Abstract
Background and Objective: Excision repair cross complementing (ERCC) group genes play important roles in the nucleotide excision repair (NER) way, which can effectively remove bulky lesions and reduce UV-caused DNA damage by environmental chemicals. Polymorphisms in ERCCs were thought to be related to prostate cancer (PCa) risk. However, it has been unclear whether this relationship is consistent. This study aimed to obtain the overall profile regarding the associations between ERCCs polymorphisms and PCa risk.
Materials and Methods: We identified relevant studies by a systematic search of PubMed, Medline, Embase, Google Scholar databases, Web of Science and Wanfang databases up to April 8, 2018. Odds ratios (ORs) with 95% confidential intervals (95%CIs) were conducted to evaluate the associations. All the statistical analyses were conducted basing on STATA 12.0 software.
Results: Finally, a total of 29 previous studies published in 17 publications were included for four polymorphisms in two DNA repair genes (ERCC2-rs1799793, ERCC2-rs238406, ERCC2-rs13181 and ERCC5-rs17655). Overall, we observed no significant connection between these four polymorphisms and PCa risk. However, after stratifying the studies by ethnicity, ERCC2-rs1799793 polymorphism was associated with an increased risk of PCa in Asian patients and the relationship was subsequently validated with the allelic model, the homozygous model and the recessive model when extracting the data of Asian patients for specific analyses (B vs. A: OR = 1.537, 95%CI: 1.240-1.906, PA< 0.001; BB vs. AA: OR = 2.089, 95%CI: 1.388-3.145, PA< 0.001 and BB vs. BA + AA: OR = 1.929, 95%CI: 1.313-2.835, PA= 0.020). Furthermore, subgroup analyses were also conducted by Hardy-Weinberg Equilibrium (HWE) and source of control, negative results were identified for ERCC2-rs238406, ERCC2-rs13181 and ERCC5-rs17655 polymorphisms (PA> 0.050).
Conclusion: To sum up, our work demonstrated that ERCC2-rs1799793 polymorphism is positively associated with PCa risk in Asian population. Further larger-scale studies with subjects of the same ethnicity and biological characteristics are required to verify these findings.
Keywords: Excision repair cross complementing (ERCC), single nucleotide polymorphism (SNP), prostate cancer(Pca), risk
Introduction
Prostate cancer (PCa) is one of the most common malignant tumors among males worldwide. Approximately 180,890 new diagnosed patients and 26,120 deaths are informed per year according to cancer statistics, 20161. A variety of risk factors such as hormones, family history and lifestyle are related to PCa. In addition, various DNA damages caused by the exposure to environmental carcinogens could also subsequently result in PCa, if left unrepaired.
To ensure genome integrity, mankind has developed advanced DNA repair mechanism in response to the insults of carcinogens during the evolution. DNA repair mechanism is a complicated biological system composed of many different pathways. A growing body of studies had uncovered the existence of the critical relationship between individuals' ability to maintain DNA stability and tumorigenesis, progression, and therapeutic responses2. Nucleotide excision repair (NER) is one of the important DNA repair mechanisms, which neutralizes the consequences of mutagenic exposure of cells3. The NER pathway consists of many proteins involved in DNA damage recognition and incision. ERCC group genes are key factors in DNA transcription and the NER pathway, and are also believed to be important in DNA repair system. It has been demonstrated that these genes are critical for genetic stability, and defects in these genes are more likely to develop cancer and present poor cancer prognosis. For example, ERCC1 gene is a key rate-limiting enzyme acting in the NER process and ERCC5 encodes a structure-specific endonuclease catalyzing 3' incision and involving the subsequent 5' incision by ERCC1 heterodimer4, 5. ERCC2 participated in DNA unwinding and was important to structurally identify the irrelevant DNA lesions, such as large adducts and thymidine dimmers6. Therefore, variants in the ERCC group could result in defects of DNA repair abilities, genomic instability, and the failure to regulate genetic transcription7, 8, which might give rise to a higher susceptibility to carcinogenesis.
To date, many studies have explored the relationship between polymorphisms in ERCC genes and PCa risk9-11. However, the results remain conflicting instead of conclusive because of the clinical heterogeneity, diverse ethnic populations and relatively small sample size in previous published studies. In addition, individual research might have insufficient power to detect a slight effect of polymorphisms on the risk of PCa. To overcome these limitations, we performed currently updated meta-analysis to further elucidate the connection of all available polymorphisms of ERCC genes and the PCa risk.
Materials and Methods
Literature screening and identification of relevant studies
We performed the complete literature search on electronic databases of PubMed, Medline, Embase, Google Scholar, Web of Science and Wanfang databases published up to March 27, 2018 to collect eligible articles on the relationship between ERCC genes polymorphisms and PCa risk by applying below MeSH terms: “xeroderma pigmentosum group OR excision repair cross complementing group 1-10 OR ERCC1-10” AND “ prostate cancer OR prostate carcinoma OR prostate tumor OR prostate malignancy OR prostate neoplasms OR prostate adenocarcinoma” AND “polymorphism OR mutation OR variant OR SNP OR genotype”. The language of eligible studies was limited to English. We reviewed all retrieved articles by reading the titles and abstracts. Then, the full text of the possibly relevant studies was examined for further suitability evaluations in our present meta-analysis. Meanwhile, to identify more eligible studies, we searched the references of related studies manually. The whole studies in the meta-analysis were firstly published in the primary literature with no reproduction in other studies.
Inclusion criteria and exclusion criteria
The publications in our meta-analysis were enrolled according to the following criteria: (1) studies that were case-control studies exploring the relationship between DNA repair gene ERCC polymorphisms and the PCa risk; (2) patients should be diagnosed by histopathology examination, and controls must be cancer-free, age- and sex-matched participants; and (3) the numbers or frequencies of alleles and genotypes in patients and controls can be extracted. The exclusion criteria were: (1) studies that are not related to ERCC polymorphisms and the PCa risk; (2) reviews, conference papers, meta-analyses, duplicated previous publications; (3) only case population; or (4) no sufficient data reported.
Quality assessment and data extraction
Two researchers independently assessed and extracted all the data from the included studies, and any divergences were solved by the discussion to achieve the agreement. Generally, the following information of each study should be extracted: first author, year of publication, study country, race, sample size, genotypic methods, allele genotype and genotype distribution for each polymorphism, and P-value of Hardy-Weinberg equilibrium (HWE) in controls.
Statistical analysis
We assessed the relationship between ERCCs polymorphisms and PCa risk by calculating crude Odds Ratios (ORs) with its 95%CI. And the significance of the pooled ORs was evaluated based on the Z-test12. In addition, Bonferroni corrections were applied to adjust the outcomes as well, with P-values less than 0.05 were considered as statistical significant13. The following five genetic models: the allele model (B vs. A), the homozygous model (BB vs. AA), the heterozygous model (BA vs. AA), the dominant model (BB + BA vs. AA), and the recessive model (BB vs. BA + AA) were employed to calculate the pooled ORs of each ERCC polymorphism (A: wild allele and B: mutated allele). Heterogeneity assumption was checked by I-square test and Q statistic test14. When the heterogeneity cannot be ignored (I-square ≥ 50% and P ≥ 0.1), the fixed-effect model was performed 15; otherwise, the random-effect model was applied16. All analyses were conducted by Stata software (Stata Corporation, College Station, TX; version 12.0).
Linkage disequilibrium (LD) analysis across populations
We extracted the datum from the 1000 genomes Project (http:// hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap3r2_B36/), containing ERCC genes polymorphisms assessed in our study. In general, populations enrolled in the project were composed of CHB (Han Chinese in Beijing, China), CEU (Utah residents with Northern and Western European ancestry from the CEPH collection), JPT (Japanese in Tokyo, Japan) and YRI (Yoruba in Ibadan, Nigeria). And we performed Haploview software to apply analyses, Moreover, for each of the above four populations, LD was assessed by I-square statistics.
Results
Study identification and characteristics of included studies
332 publications were identified after initial screening. After scoring out duplicates and screening the titles and abstracts, 305 publications were removed. Finally, 25 eligible publications were selected for further full-text review. Among these, we excluded five publications because relevant studies were less than three. We also excluded three publications because they describe the relationship of ERCCs polymorphisms with other cancer risk. Finally, 17 articles were enrolled comprising 29 case-controls studies17-33, and the researches selection process was presented in Figure S 1-4. The general demographical characteristics of all eligible publications of the ERCC2-rs1799793/rs238406/rs13181 and ERCC5-rs17655 polymorphisms were summarized in Table 1. For ERCC2-rs1799793 polymorphism, 11 eligible studies with 3,913 cases and 5,085 controls were enrolled in our meta-analysis. Five of these studies were performed in Caucasian ethnicity, two in Asian ethnicity, only one in African ethnicity and the others was mixed ethnicity (more than two descendants). Controls of five researches were hospital-based (H-B), while others were population-based (P-B). The genotype distributions of ERCC2-rs1799793 in the controls were totally consistent with HWE, except for three studies18, 24. For ERCC2-rs238406 polymorphism, four studies with 2,017 cases and 2,097 controls were selected for the present meta-analysis. Two studies were performed in Asian populations and the other two studies were conducted in Caucasian populations. The controls in three studies were P-B, and one was H-B. Additionally, the distributions of ERCC2-rs238406 polymorphisms genotype conformed to HWE, except for one study. For ERCC2-rs13181 polymorphism, we analyzed 11 studies with 4,456 cases and 4,946 controls. Four studies were in Asian ethnicity, four studies were in Caucasian ethnicity, one study was in African ethnicity and the rest of studies were mixed ethnicity. Controls in six studies were P-B, and three were H-B. As for HWE status, the genotype distributions of three studies were not in accordance with HWE18, 24. For ERCC5-rs17655 polymorphism, three studies with 1,040 cases and 1,229 controls met the inclusion criteria. They were performed in Asian, Caucasian and African ethnicity, respectively. One study was P-B, and other two studies were H-B. Additionally, the distributions of ERCC5-rs17655 polymorphism genotype were consistent with HWE, except for one studies19. We evaluated the quality of included studies by Newcastle-Ottawa Scale (NOS), and the outcomes were presented in Table S 1.
Table 1.
Characteristics of the eligible studies included in the meta-analysis.
| SNP | First Author | Year | Ethnicity | Source Of Control | Case | Control | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | AA | AB | BB | HWE | |||||
| ERCC2-rs1799793 | Rybicki et al. | 2004 | Caucasian | P-B | 230 | 269 | 72 | 180 | 218 | 39 | Y |
| Mixed | 277 | 286 | 73 | 210 | 231 | 39 | Y | ||||
| Bau et al. | 2007 | Asian | H-B | 62 | 39 | 22 | 310 | 106 | 63 | Y | |
| Agalliu et al. | 2009 | Caucasian | P-B | 545 | 575 | 120 | 527 | 528 | 166 | Y | |
| African | 106 | 31 | 7 | 65 | 15 | 2 | Y | ||||
| Mandal et al. | 2010 | Asian | P-B | 76 | 56 | 39 | 99 | 81 | 20 | Y | |
| Lavender et al. | 2010 | Mixed | H-B | 146 | 39 | 5 | 510 | 116 | 5 | Y | |
| Dhillon et al. | 2011 | Caucasian | H-B | 71 | 37 | 8 | 80 | 42 | 10 | Y | |
| Yeoh et al. | 2011 | Oceania | H-B | 71 | 37 | 8 | 80 | 42 | 10 | Y | |
| Mirecka et al. | 2014 | Caucasian | P-B | 199 | 249 | 124 | 377 | 218 | 32 | Y | |
| Fachal et al. | 2012 | Caucasian | H-B | 14 | 17 | 3 | 298 | 277 | 89 | Y | |
| ERCC2-rs238406 | Zhou et al. | 2013 | Asian | H-B | 26 | 53 | 21 | 38 | 49 | 13 | Y |
| Mirecka et al. | 2014 | Caucasian | P-B | 113 | 300 | 99 | 141 | 411 | 126 | Y | |
| Agalliu et al. | 2009 | Caucasian | P-B | 365 | 636 | 260 | 383 | 600 | 255 | Y | |
| African | 112 | 29 | 3 | 65 | 16 | 0 | Y | ||||
| ERCC2-rs13181 | Rybicki et al. | 2004 | Caucasian | P-B | 230 | 269 | 72 | 180 | 218 | 39 | Y |
| Mixed | 277 | 286 | 73 | 210 | 231 | 39 | Y | ||||
| Ritchey et al. | 2005 | Asian | P-B | 141 | 19 | 213 | 34 | N.A. | |||
| Bau et al. | 2007 | Asian | H-B | 111 | 10 | 2 | 441 | 33 | 5 | Y | |
| Agalliu et al. | 2009 | Caucasian | P-B | 505 | 575 | 153 | 480 | 571 | 177 | Y | |
| African | 87 | 48 | 11 | 50 | 28 | 5 | Y | ||||
| Mandal et al. | 2010 | Asian | P-B | 73 | 84 | 14 | 89 | 94 | 17 | Y | |
| Gao et al. | 2010 | America | H-B | 186 | 178 | 64 | 49 | 56 | 13 | Y | |
| Lavender et al. | 2010 | Mixed | H-B | 110 | 60 | 13 | 367 | 194 | 38 | Y | |
| Sobti et al. | 2012 | Asian | P-B | 62 | 67 | 21 | 67 | 69 | 14 | Y | |
| Mirecka et al. | 2014 | Caucasian | P-B | 231 | 302 | 122 | 319 | 444 | 162 | Y | |
| ERCC5-rs17655 | Hooker et al. | 2008 | African | H-B | 74 | 119 | 61 | 100 | 141 | 60 | Y |
| Berhane et al. | 2011 | Asian | H-B | 58 | 72 | 20 | 66 | 75 | 9 | Y | |
| Mirecka et al. | 2014 | Caucasian | P-B | 396 | 208 | 32 | 523 | 224 | 31 | Y | |
A: wild allele; B: mutated allele; HWE: Hardy Weinberg Equilibrium; H-B: hospital-based; P-B: population-based; Y: study conformed to HWE; N.A.: study cannot calculate HWE.
Quantitative synthesis and Subgroup analyses
Forest plots were applied to show meta-analysis results for each genetic polymorphism, and we calculated summary ORs for the pooling publications to assess the relationship between those polymorphisms and PCa risk. The results of the relationship between polymorphisms in ERCC2 and ERCC5 and PCa risk were shown in Table 2.
Table 2.
Results of the association between ERCC2 and ERCC5 polymorphisms and PCa risk.
| SNP | Comparison | Subgroup | N | PH | PZ | PA | Random | Fixed |
|---|---|---|---|---|---|---|---|---|
| rs13181 | B vs.A | Overall | 11 | 0.820 | 0.679 | 1.000 | 1.014 (0.950-1.082) | 1.014 (0.950-1.082) |
| B vs.A | Asian | 4 | 0.763 | 0.397 | 1.000 | 1.092 (0.894-1.333) | 1.090 (0.893-1.331) | |
| B vs.A | Caucasian | 4 | 0.379 | 0.709 | 1.000 | 0.986 (0.910-1.068) | 0.985 (0.911-1.065) | |
| B vs.A | Other ethnicities | 3 | 0.987 | 0.325 | 1.000 | 1.074 (0.932-1.238) | 1.074 (0.932-1.238) | |
| B vs.A | H-B | 3 | 0.833 | 0.492 | 1.000 | 1.072 (0.883-1.301) | 1.070 (0.881-1.300) | |
| B vs.A | Other | 2 | 0.874 | 0.168 | 1.000 | 1.094 (0.963-1.244) | 1.094 (0.963-1.244) | |
| B vs.A | P-B | 6 | 0.712 | 0.520 | 1.000 | 0.974 (0.897-1.056) | 0.974 (0.897-1.056) | |
| B vs.A | N | 3 | 0.874 | 0.133 | 1.000 | 1.102 (0.971-1.249) | 1.101 (0.971-1.249) | |
| B vs.A | Y | 8 | 0.847 | 0.673 | 1.000 | 0.984 (0.912-1.061) | 0.984 (0.912-1.061) | |
| BA vs.AA | Overall | 11 | 0.999 | 0.412 | 1.000 | 0.962 (0.876-1.056) | 0.962 (0.876-1.056) | |
| BA vs.AA | Asian | 4 | 0.882 | 0.785 | 1.000 | 1.039 (0.798-1.352) | 1.037 (0.797-1.350) | |
| BA vs.AA | Caucasian | 4 | 0.951 | 0.339 | 1.000 | 0.945 (0.841-1.061) | 0.945 (0.841-1.061) | |
| BA vs.AA | Other ethnicities | 3 | 0.913 | 0.756 | 1.000 | 0.970 (0.799-1.177) | 0.970 (0.799-1.177) | |
| BA vs.AA | H-B | 3 | 0.644 | 0.850 | 1.000 | 0.976 (0.753-1.266) | 0.975 (0.752-1.265) | |
| BA vs.AA | Other | 2 | 0.878 | 0.590 | 1.000 | 0.951 (0.793-1.141) | 0.951 (0.793-1.141) | |
| BA vs.AA | P-B | 6 | 0.984 | 0.542 | 1.000 | 0.964 (0.855-1.086) | 0.963 (0.855-1.086) | |
| BA vs.AA | N | 3 | 0.822 | 0.681 | 1.000 | 0.964 (0.808-1.150) | 0.964 (0.808-1.150) | |
| BA vs.AA | Y | 8 | 0.990 | 0.478 | 1.000 | 0.961 (0.861-1.073) | 0.961 (0.861-1.072) | |
| BA+BB vs. AA | Overall | 11 | 0.993 | 0.745 | 1.000 | 0.986 (0.902-1.077) | 0.985 (0.902-1.077) | |
| BA+BB vs. AA | Asian | 4 | 0.848 | 0.579 | 1.000 | 1.076 (0.836-1.385) | 1.074 (0.835-1.382) | |
| BA+BB vs. AA | Caucasian | 4 | 0.895 | 0.431 | 1.000 | 0.957 (0.857-1.068) | 0.957 (0.857-1.068) | |
| BA+BB vs. AA | Other ethnicities | 3 | 0.982 | 0.814 | 1.000 | 1.022 (0.851-1.229) | 1.022 (0.851-1.229) | |
| BA+BB vs. AA | H-B | 3 | 0.741 | 0.839 | 1.000 | 1.027 (0.804-1.312) | 1.026 (0.803-1.310) | |
| BA+BB vs. AA | Other | 2 | 0.867 | 0.805 | 1.000 | 1.022 (0.859-1.216) | 1.022 (0.859-1.216) | |
| BA+BB vs. AA | P-B | 6 | 0.939 | 0.502 | 1.000 | 0.962 (0.859-1.077) | 0.962 (0.859-1.077) | |
| BA+BB vs. AA | N | 3 | 0.838 | 0.694 | 1.000 | 1.035 (0.875-1.225) | 1.034 (0.874-1.225) | |
| BA+BB vs. AA | Y | 8 | 0.981 | 0.532 | 1.000 | 0.967 (0.872-1.073) | 0.967 (0.872-1.073) | |
| BB vs.AA | Overall | 11 | 0.466 | 0.335 | 1.000 | 1.071 (0.927-1.238) | 1.073 (0.930-1.239) | |
| BB vs.AA | Asian | 4 | 0.845 | 0.290 | 1.000 | 1.314 (0.792-2.179) | 1.313 (0.793-2.173) | |
| BB vs.AA | Caucasian | 4 | 0.124 | 0.975 | 1.000 | 1.050 (0.814-1.354) | 0.997 (0.844-1.179) | |
| BB vs.AA | Other ethnicities | 3 | 0.861 | 0.103 | 1.000 | 1.325 (0.941-1.866) | 1.327 (0.944-1.866) | |
| BB vs.AA | H-B | 3 | 0.922 | 0.352 | 1.000 | 1.241 (0.787-1.955) | 1.240 (0.788-1.953) | |
| BB vs.AA | Other | 2 | 0.954 | 0.021 | 0.420 | 1.432 (1.055-1.943) | 1.432 (1.055-1.943) | |
| BB vs.AA | P-B | 6 | 0.556 | 0.587 | 1.000 | 0.952 (0.798-1.135) | 0.953 (0.799-1.135) | |
| BB vs.AA | N | 3 | 0.991 | 0.018 | 0.360 | 1.437 (1.064-1.940) | 1.436 (1.063-1.939) | |
| BB vs.AA | Y | 8 | 0.669 | 0.824 | 1.000 | 0.981 (0.831-1.156) | 0.982 (0.833-1.157) | |
| BB vs. BA+AA | Overall | 11 | 0.357 | 0.179 | 1.000 | 1.118 (0.962-1.300) | 1.096 (0.959-1.252) | |
| BB vs. BA+AA | Asian | 4 | 0.803 | 0.325 | 1.000 | 1.274 (0.785-2.068) | 1.273 (0.787-2.061) | |
| BB vs. BA+AA | Caucasian | 4 | 0.075 | 0.489 | 1.000 | 1.096 (0.845-1.421) | 1.029 (0.883-1.200) | |
| BB vs. BA+AA | Other ethnicities | 3 | 0.796 | 0.068 | 1.000 | 1.354 (0.973-1.883) | 1.357 (0.978-1.884) | |
| BB vs. BA+AA | H-B | 3 | 0.860 | 0.245 | 1.000 | 1.289 (0.831-1.999) | 1.294 (0.838-1.998) | |
| BB vs. BA+AA | Other | 2 | 0.988 | 0.009 | 0.180 | 1.469 (1.100-1.963) | 1.469 (1.100-1.963) | |
| BB vs. BA+AA | P-B | 6 | 0.521 | 0.751 | 1.000 | 0.974 (0.828-1.145) | 0.974 (0.829-1.145) | |
| BB vs. BA+AA | N | 3 | 0.997 | 0.008 | 0.160 | 1.472 (1.107-1.958) | 1.472 (1.106-1.958) | |
| BB vs. BA+AA | Y | 8 | 0.586 | 0.947 | 1.000 | 1.003 (0.861-1.169) | 1.005 (0.863-1.170) | |
| rs1799793 | B vs.A | Overall | 11 | 0.000 | 0.072 | 1.000 | 1.259 (0.980-1.618) | 1.218 (1.138-1.303) |
| B vs.A | Asian | 2 | 0.749 | 0.000 | < 0.001 | 1.539 (1.241-1.907) | 1.537 (1.240-1.906) | |
| B vs.A | Caucasian | 5 | 0.000 | 0.426 | 1.000 | 1.215 (0.752-1.964) | 1.209 (1.113-1.313) | |
| B vs.A | Mixed | 2 | 0.244 | 0.279 | 1.000 | 1.158 (0.845-1.588) | 1.161 (0.886-1.523) | |
| B vs.A | Other ethnicities | 2 | 0.386 | 0.229 | 1.000 | 1.109 (0.935-1.315) | 1.110 (0.937-1.316) | |
| B vs.A | H-B | 5 | 0.174 | 0.024 | 0.480 | 1.191 (0.956-1.484) | 1.217 (1.026-1.445) | |
| B vs.A | Other | 2 | 0.874 | 0.168 | 1.000 | 1.094 (0.963-1.244) | 1.094 (0.963-1.244) | |
| B vs.A | P-B | 4 | 0.000 | 0.215 | 1.000 | 1.497 (0.792-2.832) | 1.285 (1.173-1.407) | |
| B vs.A | N | 3 | 0.080 | 0.071 | 1.000 | 1.198 (0.984-1.459) | 1.156 (1.027-1.301) | |
| B vs.A | Y | 8 | 0.000 | 0.229 | 1.000 | 1.264 (0.863-1.851) | 1.249 (1.150-1.358) | |
| BA vs.AA | Overall | 11 | 0.000 | 0.103 | 1.000 | 1.186 (0.966-1.455) | 1.170 (1.063-1.287) | |
| BA vs.AA | Asian | 2 | 0.030 | 0.481 | 1.000 | 1.286 (0.639-2.591) | 1.269 (0.919-1.751) | |
| BA vs.AA | Caucasian | 5 | 0.000 | 0.235 | 1.000 | 1.242 (0.869-1.777) | 1.222 (1.086-1.375) | |
| BA vs.AA | Mixed | 2 | 0.628 | 0.550 | 1.000 | 1.106 (0.798-1.532) | 1.105 (0.797-1.532) | |
| BA vs.AA | Other ethnicities | 2 | 0.422 | 0.816 | 1.000 | 0.972 (0.769-1.229) | 0.973 (0.770-1.229) | |
| BA vs.AA | H-B | 5 | 0.377 | 0.059 | 1.000 | 1.249 (0.988-1.578) | 1.244 (0.992-1.561) | |
| BA vs.AA | Other | 2 | 0.878 | 0.590 | 1.000 | 0.951 (0.793-1.141) | 0.951 (0.793-1.141) | |
| BA vs.AA | P-B | 4 | 0.000 | 0.277 | 1.000 | 1.287 (0.816-2.030) | 1.276 (1.120-1.453) | |
| BA vs.AA | N | 3 | 0.031 | 0.492 | 1.000 | 1.124 (0.805-1.569) | 1.037 (0.876-1.227) | |
| BA vs.AA | Y | 8 | 0.001 | 0.171 | 1.000 | 1.208 (0.922-1.582) | 1.239 (1.103-1.392) | |
| BA+BB vs. AA | Overall | 11 | 0.000 | 0.075 | 1.000 | 1.264 (0.977-1.636) | 1.239 (1.132-1.355) | |
| BA+BB vs. AA | Asian | 2 | 0.185 | 0.006 | 0.120 | 1.491 (1.020-2.179) | 1.490 (1.119-1.984) | |
| BA+BB vs. AA | Caucasian | 5 | 0.000 | 0.328 | 1.000 | 1.280 (0.781-2.098) | 1.268 (1.135-1.417) | |
| BA+BB vs. AA | Mixed | 2 | 0.421 | 0.381 | 1.000 | 1.152 (0.845-1.572) | 1.150 (0.842-1.570) | |
| BA+BB vs. AA | Other ethnicities | 2 | 0.384 | 0.690 | 1.000 | 1.046 (0.836-1.308) | 1.047 (0.837-1.309) | |
| BA+BB vs. AA | H-B | 5 | 0.279 | 0.028 | 0.560 | 1.255 (0.985-1.597) | 1.267 (1.026-1.564) | |
| BA+BB vs. AA | Other | 2 | 0.867 | 0.805 | 1.000 | 1.022 (0.859-1.216) | 1.022 (0.859-1.216) | |
| BA+BB vs. AA | P-B | 4 | 0.000 | 0.227 | 1.000 | 1.471 (0.786-2.752) | 1.351 (1.196-1.526) | |
| BA+BB vs. AA | N | 3 | 0.038 | 0.265 | 1.000 | 1.188 (0.878-1.606) | 1.117 (0.953-1.310) | |
| BA+BB vs. AA | Y | 8 | 0.000 | 0.191 | 1.000 | 1.279 (0.885-1.850) | 1.300 (1.166-1.450) | |
| BB vs.AA | Overall | 11 | 0.000 | 0.077 | 1.000 | 1.636 (0.948-2.824) | 1.496 (1.287-1.738) | |
| BB vs.AA | Asian | 2 | 0.376 | 0.000 | < 0.001 | 2.066 (1.367-3.123) | 2.089 (1.388-3.145) | |
| BB vs.AA | Caucasian | 5 | 0.000 | 0.508 | 1.000 | 1.423 (0.500-4.044) | 1.416 (1.183-1.695) | |
| BB vs.AA | Mixed | 2 | 0.095 | 0.446 | 1.000 | 1.675 (0.445-6.310) | 1.438 (0.667-3.100) | |
| BB vs.AA | Other ethnicities | 2 | 0.625 | 0.070 | 1.000 | 1.459 (0.965-2.206) | 1.464 (0.969-2.212) | |
| BB vs.AA | H-B | 5 | 0.260 | 0.129 | 1.000 | 1.340 (0.830-2.163) | 1.351 (0.916-1.991) | |
| BB vs.AA | Other | 2 | 0.954 | 0.021 | 0.420 | 1.432 (1.055-1.943) | 1.432 (1.055-1.943) | |
| BB vs.AA | P-B | 4 | 0.000 | 0.240 | 1.000 | 2.297 (0.574-9.200) | 1.560 (1.286-1.892) | |
| BB vs.AA | N | 3 | 0.827 | 0.003 | 0.060 | 1.499 (1.147-1.959) | 1.493 (1.141-1.954) | |
| BB vs.AA | Y | 8 | 0.000 | 0.248 | 1.000 | 1.673 (0.699-4.007) | 1.497 (1.249-1.794) | |
| BB vs. BA+AA | Overall | 11 | 0.000 | 0.087 | 1.000 | 1.533 (0.941-2.497) | 1.388 (1.203-1.603) | |
| BB vs. BA+AA | Asian | 2 | 0.127 | 0.001 | 0.020 | 1.932 (1.058-3.529) | 1.929 (1.313-2.835) | |
| BB vs. BA+AA | Caucasian | 5 | 0.000 | 0.576 | 1.000 | 1.296 (0.523-3.214) | 1.284 (1.081-1.525) | |
| BB vs. BA+AA | Mixed | 2 | 0.101 | 0.371 | 1.000 | 1.644 (0.452-5.979) | 1.413 (0.662-3.019) | |
| BB vs. BA+AA | Other ethnicities | 2 | 0.693 | 0.044 | 0.880 | 1.496 (1.008-2.222) | 1.500 (1.010-2.226) | |
| BB vs. BA+AA | H-B | 5 | 0.301 | 0.327 | 1.000 | 1.218 (0.783-1.893) | 1.205 (0.830-1.751) | |
| BB vs. BA+AA | Other | 2 | 0.988 | 0.009 | 0.180 | 1.469 (1.100-1.963) | 1.469 (1.100-1.963) | |
| BB vs. BA+AA | P-B | 4 | 0.000 | 0.245 | 1.000 | 2.077 (0.606-7.111) | 1.402 (1.166-1.686) | |
| BB vs. BA+AA | N | 3 | 0.998 | 0.003 | 0.060 | 1.462 (1.134-1.886) | 1.463 (1.134-1.887) | |
| BB vs. BA+AA | Y | 8 | 0.000 | 0.269 | 1.000 | 1.557 (0.711-3.410) | 1.355 (1.139-1.612) | |
| rs238406 | B vs.A | Overall | 4 | 0.258 | 0.290 | 1.000 | 1.063 (0.941-1.201) | 1.049 (0.960-1.146) |
| B vs.A | Caucasian | 2 | 0.611 | 0.605 | 1.000 | 1.025 (0.935-1.123) | 1.025 (0.935-1.123) | |
| B vs.A | Other ethnicities | 2 | 0.639 | 0.037 | 0.740 | 1.432 (1.023-2.005) | 1.431 (1.022-2.004) | |
| B vs.A | P-B | 3 | 0.713 | 0.534 | 1.000 | 1.029 (0.940-1.127) | 1.029 (0.940-1.127) | |
| B vs.A | Y | 3 | 0.189 | 0.179 | 1.000 | 1.168 (0.916-1.490) | 1.075 (0.967-1.195) | |
| BA vs.AA | Overall | 4 | 0.418 | 0.335 | 1.000 | 1.074 (0.928-1.243) | 1.074 (0.929-1.243) | |
| BA vs.AA | Caucasian | 2 | 0.251 | 0.525 | 1.000 | 1.040 (0.864-1.251) | 1.051 (0.901-1.226) | |
| BA vs.AA | Other ethnicities | 2 | 0.391 | 0.255 | 1.000 | 1.310 (0.824-2.083) | 1.310 (0.823-2.084) | |
| BA vs.AA | P-B | 3 | 0.517 | 0.515 | 1.000 | 1.051 (0.905-1.221) | 1.051 (0.905-1.221) | |
| BA vs.AA | Y | 3 | 0.563 | 0.137 | 1.000 | 1.137 (0.960-1.346) | 1.137 (0.960-1.346) | |
| BA+BB vs. AA | Overall | 4 | 0.300 | 0.258 | 1.000 | 1.088 (0.913-1.297) | 1.083 (0.943-1.245) | |
| BA+BB vs. AA | Caucasian | 2 | 0.307 | 0.516 | 1.000 | 1.048 (0.901-1.218) | 1.050 (0.907-1.215) | |
| BA+BB vs. AA | Other ethnicities | 2 | 0.377 | 0.102 | 1.000 | 1.456 (0.929-2.280) | 1.454 (0.928-2.280) | |
| BA+BB vs. AA | P-B | 3 | 0.570 | 0.467 | 1.000 | 1.054 (0.914-1.216) | 1.054 (0.914-1.216) | |
| BA+BB vs. AA | Y | 3 | 0.352 | 0.108 | 1.000 | 1.150 (0.959-1.379) | 1.140 (0.972-1.338) | |
| BB vs.AA | Overall | 4 | 0.234 | 0.335 | 1.000 | 1.133 (0.855-1.501) | 1.095 (0.910-1.318) | |
| BB vs.AA | Caucasian | 2 | 0.687 | 0.657 | 1.000 | 1.044 (0.863-1.264) | 1.044 (0.863-1.263) | |
| BB vs.AA | Other ethnicities | 2 | 0.728 | 0.028 | 0.560 | 2.461 (1.084-5.586) | 2.502 (1.105-5.664) | |
| BB vs.AA | P-B | 3 | 0.618 | 0.593 | 1.000 | 1.050 (0.868-1.270) | 1.053 (0.871-1.274) | |
| BB vs.AA | Y | 3 | 0.149 | 0.236 | 1.000 | 1.468 (0.755-2.856) | 1.139 (0.918-1.413) | |
| BB vs. BA+AA | Overall | 4 | 0.410 | 0.559 | 1.000 | 1.045 (0.893-1.224) | 1.048 (0.895-1.227) | |
| BB vs. BA+AA | Caucasian | 2 | 0.790 | 0.848 | 1.000 | 1.016 (0.864-1.194) | 1.016 (0.864-1.194) | |
| BB vs. BA+AA | Other ethnicities | 2 | 0.599 | 0.082 | 1.000 | 1.869 (0.899-3.889) | 1.908 (0.922-3.948) | |
| BB vs. BA+AA | P-B | 3 | 0.640 | 0.788 | 1.000 | 1.020 (0.868-1.199) | 1.022 (0.870-1.201) | |
| BB vs. BA+AA | Y | 3 | 0.237 | 0.629 | 1.000 | 1.197 (0.757-1.891) | 1.047 (0.869-1.262) | |
| rs17655 | B vs.A | Overall | 3 | 0.852 | 0.004 | 0.080 | 1.216 (1.064-1.391) | 1.216 (1.064-1.391) |
| B vs.A | H-B | 2 | 0.578 | 0.040 | 0.800 | 1.226 (1.009-1.489) | 1.226 (1.009-1.489) | |
| B vs.A | Y | 2 | 0.876 | 0.016 | 0.320 | 1.197 (1.034-1.385) | 1.197 (1.034-1.385) | |
| BA G AA | Overall | 3 | 0.889 | 0.066 | 1.000 | 1.187 (0.989-1.424) | 1.187 (0.989-1.424) | |
| BA vs.AA | H-B | 2 | 0.891 | 0.456 | 1.000 | 1.121 (0.830-1.515) | 1.121 (0.830-1.515) | |
| BA vs.AA | Y | 2 | 0.752 | 0.066 | 1.000 | 1.203 (0.988-1.466) | 1.203 (0.988-1.466) | |
| BA+BB vs. AA | Overall | 3 | 0.992 | 0.017 | 0.340 | 1.236 (1.039-1.470) | 1.236 (1.039-1.470) | |
| BA+BB vs. AA | H-B | 2 | 0.922 | 0.164 | 1.000 | 1.224 (0.921-1.626) | 1.224 (0.921-1.626) | |
| BA+BB vs. AA | Y | 2 | 0.901 | 0.028 | 0.560 | 1.234 (1.023-1.489) | 1.234 (1.023-1.489) | |
| BB vs.AA | Overall | 3 | 0.432 | 0.013 | 0.260 | 1.490 (1.082-2.051) | 1.497 (1.089-2.057) | |
| BB vs.AA | H-B | 2 | 0.222 | 0.026 | 0.520 | 1.666 (0.955-2.906) | 1.587 (1.056-2.385) | |
| BB vs.AA | Y | 2 | 0.983 | 0.074 | 1.000 | 1.369 (0.970-1.932) | 1.369 (0.970-1.932) | |
| BB vs. BA+AA | Overall | 3 | 0.363 | 0.028 | 0.560 | 1.382 (1.027-1.860) | 1.389 (1.037-1.861) | |
| BB vs. BA+AA | H-B | 2 | 0.170 | 0.043 | 0.860 | 1.576 (0.870-2.857) | 1.449 (1.012-2.075) | |
| BB vs. BA+AA | Y | 2 | 0.986 | 0.134 | 1.000 | 1.272 (0.928-1.744) | 1.272 (0.928-1.744) |
PH: P value of heterogeneity; PZ: P value of Z test; PA: adjusted P value (PA = PZ *3 polymorphisms * 5 genetic models, and PA<0.05 means statistically significant); A: wild allele; B: mutated allele; HWE: Hardy Weinberg Equilibrium; H-B: hospital-based; P-B: population-based; Y: study conformed to HWE; N: study did not conform to HWE
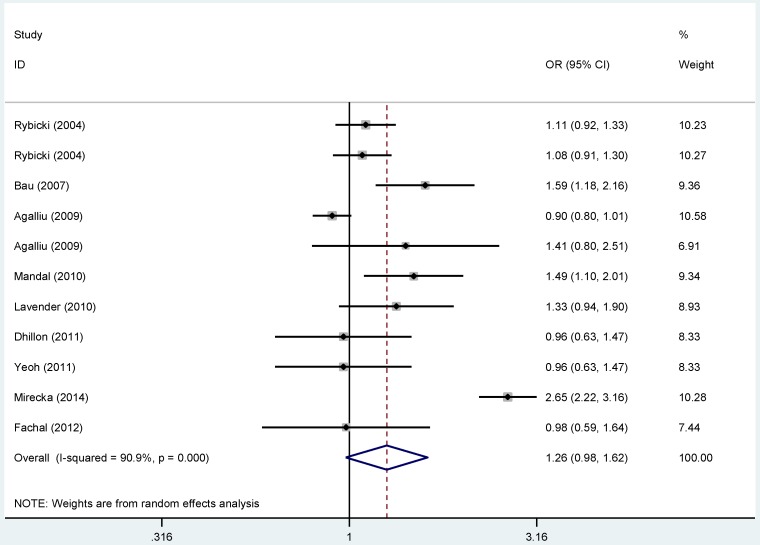
Overall, no statistical connection was observed for the relationship between the polymorphism in rs1799793 and PCa risk (Figure 1). Nevertheless, subgroup analysis based on ethnicity revealed that rs1799793 polymorphism was relevant to the increased susceptibility to Pca in Asian population in the allelic model (B vs. A: OR = 1.537, 95%CI = 1.240-1.906, PA < 0.001), the homozygous model (BB vs. AA: OR = 2.089, 95%CI: 1.388-3.145, PA < 0.001) and the recessive model (BB vs. BA+AA: OR = 1.929, 95%CI = 1.313-2.835, PA= 0.020), as showing in Figure S 16. Furthermore, we did not find any significant results between ERCC2-rs1799793 polymorphism and PCa risk for the subgroup analyses based on the source of control and HWE status.
Figure 1.
Forest plots of the association between ERCC2-rs1799793 polymorphism and the risk of prostate cancer (B vs. A). Each square indicates a study, and the area of squares is proportional to the weight of the study. The diamond represents the summary OR and 95% CI. CI= confidence interval, OR= odds ratio.
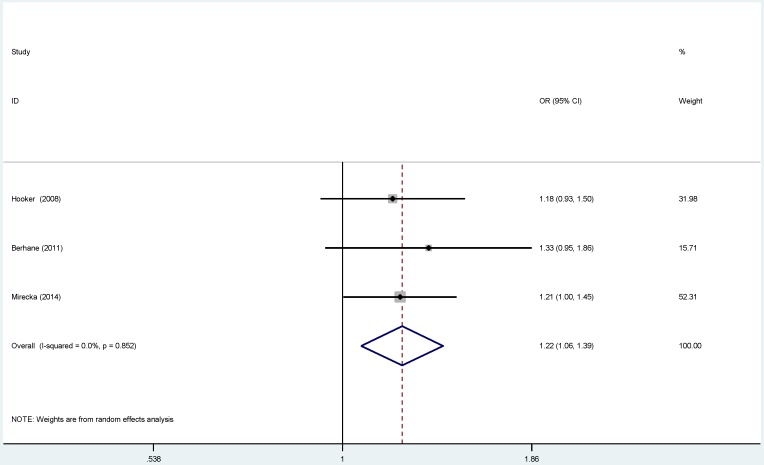
For ERCC2-rs238406, ERCC2-rs13181 polymorphisms, no significant associations were identified between gene polymorphisms and PCa risk (Figure S 5-6). Similarly, there was no significant relationship between ERCC5-rs17655 polymorphism and PCa risk (Figure 2). The subgroup analysis suggested that the ERCC2-rs238406, ERCC2-rs13181 and ERCC5-rs17655 polymorphisms were not statistically related to PCa based on the source of controls, ethnicity and HWE status.
Figure 2.
Forest plots of the association between ERCC5-rs17655 polymorphism and the risk of prostate cancer (B vs. A). Each square indicates a study, and the area of squares is proportional to the weight of the study. The diamond represents the summary OR and 95% CI. CI= confidence interval, OR= odds ratio.
Sensitivity analyses and publication bias
To better examine the influence of each study on the pooled OR, we conducted one-way sensitivity analyses by excluding each study, once at a time in every genetic model for rs1799793, rs238406, rs13181 and rs17655. The sensitivity analyses demonstrated that the ORs were not statistically influenced, which validated the stability of our data (Table S 2, Figure S 7-10). We used the Begg's funnel plots and Egger's regression tests to explore the potential publication bias for each gene polymorphism (Table S 3, Figure S 11-14). As for rs238406, rs13181 and rs17655, the shape of funnel plots and the Egger's regression tests results revealed no significant publication bias. For rs1799793, publication bias was identified in subgroup analyses of H-B and studies are not consistent with HWE, which could be graphically identified from the shape of funnel plot as well.
LD Analyses Across Populations
For a better understanding of this plot synthesis, we respectively conducted LD analysis to examine the presence of bins in the region containing these variants in each ERCC2 genes (polymorphisms including ERCC2-rs1799793, ERCC2-rs238406 and ERCC2-rs13181), and the detail information was presented in Table S 4 and Figure S 15.
Discussion
The present meta-analysis investigated the connections between four commonly studied polymorphisms (ERCC2-rs1799793/rs238406/rs13181 and ERCC5-rs17655) and the PCa risk. Finally, a total of 11,427 cases and 13,358 controls from 27 publications were enrolled in our analysis. The overall results suggested that none of these polymorphisms was connected with the risk of PCa. However, when the subgroup analysis was performed by ethnicity, the ERCC2-rs1799793 variant homozygous genotype was identified statistically related to the risk of PCa.
Among various carcinogenic factors, DNA damage could be one of the most frequently reported pathogenesis and could further result in genomic instability. Therefore, sequence variants in DNA repair genes, which are important in maintaining DNA stability, may affect DNA repair capacity and consequently increase an individual's susceptibility to PCa. Numerous studies have explored the the relationship between DNA repair gene ERCC2 polymorphisms and cancer risk. Bau et al.18 observed a significant difference in the frequency of the ERCC2-rs1799793 genotype between the PCa and control groups in Asian populations. Mandal et al.30 found that ERCC2-rs1799793 variant BB genotype was related to increased risk for PCa. Meanwhile, ERCC5 gene also plays a crucial role in the DNA repair machinery. It is generally accepted that severe autosomal recessive diseases, such as XP, CS and TTD may result from the defects of ERCC534. Several publications have reported that ERCC5 polymorphisms are related to the development of a variety of cancers, such as breast cancer35, bladder cancer36 and PCa20. Thus, it seems that deficiency in the DNA repair gene ERCC2 and ERCC5 are thought to have a central role in the modulation of PCa susceptibility.
Nevertheless, the present meta-analysis revealed no overall associations for the relationship between the ERCC2-rs1799793, ERCC2-rs238406, ERCC2-rs13181, ERCC5-rs17655 polymorphisms and the PCa risk. Our results were consistent with the findings of Agalliuet al25 but different from Mirecka et al22 who found polymorphism in ERCC2 gene appeared to be connected with the risk of PCa. Furthermore, when subgroup analysis was stratified by ethnicity, ERCC2-rs1799793 polymorphism was observed to be related to an increased risk of PCa in Asian population. Besides, stratification analyses according to the source of controls, ethnicity and HWE status, no significant results were identified for ERCC2-rs13181, ERCC2-rs238406 and ERCC5-rs17655 polymorphisms.
A comparison of the polymorphisms of ERCC2 and ERCC5 that did not present a noteworthy connection with PCa may be owing to the characteristics of low-penetrance genes. It is widely accepted that cancer etiology is polygenic and has a complex disease phenotype, thus, single genetic polymorphism is frequently inadequate to prognosticate the risk of cancer. For different carcinogens and different tumors, DNA repair may occur in a rather unspecified pattern. Although some investigators have explained the tissue-specific balance between apoptotic signals and repair effects in different tissues, the result that certain SNPs seem to appear to be in opposite risk trends at distinct carcinoma sites may be more likely owing to chance.
There are some advantages in our research that need to be point out. Firstly, as no studies had covered all the available ERCC genes polymorphisms37-39 to fully illustrate the relationship between ERCC polymorphisms and Pca, we applied a systematic document retrieval and acquire 17 qualified researches comprising 29 case-controls studies concerning the relationships between four reported ERCC genes polymorphisms and the PCa risk, which is our best advantage compared to other published articles. Secondly, quality of all the enrolled studies was assessed by Newcastle- Ottawa Scale (NOS) to avoid low quality studies affecting the overall quality. Fourthly, subgroup analysis was performed by ethnicity, source of control and HWE for the identification of the possible sources of heterogeneity. Thirdly, the data in our analysis were adjusted on the basis of the corresponding formula increasing the accuracy of the results. Lastly, the effect of each study on the pooled OR was tested using sensitivity analyses, and publication bias was evaluated by Egger's test and Begg's funnel plot.
However, there are several deficiencies in our study that should be addressed. Firstly, the number of enrolled studies for each polymorphism was insufficient, particularly the studies analyzing the rs17655 polymorphism (only three case-control studies), making it more difficult for us to find the statistical relationship between ERCC5-rs17655 and PCa risk. Secondly, histological types of PCa may be different amongst different researches. It is more reasonable to compare the relationship between ERCC polymorphism with Pca risk in the same histological type as certain polymorphisms may not be susceptibility for all Pca histological types. Thirdly, as for ethnicity subgroup analysis, the involved ethnicities were limited (only Asian, African and Caucasian population) and further studies of a larger sample size were needed to explore the influences of different ethnicities. Meanwhile, there were only two studies concerning ERCC2-rs238406 polymorphisms in Asian populations even we updated the search with more acquired databases till the lasted time, which may result in less rigorous of the conclusion. Lastly, several potentially confounding factors were not noted in this study, such as the age, smoking, alcohol consumption, lifestyles, environmental factors and so on.
In conclusion, our study demonstrates that ERCC2-rs1799793 polymorphism may be a risk factor for PCa in Asian population. Future functional researches with more ethnicities and participants are warranted to assess the more comprehensive influence of the genes encompassed in DNA repair genes and risk of PCa. And we are starting to collect clinical prostate cancer samples and paired normal control samples for further micro gene chips test, aiming to get more credible verification of the relationship between the ERCC2-rs1799793, ERCC2-rs238406, ERCC2-rs13181 and ERCC5-rs17655 polymorphisms and Pca.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
Yi Liu, Yonghui Hu, Meng Zhang, Runze Jiang and Chaozhao Liang contributed to the conception and design of the study, or acquisition of data, or analysis and interpretation of data; Yi Liu, Meng Zhang and Yonghui Hu drafted the article or revising it critically for important intellectual content; Runze Jiang and Chaozhao Liang gave the final approval of the version to be submitted.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Jalal S, Earley JN, Turchi JJ. DNA repair: From genome maintenance to biomarker and therapeutic target. Clinical Cancer Research. 2011;17:6973–84. doi: 10.1158/1078-0432.CCR-11-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedberg EC. How nucleotide excision repair protects against cancer. Nature Reviews Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 4.O'Donovan A, Davies AA, Moggs JG, West SC, Wood RD. Xpg endonuclease makes the 3|[prime]| incision in human DNA nucleotide excision repair. Nature. 1994;371:432–5. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 5.Wakasugi M, Reardon JT, Sancar A. The non-catalytic function of xpg protein during dual incision in human nucleotide excision repair. Journal of Biological Chemistry. 1997;272:16030–4. doi: 10.1074/jbc.272.25.16030. [DOI] [PubMed] [Google Scholar]
- 6.Wang LE, Wei Q. Nucleotide excision repair: DNA repair capacity, variability and cancer susceptibility.
- 7.Cheng L, Sturgis EM, Eicher SA, Spitz MR, Wei Q. Expression of nucleotide excision repair genes and the risk for squamous cell carcinoma of the head and neck. Cancer. 2002;94:393–7. doi: 10.1002/cncr.10231. [DOI] [PubMed] [Google Scholar]
- 8.Koeppel F, Poindessous V, Lazar V, Raymond E, Sarasin A, Larsen AK. Irofulven cytotoxicity depends on transcription-coupled nucleotide excision repair and is correlated with xpg expression in solid tumor cells. Clinical Cancer Research An Official Journal of the American Association for Cancer Research. 2004;10:5604–13. doi: 10.1158/1078-0432.CCR-04-0442. [DOI] [PubMed] [Google Scholar]
- 9.Berhane N, Sobti RC, Mahdi SA, Hyytinen ER, Frierson HF Jr, Sipe TW. et al. DNA repair genes polymorphism (xpg and xrcc1) and association of prostate cancer in a north indian population loss of heterozygosity and lack of mutations of the xpg/ercc5 DNA repair gene at 13q33 in prostate cancer polymorphisms of DNA repair-related genes with susceptibility and prognosis of prostate cancer. Mol Biol Rep. 2012;39:2471–9. doi: 10.1007/s11033-011-0998-5. [DOI] [PubMed] [Google Scholar]
- 10.Hyytinen ER, Frierson HF Jr, Sipe TW, Li CL, Degeorges A, Sikes RA. et al. Loss of heterozygosity and lack of mutations of the xpg/ercc5 DNA repair gene at 13q33 in prostate cancer. Prostate. 1999;41:190–5. doi: 10.1002/(sici)1097-0045(19991101)41:3<190::aid-pros6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Zhang XJ, Liu P, Zhu F. Polymorphisms of DNA repair-related genes with susceptibility and prognosis of prostate cancer. Genet Mol Res. 2014;13:4419–24. doi: 10.4238/2014.January.24.20. [DOI] [PubMed] [Google Scholar]
- 12.Cochran WG. The effectiveness of adjustment by subclassification in removing bias in observational studies. Biometrics. 1968;24:295–313. [PubMed] [Google Scholar]
- 13.Fenaroli G, Garibaldi U, Penco MA. Giochi, scommesse sulla vita, tabelle di mortalita, nascita del calcolo probabilistico, statistica e teoria delle popolazioni. Arch Hist Exact Sci. 1981;25:329–41. doi: 10.1007/BF01395659. [DOI] [PubMed] [Google Scholar]
- 14.Davey SG, Egger M. Meta-analyses of randomised controlled trials. Lancet. 1997;350:1181–2. doi: 10.1016/s0140-6736(05)63833-0. [DOI] [PubMed] [Google Scholar]
- 15.Dersimonian R, Nan L. Meta-analysis in clinical trials. Controlled Clinical Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 16.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. JNCI: Journal of the National Cancer Institute. 1959;22:719–48. [PubMed] [Google Scholar]
- 17.Zhang XJ, Liu P, Zhu F. Polymorphisms of DNA repair-related genes with susceptibility and prognosis of prostate cancer. Genetics and molecular research: GMR. 2014;13:4419–24. doi: 10.4238/2014.January.24.20. [DOI] [PubMed] [Google Scholar]
- 18.Bau D, Wu H, Cf, Lin C, Hsu C, Wang C, Wang R. et al. Association of xpd polymorphisms with prostate cancer in taiwanese patients. Anticancer Research. 2007;27:2893–6. [PubMed] [Google Scholar]
- 19.Berhane N, Sobti RC, Mahdi SA. DNA repair genes polymorphism (xpg and xrcc1) and association of prostate cancer in a north indian population. Molecular Biology Reports. 2012;39:2471–9. doi: 10.1007/s11033-011-0998-5. [DOI] [PubMed] [Google Scholar]
- 20.Hooker S, Bonilla C, Akereyeni F, Ahaghotu C, Kittles RA. Nat2 and ner genetic variants and sporadic prostate cancer susceptibility in african americans. Prostate Cancer & Prostatic Diseases. 2008;11:349–56. doi: 10.1038/sj.pcan.4501027. [DOI] [PubMed] [Google Scholar]
- 21.Huang WY, Berndt SI, Kang D, Chatterjee N, Chanock SJ, Yeager M. et al. Nucleotide excision repair gene polymorphisms and risk of advanced colorectal adenoma: Xpc polymorphisms modify smoking-related risk. Cancer Epidemiology Biomarkers & Prevention. 2006;15:306–11. doi: 10.1158/1055-9965.EPI-05-0751. [DOI] [PubMed] [Google Scholar]
- 22.Mirecka A, Paszkowska-Szczur K, Scott RJ, Górski B, Wetering TVD, Wokołorczyk D. et al. Common variants of xeroderma pigmentosum genes and prostate cancer risk. Gene. 2014;546:156–61. doi: 10.1016/j.gene.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 23.Ritchey JD, Huang WY, Chokkalingam AP, Gao YT, Deng J, Levine P. et al. Genetic variants of DNA repair genes and prostate cancer: A population-based study. Cancer Epidemiology Biomarkers & Prevention. 2005;14:1703–9. doi: 10.1158/1055-9965.EPI-04-0809. [DOI] [PubMed] [Google Scholar]
- 24.Rybicki BA, Conti DV, Moreira A, Cicek M, Casey G, Witte JS. DNA repair gene xrcc1 and xpd polymorphisms and risk of prostate cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2004;13:23–9. doi: 10.1158/1055-9965.epi-03-0053. [DOI] [PubMed] [Google Scholar]
- 25.Agalliu I, Kwon EM, Salinas CA, Koopmeiners JS, Ostrander EA, Stanford JL. Genetic variation in DNA repair genes and prostate cancer risk: Results from a population-based study. Cancer Causes & Control. 2010;21:289–300. doi: 10.1007/s10552-009-9461-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhillon VS, Yeoh E, Fenech M. DNA repair gene polymorphisms and prostate cancer risk in south australia-results of a pilot study. Urologic Oncology. 2009;29:641–6. doi: 10.1016/j.urolonc.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 27.Fachal L, Gómezcaamaño A, Peleteiro P, Carballo A, Calvocrespo P, Sánchezgarcía M. et al. Association of a xrcc3 polymorphism and rectum mean dose with the risk of acute radio-induced gastrointestinal toxicity in prostate cancer patients. Radiotherapy & Oncology Journal of the European Society for Therapeutic Radiology & Oncology. 2012;105:321–8. doi: 10.1016/j.radonc.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 28.Gao R, Price DK, Dahut WL, Reed E, Figg WD. Genetic polymorphisms in xrcc1 associated with radiation therapy in prostate cancer. Cancer Biology & Therapy. 2010;10:13–8. doi: 10.4161/cbt.10.1.12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavender NA, Komolafe OO, Benford M, Brock G, Moore JH, Vancleave TT. et al. No association between variant DNA repair genes and prostate cancer risk among men of african descent †. Prostate. 2010;70:113–9. doi: 10.1002/pros.21048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandal RK, Gangwar R, Mandhani A, Mittal RD. DNA repair gene x-ray repair cross-complementing group 1 and xeroderma pigmentosum group d polymorphisms and risk of prostate cancer: A study from north india. Dna & Cell Biology. 2010;29:183–90. doi: 10.1089/dna.2009.0956. [DOI] [PubMed] [Google Scholar]
- 31.Sobti RC, Berhane N, Melese S, Mahdi SA, Gupta L, Thakur H. et al. Impact of xpd gene polymorphism on risk of prostate cancer on north indian population. Molecular and Cellular Biochemistry. 2012;362:263–8. doi: 10.1007/s11010-011-1152-3. [DOI] [PubMed] [Google Scholar]
- 32.Yang B, Chen WH, Wen XF, Liu H, Liu F. Role of DNA repair-related gene polymorphisms in susceptibility to risk of prostate cancer. Asian Pacific Journal of Cancer Prevention Apjcp. 2013;14:5839–42. doi: 10.7314/apjcp.2013.14.10.5839. [DOI] [PubMed] [Google Scholar]
- 33.Zhou C, Xie L, Lin Y, Yang K, Mao Q, Cheng Y. Susceptibility of xpd and hogg1 genetic variants to prostate cancer. Biomedical Reports. 2013;1:679–83. doi: 10.3892/br.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fagbemi AF, Orelli B, Schärer OD. Regulation of endonuclease activity in human nucleotide excision repair. Dna Repair. 2011;10:722–9. doi: 10.1016/j.dnarep.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding DP, He XF, Zhang Y. Lack of association between xpg asp1104his and xpf arg415gln polymorphism and breast cancer risk: A meta-analysis of case-control studies. Breast Cancer Research and Treatment. 2011;129:203–9. doi: 10.1007/s10549-011-1447-9. [DOI] [PubMed] [Google Scholar]
- 36.Garciaclosas M, Malats N, Real FX, Kogevinas M, Silverman D, Tardon A. et al. Genetic variation in nucleotide excision repair (ner) and bladder cancer risk in the spanish bladder cancer study. Cancer Research. 2005;65:601–2. [Google Scholar]
- 37.He XF, Liu LR, Wei W, Liu Y, Su J, Wang SL, Association between the xpg asp1104his and xpf arg415gln polymorphisms and risk of cancer: A meta-analysis. 2014; 9: e88490. [DOI] [PMC free article] [PubMed]
- 38.Qiu L, Wang Z, Shi X, Wang Z. Associations between xpc polymorphisms and risk of cancers: A meta-analysis. European Journal of Cancer. 2008;44:2241–53. doi: 10.1016/j.ejca.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 39.Zhu ML, Wang M, Cao ZG, He J, Shi TY, Xia KQ. et al. Association between the ercc5 asp1104his polymorphism and cancer risk: A meta-analysis. Plos One. 2012;7:e36293. doi: 10.1371/journal.pone.0036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.