Abstract
Epstein-Barr virus (EBV) is an oncogenic herpes virus that is closely associated with the initiation and development of nasopharyngeal carcinoma (NPC), lymphoma and other malignant tumors. EBV encodes 44 mature miRNAs that regulate viral and host cell gene expression and plays a variety of roles in biological functions and the development of cancer. In this review, we summarized the biological functions and molecular mechanisms of Epstein-Barr virus-encoded microRNAs (EBV miRNAs) in tumor immune evasion, proliferation, anti-apoptosis, invasion, metastasis and as a potential biomarker for NPC diagnosis and prognosis. The knowledge generated by EBV miRNAs can be used for EBV miRNA-based precision cancer treatments in the near future.
Keywords: nasopharyngeal carcinoma, EBV miRNAs, immune evasion, anti-apoptosis, metastasis, biomarker
Introduction
The incidence rate of nasopharyngeal carcinoma (NPC) is low in the majority of the world, but high in southern China, Mediterranean Africa and some regions of the Middle East 1, 2. As lesions are hidden in the nasopharyngeal cavity and early symptoms are not obvious, early detection and diagnosis are difficult 3. Approximately 70%-80% of patients with NPC have had cervical lymph node metastases or even distant metastases when diagnosed, which becomes an important restrictive factor in patient survival 4, 5. There is mounting evidence that the vast majority of NPC is associated with Epstein-Barr virus (EBV) infection 6, 7.
EBV is a kind of human herpes virus that belongs to the γ subfamily. EBV, first discovered by Epstein and Barr in 1964, is the first virus recognized to cause cancer in people 8. EBV is closely associated with NPC, Burkitt lymphoma, lymphoblastoma, EBV-related gastric cancer, etc 9. The carry-infection rate of EBV is up to 90%, but only a small number of people would suffer from NPC. EBV is maintained extrachromosomally in infected normal and tumor cells and exists in latently infected cells primarily as an episome, usually results in defective virus or partial viral DNA being integrated 10. EBV expresses oncogenic proteins, such as EBNAl and LMP1 in the host cell 11. At the same time, EBV is the first virus that has been shown to encode miRNAs in host cells 12.
EBV-encoded miRNAs are transcribed as two regions according to their location: the BamH I fragment A rightward transcript (BART)-cluster 1, 2 and the BamH I fragment H rightward open reading frame 1(BHRF1)-cluster 12-15. To date, all BART miRNAs are detected in NPC. BART miRNAs contain 22 precursor miRNAs that encode 40 mature miRNAs. BHRF1 miRNAs have three precursor miRNAs, which encode four mature miRNAs 16. It has been shown that EBV-encoded miRNAs interact with their viral gene or host mRNA 3'-UTR conserved region via complete or imperfect base pairing, inducing degradation or translation inhibition of the target mRNA. There are only 44 categories of mature miRNAs, but the expression of BART miRNAs in NPC accounts for a quarter of the total miRNAs 17, 18, suggesting that EBV miRNAs play an important role in NPC pathogenesis. In this review, we will summarize the critical role of EBV-miRNAs in the development of NPC, especially in the biological functions and molecular mechanisms of immune evasion, proliferation, apoptosis, invasion and metastasis.
1. EBV miRNAs are involved in immune evasion in NPC
EBV infects nasopharyngeal mucosa epithelial cells, and the virus depends on host cells to synthesize EBV miRNAs, which can regulate viral and host cell gene expression to mask the infected cell from the host immune response and help the virus maintain long-term chronic infection. Once immune function is attenuated, the latent EBV is explosively activated, forming a recrudescent infection 19, 20. Viral gene products, such as LMP1, LMP2, EBNA1, may act as antigens to trigger host cell immune response as the cell continues to produce cytokines. Multiple groups demonstrated that viral antigens are regulated by various EBV miRNAs 21. This results in activation of the inflammatory cascade, which induces the proliferation and differentiation of immune cells that are recruited to the infected site to exert an immune effect under the guide of chemokines. During this process, EBV-miRNAs regulate the host immune response by regulating viral gene-encoded products and blocking the host immune cell activation. EBV-miRNAs can interfere with host cell immune modulation by dulling immune recognition, blocking immune signaling pathways, affecting the levels of antigen proteins and other behaviors to paralyze the host immune system.
1.1. EBV miRNAs inhibit recognition of pattern recognition receptors
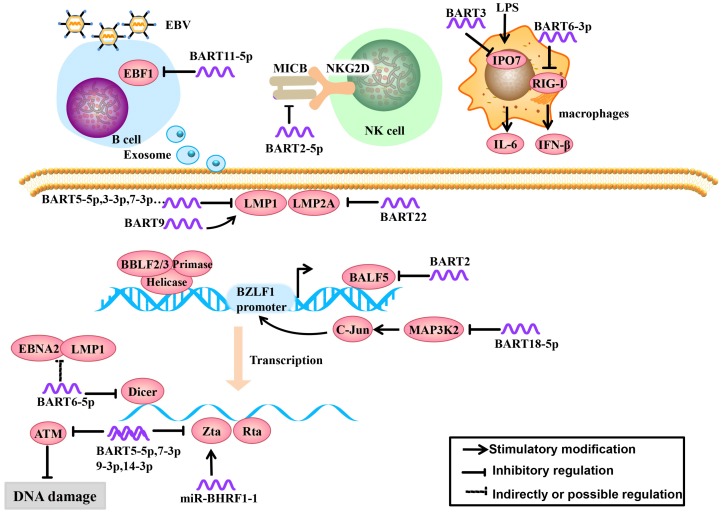
Recognition of viral pathogen-associated molecular patterns by pattern recognition receptors (PRRs) is the first step in the initiation of a host innate immune response. The RIG-I-like receptor family (including RIG-I, MDA5 and LGP2) is an important member of PRRs, which can recognize viral RNAs or replication transcripts in the cytoplasm. The viruses employ the strategy that inhibit RIG-I recognition to achieve immune evasion. A recent study demonstrated that miR-BART6-3p suppressed RIG-I-like receptor signaling-mediated IFN-β production. Furthermore, miR-BART6-3p inhibited the EBV-triggered IFN-β response and facilitated EBV infection through targeting the 3'UTR of RIG-I mRNA 22 (Figure 1).
Fig 1.
Role of EBV miRNAs in immune evasion and EBV latency. EBV miRNAs can be secreted from infected B cells to non-infected cells via exosomes. EBV miRNAs regulate lymphocyte activation and lethality, modulate infected host cell antigens and maintain EBV latent infection status to establish persistent latent infections.
1.2. EBV miRNAs regulate lymphocyte activation and lethality
EBV miRNAs can attenuate immune recognition and lymphocyte activation. To tolerate the virus after EBV infection, the host cells produce a ligand, major histocompatibility complex class I-related chain B (MICB), which is an NKG2D receptor ligand. The NKG2D receptor is located on the surface of NK cells, can function in immune recognition in infected cells and effectively activate NK cells and T cells 23. EBV-miR-BART2-5p downregulates the expression of MICB by targeting the host cell MICB ligand, which reduces host cell immune recognition and activation in infected cells 24 (Figure 1).
In the early stage of EBV infection, dormant B cells are rapidly differentiated into memory B cells driven by the germinal center. Therefore, the germinal center plays an important role in lymphocyte cell cycle and differentiation. Early B-cell factor 1 (EBF1), a B cell transcription factor, plays a key role in the formation of the germinal center 25, 26. Moreover, EBF1 targets many B cell-specific genes, including the B cell developmental regulator PAX-5 (which is necessary for maintaining B cell phenotype), B cell receptors (BCR) and Bcl-2 interacting mediator of cell death (Bim). If EBF1 function is absent, mature B cells will not function 27, 28. Ross et al. 29 found that EBV-miR-BART11-5p binds to the EBF1 3'-UTR, highlighted a potential role of BART-11-5p in the regulation of B-cell differentiation (Figure 1).
IPO7 is a nuclear receptor associated with congenital immunity. Under the stimulation of the endotoxin LPS, interfering with IPO7 expression in macrophages can reduce the secretion of IL-6. IPO7 is also associated with autoimmune regulatory factors, including a transcription factor that regulates immune tolerance, and IPO7 expression is positively correlated with T cell immune toxicity 30. Dolken et al. 31 found that EBV-miR-BART3 could target IPO7, reduce its expression and lymphocyte cytotoxicity, and protect infected cells (Figure 1).
EBV miRNAs inhibit EBV surveillance via CD8+ T cells through multiple mechanisms, such as reducing virus-specific proliferation by directly targeting TAP2, diminishing IL-12 release, and repressing the nuclear antigen EBNA1 32. These proteins are found in Burkitt lymphoma, but their interplay with EBV miRNAs in NPC remains unclear. Cytokines are a class of proteins or polypeptides that convert information in intercellular communication and immune response 33. CXCL11 binds to the lymphocyte surface chemokine receptor CXCR3, which drives immune cells migrate to infected loci and exerts immune functions 34-36. In non-Hodgkin's lymphoma, miRBHRF1-3 can restrain host cell immune chemotactic functions by inhibiting the expression of CXCL11, which inhibits inflammation and is conducive to infected cell survival 37. The inflammatory body is composed of multiple protein complexes. To date, nod-like receptor protein 3 (NLRP3) is the most well-recognized inflammatory body 38. Upon activation, NLRP3 can recognize foreign pathogens and dangerous signals; recruit and activate proinflammatory protease Caspase-1, cleaved IL-1β and IL-18 precursors, and induce anti-viral inflammation. MiR-BART15 can bind to the NLRP3 3′-UTR, downregulate its downstream cytokines and inhibit inflammation 39. Thus, the EBV BART miRNA-mediated tumor inflammation microenvironment plays an important role in immunity, though its role remains obscure in NPC 40.
1.3. EBV miRNAs modulate infected host cell antigens
EBV usually remains latent in the tumor and expresses small amounts of latent membrane proteins to evade the host immune surveillance. Some of viral proteins, including a lytic EBV protein, has been shown to bind or target the SCF complex, thereby indirectly suppressing NF-κB activation pathways that elicit innate immune responses 41-43. For example, LMP2A is an EBV-encoded protein that is a highly immunogenic virus antigen and can be specifically recognized by CD4+ and CD8+ T cells 44. Lung et al. 21 found that EBV-miR-BART22 can directly target the 3'-UTR of LMP2A, which may facilitate NPC carcinogenesis by evading the host immune response (Figure 1). EBV encodes another latent protein, LMP1, which is an oncoprotein that it considered to be an important factor in the development of NPC. LMP1 can upregulate the epithelial-mesenchymal transition (EMT) and regulate many signaling pathways, including the NF-κB, JNK-p38/SAPK, Ras-MAPK, PI3K-Akt and JAK-STAT pathways 45-50. It has been reported that miR-BART9 upregulates the expression of LMP1 in an indirect regulatory manner, such as enhancing LMP1 mRNA stability 51, maintaining high LMP1 expression levels in the host cell and promoting LMP1 oncogenic effects (Figure 1). However, LMP1 has a dual effect on NPC. In the other side, EBV-encoded BART miRNAs can target LMP1, prevent apoptosis or induction of host immune response due to excessive LMP1 accumulation. MiR-BART5-3p can target LMP1, but miR-BART5-3p is a passenger strand (not mature miRNA) and therefore does not have functional effects. MiR-BART3-3p and miR-BART7-3p also can target LMP1 but are weaker than miR-BART5-5p 52. Lo et al. initially described that EBV BART cluster I miRNAs, specifically miR-BART1-5p, miR-BART16, and miR-BART17-5p can target the LMP1 3'-UTR and regulate LMP1 expression 53. But Skalsky faired to conform the miR-BART1-5p targeting by PAR-CLIP analysis, they showed that five EBV BART miRNAs (miR-BART3, miR-BART5, miR-BART16, miRBART17, and miR-BART20) target LMP1 3'-UTRs, which uncovered a role for EBV BART miRNAs in modulating NF-κB signaling pathways (Figure 1).
1.4. EBV miRNAs maintain EBV infection status
In the latent stage and early stage of lytic replication during EBV infection, EBV miRNAs regulate the host cell cycle for the virus's own benefit by regulating EBV and host cell gene expression. To achieve a persistent latent infection, EBV inhibits its own lytic replication to prevent excessive proliferation 54. For example, the endonuclease Dicer is part of the RNase III family and specifically recognizes double-stranded RNA. Dicer not only regulates the expression of host cell and viral miRNAs but also affects the EBV infection process 55. In the study by Iizasa et al. 56, antisense oligonucleotide silencing was used in C666-1 cells to inhibit endogenous EBV miR-BART6-5p expression, which increased Dicer expression, followed by increased mRNA levels of the immediate early genes Zta and Rta and immunogen latent proteins Epstein Barr virus nuclear antigen 2 (EBNA2) and LMP1, indicating that EBV-miR-BART6-5p can target and bind to the Dicer gene and subsequently regulate the conversion of a type I/II latent infection to a type III infection (Figure 1).
When EBV enters into the early stage of lytic replication, BALF5 is highly expressed. This gene is a necessary DNA polymerase for DNA replication during the lytic process. Barth et al. showed that EBV-miR-BART2 is complementary to the BALF5 3'-UTR and EBV-miR-BART2 overexpression results in a decrease in BALF5 protein levels in the lytic phase. Mechanistically, EBV-miR-BART2 directly targets and downregulates the expression of BALF5. This was compatible with the notion that EBV miR-BART2 inhibits transition from latent to lytic viral replication 19 (Figure 1).
BHRF1 miRNAs were reported no or low expressed in NPC cells. Li et al. 57 confirmed that miR-BHRF1-1 is involved in TPA-induced EBV late lytic replication in NPC cells. MiR-BHRF1-1 can start the virus lytic replication switch gene Zta in the EBV-positive NPC cell line SUNEl to promote EBV lytic replication and complete the conversion to the viral infection state. Further study demonstrated that the EBV original pathogen miR-BHRF1-1 is involved in the control of EBV late lytic replication by directly targeting the host p53 gene 4. Specifically, the host p53 is functionally inhibited by EBV in latent stage 58, transiently elevated in the early lytic stage 59 and then declined in late lytic stage to benefit viral replication 60. MiR-BHRF1-1 expression was induced but delayed when entry into lytic cycle. This phenomenon indicates that miR-BHRF1-1 may downregulate host p53 in late lytic stage (Figure 2).
Fig 2.
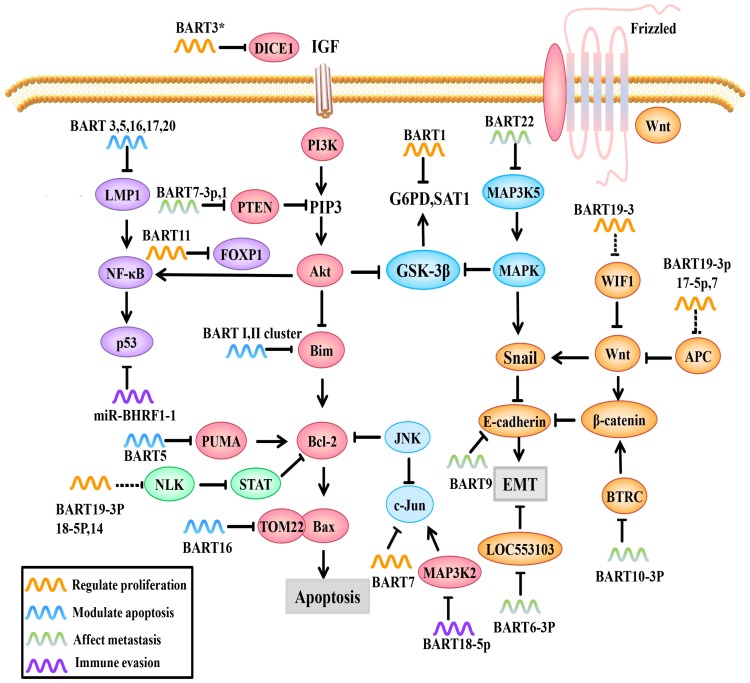
Role of EBV miRNAs in signaling pathways. EBV miRNAs can modulate viral and host cell gene expression by directly or indirectly regulating molecules and related signaling pathways.
In addition to modulating EBV lytic replication, EBV miRNAs also regulate cell cycle-related regulatory molecules in the host cell to create conditions for viral persistent latent infection. For example, MAP3K2 is a pivotal molecule in various signaling pathways. There are many transcription factors in its downstream signaling pathways, such as CREB, ATF1-2 and c-jun, that bind to the promoter region of the immediate early gene BZLF1 and promote its expression 61. BZLF1 expression in both B cells and epithelial cells can activate lytic replication 62. In a study by Qiu, et al. 63, it was confirmed that EBV-miR-BART18-5p inhibits MAP3K2 expression by targeting the MAP3K2 3'-UTR in B lymphocytes, thus inhibiting viral lytic replication during the immediately early phase, this allows EBV to achieve its long-term latent infection aim (Figure 1&2).
A recent study shows that BART5-5p, BART7-3p, BART9-3p, and BART14-3p could negatively regulate the expression of ataxia telangiectasia mutated (ATM), which is a key DNA double-strand break repair gene, by targeting its 3'-UTR. By aborogation these endogenous miR-BARTs in EBV-positive NPC cells, they further demonstrated the role of miR-BARTs in inhibiting Zta-induced lytic reactivation. Taking together, the four EBV miRNAs work together to modulate ATM activity in response to DNA damage and to maintain viral latency, contributing to the tumorigenesis of NPC 64 (Figure 1).
2. EBV miRNAs promote cell proliferation in NPC
The activation of oncogenes and the inactivation of tumor suppressor genes are common events in tumors. EBV miRNAs promote NPC cell proliferation by directly or indirectly regulating genes and related signaling pathways.
FOXP1, a transcription factor, can not only prevent the secretion of inflammatory factors by macrophages but also regulate signaling pathways important for the differentiation of mononuclear cells to macrophages. FOXP1 overexpression blocks mononuclear cell differentiation and inhibits NPC cell growth. Song et al. found that EBV-miRBART11 directly targets the FOXP1 3'-UTR and attenuates FOXP1 expression, which inhibits tumor-associated macrophage (TAM) differentiation and inflammatory factor secreting into the tumor microenvironment, inducing NPC cell proliferation (Figure 2). EBV-miR-BART11 can also promote NF-κB transcription through FOXP1 65.
EBV miRNAs can cooperate to activate the Wnt signaling pathway by downregulating Wnt inhibitory genes, such as WIF1 (targeted by miR-BART19-3p), APC (targeted by miR-BART19-3p, 17-5p, and 7), and NLK (targeted by miR-BART14, 19-3p, 18-5p), but wong et al. only use Q-PCR and western blotting to demonstrate the potential correlation, the specific mechanism still need to be clarified66-68 (Figure 2). This gives us a hypothesis that EBV miRNAs may regulate cell proliferation by indirectly regulating Wnt signaling pathways.
The DICE1 gene is highly conserved and is widely expressed in tissues. The DICE1 protein shows 92.9% identity to the carboxy-terminal half of the mouse EGF repeat transmembrane protein DBI-1. The DBI-1 protein interferes with the mitogenic response to insulin-like growth factor 1 (IGF-I) and is presumably involved in anchorage-dependent growth 69. DICE1 is a tumor suppressor that can inhibit growth and interfere with the anchorage-independent growth of IGF-IR-transformed tumor cells. Lei et al. found that EBV-miR-BART3* downregulates the expression of the NPC suppressor DICE1, which in turn promotes the growth and transformation of NPC cells 70 (Figure 2).
EBV-miR-BART7 plays multiple roles in NPC, such as promoting cell growth, clonal formation and cell cycle progression, by activating PI3K/Akt/c-Myc and c-Jun. C-Myc and c-Jun are important transcription factors in cell growth and usually highly expressed in NPC 71, 72 (Figure 2). Additionally, NPC cells transfected with miR-BART7 mimic (a synthetic miRNA function substitute) are more tolerant to cisplatin 71. Furthermore, Cai et al. found that gold PEI nanocarriers can deliver anti-miRBART7-3p to target and inhibit the function of BART7, which provides a new therapeutic strategy for targeted therapy based on EBV-miRNAs 73.
EBV miRNAs can also influence cancer cell metabolism. Ye et al. reported that EBV-miR-BART1 dramatically modulates G6PD, SAT1 and other metabolism-related genes (Figure 2). PAST1 and PHGDH are upregulated in NPC specimens and are important for tumor cell proliferation and growth, respectively 74, 75. Qiu, et al. 76 demonstrated that there are no obvious associations between tumor growth and the expression of specific BART-miRNAs. However, upon further study, it was found that the upregulation of all the BART-miRNA expression is positively correlated with tumor growth, consistent with previous observations that all the BART miRNAs are highly expressed in all of the EBV associated cancers, suggesting that dysregulated expression of the BART miRNAs potentiates tumor growth and represents a general mechanism behind EBV associated oncogenesis.
3. EBV miRNAs play a crucial role in cell apoptosis in NPC
There are three main cell apoptosis pathways. The first is the mitochondrial pathway, which is divided into the following two categories: the first class requires activation of the caspase pathway via mitochondrial CytoC release and the second type does not require caspase activation, but occurs through the release of mitochondrial apoptosis-inducing factor (AIF). The second pathway is accomplished by activating death receptors through the TNF pathway. The third pathway is the endoplasmic reticulum pathway, whereby endoplasmic reticulum stress leads to a calcium ion imbalance that can either activate caspase 9 to promote apoptosis or induce the activation of the proapoptotic Bcl-2 family proteins Bax and Bak. EBV miRNAs can regulate the expression of Caspase-3 and the apoptotic pathways to inhibit host cell apoptosis and achieve persistent infection 77.
A study by Kang et al. 78 used PAR-CLIP and luciferase assay to experimentally identify the miRNA targetome in C666 NPC cells, including FEM1B and CASZ1a (miR-BART3), OCT1 (miR-BART6), ARID2 (miR-BART8), CREBBP and SH2B3 (miR-BART16) and finally PPP3R1, PAK2 and TP53INP1 (miR-BART22). They introduced artificial miRNAs specific for pro-apoptotic miR-BART target mRNAs to phenocopy their anti-apoptotic effect in AGS gastric carcinomas cells, and found that 7 cellular genes identified as targets for anti-apoptotic EBV miR-BART (CASZ1, DICE1, SH2B3, PAK2, TP53INP1, OCT1 and CREBBP). However, they faired to test the anti-apoptotic effect in NPC cell lines, which leave to be proven. P53 upregulated modulator of apoptosis (PUMA) is a pro-apoptotic protein and member of the Bcl-2 family that has been discovered in recent years; it is a key factor that induces non-p53-dependent and p53-dependent apoptosis. PUMA is usually lowly expressed in 60% of NPC patients. Choy et al. 79 showed that miR-BART5 makes NPC cells less sensitive to proapoptotic agents, suggesting that depleting miR-BART5 or inducing PUMA expression could induce cell apoptosis. Mechanistically, EBV-miR-BART5 directly targets PUMA and maintains latent virus infection by protecting host cells from apoptosis (Figure 2). Bim is a pro-apoptotic protein that activates Bax-induced mitochondrial apoptosis through Bcl-2/Bax interactions. Marquitz et al. verified that Cluster I miRNAs (miR-BART1-5p, 4, and 5) and Cluster II miRNAs (miR-BART9, 11-3p, 11-5p, 1, and 18-3p) both bind to the Bim 3'-UTR to inhibit host cell apoptosis 80 (Figure 2).
In addition to regulating apoptotic regulators, EBV miRNAs can also affect pro-apoptotic protein receptors. For example, translocase of outer mitochondrial membrane 22 (TOM22) is a receptor for the pro-apoptotic protein Bax that is located on the mitochondrial outer membrane. During the execution phase of apoptosis, activated Bax forms a complex with TOM22 and binds to the mitochondrial outer membrane; this controls the permeability of the mitochondrial outer membrane and regulates the release of apoptotic factors into the cytosol 81. Dolken et al. showed that EBV-miR-BART16 binds to TOM22 by competitive targeting, thereby reducing Bax binding to TOM22 and inhibiting the Bax-TOM22-mediated mitochondrial apoptotic pathway 31, 82 (Figure 2).
BART 16 and 17-5p induce apoptotic resistance by targeting LMP1 in NPC (Figure 2), which plays an important role in the initial stage of tumor. LMP1 has different roles in different stages of EBV infection. In the precancerous stage, LMP1 overexpression promotes its cancer-promoting function, whereas in the advanced stage of cancer, LMP1 expression is relatively low. EBV-miR-BART 16 and 17-5p prevent cytotoxic effects by downregulating LMP1 and inducing NPC cell apoptosis resistance 83.
NF-κB is one of the most important factors in NPC progression because it regulates cell proliferation and inflammatory response. NF-κB activation promotes the release of cytokines from inflammatory cells, thus promoting normal cells to transform into malignant cells. In turn, the increased cytokines promote NF-κB activation, which forms a positive feedback loop. Moreover, NF-κB activation leads to p53 accumulation, which prevents JNK-induced apoptosis 6, 84. The BART cluster I regulates NF-κB activity by regulating LMP1 levels 53 (Figure 2).
Wan et al. established a cellular and viral miRNA profile in NPC via microdissection that included 9 upregulated EBV miRNAs (BART1-5p, BART3, BART4, BART5, BART6, BART7, BART8, BART10 and BART18-3p). The targets of these EBV miRNAs are mainly enriched in the TGF-β and Wnt signaling pathways and involved in NPC apoptosis and transformation. The target genes are mainly ARHGEF12, SH3GLB1, DHCR24, IGFBP5, ITGA2, FOXO1 and FZD4. Only BART18-3P had no predicted results. This will help us to further dissect miRNA targets and the associated regulatory network 85. However, the specific mechanisms associated with these miRNAs and targets need to be verified.
EriSeto et al. found that mutations in EBV precursor miRNAs in B cell lymphomas prevent them from being processed into mature viral miRNAs. These mutated phenotypes were observed during the early stages of primary B cell infection. The BHRF1 locus of viral miRNAs inhibits apoptosis and promotes cycle progression and proliferation, indicating that viral miRNAs promote cell transformation rather than controlling viral genes during the lytic phase. Additionally, EBV genomic variation plays an important role in EBV-related tumors 86. Shu-Jen Chen revealed extensive sequence variation by NPC tissue deep sequencing. A wide range of sequence variations were detected in all 44 EBV miRNAs, including 1-3 nucleotide mismatches compared to the reference EBV genome, most of which occurred at the 3'-end between nucleotides 17 and 24. The nucleotide mutation distribution pattern is similar to that in human brain tissues, where EBV miRNAs and cellular miRNAs share the same seed sequence. With respect to the mechanism of genomic variation, SNP and post-transcriptional modifications may promote mutations, RNA editing and template-free extension, which are common post-transcriptional modifications 17.
4. EBV miRNAs affect invasion and metastasis
Tumor metastasis is often the main cause of cancer recurrence, poor prognosis and death. EMT is a predominant way for epithelial-derived malignancies to acquire migration and invasion abilities 68. During this process, polarized epithelial cells are transformed into mesenchymal-like cells, which can enhance skeletal rearrangement and display increased migration and invasion abilities 67, 87. Thus, EMT is considered to be an initial step in tumor metastasis and plays an important role in tumor initiation and development. In EBV-related tumor cells, EBV miRNAs can regulate EMT-related molecules and signaling pathways to enhance the invasion and metastasis of tumor cells and promote tumor deterioration.
PTEN, a tumor suppressor that competes with tyrosine kinases for a common substrate, can not only promote cell apoptosis and delay cell cycle progression but can also affect other PTEN-dependent signaling pathways, such as the PI3K/Akt and Akt/GSK-3β signaling pathways, leading to high expression of EMT markers, such as Snail and β-catenin 88. In two successive reports 72, 83, Cai, L.M showed that EBV-miR-BART7-3p and EBV-miR-BART1 overexpression are positively correlated with the number of lymph nodes and the clinical stage of NPC malignancy. Interestingly, EBV-miR-BART7-3p and EBV-miR-BART1 both directly target PTEN, and therefore promote EMT (Figure 2). Further studies have shown that EBV-miR-BART7-3p targets PTEN, which leads to high Snail and β-catenin expression by regulating PI3K/Akt/GSK-3β, whereas miR-BART1 directly targets PTEN, resulting in the activation of PTEN-dependent pathways, including the PI3K-Akt, FAK-p130Cas and Shc-MAPK/ERK1/2 pathways. MiR-BART1 forces actin cytoskeleton reshaping and EMT, thereby promoting NPC invasion and metastasis. Based on bioinformatics analysis and PAR-CLIP prediction, Wong et al. predicted that several EBV-BART-miRNAs, including EBV-miR-BART9 and EBV-miR-BART18-5p, directly target the tumor suppressor PTEN, thereby promoting tumor cell invasion and metastasis abilities; however, the exact mechanism requires further verification 66, 89.
Beta-transducin repeat containing E3 ubiquitin protein ligase (BTRC) is another signaling molecule capable of regulating EMT that promotes the ubiquitination of its downstream substrates β-catenin and Snail, thereby inhibiting EMT. According to Li Guiyuan's team, EBV-miR-BART10-3p expression is negatively correlated with BTRC, EBV-miR-BART10-3p overexpression and low BTRC expression are positively correlated with poor prognoses in NPC patients. Mechanistically, EBV-miR-BART10-3p overexpression enhances the invasion and migration capacity of NPC cells by targeting BTRC (Figure 2) and regulates the expression of its downstream target genes β-catenin and Snail, ultimately promoting EMT and metastasis 90.
MAP3K5 is an important early response and apoptosis-related gene in the P38-MAPK pathway. MiR-BART22 directly targets the MAP3K5 3'-UTR and downregulates the MAP3K5 protein and the expression of the downstream target gene MAP2K4 (Figure 2). MiR-BART22 can promote proliferation and invasion in the NPC cell line 5-8F, partly by regulating MAP3K5 91.
In a recent study, EBV-miR-BART6-3p expression is negatively correlated with distant metastases and tumor recurrence. Further study showed that EBV-miR-BART6-3p exerts an inhibitory effect on EMT by targeting the lncRNA LOC553103 92, thereby reducing the invasion and metastasis abilities of these cells 92 (Figure 2).
In recent years, it has been found that EBV miRNAs can directly regulate EMT markers, induce the formation of the mesenchymal phenotype and promote EMT. Hsu, CY et al. 93 confirmed that EBV-miR-BART9 could directly target and inhibit E-cadherin expression and promote EMT (Figure 2). As a result, EBV-miR-BART9 facilitates migration and invasion in NPC. Additionally, intracellular NDRG1 is not only an inhibitory molecule in metastasis but also a marker for epithelial cell differentiation. NDRG1 is highly expressed in epithelial cells and positively correlated with cell differentiation signals 94. Moreover, NDRG1 interacts with the Wnt signaling receptor LRP6 and hinders Wnt signaling 95, 96. Multiple EBV miRNAs in combination can inhibit NDRG1 expression and promote cell dedifferentiation and growth 97. This can activate the Wnt signaling pathways and promote the transformation of epithelial cancer cells (summarized in Table 1).
Table 1.
EBV-miRNAs and its targets in NPC
| EBV-miRNAs | Targets | Function of targets | Ref |
|---|---|---|---|
| miR-BART 2-5P | MICB | Ligand of NKG2D receptor | 24 |
| miR-BART 11-5p | EBF1 | B cell proliferation and differenciation | 29 |
| miR-BART 3 | IPO7 | Immunoregulatory factor | 31 |
| miR-BART 22 miR-BART 6-3p |
LMP2A RIG-I |
Initiation of latent cycle pattern recognition receptors |
21 22 |
| miR-BART 9 | LMP1 | Immortalization of B cells | 51 |
| miR-BART 5-5p,16 | LMP1 | Immortalization of B cells | 52 |
| 3-3p, 17-5p,20 | 98 | ||
| miR-BART 6-5p | Dicer,Zta,Rta | Catalytic miRNA mature | 56 |
| EBNA2 | Lytic replication | 56 | |
| miR-BART 2 | BALF5 | DNA polymerase | 19 |
| miR-BHRF 1 | P53 | Late lytic replication | 57 |
| miR-BART 18-5p BART5-5p,7-3p, 9-3p,14-3p |
MAP3K2 ATM |
Protein phosphorylation DNA double-strand break repair gene |
63 64 |
| miR-BART 11 | FOXP1 | Cell differentiation | 65 |
| miR-BART 19-3p | WIF1 | Wnt inhibitory genes | 66 |
| miR-BART 19-3p,7,17-5p | APC | Wnt inhibitory genes | 66 |
| miR-BART19-3p,14,18-5p | NLK | STAT inhibitory genes | 99 |
| miR-BART 3* | DICE1 | Tumor suppressor | 70 |
| miR-BART 7 | C-Myc,C-Jun | Transcription factor | 71, 73 |
| miR-BART 1 | G6PD,SAT1 | Metabolism-associated enzymes | 74 |
| miR-BART 5 | PUMA | Pro-apoptotic protein | 79 |
| miR-BART I,II cluster | Bim | Pro-apoptotic protein | 80 |
| miR-BART 16 | TOM22 | Bax mitochondrial receptor | 31, 82 |
| miR-BART 1-5p,16,17-5p | LMP1 | Immortalization of B cells | 83 |
| miR-BART I cluster | LMP1 | Immortalization of B cells | 52 |
| miR-BART3# | FEM1B,CASZ1a | Pro-apoptotic protein | 78 |
| miR-BART6# | OCT1 | Pro-apoptotic protein | 78 |
| miR-BART8# | ARID2 | Pro-apoptotic protein | 78 |
| miR-BART16# | CREBBP,SH2B3 | Pro-apoptotic protein | 78 |
| EBV-miRNAs | Targets | Function of targets | Ref |
| miR-BART22# | PPP3R1,TP53INP1,PAK2 | Pro-apoptotic protein | 78 |
| miR-BART 7-3p,1 | PTEN | Tumor suppressor | 72, 83 |
| miR-BART 9,18-5p | PTEN | Tumor suppressor | 66, 89 |
| miR-BART 10-3p | BTRC | Ubiquitin of β-catenin and snail | 90 |
| miR-BART 22 miR-BART 6-3p miR-BART 9 |
MAP3K5 LOC553103 E-cadherin |
P38MAPK early response genes Suppress EMT Mediate cell aggregation |
91 92 93 |
# Identified EBV BART miRNA-mRNA targets, but its anti-apoptotic effect remains to be proven.
Ref: reference
5. EBV miRNAs are transported via exosomes and could serve as biomarkers in NPC
D. Michiel Pegtel et al. first detected a large number of BART-miRNAs in circulating non-infected non-B cells (the EBV genome was absent in these cells). BART1-5p was circulating in infected and non-infected B cells, indicating that EBV-miRNAs were transported to neighboring cells by exosomes and have gene-inhibition functions 100, 101. Canitano demonstrated that all of the EBV-miRNAs detected in LCL and 721 cells (two lymphoblastoid cell lines) were also present in exosomes purified from these cells, suggesting their delivery via exosomes 102. Gourzones et al. determined that BART miRNAs are released into the extracellular space by NPC cells via secreted exosomes, these miRNAs were detected with good selectivity in plasma samples from NPC-xenografted nude mice as well as NPC patients. 103. Meckes et al. further found that not only were BART-miRNAs secreted from NPC cell lines via exosomes, but LMP1 was also present within the exosomes. Even though the cell line C666-1 contained very few LMP1, this protein was easy to detect in exosomes. One might speculate that the release of EBV miR-BARTs via exosomes downregulates the expression of LMP1 in NPC, thus inducing signaling pathways such as AKT and ERK. More interestingly, exosomal EBV-miRNAs have different abundances compared with their intracellular levels, suggesting that some of the viral miRNAs may be selectively packaged into exosomes. Exosomal EBV miRNAs could manipulate the tumor microenvironment to enhance tumor progression. Moreover, the exosome contents and properties may be changed by these molecules 104. These biological changes are consistent with EBV exploiting the host cell miRNA machinery for its own benefit. This is also a potential mechanism for intercellular exchange and immunoregulation, although the mode of this transport remains obscure 100, 105. Other EBV-miRNAs, including miR-BART4, miR-BART7, miR-BART9, miR-BART12, miR-BART13 and miR-BART16, were found to be transported to neighboring cells by vesicles or exosomes that were free of RNases.
More than 75% of NPC patients have developed local metastases when diagnosed, which hinders effective treatments and leads to poor prognoses. Thus, it is particularly important to identify sensitive and specific biomarkers for early diagnosis and prognosis 106. Because EBV miRNAs could be delivered by exosomes, out of RNases, they are more stable in tissues and serum compared to circulating tumor DNAs. Meanwhile, EBV miRNAs present in higher abundance than cellular miRNAs, and they are easier to extract and detect with higher specificity in comparison with proteins, thus EBV miRNAs are ideal molecular markers in EBV-related cancers 107-109.
Zhang et al. established an EBV BART microRNA profile and found that three are highly expressed (miR-BART3, miR-BART7 and miR-BART13). However, only miR-BART7 and miR-BART13 were found in NPC patient serum samples. ROC curves showed that miR-BART7 and miR-BART13 showed a 90% prognostic value and the expression of them decreased after radiotherapy, indicating that miR-BART7 and miR-BART13 have the potential to be plasma biomarkers for NPC diagnosis and prognosis 110. Zhen et al. examined the expression of 4 EBV miRNAs in a large number of clinical specimens and normal controls (miR-BART1-5p, miR-BART5, miR-BART6-5p and miR-BART17-5p) and found that all four miRNAs were higher in the clinical samples than in the normal group. Specifically, miR-BART1-5p is suitable to be a diagnostic indicator (93.5% sensitivity and 100% specificity), even in cases that were falsely diagnosed as negative based on histopathological analysis, plasma EBV DNA load, and VCA-IgA and EA-IgA titers 111. Gourzones C confirmed that EBV-miR-BART17 has good sensitivity (77%) and high specificity (90%) 112.
Hirai et al. collected serum from 31 EBV-positive NPC patients and 40 non-NPC controls and examined the copy numbers of circulating miR-BART2-5p, miR-BART17-5p and miR-BART18-5p. The expression of these three BART-miRNAs was not associated with the stage or treatment after diagnosis, but miR-BART17-5p was detected in 5/11 cases with relapse or residual tumors after treatment and absent in patients with no recurrence or remnants, suggesting that miR-BART17-5p is a potential prognostic biomarker 113. Further, miR-BART17 delivery is associated with ribonucleoprotein complexes and not exosomes 112. The advantages of circulating microRNAs are their stability and abundance, which means that EBV-miRNAs have the potential to be serum biomarkers for early diagnosis and prognosis. This is especially true when the circumstance that EBV DNA is undetectable 114, while EBV-miRNAs are present in almost all patients.
In conclusion, EBV miRNAs could be transported to neighboring cells or serum by vesicles or exosomes, thus they outweigh current tumor biomarkers in many aspects, and can be promising molecular biomarkers and new targets for future drugs. But its defect is that EBV miRNAs still remain to enlarge clinical application and evaluate the effectiveness. (EBV miRNAs function as biomarkers summarized in Table 2).
Table 2.
Some EBV-miRNAs markers identified
| EBV-miRNAs | Sample number (tumor/non tumor) | Relative clinic value | Ref |
|---|---|---|---|
| miR-BART 7,13 | 89/36 | 90% prognostic value | 110 |
| miR-BART 1-5p,5,6-5p,17-5p | 215/209 | 93.5% sensitivity and 100% specificity diagnostic indicator | 111 |
| miR-BART 17 | 26/10 | 77% sensitivity and 90% specificity prognostic indicator | 112 |
| miR-BART 17-5p | 31/40 | prognostic biomarker | 113 |
6. EBV-targeted therapy in NPC
EBV causes approximately 200,000 new malignancies per year, therefore, we need to find a safe and effective means to intervene in the development of tumors. For non-metastatic NPC, radiotherapy is the most prevalent method to locally control 50-90% of tumors 115. In contrast, platinum-based chemotherapy is often used to treat advanced and lymph node metastasis in NPC. Nevertheless, local failures and distant metastases still occur, and radiotherapy and chemotherapy may cause serious side effects. There is compelling evidence that cytotoxic T lymphocyte-based immunotherapy plays an important role in EBV-related solid tumors 116-118. As EBV miRNAs play a pivotal role in immunization, could they potentially interfere in the response of EBV miRNAs to T lymphocytes? With respect to cancer prevention, the main goal is a vaccine based on EBV products, such as targeting LMP2A and EBNA1 to induce a T cell response 119. A novel adenoviral vector-based vaccine termed AdE1-LMP poly has been developed that fused multiple CD8+ T-cell epitopes from LMP1 and LMP2. This adoptive immunotherapy could extend patients' life. However, these vaccines have not yet been approved. The biggest problem is the lack of animal models. Additionally, it is difficult to find immunogen and adjuvant combination therapies. EBV DNA are currently being used for clinical early detection and screening in high-risk populations 120. For EBV-based second-generation tumor prophylaxis, we should consider EBV miRNAs as a candidate for specific biomarker and targeted therapy, due to their pivotal roles in NPC and good properties. Probably the most important quality of miRNA gene therapy in comparison with current approaches of targeting single gene is its rapid modulation of many different pathways or targets, thus improving its effectiveness greatly and inducing fewer side effects. CRISPR/Cas9 system has been adopted for genetic engineering in mammalian cells, allowing us to edit the genomes with our expect precision. In a study of BL parent-derived cells, the use of CRISPR / Cas9 to clear the EBV potential genome resulted in proliferation arrest and apoptosis without significant toxic effects on uninfected cells 121. The feasibility of CRISPR/Cas9-mediated editing of the EBV genome (EBNA-1, EBNA-3C, LMP1, and BART) has also been demonstrated in cultured NPC cell lines 122. Moreover, the CRISPR / Cas9 system is being updated at a very fast pace, Zhang Feng et al. demonstrated RNA editing by using catalytically inactive Cas13 (dCas13) to direct adenosine-to-inosine deaminase activity by ADAR2 (adenosine deaminase acting on RNA type 2) to transcripts in mammalian cells 123, 124. Perhaps we can use these highly precision techniques to target the EBV miRNA-encoding regions of the viral genome to achieve EBV-targeted therapy, and we may apply gene editing techniques to animal models.
Perspective
Approximately 20% of human tumors are associated with latent virus infection, such as EBV, during initiation and development. EBV has been shown to regulate both viral and host cell gene expression and epigenetic mechanisms. When EBV infects host cells, the virus inhibits lytic replication, prevents explosive proliferation and remains latent to avoid host immune surveillance. EBV miRNAs regulate EBV and host cell gene expression, the host cells remain in the cell cycle, which continues to provide materials and energy for viral survival and reproduction. During this latency period, EBV miRNAs promote tumor cell proliferation, attenuate apoptosis, and promote invasion and metastasis, all of which pave the way for sustained virus infection. This dynamic balance cooperates to achieve persistent latent infection for the virus and further participates in the development of cancer.
So far, 44 mature EBV miRNAs have been identified, however, many of the target genes and their mechanisms remain unclear. For example, miR-BHRF1-3 and miR-BART15 can target CXCL11 and NLRP3 respectively and play an important role in lymphoma progression. However, the role of the EBV miRNAs in the NPC tumor microenvironment remains unclear. There are currently no references associated with miR-BART4, 12, 13, 15, 20, and 21 and their targets in NPC. The same EBV miRNAs can target multiple molecules to modulate a variety of functions, and the same target molecules have different roles in different stages of viral infection. EBV genomic variation also affects NPC pathogenesis. IsomiRs are miRNAs containing heterozygous mutations affecting the length and sequence that can affect the stability of target genes, miRNAs and the translation machinery. Carol Ying-Ying Szeto demonstrated the role of miRNAs in transcript expression and miRNA genomic variation through mRNA and miRNA sequencing in the NPC cell lines HK1, C666 and immortalized NP420 cells. The NGS data showed isomiRs that are worthy of study and identified that three quarters of the miRNAs had isomiRs, providing even more evidence that the functions of these EBV-encoded miRNAs need to be studied in NPC 125. Recently, Li Guiyuan's team illuminated the characteristics of EBV genomic variations in NPC tissues using "capture-sequencing" analysis. In total, 20328 variants were found, including 19903 single base variation, 88 insert mutations and 154 deletion mutations. A large number of potentially non-synonymous mutations were found in genes encoding LMP1, BZLF1, EBNA1, BKRF2 and the miRNAs 126, 127, highlighting that the role of EBV genome variation in NPC pathogenesis warrants further study.
The secretion of EBV miRNAs from infected B cells to uninfected cells through exosomes raises a fascinating possibility that EBV-miRNAs have the potential to be a serum biomarker for early diagnosis and prognosis. In the future, we may be able to use EBV-miRNA interference methods, such as mimics, inhibitors, anti-miRNA oligonucleotides or CRISPR / Cas13 system combined with exosomes or nanocarriers to transport these molecules to specific sites for targeted therapy, for the treatment of NPC 73, 128. Thus, we believe that a better understanding of the EBV-miRNA regulatory networks in NPC will lay a solid theoretical foundation for the treatment of NPC.
Acknowledgments
This work was supported in part by grants from the National Natural Science Foundation of China (81572787, 81672683, 81672993, 81772928, 81772901 and 81702907) and the Natural Science Foundation of Hunan Province (2016JC2035, 2017SK2105, 2018JJ3704, and 2018JJ3815).
References
- 1.Wei F, Wu Y, Tang L, He Y, Shi L, Xiong F. et al. BPIFB1 (LPLUNC1) inhibits migration and invasion of nasopharyngeal carcinoma by interacting with VTN and VIM. British journal of cancer. 2018;118:233–47. doi: 10.1038/bjc.2017.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang L, Tang Y, He Y, Wang Y, Lian Y, Xiong F. et al. High Expression of LINC01420 indicates an unfavorable prognosis and modulates cell migration and invasion in nasopharyngeal carcinoma. J Cancer. 2017;8:97–103. doi: 10.7150/jca.16819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei F, Tang L, He Y, Wu Y, Shi L, Xiong F. et al. BPIFB1 (LPLUNC1) inhibits radioresistance in nasopharyngeal carcinoma by inhibiting VTN expression. Cell Death Dis. 2018;9:432. doi: 10.1038/s41419-018-0409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong Z, Yang Q, Zeng Z, Zhang W, Li X, Zu X. et al. An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma. Tumour Biology. 2016;37:3683–95. doi: 10.1007/s13277-015-4156-x. [DOI] [PubMed] [Google Scholar]
- 5.Gong Z, Zhang S, Zeng Z, Wu H, Yang Q, Xiong F. et al. LOC401317, a p53-regulated long non-coding RNA, inhibits cell proliferation and induces apoptosis in the nasopharyngeal carcinoma cell line HNE2. PloS one. 2014;9:e110674. doi: 10.1371/journal.pone.0110674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei F, Wu Y, Tang L, Xiong F, Guo C, Li X. et al. Trend analysis of cancer incidence and mortality in China. Sci China Life Sci. 2017;60:1271–1275. doi: 10.1007/s11427-017-9172-6. [DOI] [PubMed] [Google Scholar]
- 7.He Y, Jing Y, Wei F, Tang Y, Yang L, Luo J. et al. Long non-coding RNA PVT1 predicts poor prognosis and induces radioresistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell Death Dis. 2018;9:235. doi: 10.1038/s41419-018-0265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epstein MA, Barr YM, Achong BG. Studies with Burkitt's lymphoma. Wistar Inst Symp Monogr. 1965;4:69–82. [PubMed] [Google Scholar]
- 9.Khan G, Hashim MJ. Global burden of deaths from Epstein-Barr virus attributable malignancies 1990-2010. Infect Agent Cancer. 2014;9:38. doi: 10.1186/1750-9378-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammerschmidt W, Sugden B. Replication of Epstein-Barr viral DNA. Cold Spring Harb Perspect Biol. 2013;5:a013029. doi: 10.1101/cshperspect.a013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo AK, Dawson CW, Jin DY, Lo KW. The pathological roles of BART miRNAs in nasopharyngeal carcinoma. J Pathol. 2012;227:392–403. doi: 10.1002/path.4025. [DOI] [PubMed] [Google Scholar]
- 12.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J. et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–6. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 13.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–50. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R. et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barth S, Meister G, Grasser FA. EBV-encoded miRNAs. Biochimica et biophysica acta. 2011;1809:631–40. doi: 10.1016/j.bbagrm.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Zeng Z, Huang H, Huang L, Sun M, Yan Q, Song Y. et al. Regulation network and expression profiles of Epstein-Barr virus-encoded microRNAs and their potential target host genes in nasopharyngeal carcinomas. Sci China Life Sci. 2014;57:315–26. doi: 10.1007/s11427-013-4577-y. [DOI] [PubMed] [Google Scholar]
- 17.Chen SJ, Chen GH, Chen YH, Liu CY, Chang KP, Chang YS, Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PloS one; 2010. p. 5. pii: e12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marquitz AR, Raab-Traub N. The role of miRNAs and EBV BARTs in NPC. Semin Cancer Biol. 2012;22:166–72. doi: 10.1016/j.semcancer.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E. et al. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic acids research. 2008;36:666–75. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apcher S, Fahraeus R, Manoury B. Epstein-Barr virus: exploiting the immune system by interfering with defective ribosomal products. Microbes Infect. 2004;6:1212–8. doi: 10.1016/j.micinf.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Lung RW, Tong JH, Sung YM, Leung PS, Ng DC, Chau SL. et al. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia. 2009;11:1174–84. doi: 10.1593/neo.09888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Y, Qin Z, Wang J, Zheng X, Lu J, Zhang X. et al. Epstein-Barr Virus miR-BART6-3p Inhibits the RIG-I Pathway. J Innate Immun. 2017;9:574–86. doi: 10.1159/000479749. [DOI] [PubMed] [Google Scholar]
- 23.Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol. 2000;1:119–26. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- 24.Yang IV, Wade CM, Kang HM, Alper S, Rutledge H, Lackford B. et al. Identification of novel genes that mediate innate immunity using inbred mice. Genetics. 2009;183:1535–44. doi: 10.1534/genetics.109.107540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gyory I, Boller S, Nechanitzky R, Mandel E, Pott S, Liu E. et al. Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes & development. 2012;26:668–82. doi: 10.1101/gad.187328.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Treiber T, Mandel EM, Pott S, Gyory I, Firner S, Liu ET. et al. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 2010;32:714–25. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Vilagos B, Hoffmann M, Souabni A, Sun Q, Werner B, Medvedovic J. et al. Essential role of EBF1 in the generation and function of distinct mature B cell types. J Exp Med. 2012;209:775–92. doi: 10.1084/jem.20112422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maier H, Hagman J. Roles of EBF and Pax-5 in B lineage commitment and development. Semin Immunol. 2002;14:415–22. doi: 10.1016/s1044532302000763. [DOI] [PubMed] [Google Scholar]
- 29.Ross N, Gandhi MK, Nourse JP. The Epstein-Barr virus microRNA BART11-5p targets the early B-cell transcription factor EBF1. Am J Blood Res. 2013;3:210–24. [PMC free article] [PubMed] [Google Scholar]
- 30.Abramson J, Giraud M, Benoist C, Mathis D. Aire's partners in the molecular control of immunological tolerance. Cell. 2010;140:123–35. doi: 10.1016/j.cell.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 31.Dolken L, Malterer G, Erhard F, Kothe S, Friedel CC, Suffert G. et al. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe. 2010;7:324–34. doi: 10.1016/j.chom.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Albanese M, Tagawa T, Bouvet M, Maliqi L, Lutter D, Hoser J. et al. Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells. Proc Natl Acad Sci U S A. 2016;113:E6467–E75. doi: 10.1073/pnas.1605884113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masmoudi A, Toumi N, Khanfir A, Kallel-Slimi L, Daoud J, Karray H. et al. Epstein-Barr virus-targeted immunotherapy for nasopharyngeal carcinoma. Cancer Treat Rev. 2007;33:499–505. doi: 10.1016/j.ctrv.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Tensen CP, Flier J, Van Der Raaij-Helmer EM, Sampat-Sardjoepersad S, Van Der Schors RC, Leurs R. et al. Human IP-9: A keratinocyte-derived high affinity CXC-chemokine ligand for the IP-10/Mig receptor (CXCR3) J Invest Dermatol. 1999;112:716–22. doi: 10.1046/j.1523-1747.1999.00581.x. [DOI] [PubMed] [Google Scholar]
- 35.Albanesi C, Scarponi C, Sebastiani S, Cavani A, Federici M, De Pita O. et al. IL-4 enhances keratinocyte expression of CXCR3 agonistic chemokines. J Immunol. 2000;165:1395–402. doi: 10.4049/jimmunol.165.3.1395. [DOI] [PubMed] [Google Scholar]
- 36.Cole KE, Strick CA, Paradis TJ, Ogborne KT, Loetscher M, Gladue RP. et al. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J Exp Med. 1998;187:2009–21. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB. et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Research. 2008;68:1436–42. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satoh T, Kambe N, Matsue H. NLRP3 activation induces ASC-dependent programmed necrotic cell death, which leads to neutrophilic inflammation. Cell Death Dis. 2013;4:e644. doi: 10.1038/cddis.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB. et al. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J Immunol. 2012;189:3795–9. doi: 10.4049/jimmunol.1200312. [DOI] [PubMed] [Google Scholar]
- 40.Wang M, Zhao J, Zhang L, Wei F, Lian Y, Wu Y. et al. Role of tumor microenvironment in tumorigenesis. J Cancer. 2017;8:761–73. doi: 10.7150/jca.17648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang SJ, Hsiao JC, Sonnberg S, Chiang CT, Yang MH, Tzou DL. et al. Poxvirus host range protein CP77 contains an F-box-like domain that is necessary to suppress NF-kappaB activation by tumor necrosis factor alpha but is independent of its host range function. J Virol. 2009;83:4140–52. doi: 10.1128/JVI.01835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gastaldello S, Callegari S, Coppotelli G, Hildebrand S, Song M, Masucci MG. Herpes virus deneddylases interrupt the cullin-RING ligase neddylation cycle by inhibiting the binding of CAND1. J Mol Cell Biol. 2012;4:242–51. doi: 10.1093/jmcb/mjs012. [DOI] [PubMed] [Google Scholar]
- 43.Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25:6758–80. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 44.Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu Rev Immunol. 2007;25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- 45.Cahir McFarland ED, Izumi KM, Mosialos G. Epstein-barr virus transformation: involvement of latent membrane protein 1-mediated activation of NF-kappaB. Oncogene. 1999;18:6959–64. doi: 10.1038/sj.onc.1203217. [DOI] [PubMed] [Google Scholar]
- 46.Li HP, Chang YS. Epstein-Barr virus latent membrane protein 1: structure and functions. J Biomed Sci. 2003;10:490–504. doi: 10.1007/BF02256110. [DOI] [PubMed] [Google Scholar]
- 47.Soni V, Cahir-McFarland E, Kieff E. LMP1 TRAFficking activates growth and survival pathways. Adv Exp Med Biol. 2007;597:173–87. doi: 10.1007/978-0-387-70630-6_14. [DOI] [PubMed] [Google Scholar]
- 48.Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes Immun. 2011;12:399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang GD, Huang TJ, Peng LX, Yang CF, Liu RY, Huang HB. et al. Epstein-Barr Virus_Encoded LMP1 upregulates microRNA-21 to promote the resistance of nasopharyngeal carcinoma cells to cisplatin-induced Apoptosis by suppressing PDCD4 and Fas-L. PloS one. 2013;8:e78355. doi: 10.1371/journal.pone.0078355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–68. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 51.Ramakrishnan R, Donahue H, Garcia D, Tan J, Shimizu N, Rice AP. et al. Epstein-Barr virus BART9 miRNA modulates LMP1 levels and affects growth rate of nasal NK T cell lymphomas. PloS one. 2011;6:e27271. doi: 10.1371/journal.pone.0027271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verhoeven RJ, Tong S, Zhang G, Zong J, Chen Y, Jin DY. et al. NF-kappaB Signaling Regulates Expression of Epstein-Barr Virus BART MicroRNAs and Long Noncoding RNAs in Nasopharyngeal Carcinoma. J Virol. 2016;90:6475–88. doi: 10.1128/JVI.00613-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G. et al. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A. 2007;104:16164–9. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F. et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mansouri S, Pan Q, Blencowe BJ, Claycomb JM, Frappier L. Epstein-Barr virus EBNA1 protein regulates viral latency through effects on let-7 microRNA and dicer. J Virol. 2014;88:11166–77. doi: 10.1128/JVI.01785-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iizasa H, Wulff BE, Alla NR, Maragkakis M, Megraw M, Hatzigeorgiou A. et al. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J Biol Chem. 2010;285:33358–70. doi: 10.1074/jbc.M110.138362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z, Chen X, Li L, Liu S, Yang L, Ma X. et al. EBV encoded miR-BHRF1-1 potentiates viral lytic replication by downregulating host p53 in nasopharyngeal carcinoma. Int J Biochem Cell Biol. 2012;44:275–9. doi: 10.1016/j.biocel.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 58.Spruck CH 3rd, Tsai YC, Huang DP, Yang AS, Rideout WM 3rd, Gonzalez-Zulueta M. et al. Absence of p53 gene mutations in primary nasopharyngeal carcinomas. Cancer Research. 1992;52:4787–90. [PubMed] [Google Scholar]
- 59.Chang SS, Lo YC, Chua HH, Chiu HY, Tsai SC, Chen JY. et al. Critical role of p53 in histone deacetylase inhibitor-induced Epstein-Barr virus Zta expression. J Virol. 2008;82:7745–51. doi: 10.1128/JVI.02717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato Y, Tsurumi T. Noise cancellation: viral fine tuning of the cellular environment for its own genome replication. PLoS Pathog. 2010;6:e1001158. doi: 10.1371/journal.ppat.1001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klinke O, Feederle R, Delecluse HJ. Genetics of Epstein-Barr virus microRNAs. Semin Cancer Biol. 2014;26:52–9. doi: 10.1016/j.semcancer.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci U S A. 1985;82:4085–9. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiu J, Thorley-Lawson DA. EBV microRNA BART 18-5p targets MAP3K2 to facilitate persistence in vivo by inhibiting viral replication in B cells. Proc Natl Acad Sci U S A. 2014;111:11157–62. doi: 10.1073/pnas.1406136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lung RW, Hau PM, Yu KH, Yip KY, Tong JH, Chak WP. et al. EBV-encoded miRNAs target ATM-mediated response in nasopharyngeal carcinoma. J Pathol. 2018;244:394–407. doi: 10.1002/path.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song Y, Li X, Zeng Z, Li Q, Gong Z, Liao Q. et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget. 2016;7:36783–99. doi: 10.18632/oncotarget.9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong AM, Kong KL, Tsang JW, Kwong DL, Guan XY. Profiling of Epstein-Barr virus-encoded microRNAs in nasopharyngeal carcinoma reveals potential biomarkers and oncomirs. Cancer. 2012;118:698–710. doi: 10.1002/cncr.26309. [DOI] [PubMed] [Google Scholar]
- 67.Yang L, Tang Y, Xiong F, He Y, Wei F, Zhang S. et al. LncRNAs regulate cancer metastasis via binding to functional proteins. Oncotarget. 2017;9:1426–43. doi: 10.18632/oncotarget.22840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang Y, He Y, Zhang P, Wang J, Fan C, Yang L. et al. LncRNAs regulate the cytoskeleton and related Rho/ROCK signaling in cancer metastasis. Mol Cancer. 2018;17:77. doi: 10.1186/s12943-018-0825-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wieland I, Arden KC, Michels D, Klein-Hitpass L, Bohm M, Viars CS. et al. Isolation of DICE1: a gene frequently affected by LOH and downregulated in lung carcinomas. Oncogene. 1999;18:4530–7. doi: 10.1038/sj.onc.1202806. [DOI] [PubMed] [Google Scholar]
- 70.Lei T, Yuen KS, Xu R, Tsao SW, Chen H, Li M. et al. Targeting of DICE1 tumor suppressor by Epstein-Barr virus-encoded miR-BART3* microRNA in nasopharyngeal carcinoma. Int J Cancer. 2013;133:79–87. doi: 10.1002/ijc.28007. [DOI] [PubMed] [Google Scholar]
- 71.Chan JY, Gao W, Ho WK, Wei WI, Wong TS. Overexpression of Epstein-Barr virus-encoded microRNA-BART7 in undifferentiated nasopharyngeal carcinoma. Anticancer Res. 2012;32:3201–10. [PubMed] [Google Scholar]
- 72.Cai LM, Lyu XM, Luo WR, Cui XF, Ye YF, Yuan CC. et al. EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN. Oncogene. 2015;34:2156–66. doi: 10.1038/onc.2014.341. [DOI] [PubMed] [Google Scholar]
- 73.Cai L, Li J, Zhang X, Lu Y, Wang J, Lyu X. et al. Gold nano-particles (AuNPs) carrying anti-EBV-miR-BART7-3p inhibit growth of EBV-positive nasopharyngeal carcinoma. Oncotarget. 2015;6:7838–50. doi: 10.18632/oncotarget.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ye Y, Zhou Y, Zhang L, Chen Y, Lyu X, Cai L. et al. EBV-miR-BART1 is involved in regulating metabolism-associated genes in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2013;436:19–24. doi: 10.1016/j.bbrc.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 75.Fan C, Tang Y, Wang J, Xiong F, Guo C, Wang Y. et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. 2017;16:130. doi: 10.1186/s12943-017-0699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qiu J, Smith P, Leahy L, Thorley-Lawson DA. The Epstein-Barr virus encoded BART miRNAs potentiate tumor growth in vivo. PLoS Pathog. 2015;11:e1004561. doi: 10.1371/journal.ppat.1004561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kiraz Y, Adan A, Kartal Yandim M, Baran Y. Major apoptotic mechanisms and genes involved in apoptosis. Tumour Biol. 2016;37:8471–86. doi: 10.1007/s13277-016-5035-9. [DOI] [PubMed] [Google Scholar]
- 78.Kang D, Skalsky RL, Cullen BR. EBV BART MicroRNAs Target Multiple Pro-apoptotic Cellular Genes to Promote Epithelial Cell Survival. PLoS Pathog. 2015;11:e1004979. doi: 10.1371/journal.ppat.1004979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF. et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551–60. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marquitz AR, Mathur A, Nam CS, Raab-Traub N. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology. 2011;412:392–400. doi: 10.1016/j.virol.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ott M, Norberg E, Zhivotovsky B, Orrenius S. Mitochondrial targeting of tBid/Bax: a role for the TOM complex? Cell Death Differ. 2009;16:1075–82. doi: 10.1038/cdd.2009.61. [DOI] [PubMed] [Google Scholar]
- 82.Bellot G, Cartron PF, Er E, Oliver L, Juin P, Armstrong LC. et al. TOM22, a core component of the mitochondria outer membrane protein translocation pore, is a mitochondrial receptor for the proapoptotic protein Bax. Cell Death Differ. 2007;14:785–94. doi: 10.1038/sj.cdd.4402055. [DOI] [PubMed] [Google Scholar]
- 83.Cai L, Ye Y, Jiang Q, Chen Y, Lyu X, Li J. et al. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat Commun. 2015;6:7353. doi: 10.1038/ncomms8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tulalamba W, Janvilisri T. Nasopharyngeal carcinoma signaling pathway: an update on molecular biomarkers. Int J Cell Biol. 2012;2012:594681. doi: 10.1155/2012/594681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wan XX, Yi H, Qu JQ, He QY, Xiao ZQ. Integrated analysis of the differential cellular and EBV miRNA expression profiles in microdissected nasopharyngeal carcinoma and non-cancerous nasopharyngeal tissues. Oncology Reports. 2015;34:2585–601. doi: 10.3892/or.2015.4237. [DOI] [PubMed] [Google Scholar]
- 86.Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6:e1001063. doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang Y, Wang J, Lian Y, Fan C, Zhang P, Wu Y. et al. Linking long non-coding RNAs and SWI/SNF complexes to chromatin remodeling in cancer. Mol Cancer. 2017;16:42. doi: 10.1186/s12943-017-0612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morotti A, Panuzzo C, Crivellaro S, Carra G, Torti D, Guerrasio A. et al. The Role of PTEN in Myeloid Malignancies. Hematol Rep. 2015;7:5844. doi: 10.4081/hr.2015.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD. et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe. 2011;10:515–26. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yan Q, Zeng Z, Gong Z, Zhang W, Li X, He B. et al. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget. 2015;6:41766–82. doi: 10.18632/oncotarget.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen R, Zhang M, Li Q, Xiong H, Liu S, Fang W. et al. The Epstein-Barr Virus-encoded miR-BART22 targets MAP3K5 to promote host cell proliferative and invasive abilities in nasopharyngeal carcinoma. J Cancer. 2017;8:305–13. doi: 10.7150/jca.15753. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92.He B, Li W, Wu Y, Wei F, Gong Z, Bo H. et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7:e2353. doi: 10.1038/cddis.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hsu CY, Yi YH, Chang KP, Chang YS, Chen SJ, Chen HC. The Epstein-Barr virus-encoded microRNA MiR-BART9 promotes tumor metastasis by targeting E-cadherin in nasopharyngeal carcinoma. PLoS Pathog. 2014;10:e1003974. doi: 10.1371/journal.ppat.1003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ellen TP, Ke Q, Zhang P, Costa M. NDRG1, a growth and cancer related gene: regulation of gene expression and function in normal and disease states. Carcinogenesis. 2008;29:2–8. doi: 10.1093/carcin/bgm200. [DOI] [PubMed] [Google Scholar]
- 95.Liu W, Xing F, Iiizumi-Gairani M, Okuda H, Watabe M, Pai SK. et al. N-myc downstream regulated gene 1 modulates Wnt-beta-catenin signalling and pleiotropically suppresses metastasis. EMBO Mol Med. 2012;4:93–108. doi: 10.1002/emmm.201100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.White BD, Chien AJ, Dawson DW. Dysregulation of Wnt/beta-catenin signaling in gastrointestinal cancers. Gastroenterology. 2012;142:219–32. doi: 10.1053/j.gastro.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kanda T, Miyata M, Kano M, Kondo S, Yoshizaki T, Iizasa H. Clustered microRNAs of the Epstein-Barr virus cooperatively downregulate an epithelial cell-specific metastasis suppressor. J Virol. 2015;89:2684–97. doi: 10.1128/JVI.03189-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Skalsky RL, Kang D, Linnstaedt SD, Cullen BR. Evolutionary conservation of primate lymphocryptovirus microRNA targets. J Virol. 2014;88:1617–35. doi: 10.1128/JVI.02071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Webb N, Connolly G, Tellam J, Yap AS, Khanna R. Epstein-Barr virus associated modulation of Wnt pathway is not dependent on latent membrane protein-1. PloS one. 2008;3:e3254. doi: 10.1371/journal.pone.0003254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL. et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–33. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang JP, Tang YY, Fan CM, Guo C, Zhou YH, Li Z. et al. The role of exosomal non-coding RNAs in cancer metastasis. Oncotarget. 2017;9:12487–502. doi: 10.18632/oncotarget.23552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Canitano A, Venturi G, Borghi M, Ammendolia MG, Fais S. Exosomes released in vitro from Epstein-Barr virus (EBV)-infected cells contain EBV-encoded latent phase mRNAs. Cancer Lett. 2013;337:193–9. doi: 10.1016/j.canlet.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 103.Gourzones C, Gelin A, Bombik I, Klibi J, Verillaud B, Guigay J. et al. Extra-cellular release and blood diffusion of BART viral micro-RNAs produced by EBV-infected nasopharyngeal carcinoma cells. Virol J. 2010;7:271. doi: 10.1186/1743-422X-7-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meckes DG Jr, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A. 2010;107:20370–5. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li J, Zeng XH, Mo HY, Rolen U, Gao YF, Zhang XS. et al. Functional inactivation of EBV-specific T-lymphocytes in nasopharyngeal carcinoma: implications for tumor immunotherapy. PloS one. 2007;2:e1122. doi: 10.1371/journal.pone.0001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Y, Xue D, Li Y, Pan X, Zhang X, Kuang B. et al. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J Cancer. 2016;7:991–1001. doi: 10.7150/jca.14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou R, Wu Y, Wang W, Su W, Liu Y, Wang Y. et al. The roles of circRNAs in human cancer. Cancer lett. 2018;425:134–42. doi: 10.1016/j.canlet.2018.03.035. [DOI] [PubMed] [Google Scholar]
- 108.Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S. et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25. doi: 10.1186/s12943-017-0598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang Y, Xia M, Jin K, Wang S, Wei H, Fan C. et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer. 2018;17:45. doi: 10.1186/s12943-018-0796-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang G, Zong J, Lin S, Verhoeven RJ, Tong S, Chen Y. et al. Circulating Epstein-Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer. 2015;136:E301–12. doi: 10.1002/ijc.29206. [DOI] [PubMed] [Google Scholar]
- 111.Zheng XH, Lu LX, Cui C, Chen MY, Li XZ, Jia WH. Epstein-Barr virus mir-bart1-5p detection via nasopharyngeal brush sampling is effective for diagnosing nasopharyngeal carcinoma. Oncotarget. 2016;7:4972–80. doi: 10.18632/oncotarget.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gourzones C, Ferrand FR, Amiel C, Verillaud B, Barat A, Guerin M. et al. Consistent high concentration of the viral microRNA BART17 in plasma samples from nasopharyngeal carcinoma patients-evidence of non-exosomal transport. Virol J. 2013;10:119. doi: 10.1186/1743-422X-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hirai N, Wakisaka N, Kondo S, Aga M, Moriyama-Kita M, Ueno T. et al. Potential Interest in Circulating miR-BART17-5p As a Post-Treatment Biomarker for Prediction of Recurrence in Epstein-Barr Virus-Related Nasopharyngeal Carcinoma. PloS one. 2016;11:e0163609. doi: 10.1371/journal.pone.0163609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yuan H, Yang BB, Xu ZF. [The clinical value of quantitative analysis of plasma Epstein-Barr virus DNA in patients with nasopharyngeal carcinoma] Zhonghua Er Bi Yan Hou Ke Za Zhi. 2004;39:162–5. [PubMed] [Google Scholar]
- 115.Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005;365:2041–54. doi: 10.1016/S0140-6736(05)66698-6. [DOI] [PubMed] [Google Scholar]
- 116.Papadopoulos EB, Ladanyi M, Emanuel D, Mackinnon S, Boulad F, Carabasi MH. et al. Infusions of donor leukocytes to treat Epstein-Barr virus-associated lymphoproliferative disorders after allogeneic bone marrow transplantation. The N Engl J Med. 1994;330:1185–91. doi: 10.1056/NEJM199404283301703. [DOI] [PubMed] [Google Scholar]
- 117.Rooney CM, Smith CA, Ng CY, Loftin S, Li C, Krance RA. et al. Use of gene-modified virus-specific T lymphocytes to control Epstein-Barr-virus-related lymphoproliferation. Lancet. 1995;345:9–13. doi: 10.1016/s0140-6736(95)91150-2. [DOI] [PubMed] [Google Scholar]
- 118.Comoli P, Labirio M, Basso S, Baldanti F, Grossi P, Furione M. et al. Infusion of autologous Epstein-Barr virus (EBV)-specific cytotoxic T cells for prevention of EBV-related lymphoproliferative disorder in solid organ transplant recipients with evidence of active virus replication. Blood. 2002;99:2592–8. doi: 10.1182/blood.v99.7.2592. [DOI] [PubMed] [Google Scholar]
- 119.Cohen JI. Erratum: Epstein-Barr virus vaccines. Clin Transl Immunology. 2015;4:e36. doi: 10.1038/cti.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chan KCA, Woo JKS, King A, Zee BCY, Lam WKJ, Chan SL. et al. Analysis of Plasma Epstein-Barr Virus DNA to Screen for Nasopharyngeal Cancer. The N Engl J Med. 2017;377:513–22. doi: 10.1056/NEJMoa1701717. [DOI] [PubMed] [Google Scholar]
- 121.Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci U S A. 2014;111:13157–62. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.White MK, Hu W, Khalili K. The CRISPR/Cas9 genome editing methodology as a weapon against human viruses. Discov Med. 2015;19:255–62. [PMC free article] [PubMed] [Google Scholar]
- 123.Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J. et al. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–27. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ. et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–4. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Szeto CY, Lin CH, Choi SC, Yip TT, Ngan RK, Tsao GS. et al. Integrated mRNA and microRNA transcriptome sequencing characterizes sequence variants and mRNA-microRNA regulatory network in nasopharyngeal carcinoma model systems. FEBS Open Bio. 2014;4:128–40. doi: 10.1016/j.fob.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tu C, Zeng Z, Qi P, Li X, Yu Z, Guo C, Genome-wide analysis of eighteen Epstein-Barr viruses isolated from primary nasopharyngeal carcinoma biopsies. J Virol; 2017. p. 91. pii: e00301-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xiao K, Yu Z, Li X, Li X, Tang K, Tu C. et al. Genome-wide Analysis of Epstein-Barr Virus (EBV) Integration and Strain in C666-1 and Raji Cells. J Cancer. 2016;7:214–24. doi: 10.7150/jca.13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–30. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]