic differentiation
For differentiation following siRNA transfection, hAMSCs were seeded at 3,000 cells per cm2 in maintenance medium in six-well plates and incubated under standard culture conditions for 24 h before being changed to osteogenic medium containing vehicle (DMSO) or 0.1 μg/mL CytoD (Sigma-Aldrich). Osteogenic medium maintenance media supplemented with 10 nM dexamethasone, 25 μg/mL ascorbic acid, and 10 mM β-glycerophosphate. Cells were maintained for up to a maximum of 14 days, with media changed every 3 to 4 days. RNA and protein were collected at various time points (days 1, 2, 3, and 6) during the course of differentiation. CytoD or vehicle was present in osteogenic media throughout the course of the experiments.
Statistical analyses
Results are provided as mean ± standard deviation, unless specified otherwise. All data were typically collected from experiments performed in triplicate and from three distinct human or mouse donors. Data were analyzed by applying Student's t-test or one-way analysis of variance where necessary using GraphPad Prism 7 version 7.03 software: P value <0.05 was considered significant.
Results
RNA-seq reveals distinct changes in gene expression with CytoD treatment
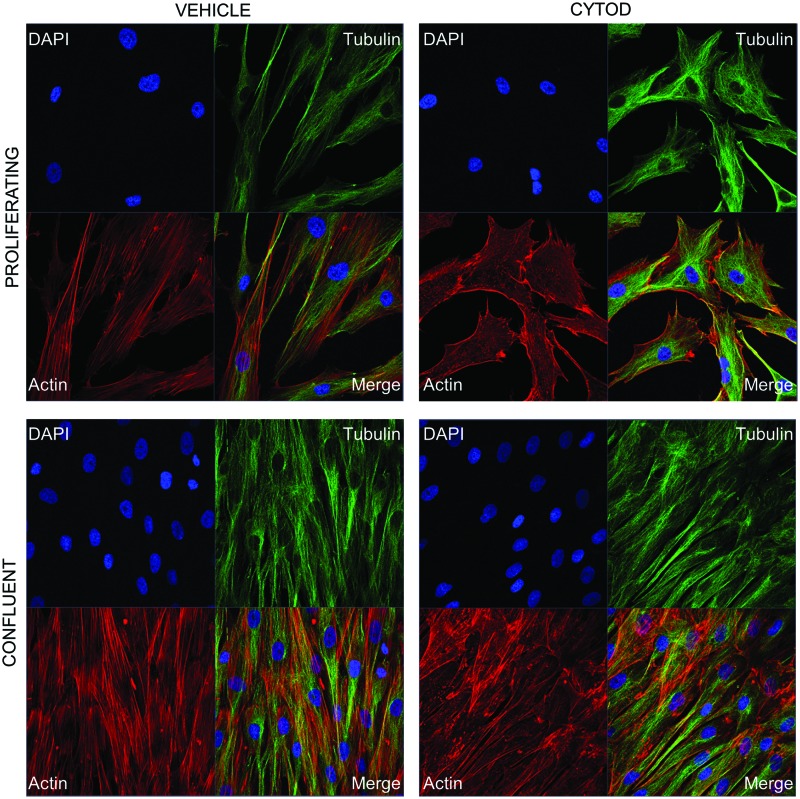
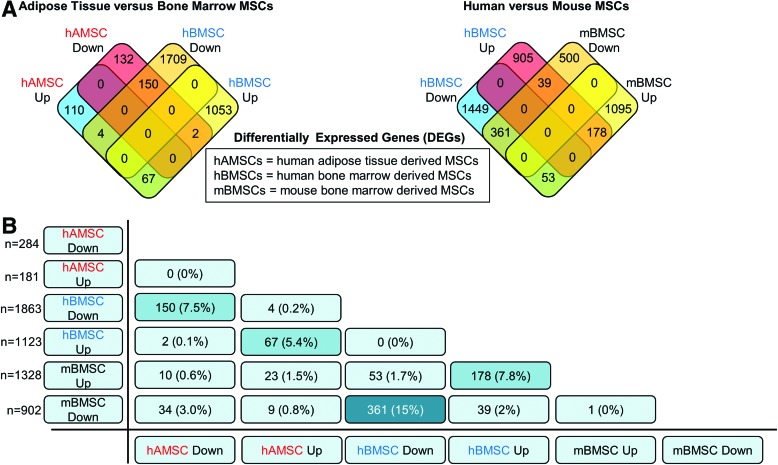
CytoD treatment of MSCs showed marked differences in cell shape and actin organization 24 h posttreatment under proliferating conditions; under confluent conditions, actin cytoskeleton was altered but cell shape was unaffected (Fig. 1). To characterize early changes in gene expression due to CytoD treatment of MSCs from different species and tissues, we performed RNA-seq on MSC cultures treated with CytoD for 24 h without osteogenic supplements. Conservative filtering of mRNAs was performed by selecting mRNAs expressed at robust levels (RPKM >0.3) with differential gene expression (fold-change >1.4) and statistically significant (P < 0.05) difference in expression values when compared between samples. A four-way Venn diagram analysis using the Venny interactive tool [17] was performed to evaluate the number of genes commonly upregulated or downregulated with CytoD treatment. Our analysis shows that genes were differentially expressed upon comparing human MSCs derived from adipose tissue and those from bone marrow: a total of 67 genes were commonly upregulated, and 150 genes were commonly downregulated in human MSCs (Fig. 2A). Comparing bone marrow-derived MSCs from human and mouse sources showed that 178 genes were commonly upregulated and 361 genes were commonly downregulated. Bone marrow-derived MSCs from mouse and human share a greater percentage of genes with similar modulations in gene expression than is shared between human MSCs from either bone marrow or adipose tissue. Additional verification by cross-table analysis (Fig. 2B) reveals a similar trend with higher percentages of genes commonly regulated between mBMSC and hBMSC (7.8% upregulated and 15% downregulated with CytoD treatment) compared to human MSCs derived from bone and adipose tissue (5.4% upregulated and 7.5% downregulated with CytoD treatment). Collectively, these data show that CytoD treatment alters the biological ground-states of undifferentiated MSCs, and importantly that CytoD has distinct cell type-specific effects on gene expression.
FIG. 1.
Morphological changes in human MSCs treated with CytoD. Immunofluorescence images of hBMSCs at 24 h posttreatment with 0.1 μg/mL CytoD or vehicle (DMSO) under proliferative (top panel) and confluent (bottom panel) conditions. Cells are stained for actin (phalloidin red), tubulin (green, Alexaflour 488), and nuclei are stained with DAPI. CytoD, cytochalasin D; DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide; hBMSCs, human bone marrow-derived MSCs; MSCs, mesenchymal stromal/stem cells. Color images available online at www.liebertpub.com/scd
FIG. 2.
Gene expression changes in mouse and human MSCs. (A) Four-way Venn diagram analysis showing number of genes upregulated or downregulated with CytoD in hAMSCs and hBMSCs. Also shown is another four-way Venn diagram analysis showing comparisons of genes regulated by CytoD in bone marrow MSCs derived from human (hBMSCs) and mouse samples (mBMSCs). (B) Cross-table analysis indicating total number (n) and percentages of genes upregulated or downregulated with CytoD across both human and mouse MSCs derived from bone marrow and adipose tissues. hAMSCs, human adipose tissue-derived MSCs; mBMSC, mouse bone marrow-derived MSCs. Color images available online at www.liebertpub.com/scd
Functional components commonly regulated with CytoD treatment
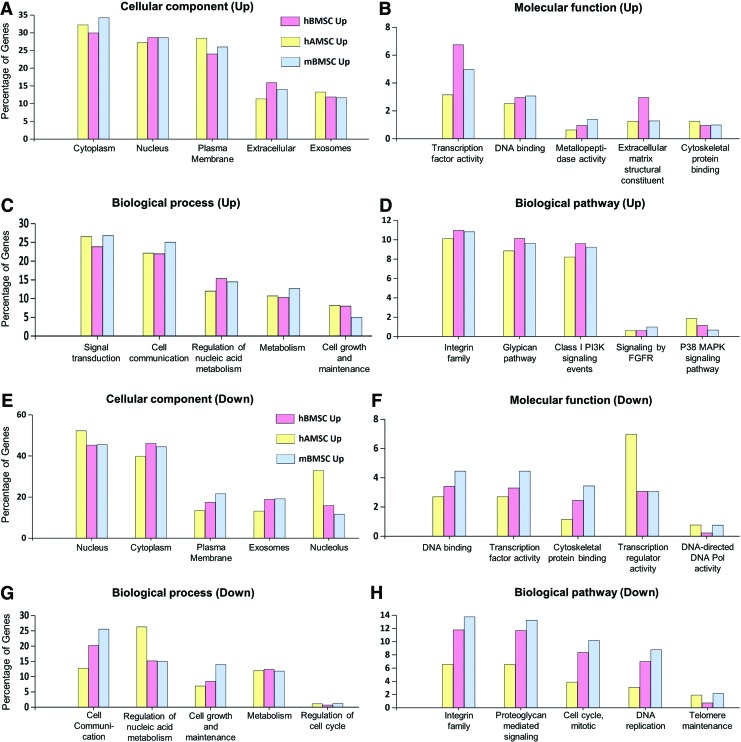
We next examined the functional role of CytoD-modulated gene sets. For this analysis, we used our CytoD-responsive gene lists (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/scd) and conducted gene ontology (GO) analysis using FunRich online software. These analyses show that the upregulated genes were mostly cytoplasmic, nuclear, and plasma membrane related. Similarly, the percentages of genes involved in biological processes such as signal transduction and cell communication were comparatively higher in CytoD treated MSCs (Fig. 3A). The percentages of upregulated genes associated with each cellular component did not vary considerably between the three groups of MSCs, suggesting that CytoD targets similar overall pathways in different types of MSCs even though different genes may be targeted depending on the tissue of origin or species. We also observed changes in a small subset of genes involved in biological processes and pathways linked to cell growth and maintenance, consistent with cell growth regulatory effects of CytoD on MSCs.
FIG. 3.
Functional factors commonly regulated with CytoD treatment. FunRich analysis charts revealing the percentages of upregulated genes and the cellular components involved (A), associated molecular functions (B), related biological process (C), and biological pathways affected (D)with CytoD treatment. Similarly, percentages of genes downregulated with CytoD and the respective functional components involved are depicted (E–H). FGFR, fibroblast growth factor receptor; MAPK, mitogen-activated protein kinase. Color images available online at www.liebertpub.com/scd
Molecular functions that were identified as subject to CytoD regulation were related to transcription factor activities, DNA binding, metallopeptidase activity, cytoskeletal protein binding, and extracellular matrix (ECM) constituents—all showing a very limited percentage of genes ranging from 1.5% to 7% (Fig. 3B). We also examined the biological processes influenced by CytoD and identified that genes linked to signal transduction and cell communication were uniformly upregulated in all three groups of MSCs (Fig. 3C). We also note equal representation of Integrin family genes and PI3K signaling genes among the three groups of MSCs, and limited percentage of genes associated with fibroblast growth factor receptor (FGFR) signaling and the p38 MAPK (mitogen-activated protein kinase) pathway (which is downstream of FGFR) were also upregulated with CytoD treatment (Fig. 3D). Among the genes downregulated, the most prominent were those involved in functions such as mitosis, cytoskeletal organization, nucleotide synthesis, and binding, DNA replication (Fig. 3E–H). This observation clearly indicates that CytoD has potent cytostatic effects. Collectively, the GO analyses reveal that a vast majority of CytoD-induced functions are linked to ECM proteins, plasma-membrane associated proteins, glycoproteins, and cell surface receptors, as well as the corresponding biological processes associated with cell communication and signal transduction. Because these processes are involved in cell differentiation, these findings suggest that the RNA-seq data uncovered pathways relevant to the osteogenic effects of CytoD on MSCs. Therefore, we proceeded to assess which genes are up- or downregulated by CytoD regardless of biological origin.
Identification of commonly upregulated genes in different types of MSCs treated with CytoD
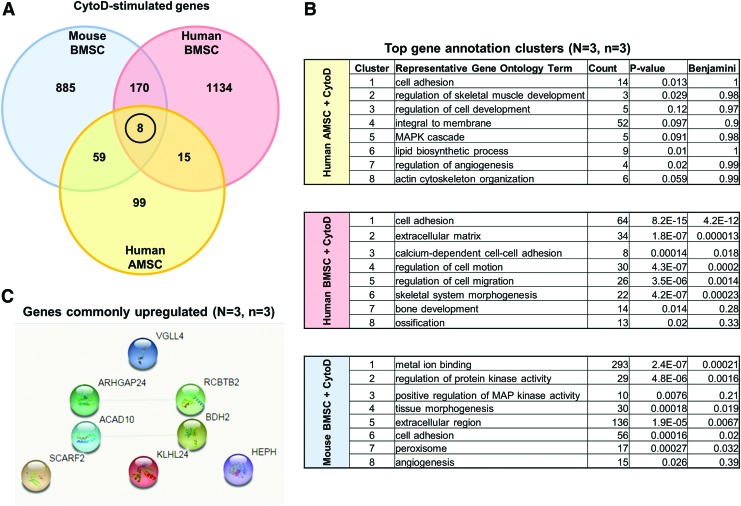
Genes that were revealed by RNA-seq analysis to be significantly upregulated with CytoD treatment of hBMSCs (pink circle), mBMSCs (blue circle), and hAMSCs (yellow circle) were compared using a three-way Venn diagram (Fig. 4A). GO analysis revealed that CytoD increases expression of genes involved in cell adhesion, skeletal tissue development, as well as angiogenesis. In hAMSCs, the representative GO terms from the top eight gene clusters revealed by Database for Annotation, Visualization and Integrated Discovery (DAVID) analysis included genes involved in cell adhesion, regulation of cell development, angiogenesis, and actin cytoskeleton organization (Fig. 4B). Similar GO terms were revealed in CytoD-treated hBMSCs including cell adhesion, ECM, calcium-dependent cell-adhesion, bone development, and ossification among others. In mBMSCs, metal ion binding, extracellular region, angiogenesis, and protein kinase activity were the top represented terms revealed among other GO terms that were in common between hAMSCs and hBMSCs.
FIG. 4.
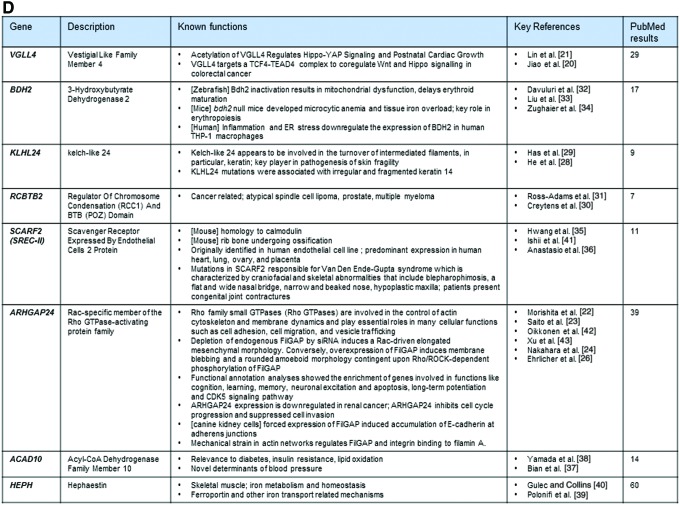
Commonly upregulated genes across species and tissue types. (A) Three-way Venn diagram of upregulated genes reveals identification of eight unique genes that are commonly upregulated across mouse and human MSCs. (B) Table derived from DAVID functional annotation tool listing top eight gene annotation cluster terms, counts, and P values. Of significant importance are representative GO terms including skeletal muscle development, bone development, ossification, tissue morphogenesis and angiogenesis. (C) STRING v10.5 network analysis illustrates that these genes are standalone and show no reported networks or connectivity between the eight genes. (D) Summary and description of previous findings of the panel of eight genes consistently modulated by CytoD independent of species or cell type. DAVID, Database for Annotation, Visualization and Integrated Discovery; GO, gene ontology. Color images available online at www.liebertpub.com/scd
Among these three gene sets, we identified a unique signature of eight genes that were commonly upregulated in all studied classes of MSCs, including VGLL4 (Vestigial Like Family Member 4), ARHGAP24 (Rac-specific member of the Rho GTPase-activating protein family), ACAD10 (Acyl-CoA Dehydrogenase Family Member 10), SCARF2 (Scavenger Receptor Expressed by Endothelial Cells 2 Protein), RCBTB2 (Regulator of Chromosome Condensation [RCC1] and BTB [POZ] Domain), BDH2 (3-hydroxybutyrate dehydrogenase 2), KLHL24 (kelch-like 24), and HEPH (Hephaestin) (Fig. 4D). STRING version 10.5 network analysis did not reveal any prominent nodes or possible regulatory relationships between these eight genes (Fig. 4C). Hence, a relatively small subset of functionally diverse genes may mediate the main biological effects of CytoD on MSCs.
Additionally, we validated the identified gene signature by performing real-time quantitative PCR (qPCR) analysis to validate the upregulation of these eight genes in CytoD-treated hAMSC. Our qPCR results confirm that CytoD promoted upregulation of the identified CytoD-responsive genes, including KLHL24, RCBTB2, SCARF2, ARHGAP24, HEPH, and BDH2 (Supplementary Fig. S1).
VGLL4 is required for osteogenic differentiation of MSCs in response to CytoD
Of the eight genes we identified, VGLL4 was of particular interest, because this gene encodes a co-regulator of the Hippo-pathway, which also includes the transcriptional co-factors Yes Associate Protein 1 (YAP1) and WW Domain Containing Transcription Regulator 1 (WWTR1/TAZ). YAP1 or WWTR1/TAZ each bind to TEA (TE for transcription enhancer factor 1 [TEF1] and trans-acting factor C [TEC1] and A for ABAA binding motif; a highly-conserved DNA-binding motif) domain transcription factors (TEADs) to control the size of organs and tissues [18]. Previous studies have suggested that YAP1, a co-factor that suppresses Runx2 function, is exported from the nucleus after CytoD stimulation [9]. Importantly, TAZ has a known function in osteogenic differentiation of MSCs [19]. In this model, loss of YAP1 in the nucleus is predicted to support Runx2-dependent induction of Osterix (SP7/OSX) in the initial stages of osteogenic differentiation of MSCs. However, YAP1 release from TEAD proteins would concomitantly also promote interactions between VGLL and TEADs [18]. Hence, increased expression of VGLL4 in all MSCs treated with CytoD could be part of a feedback loop linked to the YAP1/TAZ-TEAD-VGLL axis within the Hippo signaling pathway.
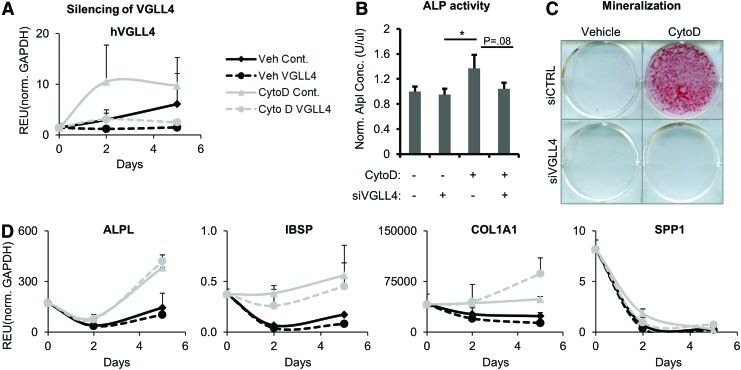
To address this hypothesis, we investigated whether silencing of VGLL4 by RNA interference affected osteogenic differentiation of hAMSCs, which typically show the most robust biological response to CytoD treatment in terms of cell morphology and gene expression. Furthermore, adipose MSCs have consistently permitted higher siRNA transfection efficiency than marrow-derived MSCs, and were therefore utilized for silencing VGLL4 mRNA. Gene expression analyses by qPCR confirmed that VGLL4 mRNA levels were depleted as expected (Fig. 5A). Osteogenic differentiation after silencing of VGLL4 modestly reduced ALP activity assayed at day 7 of osteogenic differentiation (Fig. 5B) but had rather dramatic effects on mineralization at day 10 as detected by Alizarin Red staining of calcium deposition (Fig. 5C). Further analysis by qPCR showed that silencing of VGLL4 in hAMSCs also reduced expression of the bone-related sialoprotein IBSP gene, but stimulated accumulation of COL1A1 mRNA (Fig. 5D). The latter results are consistent with the cells assuming a more fibroblastic rather than osteogenic phenotype upon CytoD treatment in the absence of VGLL4 (Fig. 1A, B), in corroboration with our previous findings showing increases in COL1A1 mRNA in CytoD-treated adipose MSCs [7]. Taken together, these results indicate that VGLL4 may contribute to CytoD effects on osteogenic differentiation of MSCs.
FIG. 5.
VGLL4 silencing and its effects on osteogenic differentiation. (A) Real-time PCR confirming siRNA knockdown of VGLL4 in hAMSCs at days 2 and 5, posttransfection. (B) ALP activity recorded at day 7 posttreatment with or without CytoD of hAMSCs transfected with siRNA VGLL4 or siRNA control. (C) Alizarin Red staining at D9 posttreatment with or without CytoD of hAMSCs transfected with siRNA VGLL4 or siRNA control. (D) Gene expression of key osteogenic genes in transfected hAMSCs assayed by real-time qPCR at multiple time points postdifferentiation with or without CytoD. ALP, alkaline phosphatase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; qPCR, quantitative PCR; REU, relative expression units; siRNA, small interfering RNA. Color images available online at www.liebertpub.com/scd
Discussion
MSCs are critical for normal skeletal development and have therapeutic potential in skeletal tissue repair. In previous studies, we have shown that CytoD alters the undifferentiated state of adipose-derived MSCs at both the phenotype and gene-expression levels and renders these cells competent for matrix mineralization [8]. Based on these and other previous findings, CytoD promoted marked increases in the osteogenic differentiation of MSCs at genetic, molecular and histological levels [7–9], irrespective of differences in culture media conditions, human versus mouse donors, or tissue source (bone marrow vs. stromal vascular fraction of adipose-tissue). In the current study, we have further explored the gene signatures of CytoD-treated MSCs from different sources (ie, bone marrow and the stromal vascular fraction of adipose tissue), two different species (human and mouse), several distinct donors, and media supplementation conditions (human platelet lysate for hAMSCs and hBMSCs vs. bovine serum for mBMSCs). Our strategy derived new information on genes that may contribute to MSC osteogenic lineage commitment and differentiation of MSCs. Our study reveals that the effects of CytoD are dependent on several biological variables that may need to be further investigated, including donor to donor variation and tissue source that were not explicitly tested in the current experimental design. Notwithstanding variation caused by these biological variables, our approach revealed a common set of genes that is co-regulated under all experimental conditions tested and that may be required for CytoD dependent effects on osteogenesis.
From a regenerative medicine perspective, it may be possible to apply a CytoD treatment strategy to tune the fate of MSCs toward enhanced osteogenesis for orthopedic applications that require bone regeneration and/or repair. Stimulation of the osteogenic gene program by CytoD in both adipose and bone marrow-derived MSCs is reflected by increased expression of classical bone markers including alkaline phosphatase (ALPL), collagen type I (COL1A1), and osteonectin (SPARC) [7,8]. Consistent with these findings, initial RNA-seq analysis of adipose tissue-derived MSCs also identified genes involved in calcium ion binding, calmodulin, collagen and other ECM markers, as well as Wnt and PI3K signaling pathways [8]. While CytoD enhances differentiation of MSCs that are also treated with standard osteogenic differentiation cocktails, one important attribute of CytoD is that treatment with this compound by itself (ie, without other inducers) can promote robust mineralization of adipose tissue derived MSCs [7].
Our current study identified eight common CytoD-responsive genes that are stimulated by CytoD in MSCs from multiple donors, different tissue sources, and distinct species (ie, VGLL4, KLHL24, RCBTB2, BDH2, SCARF2, ARHGAP24, ACAD10, and HEPH). Of these, VGLL4 represents the most interesting gene, because it is a component of the Hippo-YAP signaling pathway and targets transcription factor 4 (TCF4)/TEAD4 complex [20,21]. Indeed, loss-of-function studies show that osteogenic differentiation and mineralization of MSCs in response to CytoD treatment is inhibited by siRNA depletion of VGLL4.
Among all MSC sources, CytoD also upregulated the expression of a cytoskeletal organizing molecule, ARHGAP24, which is a member of the large group of Rho family small GTPases (Rho GTPases) involved in the control of actin cytoskeleton and membrane dynamics. These genes play essential roles in many cellular functions such as cell adhesion, cell migration, and vesicle trafficking [22,23]. Also referred to as FilGAP protein, ARHGAP24 is activated by ROCK to control cytoskeletal stiffness and prevent cell spreading [22]. In canine kidney cells, forced expression of FilGAP induced accumulation of E-cadherin at adherens junctions [24]. Also, it has been shown that mechanical strain, which promotes actin polymerization [25], causes the ARHGAP24/FilGAP protein to disassociate from filamin A on actin polymers to support cytoskeletal structure [26]. ARHGAP24 is closely related to the Rho GTPase activating protein ARHGAP18, which is also involved in MSC differentiation. ARHGAP18, which is highly expressed in bone marrow-derived MSC, supports tonic inhibition of the actin cytoskeleton [27]. It is conceivable that the activities of ARHGAP24 and ARHGAP18 are functionally linked and jointly influence osteogenic lineage commitment.
The biological roles of the other genes are less clear. KLHL24 appears to be involved in the turnover of intermediated filaments (eg, keratin) and pathogenesis of skin fragility, while KLHL24 mutations are associated with irregular and fragmented keratin 14 [28,29]. RCBTB2, which is also stimulated by CytoD, has been linked to atypical spindle cell lipoma, prostate cancer, multiple myeloma [30,31]. The CytoD responsive gene BDH2 encodes 3-hydroxybutyrate dehydrogenase 2, which is an enzyme playing a key role in iron homeostasis and transport. In zebrafish, bdh2 inactivation results in mitochondrial dysfunction and delays erythroid maturation [32]. In mice, Bdh2-null mice developed microcytic anemia and tissue iron overload, reflecting a key role in erythropoiesis [33]. Furthermore, BDH2 is negatively regulated by inflammation and endoplasmic reticulum stress in human cultured macrophage cell lines [34]. SCARF2 is known to be linked to calmodulin and related ossification processes [35]. Mutations in SCARF2 are associated with craniofacial and skeletal abnormalities [36]. ACAD10 has been linked to diabetes, insulin resistance, lipid oxidation, as well as blood pressure [37,38]. HEPH has been shown to be involved in skeletal muscle functions, iron metabolism and homeostasis, as well as with iron transport related mechanisms [39,40]. Taken together, the signatures of the eight genes that are linked to osteogenic induction by CytoD appear to have remarkably diverse functions. We conclude that our RNA-seq studies reveal important mechanistic insights into CytoD-induced osteogenesis and validate the osteogenic responses of MSCs treated with CytoD at a high level of molecular resolution.
Supplementary Material
Acknowledgments
This study received funding support from the U.S. National Institutes of Health (grant nos. R01-AR049069 to A.v.W., R01-AR066616 to J.R., and F32-AR066508 to A.D.) and the Mayo Clinic Center for Regenerative Medicine in addition to generous philanthropic gifts from William and Karen Eby. We appreciate the bioinformatic support provided by the Mayo Clinic Bioinformatics Core and especially thank Jaime Davila, Ying Li, and Jared Evans for sharing their expertise. We also thank all present and former members of our laboratories, particularly Scott Riester, Emily Camilleri, Eric Lewallen, Janet Denbeigh, Roman Thaler, Farzaneh Khani, Catalina Galeano, Martina Gluscevic, and Oksana Pichurin for sharing reagents and ideas as well as stimulating discussions throughout the course of this work.
Author Disclosure Statement
The authors indicate no conflicts of interest and have nothing to disclose.
References
- 1.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S. and Marshak DR. (1999). Multilineage potential of adult human mesenchymal stem cells. Science 284:143–147 [DOI] [PubMed] [Google Scholar]
- 2.Pittenger MF, Mosca JD. and McIntosh KR. (2000). Human mesenchymal stem cells: progenitor cells for cartilage, bone, fat and stroma. Curr Top Microbiol Immunol 251:3–11 [DOI] [PubMed] [Google Scholar]
- 3.Dudakovic A, Camilleri E, Riester SM, Lewallen EA, Kvasha S, Chen X, Radel DJ, Anderson JM, Nair AA, et al. (2014). High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J Cell Biochem 115:1816–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riester SM, Arsoy D, Camilleri ET, Dudakovic A, Paradise CR, Evans JM, Torres-Mora J, Rizzo M, Kloen P, et al. (2015). RNA sequencing reveals a depletion of collagen targeting microRNAs in Dupuytren's disease. BMC Med Genomics 8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudakovic A, Camilleri ET, Riester SM, Paradise CR, Gluscevic M, O'Toole TM, Thaler R, Evans JM, Yan H, et al. (2016). Enhancer of zeste homolog 2 inhibition stimulates bone formation and mitigates bone loss caused by ovariectomy in skeletally mature mice. J Biol Chem 291:24594–24606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dudakovic A, Camilleri ET, Xu F, Riester SM, McGee-Lawrence ME, Bradley EW, Paradise CR, Lewallen EA, Thaler R, et al. (2015). Epigenetic control of skeletal development by the histone methyltransferase EZH2. J Biol Chem 290:27604–27617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samsonraj RM, Dudakovic A, Manzar B, Sen B, Dietz AB, Cool SM, Rubin J. and van Wijnen AJ. (2017). Osteogenic stimulation of human adipose-derived mesenchymal stem cells using a fungal metabolite that suppresses the polycomb group protein EZH2. Stem Cells Transl Med 7:197–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sen B, Uzer G, Samsonraj RM, Xie Z, McGrath C, Styner M, Dudakovic A, van Wijnen AJ. and Rubin J. (2017). Intranuclear actin structure modulates mesenchymal stem cell differentiation. Stem Cells 35:1624–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sen B, Xie Z, Uzer G, Thompson WR, Styner M, Wu X. and Rubin J. (2015). Intranuclear actin regulates osteogenesis. Stem Cells 33:3065–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudakovic A, Gluscevic M, Paradise CR, Dudakovic H, Khani F, Thaler R, Ahmed FS, Li X, Dietz AB, et al. (2017). Profiling of human epigenetic regulators using a semi-automated real-time qPCR platform validated by next generation sequencing. Gene 609:28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin Y, Lewallen EA, Camilleri ET, Bonin CA, Jones DL, Dudakovic A, Galeano-Garces C, Wang W, Karperien MJ, et al. (2016). RNA-seq analysis of clinical-grade osteochondral allografts reveals activation of early response genes. J Orthop Res 34:1950–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling L, Camilleri ET, Helledie T, Samsonraj RM, Titmarsh DM, Chua RJ, Dreesen O, Dombrowski C, Rider DA, et al. (2016). Effect of heparin on the biological properties and molecular signature of human mesenchymal stem cells. Gene 576:292–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollock K, Samsonraj RM, Dudakovic A, Thaler R, Stumbras A, McKenna DH, Dosa PI, van Wijnen AJ. and Hubel A. (2017). Improved post-thaw function and epigenetic changes in mesenchymal stromal cells cryopreserved using multicomponent osmolyte solutions. Stem Cells Dev 26:828–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riester SM, Torres-Mora J, Dudakovic A, Camilleri ET, Wang W, Xu F, Thaler RR, Evans JM, Zwartbol R, et al. (2017). Hypoxia-related microRNA-210 is a diagnostic marker for discriminating osteoblastoma and osteosarcoma. J Orthop Res 35:1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Case N, Xie Z, Sen B, Styner M, Zou M, O'Conor C, Horowitz M. and Rubin J. (2010). Mechanical activation of beta-catenin regulates phenotype in adult murine marrow-derived mesenchymal stem cells. J Orthop Res 28:1531–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crespo-Diaz R, Behfar A, Butler GW, Padley DJ, Sarr MG, Bartunek J, Dietz AB. and Terzic A. (2011). Platelet lysate consisting of a natural repair proteome supports human mesenchymal stem cell proliferation and chromosomal stability. Cell Transplant 20:797–811 [DOI] [PubMed] [Google Scholar]
- 17.Oliveros JC. (2007). VENNY. An interactive tool for comparing lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html
- 18.Meng Z, Moroishi T. and Guan KL. (2016). Mechanisms of Hippo pathway regulation. Genes Dev 30:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, et al. (2005). TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 309:1074–1078 [DOI] [PubMed] [Google Scholar]
- 20.Jiao S, Li C, Hao Q, Miao H, Zhang L, Li L. and Zhou Z. (2017). VGLL4 targets a TCF4-TEAD4 complex to coregulate Wnt and Hippo signalling in colorectal cancer. Nat Commun 8:14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Z, Guo H, Cao Y, Zohrabian S, Zhou P, Ma Q, VanDusen N, Guo Y, Zhang J, et al. (2016). Acetylation of VGLL4 regulates Hippo-YAP signaling and postnatal cardiac growth. Dev Cell 39:466–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morishita Y, Tsutsumi K. and Ohta Y. (2015). Phosphorylation of serine 402 regulates RacGAP protein activity of FilGAP protein. J Biol Chem 290:26328–26338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito K, Ozawa Y, Hibino K. and Ohta Y. (2012). FilGAP, a Rho/Rho-associated protein kinase-regulated GTPase-activating protein for Rac, controls tumor cell migration. Mol Biol Cell 23:4739–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakahara S, Tsutsumi K, Zuinen T. and Ohta Y. (2015). FilGAP, a Rho-ROCK-regulated GAP for Rac, controls adherens junctions in MDCK cells. J Cell Sci 128:2047–2056 [DOI] [PubMed] [Google Scholar]
- 25.Sen B, Xie Z, Case N, Thompson WR, Uzer G, Styner M. and Rubin J. (2014). mTORC2 regulates mechanically induced cytoskeletal reorganization and lineage selection in marrow-derived mesenchymal stem cells. J Bone Miner Res 29:78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA. and Stossel TP. (2011). Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 478:260–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson WR, Yen SS, Uzer G, Xie Z, Sen B, Styner M, Burridge K. and Rubin J. (2018). LARG GEF and ARHGAP18 orchestrate RhoA activity to control mesenchymal stem cell lineage. Bone 107:172–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y, Maier K, Leppert J, Hausser I, Schwieger-Briel A, Weibel L, Theiler M, Kiritsi D, Busch H, et al. (2016). Monoallelic mutations in the translation initiation codon of KLHL24 cause skin fragility. Am J Hum Genet 99:1395–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Has C. (2017). The “Kelch” surprise: KLHL24, a new player in the pathogenesis of skin fragility. J Invest Dermatol 137:1211–1212 [DOI] [PubMed] [Google Scholar]
- 30.Creytens D, van Gorp J, Savola S, Ferdinande L, Mentzel T. and Libbrecht L. (2014). Atypical spindle cell lipoma: a clinicopathologic, immunohistochemical, and molecular study emphasizing its relationship to classical spindle cell lipoma. Virchows Arch 465:97–108 [DOI] [PubMed] [Google Scholar]
- 31.Ross-Adams H, Lamb AD, Dunning MJ, Halim S, Lindberg J, Massie CM, Egevad LA, Russell R, Ramos-Montoya A, et al. (2015). Integration of copy number and transcriptomics provides risk stratification in prostate cancer: a discovery and validation cohort study. EBioMedicine 2:1133–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davuluri G, Song P, Liu Z, Wald D, Sakaguchi TF, Green MR. and Devireddy L. (2016). Inactivation of 3-hydroxybutyrate dehydrogenase 2 delays zebrafish erythroid maturation by conferring premature mitophagy. Proc Natl Acad Sci U S A 113:E1460-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z, Ciocea A. and Devireddy L. (2014). Endogenous siderophore 2,5-dihydroxybenzoic acid deficiency promotes anemia and splenic iron overload in mice. Mol Cell Biol 34:2533–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zughaier SM, Stauffer BB. and McCarty NA. (2014). Inflammation and ER stress downregulate BDH2 expression and dysregulate intracellular iron in macrophages. J Immunol Res 2014:140728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang M, Kalinin A. and Morasso MI. (2005). The temporal and spatial expression of the novel Ca++-binding proteins, Scarf and Scarf2, during development and epidermal differentiation. Gene Expr Patterns 5:801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anastasio N, Ben-Omran T, Teebi A, Ha KC, Lalonde E, Ali R, Almureikhi M, Der Kaloustian VM, Liu J, et al. (2010). Mutations in SCARF2 are responsible for Van Den Ende-Gupta syndrome. Am J Hum Genet 87:553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bian L, Hanson RL, Muller YL, Ma L, Kobes S, Knowler WC, Bogardus C. and Baier LJ. (2010). Variants in ACAD10 are associated with type 2 diabetes, insulin resistance and lipid oxidation in Pima Indians. Diabetologia 53:1349–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada Y, Sakuma J, Takeuchi I, Yasukochi Y, Kato K, Oguri M, Fujimaki T, Horibe H, Muramatsu M, et al. (2017). Identification of polymorphisms in 12q24.1, ACAD10, and BRAP as novel genetic determinants of blood pressure in Japanese by exome-wide association studies. Oncotarget 8:43068–43079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polonifi A, Politou M, Kalotychou V, Xiromeritis K, Tsironi M, Berdoukas V, Vaiopoulos G. and Aessopos A. (2010). Iron metabolism gene expression in human skeletal muscle. Blood Cells Mol Dis 45:233–237 [DOI] [PubMed] [Google Scholar]
- 40.Gulec S. and Collins JF. (2014). Silencing the Menkes copper-transporting ATPase (Atp7a) gene in rat intestinal epithelial (IEC-6) cells increases iron flux via transcriptional induction of ferroportin 1 (Fpn1). J Nutr 144:12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishii J, Adachi H, Aoki J, Koizumi H, Tomita S, Suzuki T, Tsujimoto M, Inoue K. and Arai H. (2002). SREC-II, a new member of the scavenger receptor type F family, trans-interacts with SREC-I through its extracellular domain. J Biol Chem 277:39696–39702 [DOI] [PubMed] [Google Scholar]
- 42.Oikkonen J, Onkamo P, Jarvela I. and Kanduri C. (2016). Convergent evidence for the molecular basis of musical traits. Sci Rep 6:39707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu G, Lu X, Huang T. and Fan J. (2016). ARHGAP24 inhibits cell cycle progression, induces apoptosis and suppresses invasion in renal cell carcinoma. Oncotarget 7:51829–51839 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.